Abstract
Bioorthogonal labeling of antibodies enables the conjugation of compounds, such as small molecules or peptides, which expand targeting capacity or enhance cytotoxicity. Taking advantage of a cyclohexene sulfonamide compound that site-selectively labels Lys64 in human serum albumin (HSA), we demonstrate that domain I of HSA can be used as a fusion protein for the preparation of antibody conjugates. Trastuzumab fusions were expressed at the N-terminus of the light chain or the C-terminus of the heavy chain enabling conjugation to small molecules. Moreover, these conjugates retained HER2 binding and proved to be highly stable in human plasma. Antibody conjugation via HSA domain I fusion should therefore have broad utility for making serum-stable antibody conjugates, particularly for antibody-drug conjugates.
Graphical Abstract
INTRODUCTION
Antibody conjugates provide a route to expand antigen targeting or introduce a cytotoxic drug such as with antibody-drug conjugates (ADCs). However, many labeling strategies are non-specific resulting in heterogeneous conjugates with complex pharmacodynamic, pharmacokinetic, and safety profiles.1–3 Site-specific labeling strategies have yielded homogenous conjugates with an improved therapeutic index.4,5 Such efforts have focused on the chemical modification of engineered cysteine/selenocysteine residues, genetically-encoded unnatural amino acids, glyco-engineering, and enzymatic derivatization.
New methods for chemoselective antibody labeling offer a viable approach for generating conjugates with substantial therapeutic potential. Non-covalent antibody labeling has been successfully employed, but relies on high affinity binding to ligands or haptens.6,7 In contrast, covalent conjugation eliminates dissociation of the small molecule or peptide,8 an important distinction when preparing ADCs bearing a cytotoxic payload. Our laboratory has previously identified a peptide by phage display for conjugation of 1,3-diketone compounds to proteins via formation of an enaminone.9 Similarly, the Francis group screened a combinatorial peptide library to improve pyridoxal 5′-phosphate-mediated transamination enabling oxime or hydrazone formation with proteins, including an Fc domain.10,11 Genetically encoded protein partners, such as HaloTag,12 SNAP-Tag,13 and CLIP-Tag,14 covalently bind their respective linkers enabling the incorporation of synthetic molecules. When using such protein fusion strategies to produce therapeutic ADCs, the fusion partner should ideally be chosen to minimize immunogenicity.
Towards this goal, we have recently explored methods for the covalent attachment of chemical moieties to human serum albumin (HSA).15 A cyclohexene sulfonamide compound derived from TAK-242—an inhibitor of Toll-like receptor 4 that covalently binds Cys74716,17—was shown to rapidly modify HSA and demonstrated excellent serum stability (Figure 1A). We mapped the primary labeling site to Lys64 of HSA domain I (HSAdI) (Figure 1B). Intriguingly, labeling was site-selective despite the high abundance of surface lysine residues on HSA, which has led us to explore the use of this protein as a fusion partner for antibodies and subsequent conjugation to small molecules. We show that HSAdI fusion proteins can be readily labeled using a rhodamine-linked cyclohexene sulfonamide compound, cHx-Rho. Trastuzumab (Herceptin®) conjugates were also prepared to evaluate the robustness of such fusion proteins. Our results suggest that antibody-HSAdI fusions possess favorable properties, such as retained antigen binding and conjugate stability. Antibody conjugation using HSAdI as a fusion protein should therefore be amenable to therapeutic applications.
Figure 1.
