Abstract
This study examined the effects of buffalo oocyte extracts (BOE) on donor cells reprogramming and molecular characterisation of oocytes screened via brilliant cresyl blue (BCB) staining and comparison of gene expression profiles of developmentally important genes in blastocysts from IVF and cloned derived from BOE treated donor cells with BCB selected recipient cytoplasts. Relative abundance (RA) of OCT4 and NANOG was increased (P < 0.05) and HDAC-1, DNMT-1, and DNMT-3A decreased (P < 0.05) in extract treated cells (ETCs). This ETCs dedifferentiated into neuron-like lineage under appropriate induction condition. The RA of NASP, EEF1A1, DNMT1, ODC1 and RPS27A was increased (P < 0.05) in BCB+ oocytes, whereas ATP5A1 and S100A10 increased (P < 0.05) in BCB− oocytes. Total cell number and RA of OCT4, NANOG, SOX2, DNMT1, IGF2, IGF2R, MNSOD, GLUT1, BAX and BCL2 in cloned blastocysts derived from BCB+ oocytes with ETC more closely followed that of IVF counterparts compared to BCB+ oocytes with extract untreated cell and BCB− oocytes with ETC derived blastocysts. In conclusion, BOE influenced epigenetic reprogramming of buffalo fibroblasts making them suitable donors for nuclear transfer (NT). BCB staining can be effectively used for selection of developmentally competent oocytes for NT. The combined effects of epigenetic reprogramming of donor nuclei by BOE and higher nuclear reprogramming capacity of BCB+ oocytes improve developmentally important gene expression in cloned blastocysts. Whether these improvements have long-term effects on buffalo calves born following embryo transfer remains unknown.
Electronic supplementary material
The online version of this article (doi:10.1007/s10616-016-0057-0) contains supplementary material, which is available to authorized users.
Keywords: Oocytes extract, Reprogramming, Brilliant cresyl blue stain, Cloned blastocyst, Gene expression profile
Introduction
Somatic cell nuclear transfer (SCNT) using the zona-free approach or handmade cloning (HMC) has been studied in buffalo (Bubalus bubalis) (Sadeesh et al. 2014; Shah et al. 2008). Although some information is available on HMC in buffalo comparing in vitro culture conditions (Shah et al. 2008) examining the result of donor nucleus source (Sadeesh et al. 2016a, b), HMC is an incompetent process in buffalo (Sadeesh et al. 2014). Thus, further data are required to enable large-scale application of HMC technology to this species.
While SCNT-derived offspring have been effectively produced in many mammalian species, development to term is low (Keefer 2015; Sadeesh et al. 2016a), most cloned offspring produced by the SCNT technique show abnormalities (Rathbone et al. 2010). Following nuclear transfer (NT), the donor nucleus often fails to express early embryonic genes and establish a normal embryonic pattern of chromatin modifications. Studies have shown that low efficiency in SCNT is due to incomplete reprogramming of donor nuclei (Dean et al. 2001; Santos et al. 2003). Aiming to solve this problem, several researchers have developed favorable reprogramming of donor nuclei with some extrinsic factors before exposure to the recipient environment. Research reported transcriptional reprogramming of human and bovine nuclei increased after treatment of cells in extracts from Xenopus laevis oocytes or eggs (Alberio et al. 2005; Hansis et al. 2004). Reports showed that oocyte/egg extract treated somatic cells undergo improved reprogramming as indicated by expression of certain embryonic genes, such as octamer-binding transcription factor-4 (OCT-4) and Nanog homeobox (NANOG) (Miyamoto et al. 2009) and better development of blastocysts from extract treated cells (ETCs) as donor nuclei (Sadeesh et al. 2016a). Overall, these results propose that extract derived from eggs or oocytes is useful for understanding the mechanisms of nuclear reprogramming.
Cytoplasm quality is especially important in SCNT as overall cloning efficiency is directly related to cytoplast ability to reprogram a terminally distinguished somatic cell (Akagi et al. 2013; Tang et al. 2009). With a higher amount of cytoplasm loss connected with HMC compared to that in micromanipulation-based SCNT, cytoplasm quality becomes even more important. Thus, non-invasive oocyte quality appraisal for early selection of developmentally competent oocytes remains a major challenge for HMC. Currently, measures for selecting oocytes for use in SCNT are inadequate and in recent times the brilliant cresyl blue (BCB) test has reportedly improved in vitro development of SCNT embryos in the bovine (Su et al. 2012). Opiela et al. (2008) reported that oocytes screened with BCB staining differed in various oocyte quality markers, such as cytoplasmic volume, mitochondrial DNA copy number and relative abundance (RA) of marker genes.
In our previous study, we found higher (P < 0.05) 8- to 16-cell embryo and blastocyst development rate in HMC embryos reconstructed from BCB+ oocytes with ETCs (73.9, 32.8%, respectively) compared to in vitro fertilized (IVF) embryos (49.2, 24.2%, respectively). Explanation for these results may be that oocyte extract altered chromatin structure of donor nuclei improving reprogramming by recipient cytoplasm or the ability of the BCB stain to differentially select developmentally competent oocytes (Sadeesh et al. 2014). Nevertheless, no information is available at present regarding the molecular and cellular characteristics by which ETCs and BCB+ oocytes improved NT efficiency in buffalo species. Our previous study found no significant differences in gene expression profiles at maternal-to-zygotic transition stage (MZT; 8–16-cells) amongst cloned embryos derived from BCB+ oocytes with ETCs and IVF embryos. Whether these results in gene expression at MZT stage have any long term–effects remains unknown. These results suggest that developmental differences observed in HMC and in vitro fertilization produced fetuses and neonates are the results of aberrant gene expression during the pre-implantation stage and discrepancies in expression are indistinct or appear after the MZT stage of development.
In the present study we investigated the effect of buffalo oocytes extract (BOE) on epigenetic reprogramming of donor buffalo fibroblast cells based on gene expression using genes related to pluripotency (OCT-4, NANOG), histone deacetylation (histone deacetylase 1, HDAC-1) and methylation (DNA methyltransferase 1, DNMT-1; DNA methyltransferase 3 alpha, DNMT-3A); histone acetylation status and in vitro differentiation assays. Along with mRNA expression of certain maturation gene candidates in BCB screened oocytes, developmentally important genes in cloned blastocysts derived from ETCs and BCB screened oocytes using IVF embryos as control were compared by quantitative real–time PCR. This research may have implications for understanding molecular and cellular characteristics by which ETCs and BCB screened oocytes improved NT efficiency in buffalo species.
Materials and methods
Ethics statement, chemicals, and reagents
Experiments were conducted in accordance with the guidelines laid down by the committee for control and supervision of experiments on animals and with the approval from the institute animal ethics committee. All chemicals, reagents, culture media used in this study were of cell culture grade and purchased from Sigma Chemical Co., (St. Louis, MO, USA) unless otherwise indicated. Cell culture Petri dishes, four-well multi dishes, and six-well tissue culture plates were purchased from Nunc (Roskilde, Denmark). Membrane filters (0.2 μm) were sourced from Pall Life Sciences (Pall Corporation, Ann Arbor, MI, USA). Primers were synthesized by Sigma (P) Ltd. (Delhi, India). Reagents for molecular biology work were of biotechnology grade and purchased from Invitrogen (Carlsbad, CA, USA) and Fermentas (Thermo Fisher Scientific, Waltham, MA, USA) unless otherwise stated.
Establishment of fibroblast cell culture
Primary ear fibroblast culture of a Murrah buffalo calf was established and prepared for cloning as reported earlier (Sadeesh et al. 2014). All fibroblasts used in this study were from a single donor. Briefly, ear skin samples from a newborn Murrah buffalo calf were obtained aseptically in sterile phosphate buffered saline (PBS) with 1% pen/strep/amp (Gibco, Life Technologies, Grand Island, NY, USA) solution and transferred to the laboratory within 10 min. Tissue samples were washed six times with Dulbecco’s phosphate buffered saline (DPBS) and cut into small pieces (0.5 mm). These were transferred to 25 cm2 culture flasks and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2 mM l-glutamine, 15% FBS, 1% nonessential amino acids, 1% vitamins and 50 µg/ml gentamicin in a CO2 incubator (5% CO2 in air) at 37 °C. Cell cultures at different passages (passage 5–7) at 80–90% confluence (Fig. 1) were trypsinized with 0.25% trypsin–EDTA and washed with cell culture medium to remove traces of trypsin–EDTA. The cell pellet, obtained by centrifugation (200×g, 4 °C, 5 min), was resuspended in precooled (4 °C) cryopreservation medium (DMEM supplemented with 1% nonessential amino acids, 1% vitamins, 1% pen/strep/amp, 10% (v/v) Dimethyl sulfoxide (DMSO) and 15% FBS) stored at −80 °C overnight and then transferred directly to liquid nitrogen (−196 °C).
Fig. 1.
