ABSTRACT
Cellular RNAs with diverse chemical modifications have been observed, and N6-methyladenosine (m6A) is one of the most abundant internal modifications found on mRNA and non-coding RNAs, playing a vital role in diverse biologic processes. In humans, m6A modification is catalyzed by the METTL3-METTL14 methyltransferase complex, which is regulated by WTAP and another factor. Three groups have recently and independently reported the structure of this complex with or without cofactors. Here, we focus on the detailed mechanism of the m6A methyltransferase complex and the properties of each subunit. METTL3 is predominantly catalytic, with a function reminiscent of N6-adenine DNA methyltransferase systems, whereas METTL14 appears to be a pseudomethyltransferase that stabilizes METTL3 and contributes to target RNA recognition. The structural and biochemical characterization of the METTL3-METTL14 complex is a major step toward understanding the function of m6A modification and developing m6A-related therapies.
KEYWORDS: Epigenetics, m6A, methyltransferase, METTL3, WTAP
To date, approximately 140 types of chemical modification have been identified in RNA.1 Most modifications have been observed on rRNA and tRNA, whereas mRNA modification was considered rare. Nevertheless, several modifications have recently been identified on mRNA including N6-methyladenosine (m6A),2,3 N1-methyladenosine (m1A),4 inosine (I),5 5-methylcytosine (m5C),6 and pseudouridine (ψ).7,8 Among these, m6A is the most prevalent modification on mRNA and long noncoding RNA. In 2012, 2 groups independently identified thousands of m6A sites on mammalian RNAs,2,3 driving research examining the epitranscriptome. Several studies have characterized the m6A RNA landscape in organisms, including viruses,9,10 bacteria,11 yeast,12 and plants,13 and these studies have identified the consensus sequence RRACH (R represents purine, A is m6A and H is a non-guanine base), implying the functional importance of this modification. In fact, m6A affects multiple cellular functions,14,15 including developmental regulation, the cell cycle, fate determination,16,17 and the heat-shock stress response18 by affecting different stages of RNA metabolism such as RNA processing,19,20 stability,21 and translation efficiency.18,22,23
Analogous to dynamic chemical modifications of DNA and protein, the m6A RNA modification can be reversibly appended and removed by a methyltransferase and a demethylase (or “writer” and “eraser”), respectively (Fig. 1a). One m6A writer, the METTL3 methyltransferase (formerly called MT-A70), was first identified as part of a ∼200 kDa complex isolated from enzymatic mammalian cell nuclear extracts in 1997.24 METTL3 was grouped into the DNA m6A methyltransferase subfamily due to the conserved motif [D/N/S/H]PP[Y/F/W] (Fig. 1b) and exhibits high sequence conservation among eukaryotes including yeasts, plants, Drosophila and mammals.25 Importantly, disruption of METTL3 homologs causes severe developmental defects in yeasts and Drosophila and has a lethal phenotype in Arabidopsis and mice.16,26-28
Figure 1.
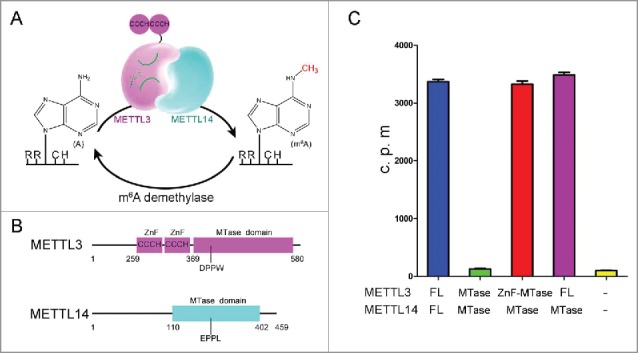
(A) Proposed model for reversible m6A methylation. m6A on RRACH is mainly appended by the METTL3-METTL14 complex, and oxidative demethylation is performed by m6A demethylase. CCCH: Cys-Cys-Cys-His type zinc finger motif (ZnF). (B) Schematic illustration of METTL3 and METTL14 domain structures. The METTL3 ZnF and METTL3 methyltransferase (abbreviation as MTase) domains are colored in magenta, and the METTL14 methyltransferase domain is colored in cyan. Detailed domain boundaries are labeled beneath the structures. (C) Comparison of the methyltransferase activity of full-length (abbreviation as FL) and truncated methyltransferase complexes. The c.p.m. of the extracted RNA was measured in a scintillation counter and averaged ( ± s.e.m.); the c.p.m. was determined from 3 replicates.