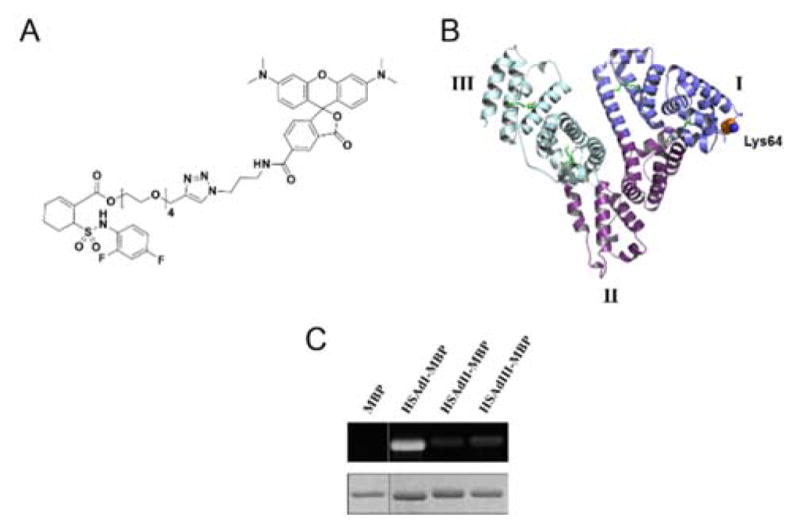
(A) Structure of rhodamine-linked cyclohexene sulfonamide compound (cHx-Rho). (B) HSA crystal structure showing the primary labeling site, Lys64 (1BJ5). HSA domains I, II, and III are indicated. (C) Fluorescent (top) and Coomassie-stained (bottom) gel of HSA-MBP domain fusion proteins labeled with 10 equivalents of cHx-Rho for 2 h.
RESULTS AND DISCUSSION
Our initial efforts were aimed at minimizing the size of the HSA fusion partner while maintaining labeling efficiency with cHx-Rho. Individual HSA domains I, II, and III were expressed with a C-terminal maltose-binding protein (MBP) to serve as a model fusion protein (Table S1). No labeling was observed with MBP alone, and only minor fluorescent bands were evident with the MBP fusion proteins of domain II (HSAdII-MBP) and domain III (HSAdIII-MBP) (Figure 1C). The fusion protein between domain I and MBP (HSAdI-MBP) was strongly labeled compared to HSAdII-MBP and HSAdIII-MBP, which is consistent with our previous analysis showing that Lys64 was predominantly modified (Figure 1B).15 Truncation of domain I to subdomain IA18 caused a substantial loss in cHx-Rho labeling (data not shown), suggesting that the conformation of HSAdI may be important for modulating reactivity.
Next, the reaction between cHx-Rho and HSAdI-MBP was further characterized. Titration of cHx-Rho from 1 to 50 equivalents over 2 h yielded conjugate in a concentration-dependent manner (Figure S1A). The nucleophilic Cys34 residue of HSA was also mutated to Ser in order to confirm that the cysteine residue does not react with cHx-Rho as well as to decrease the chances for non-specific dimerization. As expected, the C34S substitution (HSAdIC34S-MBP) had no impact on labeling efficiency (Figure S1A). A time-course study using 10 equivalents of cHx-Rho further demonstrated comparable labeling between HSAdI-MBP and HSAdIC34S-MBP at 0, 4, 8, and 24 h (Figure S1B and Table S2). The major product by ESI-MS at 24 h was +1 conjugate with only minor unmodified HSAdIC34S-MBP and +2 observed (Figure S2).
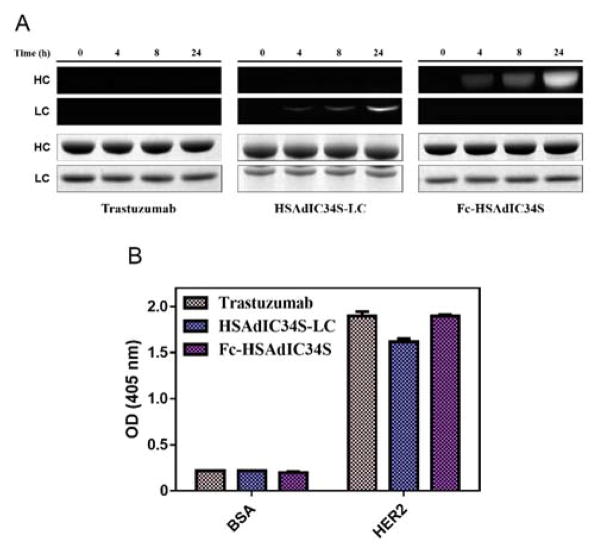
Fusion proteins between trastuzumab and HSAdIC34S were generated to investigate the utility of this approach for antibody conjugation. We chose to fuse HSAdIC34S to the N-terminus of the light chain (HSAdIC34S-LC) or C-terminus of the heavy chain Fc domain (Fc-HSAdIC34S) using a (GlySer)3 spacer. Specific antibody chains containing the fused HSAdIC34S protein were labeled with cHx-Rho, but no labeling was observed for trastuzumab (Figure 2A). This further reflects the high site-selectivity of the cyclohexene sulfonamide compound for domain I. While HSAdI conjugation reactions required longer incubation time than we previously noted for HSA,15 conjugates were prepared in good yield (Table S2). Thus, HSAdIC34S antibody fusions can be readily generated in both fusion orientations, providing a significant benefit when making antibody conjugates.