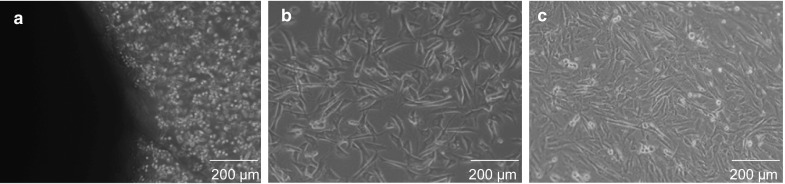
Isolation and culture of fibroblasts cells. a Primary explants outgrowth from ear skin tissue. b Subcultured fibroblast cells having 60–70% confluence. c Subcultured fibroblast cells having 80–90% confluence
Collection of oocytes and in vitro maturation
Apparently, normal ovaries of buffalo were collected from a slaughterhouse and aspiration of cumulus oocyte complexes (COCs) and in vitro maturation (IVM) of COCs were performed as described earlier (Sadeesh et al. 2014). Briefly, ovaries were washed three times with warm isotonic saline (32–37 °C) containing 400 IU/ml penicillin and 500 µg/ml streptomycin and transported to the laboratory within 4–5 h. Oocytes from follicles (2–8 mm) were aspirated with an 18 gauge needle attached to a 10 ml syringe (Sigma Chemical Co., # Z248029) loaded with aspiration medium (TCM-199 containing 0.3% BSA, 0.1 mg/ml glutamine, and 50 µg/ml gentamicin). Oocytes were washed four to six times with washing medium which consisted of TCM-199 with 10% FBS, 0.09 mg/ml sodium pyruvate, 0.1 mg/ml l-glutamine and 50 µg/ml gentamicin. The COCs with compact and unexpanded cumulus mass with equal to or greater than three layers of cumulus cells and homogeneous granular ooplasm were selected for IVM. The COCs were subject to maturation in media consisting of TCM-199 + sodium pyruvate (0.80 mM) + l-glutamine (2 mM) + 10% FBS + 5% Follicular fluid + pregnant mare’s serum gonadotropin (PMSG, 20 IU/ml) + human chorionic gonadotropin (hCG, 10 IU/ml), + gentamicin (50 µg/ml). Medium pH was adjusted to 7.4 and filtered through a 0.2 µm membrane filter immediately before use. The COCs were washed several times with IVM medium and groups of 10–15 COCs were placed independently in 100 µl droplets of IVM medium covered with sterilized mineral oil in 35 mm Petri dishes and cultured for 24 h under 5% CO2 at 38.5 °C.
Oocytes extract preparation
Matured metaphase II oocytes extract preparation was carried out as described earlier (Sadeesh et al. 2014). Briefly, COCs with expanded cumulus were transferred to a 1.5 ml micro centrifuge tube containing 500 µl hyaluronidase (0.5 mg/ml) in T2 (where T denotes HEPES modified TCM–199 supplemented with 2.0 mM l-glutamine, 0.2 mM sodium pyruvate, 50 µg/ml gentamicin and number denotes % FBS) and incubated for 1 min at 38.5 °C followed by vortexing (2–3 min). Denuded oocytes with uniformly granular cytoplasm were selected and incubated in pronase (2.0 mg/ml in T10) for 3–5 min at 38.5 °C. When the zona pellucida was digested, oocytes were transferred into T20 to deactivate the pronase. Oocytes with totally digested zona pellucida were washed twice with extraction buffer [50 mM KCl, 5 mM MgCl2, 5 mM ethylene glycol tetraacetic acid (EGTA), 2 mM β-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride (PMSF, protease inhibitor cocktail) and 50 mM HEPES, pH 7.6] containing an energy regeneration system (ERS: 1 mM ATP, 10 mM phosphocreatine, 25 µg/ml creatine kinase, pH 7.4). Nearly 400 zona-free oocytes were transferred into 10 µl of ERS in a 0.2 ml Eppendorf tube and centrifuged at 20,879×g for 20 min at 4 °C. The supernatant was used as the extract. The procedure was repeated to prepare enough extract, supplemented with 2% glycerol and stored at −80 °C.
SDS-PAGE and silver staining
Small portions of BOE from each batch were suspended in the SDS-lysis buffer and the lysed samples were subjected to SDS-PAGE. The protein bands were visualized by staining the gels with Silver Stain Kit (ThermoScientific USA) following the manufacturer’s instructions.
Permeabilization of cell membranes, extract treatment and donor cell preparation
Donor cell pre-treatment with BOE and donor cell preparations for HMC were done as described earlier (Sadeesh et al. 2014) with slight modifications. Briefly, donor cell suspensions were washed twice in DPBS (Ca2+, Mg2+ free) by spinning down at 10 × g for 5 min and supernatant was discarded carefully. Cells were suspended in 500 µl DPBS (Ca2+, Mg2+ free) containing 1.5 U/ml streptolysin-O at 38.5 °C for 30 min. Aliquots of permeated cells (about 1 × 106 cells/aliquot) were resuspended in 10 µl oocyte extract and incubated at 38.5 °C for 30 min. The control group was resuspended in 10 µl of ERS instead of oocyte extract. After these treatments, a part of the extract-treated cells (ETCs) was centrifuged at 39×g for 5 min and resuspended in DMEM containing 10% FBS, penicillin, streptomycin and 2 mM CaCl2 (Resealing media). Cells were then cultured for 2 h at 38.5 °C in 5% CO2 in air for membrane resealing. Membrane resealing was assessed by uptake of the membrane-impermeant DNA staining dye, propidium iodide (1 mg/ml), by plated cells. After resealing, floating cells were removed and the remaining cells or extract untreated cells (EUTCs, control) were cultured in DMEM supplemented with 2 mM l-glutamine, 15% FBS, 1% non-essential amino acids, 1% vitamins and 50 μg/ml gentamicin in a CO2 incubator (5% CO2 in air) at 37 °C. Morphological features, shape and size of cells and tendency to form the attachment to the culture flask were recorded at 24 h interval and medium was replaced every three days. Cell viability was monitored by standard protocol of exclusion of tryplan blue dye and cells were counted using a haemocytometer. Cells were cultured up to one week in above mentioned DMEM medium. Immediately before use, cell cultures were dispersed to single cells by treatment for 5 min by 0.25% trypsin-EDTA, subsequently washed by centrifugation and resuspended in T20 media for use as nucleus donor cells.
Immunodetection of H3K9ac
Cells were immunostained with antibodies against acetyl-histone H3 lysine 9 (Abcam, Cambridge, UK). Cells were washed briefly with PBS, fixed with 4% paraformaldehyde in PBS for 20 min, permeabilized with 0.1% Triton X-100 in PBS for 20 min, blocked in 2% bovine serum albumin (BSA) in PBS for 15 min, and incubated with primary antibodies (rabbit polyclonal to histone H3 acetyl K9) for 1 h. Cells were washed three times in PBS for 5 min and incubated for 2 h with 1:200 diluted fluorescein isothiocyanate-labelled secondary antibodies (goat anti-mouse IgG; BA1101, BOSTER, Pleasanton, CA, USA). Then, DNA was stained with a propidium iodide solution (10 µg/ml). As a negative control, immunostaining was performed without primary antibodies. Fluorescence was detected using a fluorescence microscope (Nikon, Tokyo, Japan).
In vitro induced differentiation
For neuron-like differentiation, cultured cells were detached by trypsinization, centrifuged and then seeded in complete growth medium supplemented with 10 μM all-trans retinoic acid. Cells were cultured for 2 weeks in retinoic acid; with the medium replaced every 2 days and differentiation of cells was assessed morphologically. The differentiated cells were identified by NESTIN-specific RT-PCR.
Brilliant cresyl blue staining and IVM of COCs
For the selection of developmentally competent oocytes as recipient cytoplasts, BCB staining of collected COCs was performed as described earlier (Sadeesh et al. 2014). To carry out the BCB test, immediately after collection oocytes were washed three times in DPBS modified by adding 0.4% BSA (mDPBS). Oocytes were exposed to 26 μM BCB diluted in mDPBS for 90 min at 38.5 °C in a humidified air atmosphere. Oocytes were then transferred to mDPBS and washed twice. On the basis of coloration, oocytes were divided into two groups, those with or without blue coloration of cytoplasm were named as BCB+ and BCB−, respectively. To achieve accurate data, oocytes of uncertain staining status were discarded. Further COCs from BCB+ and BCB− groups were subjected to maturation as described earlier (Sadeesh et al. 2014).
Handmade cloning
Recipient cytoplast preparations (cumulus or zona removal and manual enucleation) from BCB screened in vitro matured oocytes and the procedures for HMC were performed using standard protocols as described earlier (Shah et al. 2008). In vitro culture of NT embryos was performed according to the methods described previously (Sadeesh et al. 2014). Randomly collected blastocysts derived from ETCs/EUTCs with BCB screened oocytes were stained with Hoechst 33342 for 1 h and the number of nuclei was counted under the fluorescent microscope.
Sperm capacitation, in vitro fertilization and in vitro culture
In all experiments, frozen-thawed semen from the same bull was used. Spermatozoa were capacitated using Brackett and Oliphant (BO; 1975) medium and procedures for in vitro fertilization and in vitro culture of IVF embryos were performed according to the methods described previously (Sadeesh et al. 2014). Randomly collected IVF blastocysts were stained with Hoechst 33342 for 1 h and the number of nuclei was counted under a fluorescent microscope.
Isolation of total RNA, complementary DNA synthesis, and RT-PCR analysis
Total RNA was extracted from a portion of ETCs, EUTCs and reprogrammed cells using a cell to cDNA kit (Ambion Inc, Austin, TX, USA) according to the manufacturer’s protocol. A representative number of expanded COCs (n = 50, all oocytes collected from the same batch) from BCB+ and BCB− group after 21 h maturation was denuded with hyaluronidase as described earlier (Sadeesh et al. 2014). Denuded oocytes were washed three times with PBS by centrifugation for 5 min at 4 °C at 1200×g in 0.2 ml centrifuge tubes and treated using a cell to cDNA kit (Ambion Inc) to prepare the oocyte lysates, DNase digestion of prepared lysates and reverse transcription was conducted according to manufacturer’s instructions. Morphologically normal cloned blastocyst stage embryos from BCB screened oocytes with ETCs/EUTCs and IVF blastocysts were collected at 168–192 h post-activation and 168–192 h post-insemination, respectively, and separately treated using a cell to cDNA kit (Ambion Inc) according to the manufacturer’s protocol. Ten embryos at blastocyst stage were analyzed in each group in each trial. All blastocysts were from the same batch.
