ABSTRACT
HuR influences gene expression programs and hence cellular phenotypes by binding to hundreds of coding and noncoding linear RNAs. However, whether HuR binds to circular RNAs (circRNAs) and impacts on their function is unknown. Here, we have identified en masse circRNAs binding HuR in human cervical carcinoma HeLa cells. One of the most prominent HuR target circRNAs was hsa_circ_0031288, renamed CircPABPN1 as it arises from the PABPN1 pre-mRNA. Further analysis revealed that HuR did not influence CircPABPN1 abundance; interestingly, however, high levels of CircPABPN1 suppressed HuR binding to PABPN1 mRNA. Evaluation of PABPN1 mRNA polysomes indicated that PABPN1 translation was modulated positively by HuR and hence negatively by CircPABPN1. We propose that the extensive binding of CircPABPN1 to HuR prevents HuR binding to PABPN1 mRNA and lowers PABPN1 translation, providing the first example of competition between a circRNA and its cognate mRNA for an RBP that affects translation.
KEYWORDS: Cell proliferation, circular RNA, circPABPN1, endogenous competing RNA, HuR, PABPN1, RNA-binding Protein, translation
Introduction
HuR is an extensively studied RNA-binding protein (RBP) that regulates protein expression patterns by associating with a wide range of RNAs.1,2 The best-known HuR targets are mRNAs such as those that encode TP53, VHL, MYOD, MYC, CCND1 (cyclin D1), CDKN1A (p21), HIF1A, and BCL2; HuR typically binds to U-rich stretches within their 3′-untranslated regions (UTRs).1-3 HuR often increases the half-life of a target mRNA, but it can also lower the stability of other target mRNAs, and similarly it can promote the translation of some mRNAs and suppress the translation of others.4 In addition, it can modulate the splicing of several target pre-mRNAs and enhance the nuclear export of some mRNAs (reviewed in Ref. 5). HuR can also interact with several noncoding (nc)RNAs. It was found to associate with the microRNA let-7,6,7 although the consequences of this interaction are unclear, and with several long noncoding (lnc)RNAs, including LINCRNAP21, HOTAIR, and RMRP, influencing their stability or subcellular localization.6,8,9 However, whether or not HuR binds to circular RNAs (circRNAs) and the functional consequences of these interactions have not been studied.
Circular RNAs had been observed for several decades, but their high abundance and function have only begun to be appreciated with the advent of new sequencing and molecular biology techniques. These heterogeneous ncRNAs are often expressed in a tissue-specific manner and generally arise from the canonical spliceosomal machinery through head-to-tail backsplicing.10-13 Several studies have shown that circRNAs can interact with and sponge microRNAs. For example, the circRNA CiRS-7 contains multiple sites for miR-7, enabling it to sequester miR-7 and thereby decreasing its availability to target mRNAs bearing miR-7 sites; likewise, circSRY sponges miR-138, and circITCH sponges miR-7, miR-17, and miR-214.10,12,14,15 These examples indicate that circRNAs can suppress the impact of microRNAs as negative regulators of mRNA stability or translation. Besides microRNAs, circRNAs can also bind RBPs, regulate their availability in the cell, and influence the post-transcriptional fates (e.g., stability or translation) of RBP-interacting mRNAs.16,17
In this study, we sought to investigate systematically HuR-containing circRNA ribonucleoprotein complexes (circRNPs). First, we employed RNP immunoprecipitation (RIP) analysis followed by identification of circRNAs using microarrays, and second, we utilized bioinformatic analysis of HuR-interacting RNAs from HuR crosslinking immunoprecipitation (CLIP) data sets transcriptome-wide. One of the top HuR target circRNAs was examined in depth. As this circRNA was derived from PABPN1 mRNA, itself a target of HuR, it was renamed CircPABPN1. HuR did not affect CircPABPN1 levels, but, unexpectedly, CircPABPN1 significantly suppressed HuR binding to PABPN1 mRNA and inhibited PABPN1 translation. This regulatory paradigm is the first example of a circRNA (CircPABPN1) that modulates translation of its cognate mRNA (PABPN1 mRNA) by competing with and thereby reducing the availability of a translational activator (HuR).
Results
Identification of HuR target circRNAs
To investigate HuR-circRNA interactions, we first isolated RNP complexes containing HuR from human cervical carcinoma HeLa cells. Isolation was carried out by RIP (RNP immunoprecipitation) analysis using an anti-HuR antibody in conditions that preserved endogenous HuR-RNA complexes. Control IgG IP samples were processed in parallel. The RNA isolated from RIP complexes was treated with RNase R to degrade all linear RNA molecules, and the remaining RNA was used for reverse transcription (RT) and circular RNA microarray analysis (Fig. 1A). The specificity of the IP was assessed by Western blot analysis, which revealed a unique HuR band detected in HuR IP, but not in IgG IP (Fig. 1B). Microarray analysis after RIP (RIP-chip) identified the circRNAs associated with HuR as those enriched in HuR IP relative to those found in control IgG IP. A partial list of circRNAs that were most highly enriched in HuR IP is shown (Fig. 1C); the list of all HuR target circRNAs identified by RIP-chip is shown in Table S1. These data indicate that several circRNAs were enriched in HuR IP samples, supporting the existence of HuR-circRNA complexes.