Figure 2.
Antibody fusion conjugation. (A) Time-course study of trastuzumab, HSAdIC34S-LC, and Fc-HSAdIC34S labeling with 10 equivalents of cHx-Rho. Top, fluorescent gel. Bottom, Coomassie-stained gel. Heavy (HC) and light (LC) chains are indicated. (B) HER2 binding by ELISA for trastuzumab and conjugates.
To further interrogate the practicality of using HSAdIC34S antibody fusions for therapeutic purposes, binding to HER2 was evaluated to ensure that conjugates retained antigen recognition. Neither trastuzumab nor the trastuzumab fusion conjugates bound to BSA, but binding was observed between the antibodies with HER2 at sub-saturating concentrations by ELISA (Figure 2B). Hence, antigen binding was not compromised with either conjugate. A marginal decrease in binding by HSAdIC34S-LC was noted, which correlates with a previous study of Zybodies that attached an Ang2-binding peptide to the N-terminus of the trastuzumab light chain resulting in a minor ~2-fold decrease in HER2 affinity.19 Future studies will be needed to validate binding between other antibody/antigen pairs as the fusion protein may potentially occlude important interactions depending on fusion orientation. Nonetheless, both trastuzumab conjugates demonstrated excellent antigen binding despite fusing and labeling the HSAdIC34S protein.
HER2 recognition by Fc-HSAdIC34S conjugate was also confirmed through detection of cHx-Rho fluorescence using flow cytometry (Figure S3). Binding was seen with the HER2+ breast cancer cell line SKBR-3 but not with HER2- MDA-MB-468 cells. Dose-dependent antigen binding by antibody fusion conjugate was observed on SKBR-3 cells with saturation occurring at ~25 nM (Figure S4). Trastuzumab, Fc-HSAdIC34S, and cHx-Rho-conjugated Fc-HSAdIC34S binding to SKBR-3 cells was also detected with APC-conjugated secondary antibody against the constant domain of the kappa light chain (Cκ) (Figure S5). This comparison showed similar binding at 5 nM between all three antibodies, verifying that neither the HSAdI fusion nor cHx-Rho conjugation has detrimental effects on antigen engagement. Binding to FcγRIIIa was only modestly decreased suggesting that Fc-HSAdIC34S fusions are capable of antibody-dependent cellular cytotoxicity (ADCC) (Figure S6), however, the antibody isotype and point of fusion will need to be carefully considered for each application.20
Taking advantage of the intrinsic fluorescence provided by cHx-Rho conjugation, we performed kinetic characterization of trastuzumab and Fc-HSAdIC34S labeling by flow cytometry (Figure 3A). Conjugation reactions were performed with cyclohexene sulfonamide compound ranging from 5 to 50 equivalents over 24 h. Plotting mean fluorescence intensity (MFI) values over time allowed pseudo-first order kinetic analysis. The rate for Fc-HSAdIC34S labeling was determined to be ~1 M−1s−1, providing nearly two orders of magnitude rate enhancement relative to trastuzumab conjugation (Table S3). Antibody fusion labeling proceeded at a rate comparable to uncatalyzed oxime ligation,21,22 which has been used to readily prepare ADCs.4,23,24
Figure 3.