A polymerase chain reaction (PCR) was carried out in a 50 μl final volume containing 45 μl platinum PCR supermix (Invitrogen) and 5 μl of primer (200 nM each) and template DNA solution. A set of reaction without template cDNA was used as negative control for PCR. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the reference gene. The primer sequences of target genes of pluripotency (OCT-4, NANOG), histone deacetylation (HDAC1), methylation (DNMT1, DNMT3A), neural differentiation (NESTIN), maturation (nuclear autoantigenic sperm protein, NASP; elongation factor 1-alpha 1, EEF1A1; ornithine decarboxylase 1, ODC1; 40S Ribosomal protein S27a, RPS27A; ATP synthase subunit alpha, ATP5A1; S100 calcium-binding protein A10, S100A10,), and development-related [SRY (sex determining region Y)-box 2, SOX 2; insulin-like growth factor 2, IGF2; insulin-like growth factor 2 receptor, IGF2R; manganese superoxide dismutase, MnSOD; glucose transporter 1, GLUT1; BCL2 Associated X, BAX; B cell lymphoma 2, BCL2] are shown in Table 1. The primers all span introns. The PCR conditions were the same for all genes of interest, except for the annealing temperature (Table 1), as 94 °C for 2 min (initial denaturation), denaturation at 94 °C for 30 s, elongation at 72 °C for 1 min (35 cycles). The amplified DNA fragments were resolved on 2% agarose gel containing 0.5 μg/ml ethidium bromide against a 50-bp ladder and visualized under a gel documentation system (Alpha Imager, Alpha Innotech, San Leandro, CA, USA).
Table 1.
Details of primer sequence
| Gene name | Sequence (5′–3′) | Fragment size (bp) | Annealing temp (°C) | GenBank accession no. | |
|---|---|---|---|---|---|
| GAPDH | Forward | CCTGCCAAGTATGATGAGA | 131 | 53 | GU324291 |
| Reverse | GAAGGTAGAAGAGTGAGTGT | ||||
| OCT4 | Forward | GACAAGGAGAAGCTGGAG | 76 | 54 | JF898834 |
| Reverse | GCAAATTGTTCAAGGTCTTTC | ||||
| NANOG | Forward | GGGAAGGGTAATGAGTCCAA | 182 | 56 | DQ487022 |
| Reverse | AGCCTCCCTATCCCAGAAAA | ||||
| SOX2 | Forward | CATGGCAATCAAAATGTCCA | 215 | 54 | DQ126150 |
| Reverse | AGACCACGGAGATGGTTTTG | ||||
| HDAC1 | Forward | ATCGGTTAGGTTGCTTCAATCTG | 168 | 58 | BT030718.1 |
| Reverse | GTTGTATGGAAGCTCATTAGGGA | ||||
| DNMT1 | Forward | ATAAGTAAGATAGTGGTTGAGTTC | 100 | 55 | NM_182651 |
| Reverse | TTGAGCATACAAGGAGGAA | ||||
| DNMT3A | Forward | GACAAGAATGCCACCAAAGC | 134 | 55 | AY271298 |
| Reverse | ATCCACCAAGACACAATGCG | ||||
| NESTIN | Forward | ACCTGCTGTACATCGGCTTT | 307 | 60 | X93604 |
| Reverse | GAGGATGGTGAAGACGGAGA | ||||
| NASP | Forward | AATGAGGAGGAGGAGATTG | 193 | 56 | BT006757 |
| Reverse | GTTAAGGCAGGACTGGGAA | ||||
| EEF1A1 | Forward | TTGGTAAGAGTTGCTCATAAG | 168 | 53 | NM_174535 |
| Reverse | TTGTTCATCTGTCCTAATCTAG | ||||
| ODC1 | Forward | TGTAGATACCACTCTTGTAGC | 105 | 50 | NM_174130 |
| Reverse | CTCACATAGACATCTCAGAACTA | ||||
| RPS27A | Forward | CAA GAAGAACAAGCATAAGAGAA | 123 | 53 | XM_005212558 |
| Reverse | TCCAGCACCACATTCATC | ||||
| ATPSA1 | Forward | GCCTCTAACTCTCGTCTG | 133 | 51 | NM_174684 |
| Reverse | GCAATACCATCACCAATACTT | ||||
| S100A10 | Forward | AAATGCCGTCTCAAATGG | 182 | 54 | NM_174650 |
| Reverse | GGTCCAGGTCCTTCATTA | ||||
| IGF2 | Forward | CTTCAGCCGACCATCCAG | 107 | 55 | FJ032306 |
| Reverse | GGGGTGGCACAGTAAGTC | ||||
| IGF2R | Forward | GCAGATTTATTTCTTCTCCCAC | 186 | 55 | J03527 |
| Reverse | CACTCAAACTCGTAGAAGCA | ||||
| MNSOD | Forward | GGGCCATATCAATCACAG | 193 | 55 | JN687584 |
| Reverse | GTCCCTGCTCCTTATTG | ||||
| GLUT1 | Forward | CCGTCTCTTCCTATCCAA | 194 | 56 | HQ434959 |
| Reverse | GGTCTTCTTGAATAGTGAGTT | ||||
| BAX | Forward | ATCCACCAAGAAGCTGAG | 82 | 52 | HE661581 |
| Reverse | CTGCGATCATCCTCTGTA | ||||
| BCL2 | Forward | GGCCCCTGTTTGATTTCT | 97 | 56 | U92434 |
| Reverse | CTTATGGCCCAGATAGGC | ||||
Real-time PCR for relative quantification of target genes
Real-time PCR (Applied Biosystems 7500 Real-Time PCR system, Foster City, CA, USA) was performed using SYBR Green qPCR supermix (Invitrogen SYBR Green qPCR supermix) as a double-standard DNA-specific fluorescent dye to assess gene expression of OCT4, NANOG, HDAC1, DNMT1, DNMT3A in ETCs or EUTCs, NASP, EEF1A1, DNMT1, ODC1, RPS27A, ATP5A1 and S100A10 in BCB screened oocytes and OCT4, NANOG, SOX2, DNMT1, DNMT3A, IGF2, IGF2R, MnSOD, GLUT1, BAX and BCL2 in blastocysts. Validation studies were performed using GAPDH as the internal reference gene and GAPDH expression was consistent between various treatment groups (ETCs vs. EUTCs, BCB+ oocytes vs. BCB− oocytes and NT blastocysts vs. IVF blastocysts). All reactions were run in triplicate and three biological replicates were carried out. Amplification was carried out in 25 μl volume reaction mixture containing 12.5 μl SYBR Green qPCR supermix, 1 μl primer (10 pM each forward and reverse primer), 2 μl cDNA template and 9.5 μl nuclease free water. Samples not exposed to reverse transcriptase were used as negative controls. For PCR, samples were activated at 95 °C for 10 min, followed by 40 cycles of denaturion at 95 °C for 45 s, then annealing at the specific primer annealing temperature (Table 1) for 30 s and extension at 72 °C for 30 s. The comparative CT method was used for relative quantification of target gene expression levels. Quantification was normalized to the internal control GAPDH gene. Within the exponential phase of the amplification curve, each cycle doubled the amplified product. The ΔCT value was determined by subtracting the GAPDH CT value for each sample from the target gene CT value. Calculation of ΔΔCT value involved using the highest sample method ΔCT as an arbitrary constant to subtract from all other ΔCT samples values. Fold changes in relative mRNA expression of target genes were determined using the formula 2−ΔΔCT.
Statistical analysis
Data were analyzed using GraphPad Prism (version 6.05). Experimental results are presented as mean ± SEM. Data were subject to analysis of variance and the Tukey test was used to compare means (P < 0.05, considered statistically significant).
Results
Characterization of cell extracts from buffalo matured metaphase II oocytes
Only matured metaphase II (MII) oocytes with normal morphological appearance were selected for preparing cell extracts. The maturation rate was 78.08% (406/520; n = 3). Extraction of oocyte components was achieved by disruption with high-speed centrifugation. The resulting solutions had liquid droplets and cell debris which was originally present in oocytes. The protein concentration of matured MII extracts from buffalo oocytes was 1.92 ± 0.75 mg/ml. Protein band patterns of SDS-lysed oocyte extract showed various sized proteins ranging from 20 to 120 kDa (shown in supplementary Fig. 1). These results suggest protein components were successfully extracted from buffalo matured MII oocytes.