Figure 1.

Transcriptome-wide identification of HuR-associated circRNAs. (A,B) Schematic of the strategy used to identify globally HuR-interacting circRNAs in HeLa cells (A). Following cell lysis, RIP analysis was carried out using anti-HuR or control IgG antibodies; the presence of HuR in the IP samples was assessed by Western blot analysis [(B); HC, heavy IgG chain]. Total RNA was isolated and digested with RNase R, and circRNAs present in the sample were identified using circRNA microarrays (Arraystar). (C) Partial list of circRNAs highly enriched in HuR IP relative to IgG IP as identified in microarrays (n = 3).
HuR associates with CircPABPN1
The interactions detected by RIP-chip analysis were verified by RIP followed by RT-qPCR analysis in HeLa cells (Fig. 2A). The circRNA hsa_circ_0031288 was one of the most highly enriched HuR targets (Fig. 2A). This circRNA originates from Poly(A)-binding protein nuclear 1 (PABPN1) pre-mRNA and thus we named it CircPABPN1 (Fig. S1A). Further analyses included specific CircPABPN1 RT-qPCR amplification, visualization on agarose gels (with no amplification in -RT reactions, Fig. 2B), and sequencing of the amplified PCR product to verify the specific circRNA junction (Fig. 2C). Digestion of total RNA with RNase R to degrade linear RNAs followed by RT-qPCR analysis indicated that CircPABPN1 was protected from RNase R digestion, while GAPDH and PABPN1 mRNAs were degraded (Fig. 2D).
Figure 2.

Validation of HuR target circRNAs. (A) RT-qPCR analysis of HeLa circRNAs enriched in HuR IP samples compared with IgG samples; RNA levels were normalized to the background levels of GAPDH mRNA in each IP sample. (B) Example of PCR product validation; hsa_circ_0031288(CircPABPN1) was visualized on ethidium bromide-stained agarose gels to confirm the specificity of the circRNA amplification band (RT, reverse transcription). (C) Validation of circRNA PCR product from (B) by DNA sequencing analysis. All of the circRNAs in (A) were verified on agarose gels and many were further verified by sequencing. (D) RT-qPCR analysis to measure GAPDH mRNA, PABPN1 mRNA, and hsa_circ_0031288 (CircPABPN1) with or without RNase R treatment; RNA levels in RNase R-treated samples were compared with mock (untreated) samples (using identical input RNA amounts). (E) Western blot analysis of HuR levels in the samples pulled down using control biotinylated GAPDH (3′UTR) and linear biotinylated CircPABPN1. ‘Input’, HuR levels in the samples used for pulldown analysis. (F) Western blot analysis of HuR levels in pulldown assays using a biotinylated antisense oligomer targeting the junction of CircPABPN1 or a control biotinylated oligomer. Data in (A,D) are the means and ± SEM from three independent experiments. Data in (B,E,F) are representative of three independent experiments.
We further confirmed the interaction of HuR and CircPABPN1 by using biotin pulldown analyses. First, we generated linear biotinylated segments of CircPABPN1, incubated them with HeLa whole-cell lysates, and used streptavidin-coated beads to identify proteins bound to the biotinylated RNA. The biotinylated CircPABPN1 segment associated with HuR, while biotinylated GAPDH RNA segments did not (Fig. 2E). Second, pulldown analysis of the endogenous CircPABPN1 using antisense biotinylated oligomers complementary to the junction sequence of CircPABPN1 was followed by pulldown using streptavidin beads and Western blot analysis; HuR was detected in CircPABPN1 pulldown, but not in control pulldown samples (Fig. 2F).