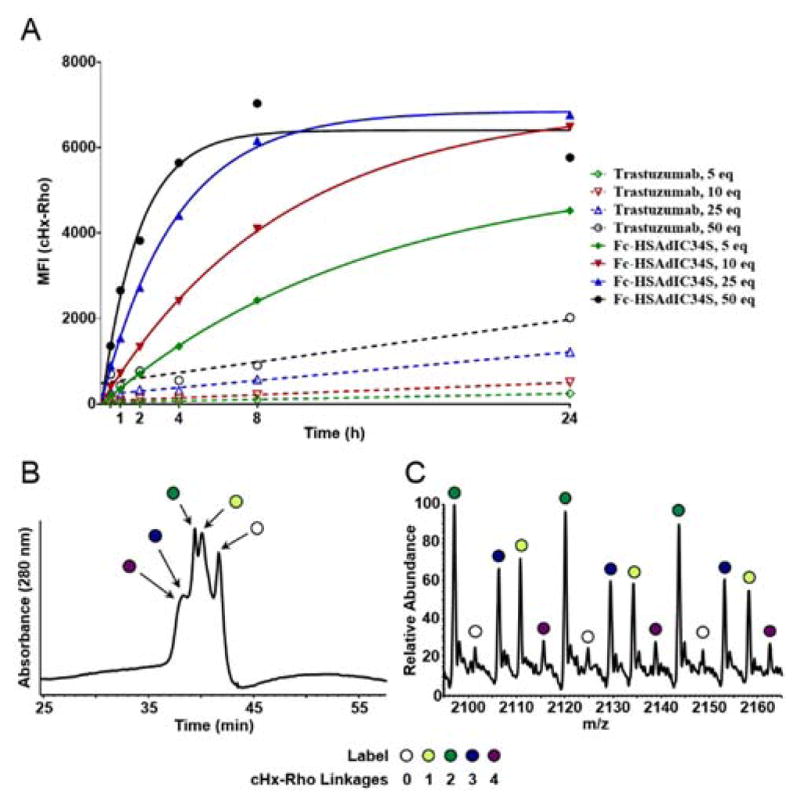
Antibody fusion conjugate analysis. (A) Kinetics of cHx-Rho labeling for trastuzumab and Fc-HSAdIC34S by flow cytometry. Conjugations were conducted using 5, 10, 25, and 50 equivalents of cHx-Rho. After 0.5, 1, 2, 4, 8, and 24 h, reactions were quenched and analyzed by HER2-dependent staining of SKBR-3 cells using cHx-Rho fluorescence detection and fitted to pseudo-first order association kinetics. (B–C) HIC-HPLC (left) and ESI-HRMS spectrum (right) of Fc-HSAdIC34S after treatment with 10 equivalents of cHx-Rho for 24 h.
Nevertheless, some trastuzumab labeling was detected indicating minor non-specific conjugation with cHx-Rho, particularly at high equivalents and long incubation periods (Figure 3A). Such non-specific labeling (e.g. other lysine residues and/or the N-termini) was not completely unexpected, as we had previously observed slight over-conjugation with full-length HSA, albeit to a lesser degree given the faster labeling kinetics.15 Examination of the 10 equivalents cHx-Rho at 24 h condition by HIC-HPLC revealed +2 and +1 as the dominant species (Figure 3B). A minor amount of unmodified and +3/+4 species was also detected, consistent with our observations with HSAdIC34S-MBP (Figure S2). Here, high equivalents and extended conjugation times also favored over-conjugation of the antibody fusion (Figure S7). These products were confirmed by ESI-HRMS (Figure 3C, Figure S8, Figure S9, and Table S4). Conjugation with more hydrophobic payloads (e.g. maytansines, auristatins, or pyrrolobenzodiazepines commonly used in ADCs) is anticipated to provide sufficient resolution by HIC to readily purify homogenous antibody fusion conjugates.