Expression of pluripotency, histone deacetylase, and DNA methyltransferase-related genes in ETCs and ETUCs
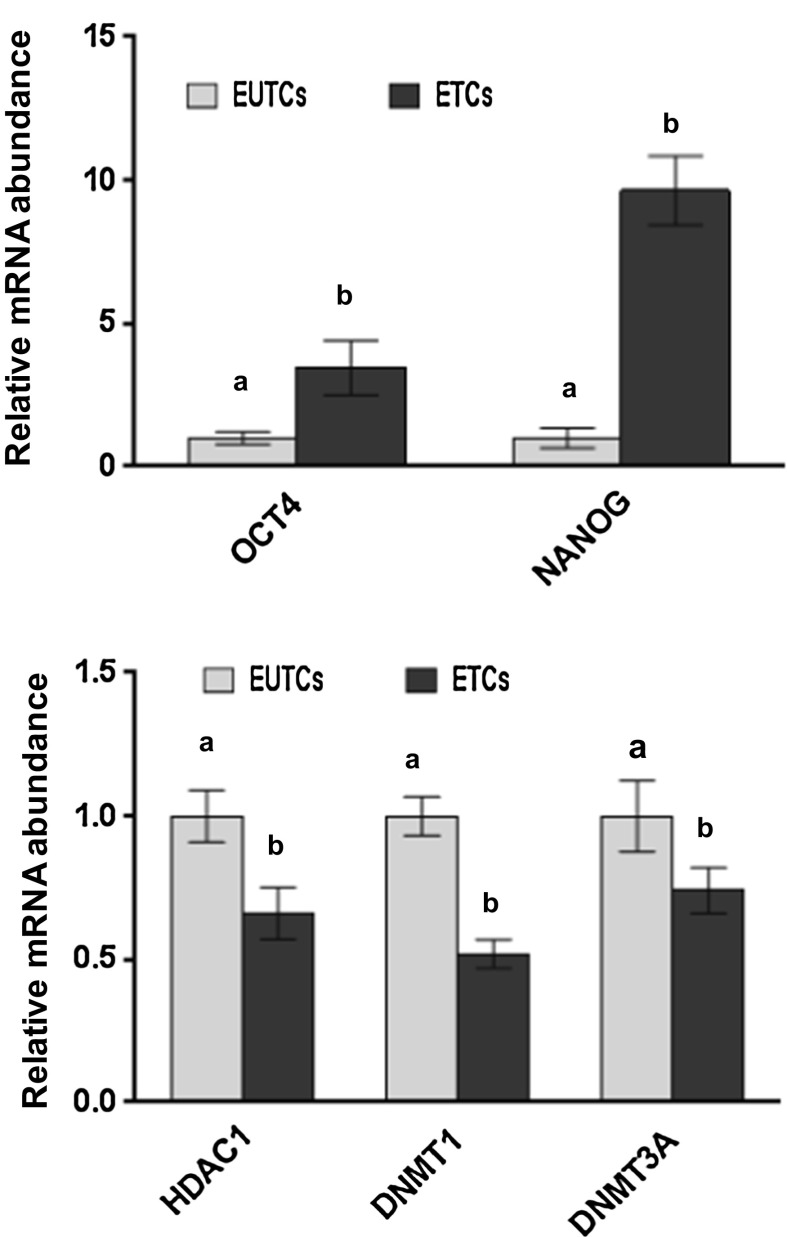
Buffalo fibroblast cells were exposed to BOE for one week and expression of pluripotent marker genes, OCT4 and NANOG, in ETCs were analyzed for assessing changes in the epigenetic state compared with EUTCs as a control. Changes in relative expression of these transcriptional factors were determined by real-time quantitative PCR (Fig. 2). The NANOG and OCT4 genes were differentially expressed in ETCs after one week of culture. The results revealed the expression of NANOG and OCT4 genes was increased (P < 0.05) in ETCs compared with EUTCs. Expression levels of genes related to histone deacetylation (HDAC1) and DNA methylation (DNMT1 and DNMT3A) were also compared for ETCs and EUTCs. Relative abundance of HDAC1, DNMT1 and DNMT3A was reduced (P < 0.05) in ETCs compared to EUTCs.
Fig. 2.
Relative expression profile of pluripotency (OCT4, NANOG), histone deacetylase (HDAC1), and DNA methyltransferase-related (DNMT1, DNMT3A) genes in ETCs and ETUCs. The data are representative of three (n = 3) independent experiments. The standard error is indicated by error bars. Bars with different superscripts differ (P < 0.05)
RT-PCR analysis of expression of pluripotency, histone deacetylase, and DNA methyltransferase-related genes in ETCs and ETUCs is shown in supplementary Fig. 2.
BOE mediated histone acetylation of H3K9ac
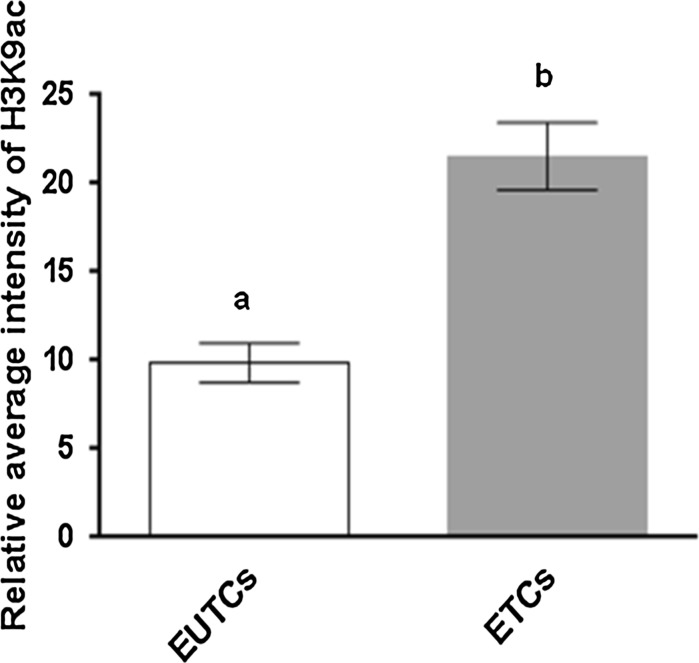
Levels of acetylated histone H3K9ac in fibroblasts exposed to BOE were determined semi-quantitatively (Fig. 3) and the H3K9ac signal in BOE treated cells was greater (P < 0.05) than in the control group. Qualitatively results of immunocytochemistry revealed the presence of acetylated histone H3K9ac in living cells, BOE influenced H3K9ac acetylation levels, and negative control experiments with primary antibody omitted showed no results in both groups (Fig not shown).
Fig. 3.
Effect of fibroblast cells treatment with BOE on the level of H3K9ac in these cells. Presented as mean ± SEM. N = 3 number of individual samples from each group over three (n = 3) independent experiments were counted. Bars with different superscripts were significantly different (P < 0.05)
In vitro differentiation potential of reprogrammed cells
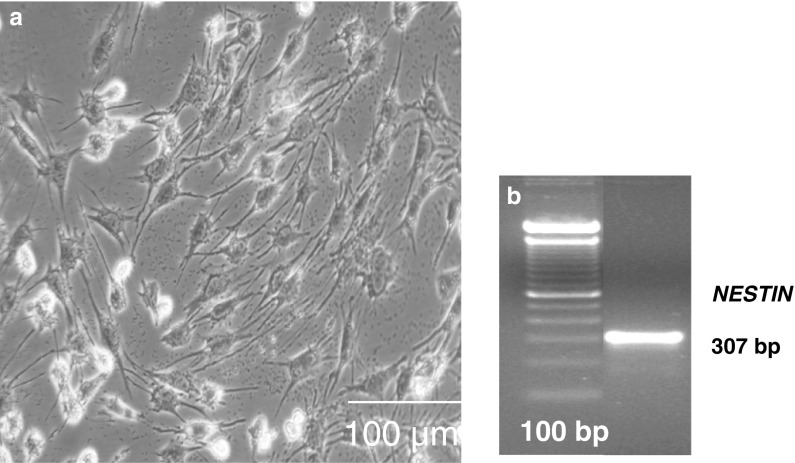
When reprogrammed cells were cultured under the neurogenic condition, neuron-like differentiation of reprogrammed cells was observed after induction with retinoic acid after 2 weeks. RT-PCR based studies showed the strong expression of NESTIN in neuron-like differentiated cells (Fig. 4). Cells maintained in regular control medium did not change their morphology and negative for NESTIN expression (Fig not shown).
Fig. 4.
In vitro differentiation potential of reprogrammed cells. a Neuron-like differentiated cells, b NESTIN for neurogenesis
Effect of BCB screening on maturation rates of COCs
Clear representative images of BCB+ and BCB− oocytes after staining with BCB are shown in Fig. 5. Out of 2257 COCs examined in the present study, the percentage of BCB+ oocytes (1336, 59.2%) was higher (P < 0.05) than that of BCB− oocytes (653, 28.9%) and oocytes that were discarded (268, 11.8%). COC maturation assessed by cumulus expansion (Fig. 6) was higher (P < 0.05) in BCB+ (82.3%) than in BCB− (52.8%) oocytes. The percentage of oocytes with a clear polar body extrusion was higher (P < 0.05) for BCB+ oocytes (72.4%) than that for BCB− oocytes (46.6%).
Fig. 5.
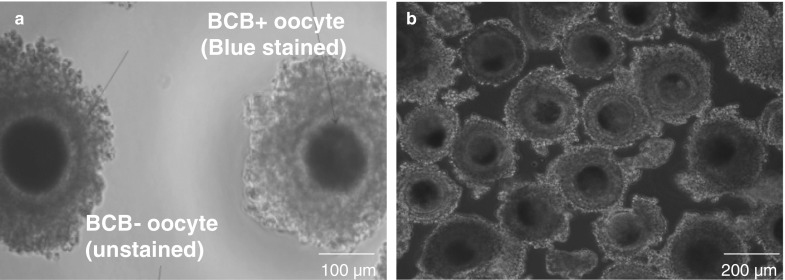
Oocytes with different glucose-6-phosphate dehydrogenase (G6PDH) activity. a Brilliant cresyl blue (BCB)+ (Blue due to low G6PDH activity) and BCB− (colorless due to high G6PDH activity) COC, b COCs nonexposed to BCB stain
Fig. 6.