Taken together, these data indicate that endogenous HuR can bind to cellular circRNAs and reveal strong binding of CircPABPN1 to HuR, as determined using microarrays and RT-qPCR analyses, sequencing of circRNA junctions, and biotinylated-RNA pulldown analysis.
circRNA-wide mapping of HuR binding sites
Although HuR associated with many circRNAs, we were only able to investigate a small subset of circRNAs given the limited set of probes on the array platform (∼5,000), while the whole circRNA-ome is believed to consist of >100,000 circRNAs (>15,000 in HeLa cells), according to circBase.18 Thus, we sought to expand our search for HuR target circRNAs by conducting a transcriptome-wide analysis. We utilized publicly available CLIP data sets of HuR binding sites (Fig. 3A).1,2,19 Since CLIP data sets do not distinguish HuR binding sites present in linear RNA from those in circular RNAs, we created a comprehensive computational map of HuR binding sites on circRNAs. This map showed that HuR can potentially bind to a large number of circRNAs reported in circBase: ∼78,845 (∼56%) of circRNAs annotated, including 455 circRNAs that can be targeted at both the junction and outside of the junction, and 95 circRNAs that can be targeted only at the junction (Fig. 3B). The complete list of potential HuR target circRNAs is shown in Table S2. A subset of HuR target circRNAs was validated by RT-qPCR analysis employing specific circRNA primers. Several transcripts were highly enriched in HuR IP compared to IgG IP samples (Fig. 3C), including a subset of potential HuR targets predicted to have binding sites on or near the junction of the circRNA (Fig. 3D). These data indicate that HuR target circRNAs can be identified from pre-existing data sets.
Figure 3.
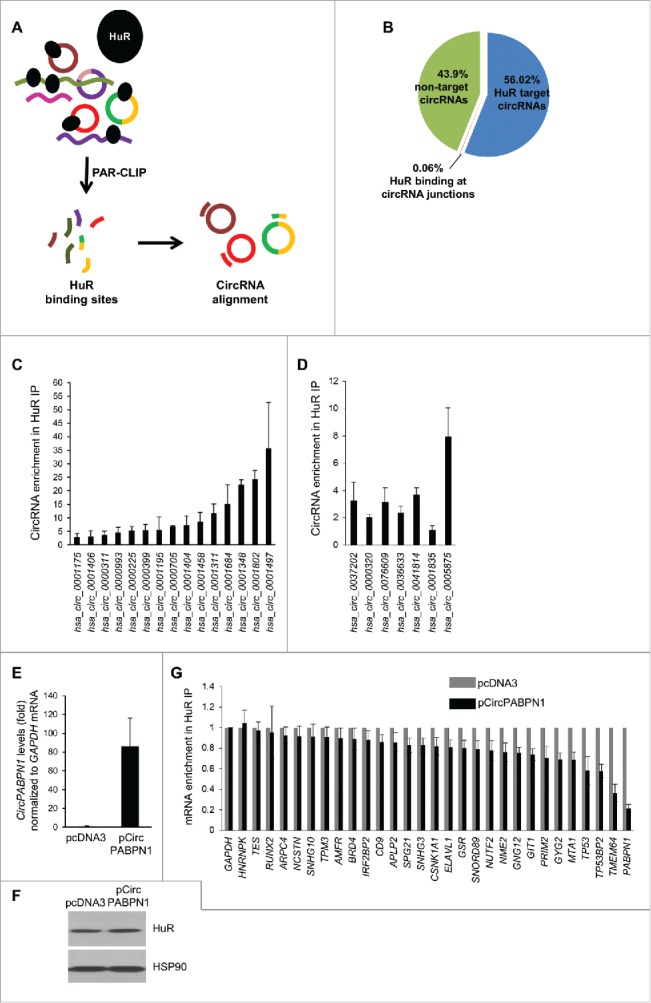
Transcriptome-wide identification and validation of HuR target circRNAs. (A) Schematic representation of the strategy used to identify globally HuR-interacting circRNAs using publicly available CLIP data sets (Methods). (B) Among the annotated circRNAs (CircBase, Aug 4 2015), ∼56% potentially contained HuR target sites, 44% did not, and for <1% HuR binding sites were at the junction. Among the putative target, most HuR binding sites were found within the body, and a few spanned the junction. (C, D) Partial validation of HuR target circRNAs found in panel (B) by RIP (HuR IP relative to IgG IP) followed by RT-qPCR analysis; circRNAs with HuR sites in the body (C) and the junction (D) were studied. (E-G) HeLa cells were transfected with either pcDNA3 or with pCircPABPN1; 48 h later, the levels of CircPABPN1 were assessed by RT-qPCR analysis (normalized to GAPDH mRNA) (E), the levels of HuR by Western blot analysis (loading control HSP90) (F), and the interaction of HuR with target mRNAs by RIP followed by RT-qPCR analysis (normalized to GAPDH mRNA) (G). Data in (C-E,G) represent the means ± SEM from three independent experiments; data in (F) are representative of three repeats.
We investigated if HuR globally affected the levels of target circRNAs by silencing HuR in HeLa cells and examining changes in the abundance of circRNA levels by RT-qPCR analysis of a subset of circRNAs. As shown in Fig. S2, HuR silencing did not generally alter the levels of target circRNAs Fig. S2, although a few circRNAs were slightly less abundant (e.g., hsa_circ_0001406 and hsa_circ_0001404) and others more abundant (e.g., hsa_circ_0005875) (Fig. S2C, D). These results suggest that HuR abundance may not broadly affect target circRNA levels.