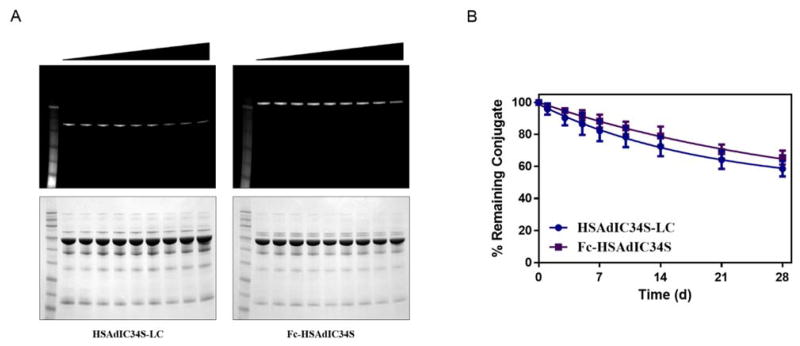
Lastly, plasma stability was examined for the HSAdIC34S-LC and Fc-HSAdIC34S cHx-Rho conjugates. In particular, the efficacy and therapeutic window of ADCs in preclinical mouse models was directly related to the stability of the conjugate,25 emphasizing the importance of the conjugation strategy. HSAdIC34S fusion conjugate stability was evaluated in human plasma over 28 days (Figure 4). Both HSAdIC34S-LC and Fc-HSAdIC34S retained the majority (~2/3) of their fluorescent conjugate during the 4 week incubation. Stability of these antibody fusion conjugates is similar to that of site-specific THIOMAB conjugates.25,26 In fact, comparable stability has been reported for HC-A114C THIOMAB drug conjugates, which were also shown to have improved therapeutic index to conventionally produced heterogeneous ADCs.27 Therefore, HSAdIC34S antibody fusion proteins provide a viable strategy for producing serum-stable antibody conjugates.
Figure 4.
Stability of trastuzumab-HSAdIC34S proteins conjugated to cHx-Rho in human plasma. Time-points were taken at T = 0, 1, 3 5, 7, 10, 14, 21, and 28 days. (A) Fluorescent (top) and Coomassie-stained (bottom) gel of HSAdIC34S-LC and Fc-HSAdIC34S conjugates in plasma. (B) Quantification of remaining antibody conjugate.
Conclusions
A fusion protein derived from HSA domain I has been developed for antibody conjugation. The preferential reactivity of our cyclohexene sulfonamide compound with Lys64 has made it possible to minimize the protein while still maintaining the ability to be modified. We have bioorthogonally labeled MBP fusion proteins as well as both N- and C-terminal trastuzumab fusion proteins of the light and heavy chains, respectively, with cHx-Rho. Trastuzumab-HSAdIC34S proteins and their corresponding conjugates bound HER2 comparably relative to trastuzumab. Kinetic analysis identified conditions that preferentially modify the HSAdIC34S fusion partner. Furthermore, antibody conjugates were also stable in human plasma. Fusions to other proteins should similarly allow installation of synthetic molecules to provide stable conjugates.
Cyclohexene sulfonamide compounds containing diverse chemical moieties are anticipated to impart new functionalities to HSAdI fusions. One significant advantage of this fusion protein is the ability to site-selectively modify lysine instead of using non-specific lysine labeling strategies such as N-hydroxysuccinimide esters. Additionally, we have previously shown that HSA can be simultaneously labeled with the cyclohexene sulfonamide compound at Lys64 and with a methylsulfonyl phenyloxadiazole compound at Cys34,15 suggesting that the HSAdI fusion protein could be even further modified in a site-specific and bioorthogonal manner. Given the scope of potential uses for this fusion partner, we expect that our conjugation method will provide a versatile platform for generating novel therapeutic conjugates.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (DP1CA174426 to C. F. B. III, U01 CA174844 to C. R. and W. R. R., and R01 CA181258 to C. R.). We thank Shannon Sirk for graciously cloning the trastuzumab expression vectors that enabled construction of the antibody fusion vectors used in this work and Pablo Martinez Acedo (Proteomics and Mass Spectrometry Core, The Scripps Research Institute, Scripps Florida) for supporting the mass spectrometry analyses.
Footnotes
Notes
The authors declare no competing financial interests.