Recently, a second methyltransferase, METTL14, was identified as the other writer complex component.29-32 Knockdown of METTL14 leads to decreased m6A levels in human cell lines,30,32 and the METTL14 methyltransferase domain is phylogenetically close to that of METTL3.33 Interestingly, METTL3 and METTL14 were found to be associated in a global proteomic profiling and co-immunoprecipitation experiment.31,34 Consistent with in vivo observations, recombinant METTL3 and METTL14 form a stable heterodimer in vitro.29,32 Individually, METTL3 and METTL14 exhibit nearly undetectable methyltransferase activity, but the METTL3-METTL14 complex displays synergistic function. Why does the m6A methyltransferase complex contain 2 conserved methyltransferase components, and what are the roles of each subunit in the complex? We recently reported crystal structures of the METTL3-METTL14 methyltransferase domains complex alone and bound to S-adenosylmethionine (AdoMet) or S-adenosylhomocysteine (AdoHcy).35 Two other groups independently published nearly identical structures of the complex.36,37 On the basis of these structures, we suggest that METTL3 plays a catalytic role in the complex, whereas METTL14 is a pseudomethyltransferase that stabilizes METTL3 and contributes to RNA binding. Here, we focus on the detailed mechanism of the m6A methyltransferase complex and the properties of each component.
METTL3 primarily serves a catalytic role
The overall structures of both the METTL3 and METTL14 methyltransferase domains resemble dozens of class I DNA N6 methyltransferase via DALI analysis.38 Most of these contain one catalytic motif [D/N/S/H]PP[Y/F/W] located centrally in the methyltransferase domain.33 Interestingly, in the AdoMet-bound METTL3-METTL14 complex structure, a single AdoMet molecule was positioned in the catalytic pocket of METTL3 but was not observed in METTL14. The AdoMet molecule is adjacent to the most conserved DPPW motif and coordinated by Asp in this motif via a hydrogen-bonding interaction. The mutation that substitutes Asp with Ala completely abolished AdoMet binding and enzyme activity in vitro, reinforcing in vivo mutagenesis studies, in which a mutated METTL3 (DPPW replaced by APPA) altered circadian clock speed in mammalian cells.39 Additionally, an IME4 (METTL3 homolog in Saccharomyces cerevisiae) mutant encoding a D384A mutation displayed meiotic defects.28 Together with these data, we speculate that METTL3 primarily plays a catalytic role in the complex using a similar mechanism to DNA N6-methyladenine transfer of a methyl group to a target adenosine: the aromatic residue [Y/F/W] stacks with the target base via π–π interactions, and the side chain of the polar residue [D/N/S/H] and the carbonyl oxygen of the proline donate 2 hydrogen bonds to the 6-amino group of adenine, priming the SN2 chemical reaction by increasing its negative charge.
Sequence analysis indicated that the N-terminus of METTL3 contains 2 Cys-Cys-Cys-His (CCCH)-type zinc finger (ZnF) motifs common in RNA-binding proteins (Fig. 1b).33 The crystal structures of the METTL3-METTL14 complex contain only methyltransferase domains without the ZnF motifs, highlighting the flexibility of these regions. However, the crystallized truncation showed no detectable methyltransferase activity, whereas truncations containing ZnF motifs exhibited comparable activity to the full-length complex, suggesting a critical role for ZnF motifs (Fig. 1c). In contrast, deleting the N-terminal and C-terminal motifs of METTL14 had negligible effects. Several CCCH-type ZnF protein structures in complex with target RNAs have exhibited direct interactions between the ZnF motif and RNA.40,41 Accordingly, we hypothesized that the METTL3 ZnF motifs are necessary for methylation because they enhance interaction with substrate RNA. In the future, the appearance of these motifs and how they precisely recognize RNA sequences await to be investigated.
Is METTL14 a pseudomethyltransferase?