CircPABPN1 suppresses HuR binding to PABPN1 mRNA and lowers PABPN1 production
To investigate the impact of HuR on a specific target circRNA, we focused on CircPABPN1, which showed the most robust interaction with HuR in validation experiments (Fig. 2). In agreement with the binding results (Fig. 2A,E,F), RIP followed by sequencing analysis (downloaded from the UCSC browser, Supplemental text) identified a large number of binding tags/sites spanning the single exon which forms CircPABPN1 (Fig. S1B-D). First, we asked if HuR influenced CircPABPN1 levels and found that silencing HuR did not affect CircPABPN1 abundance (Fig. S2B). We then asked the converse question, whether CircPABPN1 had an impact on HuR levels or function. We overexpressed CircPABPN1 (Methods), verified the increase in CircPABPN1 levels in cells by RT-qPCR analysis (Fig. 3E), and observed that this intervention did not modify HuR levels (Fig. 3F). However, HuR binding to several target mRNAs did decline in cells that overexpressed CircPABPN1, as assessed by RIP followed by RT-qPCR analysis. Interestingly, among the target mRNAs examined (chosen from HuR RIP-chip analysis20) HuR binding to PABPN1 mRNA was the most strikingly impaired interaction (Fig. 3G). In fact, PABPN1 mRNA is one of the major targets of HuR, as determined by RIP followed by RT-qPCR analysis (Fig. S3).
Given that CircPABPN1 was capable of suppressing HuR binding to PABPN1 mRNA, we hypothesized that CircPABPN1 might affect the cellular levels of PABPN1. To investigate this possibility, we first tested if HuR regulates PABPN1 abundance. As shown (Fig. 4A, left), silencing HuR in HeLa cells led to a potent reduction in PABPN1 levels, as shown by Western blot analysis. We then tested the effect of overexpressing CircPABPN1 on the levels of PABPN1; as shown (Fig. 4A, right), this intervention also reduced PABPN1 abundance. Together, these data suggested that CircPABPN1 suppressed HuR binding to PABPN1 mRNA, consequently lowering PABPN1 levels.
Figure 4.

CircPABPN1 suppresses PABPN1 translation. (A) 48 h after transfecting HeLa cells with either control siRNA or HuR siRNA (left) or with pcDNA3 or pCircPABPN1 (right), the levels of PABPN1, HuR, and the loading control HSP90 were assessed by Western blot analysis. Following quantification of the bands on Western blots, the relative signal intensities were represented as the means ± SEM from three independent experiments. *, P <0.05 (Student's t-test)., (B, C) HeLa cells prepared as in (A) were size-separated through sucrose gradients into 12 fractions (arrow, direction of sedimentation). Unbound RNA was in fractions 1 and 2; 40S, 60S, and 80S were in fractions 3–5; and low- and high-molecular-weight polysomes (LMWP and HMWP) were in fractions 6–8 and 9–12, respectively (B right, C right) and Methods. After isolating RNA from each fraction, the relative distribution (%) of PABPN1 and GAPDH mRNAs on the sucrose gradients was quantified by RT-qPCR analysis (B left, C left). (D) Proposed model whereby CircPABPN1 sequesters HuR away from PABPN1 mRNA, in turn suppressing PABPN1 mRNA translation.
CircPABPN1 suppresses PABPN1 mRNA translation
To determine the mechanism through which CircPABPN1 elicited the HuR-dependent control of PABPN1 production, we tested PABPN1 mRNA levels and translation after overexpressing CircPABPN1 and after silencing HuR. Given that neither intervention influenced PABPN1 mRNA levels significantly (Fig. S4A), we postulated that CircPABPN1 and HuR might influence PABPN1 mRNA translation. We tested this possibility by analyzing the sizes of PABPN1 mRNA polysomes in cells expressing different levels of HuR and CircPABPN1. Representative sucrose gradient profiles (Fig. 4B right, Fig. 4C right) indicated that neither CircPABPN1 overexpression nor HuR silencing affected the global distribution pattern of polysomes. RNA was isolated from each fraction – nonpolysomal fractions (1 and 2), fractions containing ribosomal subunits and monosomes (3 and 4), and fractions containing low- (5–8) and high-molecular-weight (9–11) polysomes (LMW and HMW polysomes, respectively). After RT, the relative distribution of PABPN1 mRNA was measured by qPCR analysis. To control for overall translation, the levels of GAPDH mRNA, encoding a housekeeping protein, were also calculated from the same fractions.