Experimental Procedures; Supplementary Tables; Supplementary Figures. HSA domain sequences; MBP fusion labeling; antibody, antibody fusion, and antibody fusion conjugate binding by flow cytometry; Fc-HSAdIC34S labeling kinetics; hydrophobic interaction chromatography; mass spectrometry analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hamblett KJ, Senter PD, Chace DF, Sun MM, Lenox J, Cerveny CG, Kissler KM, Bernhardt SX, Kopcha AK, Zabinski RF, Meyer DL, Francisco JA. Effects of drug loading on the antitumor activity of a monoclonal antibody drug conjugate. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:7063–70. doi: 10.1158/1078-0432.CCR-04-0789. [DOI] [PubMed] [Google Scholar]
- 2.Boswell CA, Mundo EE, Zhang C, Bumbaca D, Valle NR, Kozak KR, Fourie A, Chuh J, Koppada N, Saad O, Gill H, Shen BQ, Rubinfeld B, Tibbitts J, Kaur S, Theil FP, Fielder PJ, Khawli LA, Lin K. Impact of drug conjugation on pharmacokinetics and tissue distribution of anti-STEAP1 antibody-drug conjugates in rats. Bioconjugate chemistry. 2011;22:1994–2004. doi: 10.1021/bc200212a. [DOI] [PubMed] [Google Scholar]
- 3.Jeffrey SC, Burke PJ, Lyon RP, Meyer DW, Sussman D, Anderson M, Hunter JH, Leiske CI, Miyamoto JB, Nicholas ND, Okeley NM, Sanderson RJ, Stone IJ, Zeng W, Gregson SJ, Masterson L, Tiberghien AC, Howard PW, Thurston DE, Law CL, Senter PD. A potent anti-CD70 antibody-drug conjugate combining a dimeric pyrrolobenzodiazepine drug with site-specific conjugation technology. Bioconjugate chemistry. 2013;24:1256–63. doi: 10.1021/bc400217g. [DOI] [PubMed] [Google Scholar]
- 4.Agarwal P, Bertozzi CR. Site-specific antibody-drug conjugates: the nexus of bioorthogonal chemistry, protein engineering, and drug development. Bioconjugate chemistry. 2015;26:176–92. doi: 10.1021/bc5004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panowksi S, Bhakta S, Raab H, Polakis P, Junutula JR. Site-specific antibody drug conjugates for cancer therapy. mAbs. 2014;6:34–45. doi: 10.4161/mabs.27022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dengl S, Sustmann C, Brinkmann U. Engineered hapten-binding antibody derivatives for modulation of pharmacokinetic properties of small molecules and targeted payload delivery. Immunological reviews. 2016;270:165–77. doi: 10.1111/imr.12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McEnaney PJ, Parker CG, Zhang AX, Spiegel DA. Antibody-recruiting molecules: an emerging paradigm for engaging immune function in treating human disease. ACS chemical biology. 2012;7:1139–51. doi: 10.1021/cb300119g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dengl S, Hoffmann E, Grote M, Wagner C, Mundigl O, Georges G, Thorey I, Stubenrauch KG, Bujotzek A, Josel HP, Dziadek S, Benz J, Brinkmann U. Hapten-directed spontaneous disulfide shuffling: a universal technology for site-directed covalent coupling of payloads to antibodies. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2015;29:1763–79. doi: 10.1096/fj.14-263665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka F, Fuller R, Asawapornmongkol L, Warsinke A, Gobuty S, Barbas CF., 3rd Development of a small peptide tag for covalent labeling of proteins. Bioconjugate chemistry. 2007;18:1318–24. doi: 10.1021/bc070080x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witus LS, Moore T, Thuronyi BW, Esser-Kahn AP, Scheck RA, Iavarone AT, Francis MB. Identification of highly reactive sequences for PLP-mediated bioconjugation using a combinatorial peptide library. Journal of the American Chemical Society. 2010;132:16812–7. doi: 10.1021/ja105429n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheck RA, Francis MB. Regioselective labeling of antibodies through N-terminal transamination. ACS chemical biology. 2007;2:247–51. doi: 10.1021/cb6003959. [DOI] [PubMed] [Google Scholar]