Although the methyltransferase domains of METTL3 and METTL14 share approximately 22% sequence identity and an almost identical topological structure, 3 pieces of data suggested that METTL14 is a pseudomethyltransferase in the complex. First, in the crystal structures, neither AdoMet nor AdoHcy is present in the METTL14 pocket. Second, the METTL3-METTL14 complex binds to the ligand in a 1:1 stoichiometric ratio as measured by isothermal titration calorimetry (ITC). Finally, there are moderately conserved EPPL sequences in METTL14 corresponding to the catalytic motif of METTL3, but the substitution of Glu to Ala had little effect on ligand binding and enzymatic activity.35-37 The structure of METTL14 offers a possible explanation. METTL14 superimposes well onto METTL3 except for 3 loops with distinct conformations, which are referred to as gate loop1 (residues 192–211 in METTL14), interface loop (residues 265–284 in METTL14) and gateloop2 (residues 318–328 in METTL14).(Fig. 2a) Gate loop1 and gateloop2 of METTL3 contribute to coordination with AdoMet, constituting part of the catalytic center. Interestingly, both METT14 gate loops are longer than those of METTL3. For METTL14, 7 residues of gate loop1 adopt a short helical conformation, resulting in a possible obstacle to ligand entry. (Fig. 2a) Gate loop 2 shows significant inward movement, which likely leads to closure of the ligand-binding pocket.
Figure 2.
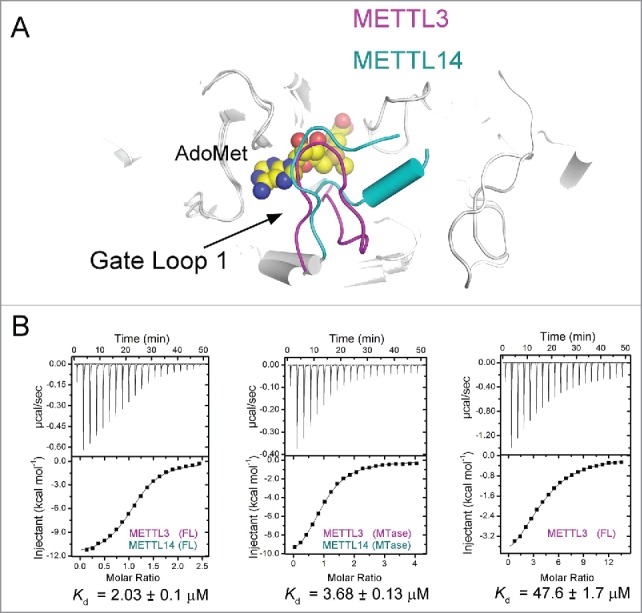
(A) Superposition of AdoMet-bound METTL3 and METTL14. AdoMet is shown in space-filling view. Gate loop 1 in METTL3 and METTL14 is colored in magenta and cyan, respectively. (B) Measurement of binding affinity between AdoMet and the METTL3–METTL14 complex using ITC. The first data point was removed from the analysis. The dissociation constant (Kd) of the wild type was approximately 2.0 μM. The AdoMet binding affinity of the methyltransferase domain complex was comparable to wild type. METTL3 alone exhibited significantly reduced AdoMet binding activity.
What is the function of METTL14 in the complex? We observed a positively charged groove between METTL3 and METTL14 adjacent to AdoMet using surface electrostatic potential analysis. At least 6 positively charged residues from METTL14—R245, R249, R254, R255, K297 and R298—are located in this groove. A complex with double mutations (K297E and R298E) showed moderately reduced RNA binding affinity and methyltransferase activity.35 Consistent with this observation, other groups have biochemically shown that R298P in METTL14 alters the sequence specificity for the RNA substrate.37 We further modeled the RNA-binding state of the complex using SAXS data. In the model, the RNA substrate appeared in the positively charged groove, indicating that METTL14 might contribute to RNA interaction.
Moreover, METTL3 and METTL14 are closely associated through an extensive buried interface, which explains the high stability of the complex. Indeed, without METTL14, METTL3 binds to AdoMet alone with a dissociation constant of approximately 47μM, 20-fold weaker than the wild-type complex (Fig. 2b). A crystallized truncation of the METTL3-METTL14 complex had a slight effect on AdoMet binding activity, suggesting that METTL14 stabilizes METTL3 to enhance AdoMet binding activity. We hypothesize that METTL14 is a pseudomethyltransferase in the complex that primarily plays a stabilization role and provides a platform with METTL3 for RNA recognition. However, we cannot exclude the possibility that METTL14 exhibits methyltransferase activity after binding additional factors. METTL14 is not well conserved; in plants, METTL14 homologues contain the DPPW motif instead of the EPPL motif.33 To date, METTL14 in plants is not known to associate with METTL3, leading us to speculate that there is a different m6A writer system in plants.