As shown in Fig. 4B (left) and Fig. 4C (left), PABPN1 mRNA levels were low in the non- and low-translating part of the gradient (fractions 1–5), and was abundant in the polysomal fractions, indicating that PABPN1 mRNA was actively translated. Importantly, overexpression of CircPABPN1 caused a leftward shift in the distribution of PABPN1 mRNA, indicating that PABPN1 mRNA associated with smaller polysomes or with no polysomal components, and suggesting that CircPABPN1 suppressed the translation of PABPN1 (Fig. 4B, left). Likewise, silencing HuR caused a leftward shift of PABPN1 mRNA on the gradient (Fig. 4C, left), consistent with reduced PABPN1 translation. In both treatment groups, the distribution of the control GAPDH mRNA was essentially unchanged, supporting the view that the changes in PABPN1 translation were specifically affected by overexpressing CircPABPN1 and by silencing HuR (Fig. 4B,C). Together, these data support the notion that HuR binds to PABPN1 mRNA and promotes the translation of PABPN1, while CircPABPN1, which competes with PABPN1 mRNA for binding to HuR, suppresses PABPN1 translation.
In sum, HuR binds to circRNAs, as revealed by RIP-chip, bioinformatic, in vitro binding, and RT-qPCR analyses, but it does not seem to influence abundance of the target circRNAs tested. One specific HuR target, CircPABPN1, prevented HuR binding to its cognate transcript, PABPN1 mRNA, in turn leading to a suppression of PABPN1 translation (Fig. 4D).
Discussion
Recent surveys have found that RBPs can potentially bind numerous circRNAs.21,22 Here, we identified HuR-associated circRNAs using RIP-chip and RIP-RT-qPCR analyses, bioinformatic prediction, and in vitro binding assays (Figs. 1-3, Table S2). Other RBPs, including AUF1/HNRNPD1 and LIN28, may also form complexes similar to those identified for HuR, but such complexes await experimental confirmation.22 IMP3 (IGF2BP3) was recently reported to form circRNPs with a subset of circRNAs, further supporting the idea that these complexes are common in human cells.23
We hypothesized that HuR might influence circRNA levels since HuR alters the turnover of several coding and noncoding RNAs.2 However, HuR silencing did not affect the abundance of most target circRNAs (Fig. S2). We then considered the possibility that circRNAs might function like lncRNAs, which form RNP complexes (lncRNPs) that might modulate the functions of the proteins with which they interact. Some lncRNPs can affect gene expression at the transcriptional level; for example, lncRNA ANRIL represses p16INK4 transcription by interacting with the PRC2 complex at the INK4 locus,24 and lncRNA GAS5 binds glucocorticoid receptors (GCRs) and thus acts as a decoy to prevent the transcriptional activity of GCRs.25 Other lncRNPs can affect gene expression post-transcriptionally,26 including the DNA damage-inducible lncRNA gadd7, which associates with the RBP TDP-43 and interferes with the ability of TDP-43 to stabilize CDK6 mRNA,27 and LincRNA-RoR, which interacts with the RBP HNRNPI, an enhancer of TP53 production, and thus represses TP53 translation.28
Similar to these examples, HuR-circRNA complexes (HuR circRNPs) may also influence the function of HuR by altering its ability to bind other target RNAs. In this regard, HuR was found to interact extensively with circRNAs, as determined by RIP-chip and CLIP analyses. HuR binding to target mRNAs is modulated by HuR phosphorylation by kinases like CHEK2, PKC, and JAK3, and by methylation through CARM1.5 As these post-translational modifications affect the subcellular localization of HuR and its association with target RNAs, it will be important to investigate if they also affect the levels and function of HuR-circRNA complexes.
HuR function is also elicited by competing with or recruiting other trans-acting factors (such as RBPs, miRNAs, or lncRNAs) to target mRNAs.4,29,30 In a recent example, lncRNA OIP5-AS1/cyrano displayed strong affinity for HuR and prevented HuR binding to a subset of mRNAs encoding proliferative proteins, in turn reducing cell growth.31 However, in other cases, the ncRNA and the RBP may compete for binding to the shared target mRNA; for example, 7SL and HuR competed for binding to TP53 mRNA, and thus 7SL was capable of preventing the HuR-mediated promotion of TP53 translation.30 Given that the main HuR target identified in this study, CircPABPN1, is not very abundant (∼12 copies per cell in HeLa cells, Fig. S4B), it is not immediately apparent how it might seize the plentiful protein HuR (∼1,300 copies in the cytoplasm of HeLa cells) away from target mRNAs. This ability is likely explained in part by the fact that most of the body of CircPABPN1 (134 nt out of 152 nt) represents a continuous HuR target RNA (Fig. S1) thus potentially capable of accommodating dozens of HuR molecules on a single CircPABPN1 molecule. If one then considers that each HuR site on CircPABPN1 can allow binding of HuR multimers32,33 and that a fraction of cellular HuR may be phosphorylated at residues that inhibit HuR binding to RNA, CircPABPN1 might be able to associate with a sizeable pool of cytoplasmic HuR.