- 12.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS chemical biology. 2008;3:373–82. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 13.Juillerat A, Gronemeyer T, Keppler A, Gendreizig S, Pick H, Vogel H, Johnsson K. Directed evolution of O6-alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo. Chemistry & biology. 2003;10:313–7. doi: 10.1016/s1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- 14.Gautier A, Juillerat A, Heinis C, Correa IR, Jr, Kindermann M, Beaufils F, Johnsson K. An engineered protein tag for multiprotein labeling in living cells. Chemistry & biology. 2008;15:128–36. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Asano S, Patterson JT, Gaj T, Barbas CF., 3rd Site-selective labeling of a lysine residue in human serum albumin. Angewandte Chemie (International ed in English) 2014;53:11783–6. doi: 10.1002/anie.201405924. [DOI] [PubMed] [Google Scholar]
- 16.Takashima K, Matsunaga N, Yoshimatsu M, Hazeki K, Kaisho T, Uekata M, Hazeki O, Akira S, Iizawa Y, Ii M. Analysis of binding site for the novel small-molecule TLR4 signal transduction inhibitor TAK-242 and its therapeutic effect on mouse sepsis model. British journal of pharmacology. 2009;157:1250–62. doi: 10.1111/j.1476-5381.2009.00297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada M, Ichikawa T, Ii M, Sunamoto M, Itoh K, Tamura N, Kitazaki T. Discovery of novel and potent small-molecule inhibitors of NO and cytokine production as antisepsis agents: synthesis and biological activity of alkyl 6-(N-substituted sulfamoyl)cyclohex-1-ene-1-carboxylate. Journal of medicinal chemistry. 2005;48:7457–67. doi: 10.1021/jm050623t. [DOI] [PubMed] [Google Scholar]
- 18.Curry S, Mandelkow H, Brick P, Franks N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nature structural biology. 1998;5:827–35. doi: 10.1038/1869. [DOI] [PubMed] [Google Scholar]
- 19.LaFleur DW, Abramyan D, Kanakaraj P, Smith RG, Shah RR, Wang G, Yao XT, Kankanala S, Boyd E, Zaritskaya L, Nam V, Puffer BA, Buasen P, Kaithamana S, Burnette AF, Krishnamurthy R, Patel D, Roschke VV, Kiener PA, Hilbert DM, Barbas CF., 3rd Monoclonal antibody therapeutics with up to five specificities: functional enhancement through fusion of target-specific peptides. mAbs. 2013;5:208–18. doi: 10.4161/mabs.23043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters C, Brown S. Antibody-drug conjugates as novel anti-cancer chemotherapeutics. Biosci Rep. 2015:35. doi: 10.1042/BSR20150089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wendeler M, Grinberg L, Wang X, Dawson PE, Baca M. Enhanced catalysis of oxime-based bioconjugations by substituted anilines. Bioconjugate chemistry. 2014;25:93–101. doi: 10.1021/bc400380f. [DOI] [PubMed] [Google Scholar]
- 22.Dirksen A, Dawson PE. Rapid oxime and hydrazone ligations with aromatic aldehydes for biomolecular labeling. Bioconjugate chemistry. 2008;19:2543–8. doi: 10.1021/bc800310p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, Wang RE, Wang F. Antibody-drug conjugates for non-oncological indications. Expert opinion on biological therapy. 2016;16:591–3. doi: 10.1517/14712598.2016.1161753. [DOI] [PubMed] [Google Scholar]
- 24.Hallam TJ, Wold E, Wahl A, Smider VV. Antibody conjugates with unnatural amino acids. Molecular pharmaceutics. 2015;12:1848–62. doi: 10.1021/acs.molpharmaceut.5b00082. [DOI] [PubMed] [Google Scholar]
- 25.Shen BQ, Xu K, Liu L, Raab H, Bhakta S, Kenrick M, Parsons-Reponte KL, Tien J, Yu SF, Mai E, Li D, Tibbitts J, Baudys J, Saad OM, Scales SJ, McDonald PJ, Hass PE, Eigenbrot C, Nguyen T, Solis WA, Fuji RN, Flagella KM, Patel D, Spencer SD, Khawli LA, Ebens A, Wong WL, Vandlen R, Kaur S, Sliwkowski MX, Scheller RH, Polakis P, Junutula JR. Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nature biotechnology. 2012;30:184–9. doi: 10.1038/nbt.2108. [DOI] [PubMed] [Google Scholar]
- 26.Patterson JT, Asano S, Li X, Rader C, Barbas CF., 3rd Improving the serum stability of site-specific antibody conjugates with sulfone linkers. Bioconjugate chemistry. 2014;25:1402–7. doi: 10.1021/bc500276m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, Chen Y, Simpson M, Tsai SP, Dennis MS, Lu Y, Meng YG, Ng C, Yang J, Lee CC, Duenas E, Gorrell J, Katta V, Kim A, McDorman K, Flagella K, Venook R, Ross S, Spencer SD, Lee Wong W, Lowman HB, Vandlen R, Sliwkowski MX, Scheller RH, Polakis P, Mallet W. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nature biotechnology. 2008;26:925–32. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.