WTAP and other regulators
Wilms' tumor 1-associating protein (WTAP) was identified as another component of the human m6A methyltransferase complex. WTAP plays an important role in METTL3-METTL14 complex localization to the nuclear speckle.31 WTAP knockdown significantly decreased global m6A levels in human cell lines, indicating its importance in generating the distinct landscape of mRNA methylation at 5′ and internal sites.30 In zebrafish, depletion of the WTAP homolog caused apoptosis and tissue differentiation defects.31 Similar results were observed in Arabidopsis. The 37 kDa FKBP12-intereaction protein (FIP37), a homolog of WTAP, mediates m6A modification to control shoot stem cell fate.42However, WTAP has no effect on enzymatic activity in vitro other than to change the preferred RNA substrate of the METTL3-METTL14 complex. Because domain information is not available, how WTAP regulates m6A modification cannot be easily elucidated. Furthermore, additional regulators such as KIAA1429 involved in mRNA methylation have been identified by proteomic screening.30 Further investigation into their regulation of the m6A writer complex will shed light on RNA epigenetics.
Future perspectives
These structures and biochemical data provide new insights into the m6A mechanism in which METTL3 primarily serves as a catalytic subunit and METTL14 functions as a stabilizer. Together, the subunits provide an RNA binding platform. However, the absence of an RNA-bound structure precludes our understanding of how the writer complex specifically targets the RRACH sequence and of why only a small portion of RRACH motifs contain an m6A modification.43 Therefore, determination of the METTL3-METTL14 complex structure in the presence of RNA substrate and/or including ZnF motifs is a major goal. Some m6A modifications are relevant to cancer and infectious diseases: METTL3 deletion leads to significantly reduced cell invasion in cancer cells and HIV-1 virus infectivity in T cells.10,44-46 Thus, on the basis of these structures, the development of an inhibitor-specific target for m6A writers is one of the next important tasks.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the research associates at the Center for Protein Research, Huazhong Agricultural University, for technical support.
Funding
This work was supported by funds from the Ministry of Science and Technology (grant 2015CB910900), the Fok Ying-Tong Education Foundation (grant 151021), the Fundamental Research Funds for the Central Universities (Program No. 2014PY026, No. 2015PY219, and No. 2014JQ001), Project J1103510 supported by National Natural Science Foundation of China, and the Huazhong Agricultural University Scientific & Technological Self-innovation Foundation (Program No. 2013RC013).
References
- 1.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al.. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res 2013; 41:D262-7; PMID:23118484; http://dx.doi.org/ 10.1093/nar/gks1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012; 149:1635-46; PMID:22608085; http://dx.doi.org/ 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al.. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012; 485:201-6; PMID:22575960; http://dx.doi.org/ 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 4.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al.. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016; 530:441-6; PMID:26863196; http://dx.doi.org/ 10.1038/nature16998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slotkin W, Nishikura K. Adenosine-to-inosine RNA editing and human disease. Genome Med 2013; 5:105; PMID:24289319; http://dx.doi.org/ 10.1186/gm508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 2012; 40:5023-33; PMID:22344696; http://dx.doi.org/ 10.1093/nar/gks144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al.. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014; 159:148-62; PMID:25219674; http://dx.doi.org/ 10.1016/j.cell.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014; 515:143-6; PMID:25192136; http://dx.doi.org/ 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beemon K, Keith J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J Mol Biol 1977; 113:165-79; PMID:196091; http://dx.doi.org/ 10.1016/0022-2836(77)90047-X [DOI] [PubMed] [Google Scholar]
- 10.Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE. Rana TM Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 2016; 1:16011; PMID:27572442; http://dx.doi.org/ 10.1038/nmicrobiol.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng X, Chen K, Luo GZ, Weng X, Ji Q, Zhou T, He C. Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res 2015; 43:6557-67; PMID:26068471; http://dx.doi.org/ 10.1093/nar/gkv596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, et al.. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 2013; 155:1409-21; PMID:24269006; http://dx.doi.org/ 10.1016/j.cell.2013.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J, et al.. Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun 2014; 5:5630; PMID:25430002; http://dx.doi.org/ 10.1038/ncomms6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol 2014; 15:313-26; PMID:24713629; http://dx.doi.org/ 10.1038/nrm3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet 2014; 15:293-306; PMID:24662220; http://dx.doi.org/ 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- 16.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al.. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 2015; 347:1002-6; PMID:25569111; http://dx.doi.org/25683224 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- 17.Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, Wu Y, Lv Y, Hao J, Wang L, et al.. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 2015; 16:289-301; PMID:25683224; http://dx.doi.org/ 10.1016/j.stem.2015.01.016 [DOI] [PubMed] [Google Scholar]