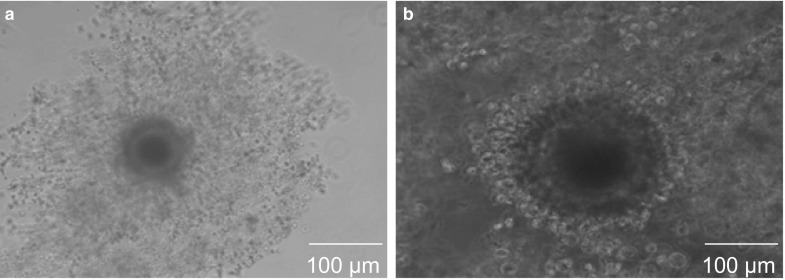
Oocytes with different glucose-6-phosphate dehydrogenase activity after in vitro maturation. a BCB+, b BCB−
Expression of selected maturation candidate genes in BCB screened oocytes
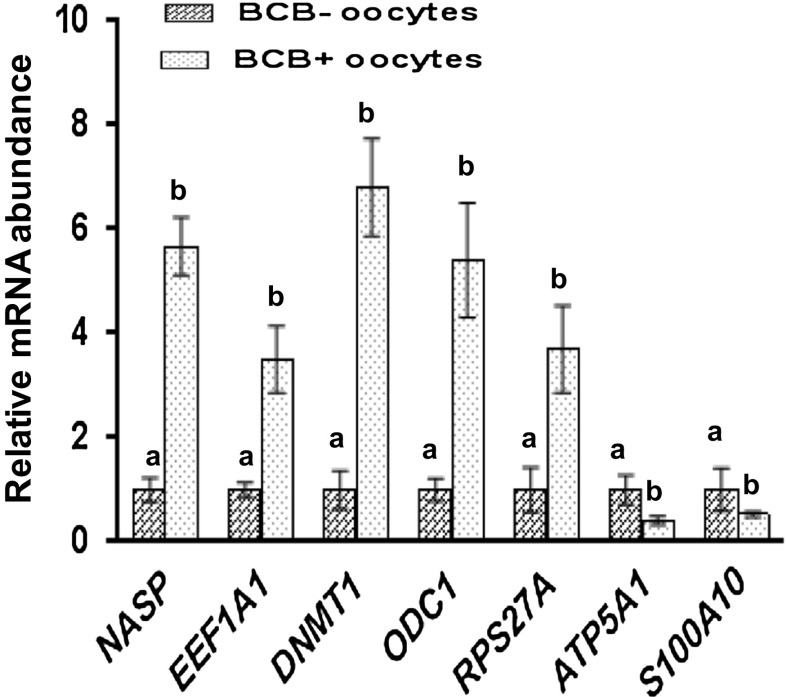
Seven genes were analyzed by quantitative real-time PCR between BCB+ and BCB− oocytes. Relative transcript abundance was significantly different for seven genes between BCB+ and BCB− oocytes (Fig. 7). The expression of five genes NASP, EEF1A1, DNMT1, ODC1and RPS27A, showed greater (P < 0.05) transcript abundance in BCB+ than in BCB− oocytes, whereas expression of two genes ATP5A1 and S100A10, was increased (P < 0.05) in BCB− oocytes than in BCB+ oocytes. RT-PCR analysis of expression of selected maturation candidate genes in BCB screened oocytes is shown in supplementary Fig. 3.
Fig. 7.
Relative expression profile of selected maturation candidate genes (NASP, EEF1A1, DNMT1, ODC1, RPS27A, ATPSA1 and S100A10) in BCB selected oocytes. N = 50 in vitro matured oocytes from BCB+ and BCB− group over three (n = 3) independent experiments. The standard error is indicated by error bars. Bars with different superscripts were significantly different (P < 0.05)
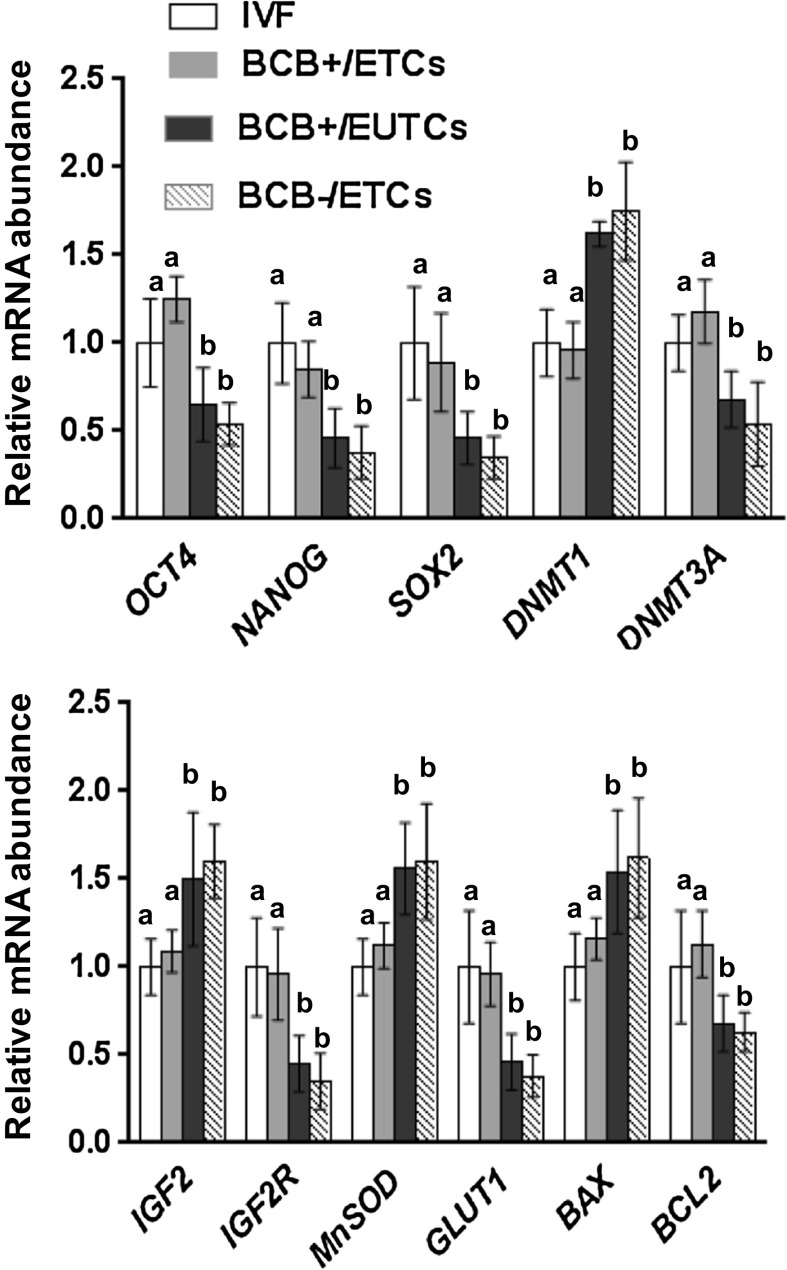
Cloned blastocysts derived from reprogrammed donor cells and BCB screened oocytes differentially express developmentally important selected candidate genes
A total number of 16 blastocysts were used for determination of total cell number. Average total cell numbers for blastocysts generated using BCB+/ETCs (217.42 ± 12.63) was greater (P < 0.05) than for BCB+/EUTCs (173.34 ± 3.42), BCB−/ETCs (140.62 ± 3.68) and BCB−/EUTCs (128.52 ± 2.86). Total cell number did not show difference (P > 0.05) between blastocysts from NT (BCB+/ETCs) and IVF (217.42 ± 12.63 vs. 194.12 ± 8.56, respectively).
The results of real-time quantitative PCR analysis of gene transcripts at blastocyst stages in NT and IVF is shown in Fig. 8. Relative mRNA abundance of OCT4, NANOG and SOX2 was similar in IVF and BCB+/ETCs blastocysts and was greater (P < 0.05) than in BCB+/EUTCs, BCB−/ETCs and BCB−/EUTCs blastocysts. Among epigenetic-related genes, expression of DNMT1 was similar in IVF and BCB+/ETCs blastocysts, was decreased (P < 0.05) than in BCB+/EUTCs, BCB−/ETCs, and BCB−/EUTCs blastocysts, whereas transcript DNMT3A was not significantly different between IVF and BCB+/ETCs blastocysts. Transcripts IGF2 and IGF2R was not significantly different among BCB+/ETCs and IVF blastocysts. For the oxidative stress-related gene, expression of MnSOD was similar in IVF and BCB+/ETCs blastocysts and was decreased (P < 0.05) compared to BCB+/EUTCs, BCB−/ETCs and BCB−/EUTCs blastocysts. For the glucose transport related gene, expression of GLUT1 was similar in IVF and BCB+/ETCs blastocyst and was significantly greater than in BCB+/EUTCs, BCB−/ETCs and BCB−/EUTCs blastocysts. Relative mRNA abundance of BAX was similar in IVF and BCB+/ETCs blastocysts and was decreased (P < 0.05) compared to BCB+/EUTCs, BCB−/ETCs and BCB−/EUTCs blastocysts. Expression of BCL2 was similar in IVF and BCB+/ETCs blastocysts and was greater (P < 0.05) than in BCB+/EUTCs, BCB−/ETCs and BCB−/EUTCs blastocysts. Data of selected candidate genes expression in BCB−/EUTCs blastocysts is not shown. RT-PCR analysis of expression of developmentally important selected candidate genes in blastocysts is shown in supplementary Fig. 4.
Fig. 8.
Relative expression profile of developmentally important selected candidate genes (OCT4, NANOG, SOX2, DNMT1, DNMT3A, IGF2, IGF2R, MnSOD, GLUT1, BAX, and BCL2) in IVF, BCB+/ETCs, BCB+/EUTCs and BCB−/ETCs blastocysts. N = 10 embryos from each group over three (n = 3) independent experiments. Bars with different superscripts differ (P < 0.05)
Discussion
Functional cell-free extracts were derived from buffalo matured oocytes, a 10 μl droplet of the extract derived from roughly 400 matured oocytes was enough to partially reprogram more than 1 × 106 cells. This processing ability implies that buffalo oocyte extracts may be used to identify reprogramming factors, as the Xenopus system (Gonda et al. 2003), and several reprogramming factors have been identified in Xenopus system, for example, ISWI, FRGY2a/b, BRG1, NPM, and DJ-1(Miyamoto et al. 2011).