It remains unclear why CircPABPN1 overexpression reduces HuR-PABPN1 mRNA complexes selectively, while other mRNAs remain bound to HuR (Fig. 3G). We hypothesized that perhaps CircPABPN1 and PABPN1 mRNA compete for HuR during splicing, but at present there are no clear data supporting this idea. Instead, it is plausible that both CircPABPN1 and PABPN1 mRNA are in physical proximity in a specific subcellular compartment and maintain a balanced interaction with HuR locally. When the levels of CircPABPN1 rise in this hypothetical compartment, PABPN1 mRNA is selectively depleted of HuR. The cellular space and/or mediators of such regulatory paradigm are not yet known. As CircPABPN1 is expressed in transformed and untransformed cells from a broad range of cell types and tissues (not shown), this mechanism could potentially influence HuR function widely.
In summary, HuR binds numerous circRNAs in HeLa cells. The circRNA showing the most abundant association with HuR, CircPABPN1, suppressed HuR binding to PABPN1 mRNA without influencing HuR levels or PABPN1 mRNA levels. Interestingly, by preventing HuR binding to PABPN1 mRNA, CircPABPN1 suppressed PABPN1 translation and lowered cell proliferation (Fig. S4C). We propose that other circRNAs might affect selectively the expression of certain mRNAs at similar post-transcriptional levels.
Methods
Cell culture, cloning, and transfections
HeLa cells were cultured in Dulbecco's modified essential medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (Gibco), antibiotics and antimycotics (Life Technologies) and transfected with pcDNA3.0 or pCircPABPN1 (2 μg) or with control siRNAs using Lipofectamine-RNAiMAX or Lipofectamine 2000, and collected 48 to 72 h later. Cloning for CircPABPN1 overexpression was performed as described.13,15 Briefly, genomic DNA was isolated using DNeasy Blood & Tissue Kit (69504, Qiagen) and 25 ng of genomic DNA was used to amplify ∼500-bp genomic sequences upstream and downstream of the PABPN1 (NM_004643.3) exon 6 to generate hsa_circ_0031288 using primers ATATATAAGCTTACCTAAATGTCTTCAGAGGCCA and ATATATCTCGAGGACAGAAGTGAAGCAAGGCA. The PCR product was cloned into the HindIII and XhoI sites of pcDNA3.0. SiRNAs used in transfection were AATTCTCCGAACGTGTCACGT (control siRNA), and a mixture of 4 siRNAs targeting HuR mRNA: AAGAGGCAATTACCAGTTTCA, AAGTGCAAAGGGTTTGGCTTT, AATCTTAAGTTTCGTAAGTTA and TTCCTTTAAGATATATATTAA (Qiagen).
Western blot analysis
Whole-cell lysates were prepared as described,17 using RIPA buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS, 1 mM dithiothreitol) containing protease inhibitor. The protein lysates were separated through 4–20% Mini-PROTEAN® TGX™ Gel (Bio-Rad) and transferred to nitrocellulose membrane using Trans-Blot® Turbo™ Transfer System (Bio-Rad). Incubations with primary antibodies to detect PABPN1 (PABP2) (14154, Cell Signaling), HuR (sc-5261, Santa Cruz Biotech), HSP90 (sc-13119), and GAPDH (sc-32233) were followed by incubations with appropriate secondary antibodies conjugated with HRP (GE Healthcare). Signals were developed using Enhanced Chemiluminescence (ECL).
RNA isolation, RNase R digestion, sequencing, RIP analysis, and circRNA microarrays
Total RNA from HeLa cells was isolated using TRIzol (15596–026, Thermo Fisher Scientific) following the manufacturer's protocol. One µg total RNA was either left untreated (mock) or treated with 10 units of RNase R (RNR07250, Epicentre) in the presence of 1× RNase R buffer, 20 units of RiboLock RNase Inhibitor (Thermo Scientific), and incubated for 30 min at 37°C. The digested RNA was isolated using acid phenol-chloroform (5:1) and ethanol precipitation. Reverse transcription (RT) was performed following the manufacturer's protocol using Maxima Reverse Transcriptase (EP0741, Thermo Fisher Scientific), and 150 ng of random hexamers (11034731001, Roche). The reaction mixtures were incubated for 10 min at 25°C followed by 30 min at 50°C and 5 min at 85°C to inactivate the RT enzyme. For qPCR analysis, 0.1 µl of cDNA was used with 250 nM of gene-specific primers (Table S2) and KAPA SYBR® FAST qPCR Kits (ABI Prism) (KK4605, KAPA Biosystems). RT-qPCR analysis was performed on Applied Biosystems 7300, 7900 and QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific) with a cycle setup consisting of 3 min at 95°C and 40 cycles of 5 sec at 95°C plus 20 sec at 60°C. The relative expression levels were calculated after normalization to GAPDH mRNA using 2−ΔΔCt method. RT-qPCR product for hsa_circ_0031288 was size-separated in ethidium bromide-stained 2% agarose gels and visualized on an ultraviolet transilluminator.