- 18.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015; 526:591-4; PMID:26458103; http://dx.doi.org/ 10.1038/nature15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al.. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell 2016; 61:507-19; PMID:26876937; http://dx.doi.org/ 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 20.Adhikari S, Xiao W, Zhao YL, Yang YG. m(6)A: Signaling for mRNA splicing. RNA Biol 2016; 13:756-9; PMID:27351695; http://dx.doi.org/ 10.1080/15476286.2016.1201628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al.. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014; 505:117-20; PMID:24284625; http://dx.doi.org/ 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015; 161:1388-99; PMID:26046440; http://dx.doi.org/ 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015; 163:999-1010; PMID:26593424; http://dx.doi.org/ 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997; 3:1233-47; PMID:9409616 [PMC free article] [PubMed] [Google Scholar]
- 25.Bujnicki JM, Feder M, Radlinska M, Blumenthal RM. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. J Mol Evol 2002; 55:431-44; PMID:12355263; http://dx.doi.org/ 10.1007/s00239-002-2339-8 [DOI] [PubMed] [Google Scholar]
- 26.Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc Natl Acad Sci U S A 2011; 108:14855-60; PMID:21873203; http://dx.doi.org/ 10.1073/pnas.1111577108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008; 20:1278-88; PMID:18505803; http://dx.doi.org/ 10.1105/tpc.108.058883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res 2002; 30(20):4509-18; PMID:12384598; http://dx.doi.org/ 10.1093/nar/gkf573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 2014; 16:191-8; PMID:24394384; http://dx.doi.org/ 10.1038/ncb2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al.. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep 2014; 8:284-96; PMID:24981863; http://dx.doi.org/ 10.1016/j.celrep.2014.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al.. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 2014; 24:177-89; PMID:24407421; http://dx.doi.org/ 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al.. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014; 10:93-5; PMID:24316715; http://dx.doi.org/ 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer LM, Zhang D, Aravind L. Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification. BioEssays 2016; 38:27-40; PMID:26660621; http://dx.doi.org/ 10.1002/bies.201500104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, et al.. A census of human soluble protein complexes. Cell 2012; 150:1068-81; PMID:22939629; http://dx.doi.org/ 10.1016/j.cell.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al.. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 2016; 534:575-8; PMID:27281194; http://dx.doi.org/ 10.1038/nature18298 [DOI] [PubMed] [Google Scholar]
- 36.ledz´ PS, Jinek M. Structural insights into the molecular mechanism of the m6A writer complex. Elife 2016; 5:e18434; PMID:27627798; http://dx.doi.org/27373337 10.7554/eLife.18434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell 2016; 63:306-17; PMID:27373337; http://dx.doi.org/ 10.1016/j.molcel.2016.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res 2010; 38:W545-9; PMID:20457744; http://dx.doi.org/ 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al.. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013; 155:793-806; PMID:24209618; http://dx.doi.org/ 10.1016/j.cell.2013.10.026 [DOI] [PubMed] [Google Scholar]
- 40.Murn J, Teplova M, Zarnack K, Shi Y, Patel DJ. Recognition of distinct RNA motifs by the clustered CCCH zinc fingers of neuronal protein Unkempt. Nat Struct Mol Biol 2016; 23:16-23; PMID:26641712; http://dx.doi.org/27396363 10.1038/nsmb.3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 2004; 11:257-64; PMID:14981510; http://dx.doi.org/27396363 10.1038/nsmb738 [DOI] [PubMed] [Google Scholar]
- 42.Shen L, Liang Z, Gu X, Chen Y, Teo ZW, Hou X, Cai WM, Dedon PC, Liu L, Yu H. N(6)-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev Cell 2016; 38:186-200; PMID:27396363; http://dx.doi.org/ 10.1016/j.devcel.2016.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu N, Pan T. N6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol 2016; 23:98-102; PMID:26840897; http://dx.doi.org/27371828 10.1038/nsmb.3162 [DOI] [PubMed] [Google Scholar]
- 44.Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife 2016; 5:e15528; PMID:27371828; http://dx.doi.org/ 10.7554/eLife.15528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 2016; 62:335-45; PMID:27117702; http://dx.doi.org/ 10.1016/j.molcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM, et al.. Posttranscriptional m(6)A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe 2016; 19:675-85; PMID:27117054; http://dx.doi.org/ 10.1016/j.chom.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]