One of the most interesting reprogramming events in this study was histone acetylation of H3K9ac in buffalo fibroblasts. Histone acetylation has an important role in reprogramming and may be associated with pluripotency of nuclei (Yamanaka et al. 2009). In this study, H3K9ac increased (P < 0.05) after buffalo fibroblasts were pre-treated with BOE. Therefore, we inferred that nuclear reprogramming of somatic cells by oocyte cytoplasm was inhibited by their own histone deacetylases (HDACs). In previous studies, treatment with HDAC inhibitors, such as Trichostatin A and Scriptaid, improved pluripotency of somatic cells (Wang et al. 2011a, b). However, increased histone acetyltransferase activity or inhibited HDAC activity was required to increase histone acetylation (Murko et al. 2010). Lysine deacetylation in the tails of core histones was controlled by HDACs, for example, HDAC1, which participated in the removal of acetyl moieties from histone tails (Murko et al. 2010). Therefore, we further examined the effects of BOE treatment on gene expression of HDAC1 in buffalo fibroblasts. Interestingly, expression levels of HDAC1 decreased after BOE treatment. Similarly, cytoplasmic extracts of Xenopus eggs improved acetylation levels in mammalian somatic cells (Allegrucci et al. 2011). Interestingly, changes in H3K9ac staining after BOE treatment showed the likeness to epigenetic modifications induced using amphibian oocyte extract (Bian et al. 2009; Rathbone et al. 2010). These findings indicate that exposure to BOE alters some epigenetic modifications (DNA demethylation and histone acetylation) of buffalo somatic cells.
Data of the present study show that expression of pluripotency-related genes (OCT4 and NANOG) were reactivated in buffalo somatic cells after BOE treatment. Therefore, the expression of these pluripotent related genes may convert somatic cells to a partially dedifferentiated state after treatment with extracts. These results suggest that reprogramming of somatic nuclei by oocyte extracts is a gradual process which requires the expression of pluripotent factors before cells begin to enter a stable self-sustaining pluripotent state. In our study, high expression of OCT4 and NANOG in ETCs may result from successful reactivation of the OCT4 and NANOG genes in ETCs. This may be due to promoter regions of OCT4 and NANOG being significantly demethylated after BOE treatment. Thus, we further investigated the expression patterns of DNMT1 and DNMT3A in buffalo fibroblasts after BOE treatment and we found that methyltransferase expression levels were decreased (P < 0.05) after BOE treatment, especially DNMT1 expression. We inferred that some reprogramming factors in oocyte cytoplasm changed methyltransferase activity and methylation binding proteins in somatic cells which altered the epigenetic reprogramming pattern. However, further research is needed to examine the global levels of DNA methylation in BOE treated cells using untreated cells as controls before meaningful conclusions can be drawn in this species.
From our study and observation after the exposition of reprogrammed cells to neurogenic induction condition in vitro, cells alter their morphology and convert into neuron-like differentiation. Laboratories have reported neuronal differentiation of oocyte extract treated fibroblasts (Bui et al. 2013; Pang et al. 2015). Our results indicate that BOE mediated the dedifferentiation of terminally differentiated buffalo fibroblasts and enabled the reprogrammed cells to undergo redifferentiation.
Our data suggest that matured oocyte extracts could provide the necessary regulatory components for inducing somatic cell nuclear reprogramming in buffalo species. Prolonged exposure of somatic cells to reprogramming factors present in oocyte extracts provided enough time for reprogramming. Therefore, the reprogramming of somatic cell nuclei to a lower state using BOE may be one reason behind the better developmental potential of cloned embryos reconstructed from BCB+ oocytes with ETCs in our previous studies.
Earlier, we reported an increased (P < 0.05) blastocyst development rate in cloned embryos reconstructed from BCB+ oocytes with ETCs. However whether the BCB test is an effective method to isolate oocytes with high growth/maturation potential is unknown in buffalo species. To address this gap in knowledge we compared seven maturation gene candidates in BCB selected buffalo oocytes. Expression profiles of seven maturation gene candidates were expressed differentially between BCB+ and BCB− oocytes. Expressed genes in BCB+ oocytes are associated with regulation of the cell cycle (NASP, ODC1), transcription (DNMT1) and translation (EEF1A1, RPS27A), whereas BCB− oocytes were enriched with genes involved in ATP synthesis (ATP5A1) and calcium ion binding (S100A10).
Various factors involved in cell cycle regulation are more evident in BCB+ than in BCB− oocytes. Among these cell cycle regulators, NASP was chiefly recognized as a nuclear-associated protein in rabbit testis (Welch et al. 1990). NASP was one of the genes with increased expression in very fast moving bovine oocytes which showed higher blastocyst rate compared with the slow groups following dielectrophoretic separation (Dessie et al. 2007). Elongation factor 1A is part of the eukaryotic translational apparatus and a GTP-binding protein that catalyses binding aminoacyl tRNAs to the ribosome (Tatsuka et al. 1992). Attainment of high developmental capacity in mammalian oocytes is dependent on high rates of RNA and protein synthesis, imprinting processes and biogenesis of organelles such as mitochondria (Eichenlaub-Ritter and Peschke 2002). Consistent with this, oocytes with better developmental potential (BCB+) showed superior mRNA transcript abundance for EEF1A1 and RPS27A which corresponds with the members of translation connected genes. Smits et al. (2011) identified ODC1 as a developmentally important gene in the equine embryo. The ODC1 gene showed greater (P < 0.05) expression for in vivo derived blastocysts compared with in vitro produced blastocysts. In the present study greater (P < 0.05) abundance of ODC1 in BCB+ oocytes was observed than in BCB− oocytes. Expression of DNMT1 was also found to be highly over expressed in BCB+ buffalo oocytes compared with BCB− oocytes which are similar to BCB+ ovine oocytes (Torner et al. 2008). Modifications in mitochondrial distribution, DNA replication, copy number and transcripts may point to overall debilitation for the mitochondria and influence the ability of embryos to scavenge free radicals and prompt an oxidative stress response which contributes to impaired development (EI-Shourbagy et al. 2006; May-Panloup et al. 2005; Van Blerkom 2004). ATP5A1 is a nuclear-encoded gene whose protein contributes to the overall function of ATP synthase and it is the universal enzyme for cellular ATP synthesis (Pedersen 1994). It has been reported that null mutations in the three-subunit of the mitochondrial ATP synthase gene in Drosophila led to embryonic death (Kidd et al. 2005). The S100A10, a member of the S100 family also known as calpactin I or p11, provinces as a mediator in the calcium-dependent signaling pathway. Significantly greater mRNA transcript abundances for ATP5A1 and S100A10 in incompetent BCB− oocytes than competent BCB+ oocytes in bovine have been reported (Torner et al. 2008). The above-mentioned data suggest that BCB+ oocytes have better stores of the cell cycle, transcription and protein biosynthesis transcripts which could be used for resuming meiosis and supporting maternal to zygotic transition. The BCB test is an effective method to isolate oocytes with high growth/maturation potential in buffalo species. These results and our previous study suggest that BCB staining can be effectively used for selection of developmentally competent oocytes for NT in buffalo species because BCB+ oocytes yielded higher nuclear reprogramming capacity than BCB− oocytes. This may be another reason behind the improved developmental potential of handmade cloned embryos derived from BCB+ oocytes with ETCs as previous studies from our laboratory have reported (Sadeesh et al. 2014).
Qualitative or morphological assessment alone is insufficient to provide an accurate and efficient estimation of embryo quality and embryonic developmental potential (Jousan et al. 2008; Wrenzycki et al. 2004). Aberrant expression of genes during early embryonic developmental stages is considered an important cause of embryonic losses and fetal abnormalities that continue during gestation and parturition resulting in abnormal offspring syndrome (Arnold et al. 2008; Walker et al. 1996). Evaluation of gene expression profiles in our previous study suggests that no differences in gene expression profiles among morphologically dissimilar embryos at MZT may contribute to the low variation observed in potential to develop to the blastocyst stage. These results support the notion that if developmental differences observed in NT and in vitro fertilization produced fetuses and neonates are the results of aberrant gene expression during the pre-implantation stage, discrepancies in expression are indistinct or appear after the MZT stage of development. Thus, to identify differences in relative abundance of transcripts of developmentally important genes between cloned embryos derived from BCB screened oocytes with ETCs and IVF embryos after MZT stage, the expression patterns of several developmentally important genes were compared at the blastocyst stage between these two groups.