Forward and reverse primers (Table S2) were used to sequence individually the amplified PCR products (MCLAB, USA). Divergent primers were designed using the CircInteractome tool.22 RT-qPCR products were visualized on agarose gels and select products were sequenced (MCLAB, USA). Endogenous circRNAs associated with HuR were identified by RIP analysis as described34 extracted using TRIzol, and identified using microarrays (Arraystar).
Polysome analysis
Polysome profiling was performed as previously described.17 Briefly, 72 h after transfection with Ctrl siRNA, HuR siRNA, pcDNA3 or pcDNA3_circ_0031288, HeLa cells were incubated with cycloheximide (Calbiochem; 100 μg/ml, 15 min) and lysed in PEB (polysome extraction buffer). After the lysate was separated through 10% to 50% sucrose gradients, 12 fractions were collected for further analysis. The distribution of GAPDH and PABPN1 mRNAs was quantified by RT-qPCR analysis and plotted as a percentage of the specific mRNA in each fraction relative to the total amount of that mRNA in the gradient.
HuR RIP analysis and identification of HuR binding sites on CircPABPN1
The association of HuR with endogenous circular RNAs in HeLa cells was analyzed by RIP analysis (immunoprecipitation (IP) of ribonucleoprotein (RNP) complexes), as described.34 Briefly, cytoplasmic lysates of HeLa cells were prepared in polysome extraction buffer (PEB; 20 mM Tris-HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2 and 0.5% NP-40) containing protease and RNase inhibitors. The supernatants were incubated with protein A sepharose beads coated with HuR antibody (sc-5261, Santa Cruz Biotech.) or control IgG (Santa Cruz Biotech.) antibodies for 2 h at 4°C. After three washes with ice-cold NT2 buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40), bound RNA was extracted from the beads using TRIzol and subjected to circRNA microarray analysis (Arraystar).
The CircInteractome tool22 was used to find HuR target circRNAs based on their sequence match with the HuR CLIP tags from previous publications1,2,19 (Table S3). HuR PAR-CLIP tags from mild MNase-treated samples19 were downloaded from GEO (GSM714639 and GSM714640) and UCSC (hg19) browser tracks were created from the raw sequenced tags (Fig. S1C).
Biotin pulldown assay
For antisense oligo pulldown of hsa_circ_0031288/circPABPN1, HeLa cells were lysed in PEB buffer containing protease inhibitors (Roche) and RNase inhibitor (Thermo Fisher). Lysates were incubated with 100 pmol of biotinylated oligomer TCGTTTTGGGATCACCTGTAGACGCGACCC complementary to the junction sequence of hsa_circ_0031288 in 1× TENT buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA at pH 8.0, 250 mM NaCl, 0.5% [v/v] Triton X-100) and protease and RNase inhibitors for 1 h at 25°C with rotation. Biotinylated oligomer GCTGGTAGAGGGAGCAGATG was used in control pulldown reactions. Streptavidin-coupled Dynabeads (50 µl; 11206D, Invitrogen) were added and incubated for 30 min at room temperature with rotation, and then complexes were isolated after three washes with ice-cold 1X TENT buffer. RNA was isolated using TRIzol, and HuR was detected in the pulldown by Western blot analysis.17
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell 2011; 43:340-52; PMID:21723171; http://dx.doi.org/ 10.1016/j.molcel.2011.06.008 [DOI] [PubMed] [Google Scholar]
- 2.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell 2011; 43:327-39; PMID:21723170; http://dx.doi.org/ 10.1016/j.molcel.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem 2008; 389:243-55; PMID:18177264; http://dx.doi.org/ 10.1515/BC.2008.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srikantan S, Gorospe M. HuR function in disease. Front Biosci (Landmark Ed) 2012; 17:189-205; PMID:22201738; http://dx.doi.org/ 10.2741/3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grammatikakis I, Abdelmohsen K, Gorospe M. Posttranslational control of HuR function.Wiley Interdiscip Rev RNA; 2017; 8(1); PMID: 27307117; http://dx.doi.org/23178169 10.1002/wrna.1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol 2013; 425:3723-30; PMID:23178169; http://dx.doi.org/ 10.1016/j.jmb.2012.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu YC, Chang SH, Hafner M, Li X, Tuschl T, Elemento O, Hla T. ELAVL1 modulates transcriptome-wide miRNA binding in murine macrophages. Cell Rep 2014; 9:2330-43; PMID:25533351; http://dx.doi.org/ 10.1016/j.celrep.2014.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012; 47:648-55; PMID:22841487; http://dx.doi.org/ 10.1016/j.molcel.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noh JH, Kim KM, Abdelmohsen K, Yoon JH, Panda AC, Munk R, Kim J, Curtis J, Moad CA, Wohler CM, et al.. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev 2016; 30:1224-39; PMID:27198227; http://dx.doi.org/ 10.1101/gad.276022.