Appropriate expression of pluripotency-related genes such as OCT4, NANOG, and SOX2, which are expressed mainly in the inner cell mass of blastocysts is necessary for successful embryonic development. We found expression of these three pluripotency genes was significantly reduced in cloned blastocysts derived from BCB− oocytes with ETCs/EUTCs than in IVF blastocysts and blastocysts reconstructed by BCB+ oocytes with ETCs/EUTCs. However, OCT4, NANOG, and SOX2 expression in cloned blastocysts reconstructed by BCB+ oocytes with ETCs were similar to those in IVF blastocysts. This finding contradicts the report of Zhang et al. (2014), who found OCT4 and SOX2 transcript levels to be higher in IVF than in SCNT bovine embryos. This divergence among experiments may be due to different protocols used to produce and culture embryos, different methods to assess gene expression profile or species-specific variation. Whether the change in expression of important genes is enough to stimulate pluripotent embryonic cells to distinguish into those of trophoblast cell lineage needs to be studied further. Nevertheless, BCB+/ETCs blastocysts may also be superior to BCB−/ETCs blastocysts due to a lower magnitude of aberrance in the expression of these three genes. Expression of DNMT1 and DNMT3A in NT blastocysts suggests that differential DNMT1 and DNMT3A mRNA levels in BCB+/EUTCs and BCB−/ETCs or EUTCs blastocysts have serious implications for pre-implantation development and result in reduced blastocyst formation from these NT embryos. Whether the consistently low efficiency of NT is related to the inability of a somatic nucleus to undergo normal changes in methylation as indicated by increased levels of DNMT1 or to the lack of de novo synthesis by low DNMT3A expression remains unclear in buffalo. Thus, further research needs to be carried out on the global levels of DNA methylation and histone acetylation in NT embryos using IVF embryos as controls before meaningful conclusions can be drawn in this species. At the blastocyst stage, the RA of IGF2 was significantly increased and IGF2R was significantly decreased in NT embryos derived from BCB− oocytes compared with IVF embryos. Our finding is consistent with Han et al. (2003) who reported that IGF2R expression was significantly down-regulated and IGF2 up-regulated in cloned embryos rather than in IVF embryos. Alteration in mRNA expression of IGF2 and IGF2R between NT- and IVF-derived blastocysts may be associated with inappropriate genomic imprinting. However, IGF2 and IGF2R expression was nonsignificant between blastocyst from BCB+ oocytes with ETCs and IVF counterpart due to successful reprogramming of imprinted genes of donor nuclei in this cloned embryo similar to IVF embryos. MnSOD is involved in promoting cellular differentiation (St Clair et al. 1994), higher expression of MnSOD in NT blastocysts (BCB+/EUTCs, BCB−/ETCs or EUTCs) in our studies could be due to inadequate dedifferentiation of the donor genome in NT embryos. Sadeesh et al. (2016c) reported a significant difference in expression of GLUT1 between NT and in vitro fertilization derived blastocysts. The mouse model suggests that for in vivo embryos GLUT1 decreased 50% in in vitro blastocysts (Morita et al. 1990). These reports clearly suggest that viable embryos have higher GLUT1 expression. In our study, relative expression of GLUT1 between two types of embryos at blastocyst stage support the notion that cloned embryos reconstructed with oocyte extract treated donor cells and selection of recipient cytoplasts through BCB staining are identical to IVF embryos regarding the ability to develop to term. Decreased glucose transporter expression triggers BAX-dependent apoptosis in murine blastocysts (Chi et al. 2000). Hence, it is possible that, in our study, the identical level of GLUT1 expression observed between these two embryos is related to similar levels of apoptotic incidence in both embryos at the blastocyst stage. Along with cell number of blastocysts, apoptosis is a measure for evaluation of blastocyst quality. We found the pro-apoptotic gene BAX was decreased in BCB+ blastocysts compared with BCB− blastocysts. BAX is a positive regulator of apoptosis, so the lower expression of BAX may contribute to reduced apoptosis of cells in BCB+ blastocysts compared with those in BCB− blastocysts. However, no difference was observed in BAX expression, among NT blastocysts from BCB+/ETCs and IVF counterpart. Furthermore, our results show, lower apoptosis was further indicated by higher expression of BCL2 in BCB+ and IVF blastocysts than in BCB− blastocysts.
The expression profile of genes as indicators of developmental competence in buffalo NT-derived blastocysts from BCB+ oocytes with ETCs was similar to IVF blastocysts which in turn, were higher than in blastocysts from BCB+ oocytes with EUTCs and BCB− oocytes with ETCs/EUTCs. Therefore, cloned blastocysts derived from BOE treated cells and BCB selected oocytes may be a useful marker of embryonic developmental potential and an additional criterion in the selection of good quality NT embryos for transfer or freezing. The mechanism(s) through which these embryos are able to develop faster and acquire better quality may be that these embryos possess all the necessary gene products and proteins required for further development, whereas the absence of many of these gene products and proteins in BCB− oocytes with ETC/EUTCs embryos could be related to higher developmental arrest or apoptosis. However, the low developmental efficiency of NT embryos may not be large because of lack of nuclear reprogramming during early embryo development but may be potentially caused by abnormal gene reprogramming during post-implantation fetal or placental development (Smith et al. 2005). Studies addressing these questions with a variety of different molecular and cellular approaches could bring about importantly needed information in the future.
Conclusion
Buffalo oocyte extracts (BOE) have the ability to convert buffalo somatic cells into an undifferentiated state following a brief period of culture. The strategy of epigenetic remodeling by treatment of BOE may prove a useful tool for reprogramming of somatic cell nuclei. We need to exploit the nuclear reprogramming of differentiated cells into an undifferentiated state induced by using oocyte extracts. Further, studies to select extract-treated cells for SCNT are required for optimized exploitation of the reprogramming ability of the extract. The G6PDH activity of buffalo oocytes is correlated with expression of developmentally important transcripts. Selection of developmentally competent oocytes through BCB staining for NT using ETCs may improve the gene expression profile of developmentally important genes in cloned blastocysts comparable to that of IVF-derived blastocysts. This improvement may be due to the combined effects of epigenetic reprogramming of donor nuclei by BOE and higher nuclear reprogramming capacity of BCB+ oocytes. Whether this improvement will have long-term effects on buffalo calves born after as a result of embryo transfer remains unknown. However, it is likely that early adaptations of the pre-implantation embryo to its environment persist during fetal and post-natal development.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Funding support from Indian Council of Agricultural Research (ICAR), New Delhi is gratefully acknowledged. The authors would like to thank Dr. Inderjeet Singh, Director, ICAR-Central Institute for Research on Buffaloes, for providing the necessary facilities for conducting this research. We are also thankful to Dr. PS Yadav, ICAR-CIRB, for his scientific guidance and Dr. Dana Thomsen (The University of Adelaide, Adelaide, Australia) for reviewing this manuscript.
Authors’ contribution
SEM conceived the study, carried out the experiments and drafted the manuscript. FS assisted with sample analysis. MK participated in study design. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- Akagi S, Geshi M, Nagi T. Recent progress in bovine somatic cell nuclear transfer. Anim Sci J. 2013;84:191–199. doi: 10.1111/asj.12035. [DOI] [PubMed] [Google Scholar]
- Alberio R, Johnson A, Stick R, Campbell KHS. Differential nuclear remodeling of mammalian somatic cells by Xenopus laevis oocyte and egg cytoplasm. Exp Cell Res. 2005;307:131–141. doi: 10.1016/j.yexcr.2005.02.028. [DOI] [PubMed] [Google Scholar]
- Allegrucci C, Rushton MD, Dixon JE, Sottile V, Shah M, Kumari R, et al. Epigenetic reprogramming of breast cancer cells with oocyte extract. Mol Cancer. 2011;10:7. doi: 10.1186/1476-4598-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold DR, Fortier AL, Lefebvre R, Miglino MA, Pfarrer C, Smith LC. Placental insufficiencies in cloned animals- a workshop report. Placenta. 2008;29:108–110. doi: 10.1016/j.placenta.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Bian Y, Alberio R, Allegrucci C, Campbell KH, Johnson AD. Epigenetic marks in somatic chromatin are remodelled to resemble pluripotent nuclei by amphibian oocyte extract. Epigenetics. 2009;4:194–202. doi: 10.4161/epi.4.3.8787. [DOI] [PubMed] [Google Scholar]
- Bracket BG, Oliphant G. Capaciation of rabbit spermatozoa in vitro. Biol Reprod. 1975;12:260–274. doi: 10.1095/biolreprod12.2.260. [DOI] [PubMed] [Google Scholar]
- Bui HT, Kwon DN, Kang MH, Oh MH, Park MR, Park WJ, et al. Epigenetic reprogramming in somatic cells induced by extract from germinal vesicle stage pig oocytes. Development. 2013;140:4330–4340. doi: 10.1242/dev.092239. [DOI] [PubMed] [Google Scholar]
- Chi MM, Pingsterhaus J, Carayannopoulos M, Moley KH (2000) Decreased glucose transporter expression triggers BAX-dependent apoptosis in the murine blastocyst. J Biol Chem 275:40252–40257 [DOI] [PubMed]
- Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, WolF E, et al. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci USA. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessie SW, Rings F, Hölker M, Gilles M, Jennen D, Tholen E, et al. Dielectrophoretic behavior of in vitro derived bovine metaphase II oocytes and zygotes and its relation to in vitro embryonic developmental competence and mRNA expression pattern. Reproduction. 2007;133:931–946. doi: 10.1530/REP-06-0277. [DOI] [PubMed] [Google Scholar]
- Eichenlaub-Ritter U, Peschke M. Expression in in vivo and in vitro growing and maturing oocytes: focus on regulation of expression at the translational level. Hum Reprod Update. 2002;8:21–41. doi: 10.1093/humupd/8.1.21. [DOI] [PubMed] [Google Scholar]
- El-Shourbagy SH, Spikings EC, Freitas M, St John JC. Mitochondria directly influence fertilisation outcome in the pig. Reproduction. 2006;131:233–245. doi: 10.1530/rep.1.00551. [DOI] [PubMed] [Google Scholar]
- Gonda K, Fowler J, Katoku-Kikyo N, Haroldson J, Wudel J, Kikyo N. Reversible disassembly of somatic nucleoli by the germ cell proteins FRGY2a and FRGY2b. Nat Cell Biol. 2003;5:205–210. doi: 10.1038/ncb939. [DOI] [PubMed] [Google Scholar]
- Han DW, Song SJ, Uhum SJ, Do JT, Kim NH, Chung KS, et al. Expression of IGF2 and IGF receptor mRNA in bovine nuclear transferred embryos. Zygote. 2003;11:245e52. doi: 10.1017/S0967199403002296. [DOI] [PubMed] [Google Scholar]
- Hansis C, Barreto G, Maltry N, Niehrs C. Nuclear reprogramming of human somatic cells by Xenopus egg extract requires BRG1. Curr Biol. 2004;14:1475–1480. doi: 10.1016/j.cub.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Jousan FD, De Castro LA, Brad AM, Roth Z, Hansen PJ. Relationship between group II caspase activity of bovine pre-implantation embryos and capacity for hatching. J Reprod Dev. 2008;54:217–220. doi: 10.1262/jrd.19175. [DOI] [PubMed] [Google Scholar]
- Keefer CL. Artificial cloning of domestic animals. Proc Natl Acad Sci USA. 2015;112:8874–8878. doi: 10.1073/pnas.1501718112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd T, Shumays RA, Katzen A, Sisson JC, Jiménez G, Pinchin S, et al. The ε-subunit of mitochondrial ATP synthase is required for normal spindle orientation during the Drosophila embryonic divisions. Genetics. 2005;170:697–708. doi: 10.1534/genetics.104.037648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May-Panloup P, Chretien MF, Jacques C, Vasseur C, Malthiery Y, Reynier P. Low oocyte mitochondrial DNA content in ovarian insufficiency. Hum Reprod. 2005;20:593–597. doi: 10.1093/humrep/deh667. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Tsukiyama T, Yang Y, Li N, Minami N, Yamada M, et al. Cell-Free extracts from mammalian oocytes partially induces nuclear reprogramming in somatic cells. Biol Reprod. 2009;80:935–943. doi: 10.1095/biolreprod.108.073676. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Nagai K, Kitamura N, Nishikawa T, Ikegami H, Binh NT. Identification and characterization of an oocyte factor required for development of porcine nuclear transfer embryos. Proc Natl Acad Sci. 2011;108:7040–7045. doi: 10.1073/pnas.1013634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Tsutsumi O, Hosoya I, Taketani Y, Oka Y, Kato T. Expression and possible function of glucose transporter protein GLUT1 during pre-implantation mouse development from oocytes to blastocysts. Biochem Biophys Res Commun. 1990;188:8–15. doi: 10.1016/0006-291X(92)92342-U. [DOI] [PubMed] [Google Scholar]
- Murko C, Lagger S, Steiner M, Seiser C, Schoefer C, Pusch O. Expression of class I histone deacetylases during chick and mouse development. Int J Dev Biol. 2010;54:1527–1537. doi: 10.1387/ijdb.092971cm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opiela J, Katska-Ksiazkiewicz L, Lipiński D, Słomski R, Bzowska M, Ryńska B. Interactions among activity of glucose-6-phosphate dehydrogenase in immature oocytes, expression of apoptosis-related genes BC-2 and BAX, and developmental competence following IVP in cattle. Theriogenology. 2008;69:546–555. doi: 10.1016/j.theriogenology.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Pang R, Zhu X, Geng J, Zhang Y, Wang Q, He J, et al. In vitro and in vivo analysis of human fibroblast reprogramming and multipotency. Cell Mol Biol Lett. 2015;20:404–417. doi: 10.1515/cmble-2015-0024. [DOI] [PubMed] [Google Scholar]
- Pedersen PL. ATP synthase: the machine that makes ATP. Curr Biol. 1994;4:1138–1141. doi: 10.1016/S0960-9822(00)00257-8. [DOI] [PubMed] [Google Scholar]
- Rathbone AJ, Fisher PA, Lee JH, Craigon J, Campbell KHS. Reprogramming of ovine somatic cells with Xenopus laevis oocyte extract prior to SCNT improves live birth rate. Cell Reprogr. 2010;12:609–616. doi: 10.1089/cell.2010.0015. [DOI] [PubMed] [Google Scholar]
- Sadeesh EM, Meena K, Balhara S, Yadav PS. Expression profile of developmentally important genes between handmade cloned buffalo embryos produced from reprogramming of donor cell with oocytes extract and selection of recipient cytoplast through brilliant cresyl blue staining and in vitro fertilized embryos. J Assist Reprod Genet. 2014;31:1541–1552. doi: 10.1007/s10815-014-0316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeesh EM, Meena K, Fozia S, Yadav PS. A comparative study on efficiency of adult fibroblasts and amniotic fluid-derived stem cells for production of handmade cloned buffalo (Bubalus bubalis) embryos. Cytotechnology. 2016;68:593–608. doi: 10.1007/s10616-014-9805-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeesh EM, Shah F, Kataria M, Yadav PS. A comparative study on expression profile of developmentally important genes during pre-implantation stages in buffalo hand-made cloned embryos derived from adult fibroblasts and amniotic fluid derived stem cells. Cytotechnology. 2016;68:1447–1461. doi: 10.1007/s10616-015-9904-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeesh EM, Shah F, Yadav PS. Differential developmental competence and gene expression patterns in buffalo (Bubalus bubalis) nuclear transfer embryos reconstructed with fetal fibroblasts and amnion mesenchymal stem cells. Cytotechnology. 2015;68:1827–1848. doi: 10.1007/s10616-015-9936-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Zakhartchenko V, Stojkovic M. Epigenetic marking correlates with developmental potential in cloned bovine pre-implantation embryos. Curr Biol. 2003;3:1116–1121. doi: 10.1016/S0960-9822(03)00419-6. [DOI] [PubMed] [Google Scholar]
- Shah RA, George A, Singh MK, Kumar D, Chauhan MS, Manik R, et al. Handmade cloned buffalo (Bubalus bubalis) embryos: comparison of different media and culture systems. Cloning Stem Cells. 2008;10:435–442. doi: 10.1089/clo.2008.0033. [DOI] [PubMed] [Google Scholar]
- Smith SL, Everts RE, Tian XC, Du F, Sung LY, et al. Global gene expression profiles reveal significant nuclear reprogramming by the blastocyst stage after cloning. Proc Natl Acad Sci USA. 2005;102:17582–17587. doi: 10.1073/pnas.0508952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits K, Goossens K, Van Soom A, Govaere J, Hoogewijs M, Peelman LJ. In vivo-derived horse blastocysts show transcriptional upregulation of developmental important genes compared with in vitro-produced horse blastocysts. Reprod Fertil Dev. 2011;23:364–375. doi: 10.1071/RD10124. [DOI] [PubMed] [Google Scholar]
- St Clair DK, Oberley TD, Muse KE, St Clair WH. Expression of manganese superoxide dismutase promotes cellular differentiation. Free Radic Biol Med. 1994;16:275–282. doi: 10.1016/0891-5849(94)90153-8. [DOI] [PubMed] [Google Scholar]
- Su J, Wang Y, Li R, Peng H, Hua S, Li Q, et al. Oocytes selected using BCB staining enhances nuclear reprogramming and the in vivo development of SCNT embryos in cattle. Plos ONE. 2012;7:12e36181. doi: 10.1371/annotation/01ebf0b5-ccd3-494d-b577-170981f7bc36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Wang Y, Zhang D, Gao Y, Ma Y, Yin B, et al. Reprogramming donor cells with oocyte extracts improves in vitro development of nuclear transfer embryos. Anim Reprod Sci. 2009;15:1–9. doi: 10.1016/j.anireprosci.2008.10.018. [DOI] [PubMed] [Google Scholar]
- Tatsuka M, Mitsui H, Wada M, Nagata A, Nojima H, Okayama H. Elongation factor-1 alpha gene determines susceptibility to transformation. Nature. 1992;359:333–336. doi: 10.1038/359333a0. [DOI] [PubMed] [Google Scholar]
- Torner H, Ghanem N, Ambros C, Holker M, Tomek W. Molecular and sub cellular characterization of oocytes screened for their developmental competence based on glucose-6-phosphate dehydrogenase activity. Reproduction. 2008;135:197–212. doi: 10.1530/REP-07-0348. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J. Mitochondria in human oogenesis and pre-implantation embryogenesis: engines of metabolism, ionic regulation and developmental competence. Reproduction. 2004;128:269–280. doi: 10.1530/rep.1.00240. [DOI] [PubMed] [Google Scholar]
- Walker SK, Hartwich KM, Seamark RF. The production of unusually large offspring following embryo manipulation: concepts and changes. Theriogenology. 1996;45:111–120. doi: 10.1016/0093-691X(95)00360-K. [DOI] [Google Scholar]
- Wang LJ, Zhang H, Wang YS, Xu WB, Xiong XR, Li YY, et al. Scriptaid improves in vitro development and nuclear reprogramming of somatic cell nuclear transfer bovine embryos. Cell Reprogr. 2011;13:431–439. doi: 10.1089/cell.2011.0024. [DOI] [PubMed] [Google Scholar]
- Wang YS, Xiong XR, An ZX, Wang LJ, Liu J, Quan FS, et al. Production of cloned calves by combination treatment of both donor cells and early cloned embryos with 5-aza-2(/)-deoxycytidine and trichostatin A. Theriogenology. 2011;75:819–825. doi: 10.1016/j.theriogenology.2010.10.022. [DOI] [PubMed] [Google Scholar]
- Welch JE, Zimmerman LJ, Joseph DR, O’Rand MG. Characterization of a sperm-specific nuclear auto antigenic protein. I. Complete sequence and homology with the Xenopus protein, N1/N2. Biol Reprod. 1990;43:559–568. doi: 10.1095/biolreprod43.4.559. [DOI] [PubMed] [Google Scholar]
- Wrenzycki C, Herrmann D, Lucas-Hahn A, Lemme E, Korsawe K, Niemann H. Gene expression patterns in in vitro produced and somatic nuclear transfer derived pre-implantation bovine embryos: relationship to the large offspring syndrome. Anim Reprod Sci. 2004;82–83:593–603. doi: 10.1016/j.anireprosci.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Sugimura S, Wakai T, Kawahara M, Sato E. Acetylation level of histone H3 in early embryonic stages affects subsequent development of miniature pig somatic cell nuclear transfer embryos. J Reprod Dev. 2009;55:638–644. doi: 10.1262/jrd.20245. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang Y, Sang Y, Zhang Y, Hua S. Combination of S-adenosylhomocysteine and scriptaid, a non-toxic epigenetic modifying reagent, modulates the reprogramming of bovine somatic-cell nuclear transfer embryos. Mol Reprod Dev. 2014;81:87–97. doi: 10.1002/mrd.22287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.