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384-8; PMID:23446346; http://dx.doi.org/ 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 11.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141-57; PMID:23249747; http://dx.doi.org/ 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333-8; PMID:23446348; http://dx.doi.org/ 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 13.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014; 56:55-66; PMID:25242144; http://dx.doi.org/ 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 14.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget 2015; 6:6001-13; PMID:25749389; http://dx.doi.org/ 10.18632/oncotarget.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al.. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016; 7:11215; PMID:27050392; http://dx.doi.org/ 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hentze MW, Preiss T. Circular RNAs: splicing's enigma variations. EMBO J 2013; 32:923-5; PMID:23463100; http://dx.doi.org/ 10.1038/emboj.2013.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panda AC, Abdelmohsen K, Martindale JL, Di Germanio C, Yang X, Grammatikakis I, Noh JH, Zhang Y, Lehrmann E, Dudekula DB, et al.. Novel RNA-binding activity of MYF5 enhances Ccnd1/Cyclin D1 mRNA translation during myogenesis. Nucleic Acids Res 2016; 44:2393-408; PMID:26819411; http://dx.doi.org/ 10.1093/nar/gkw023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014; 20:1666-70; PMID:25234927; http://dx.doi.org/ 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods 2011; 8:559-64; PMID:21572407; http://dx.doi.org/ 10.1038/nmeth.1608 [DOI] [PubMed] [Google Scholar]
- 20.López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA 2004; 101:2987-92; PMID:14981256; http://dx.doi.org/ 10.1073/pnas.0306453101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014; 42:D92-97; PMID:24297251; http://dx.doi.org/ 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteracto me: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 2016; 13:34-42; PMID:26669964; http://dx.doi.org/ 10.1080/15476286.2015.1128065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider T, Hung LH, Schreiner S, Starke S, Eckhof H, Rossbach O, Reich S, Medenbach J, Bindereif A. CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci Rep 2016; 6:31313; PMID:27510448; http://dx.doi.org/ 10.1038/srep31313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011; 30:1956-62; PMID:21151178; http://dx.doi.org/ 10.1038/onc.2010.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA Gas5 Is a Growth Arrest- and Starvation-Associated Repressor of the Glucocorticoid Receptor. Sci Signal 2010; 3; PMID:20124551; http://dx.doi.org/ 10.1126/scisignal.2000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, et al.. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun 2013; 4:2939; PMID:24326307; http://dx.doi.org/ 10.1038/ncomms3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Li D, Zhang W, Guo M, Zhan Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J 2012; 31:4415-27; PMID:23103768; http://dx.doi.org/ 10.1038/emboj.2012.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang A, Zhou NJ, Huang JG, Liu Q, Fukuda K, Ma D, Lu ZH, Bai CX, Watabe K, Mo YY. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013; 23:340-50; PMID:23208419; http://dx.doi.org/ 10.1038/cr.2012.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 2009; 23:1743-8; PMID:19574298; http://dx.doi.org/ 10.1101/gad.1812509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelmohsen K, Panda AC, Kang MJ, Guo R, Kim J, Grammatikakis I, Yoon JH, Dudekula DB, Noh JH, Yang X, et al.. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res 2014; 42:10099-111; PMID:25123665; http://dx.doi.org/ 10.1093/nar/gku686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Abdelmohsen K, Yang XL, De S, Grammatikakis I, Noh JH, Gorospe M. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res 2016; 44:2378-92; PMID:26819413; http://dx.doi.org/ 10.1093/nar/gkw017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toba G, White K. The third RNA recognition motif of Drosophila ELAV protein has a role in multimerization. Nucleic Acids Res 2008; 36:1390-9; PMID:18203745; http://dx.doi.org/ 10.1093/nar/gkm1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheiba RM, Ibáñez de Opakua A, Díaz-Quintana A, Cruz-Gallardo I, Martínez-Cruz LA, Martínez-Chantar ML, Blanco FJ, Díaz-Moreno I. The C-terminal RNA binding motif of HuR is a multi-functional domain leading to HuR oligomerization and binding to U-rich RNA targets. RNA Biol 2014; 11:1250-61; PMID:25584704; http://dx.doi.org/ 10.1080/15476286.2014.996069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penalva LO, Tenenbaum SA, Keene JD. Gene expression analysis of messenger RNP complexes. Methods Mol Biol 2004; 257:125-34; PMID:14770002; http://dx.doi.org/ 10.1385/1-59259-750-5:125 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


