ABSTRACT
Toxin-antitoxin (TA) systems are gene modules that appear to be horizontally mobile across a wide range of prokaryotes. It has been proposed that type I TA systems, with an antisense RNA-antitoxin, are less mobile than other TAs that rely on direct toxin-antitoxin binding but no direct comparisons have been made. We searched for type I, II and III toxin families using iterative searches with profile hidden Markov models across phyla and replicons. The distribution of type I toxin families were comparatively narrow, but these patterns weakened with recently discovered families. We discuss how the function and phenotypes of TA systems as well as biases in our search methods may account for differences in their distribution.
KEYWORDS: Antisense RNA, horizontal gene transfer, post-segregational killing, toxin-antitoxin systems
Abbreviations
- TA
toxin-antitoxin
- PSK
post-segregational killing
- HGT
horizontal gene transfer
Introduction
Toxin-antitoxin (TA) systems are 2 gene modules comprised of a toxic protein and an antitoxin.1,2 There are 6 types of TA systems grouped according to the mechanism of the antitoxin and whether it is a protein or an RNA molecule. The best described are type I and type II systems (Fig. 1). Type I systems have an antisense RNA antitoxin that binds the toxin mRNA3,4 occluding the binding sites that are necessary for translation5-8 or inducing RNase degradation9 (Fig. 1). The toxins are typically small hydrophobic proteins that destabilize cellular membranes, though some nucleases have been found8 (Table 1). Type II systems have a protein antitoxin that directly binds and inhibits the protein toxin. The antitoxin also represses transcription from the operon but does so more effectively when bound by the toxin (Fig. 1).10 The toxin can have a range of functions, usually inhibiting DNA replication or translation.1,11
Figure 1.
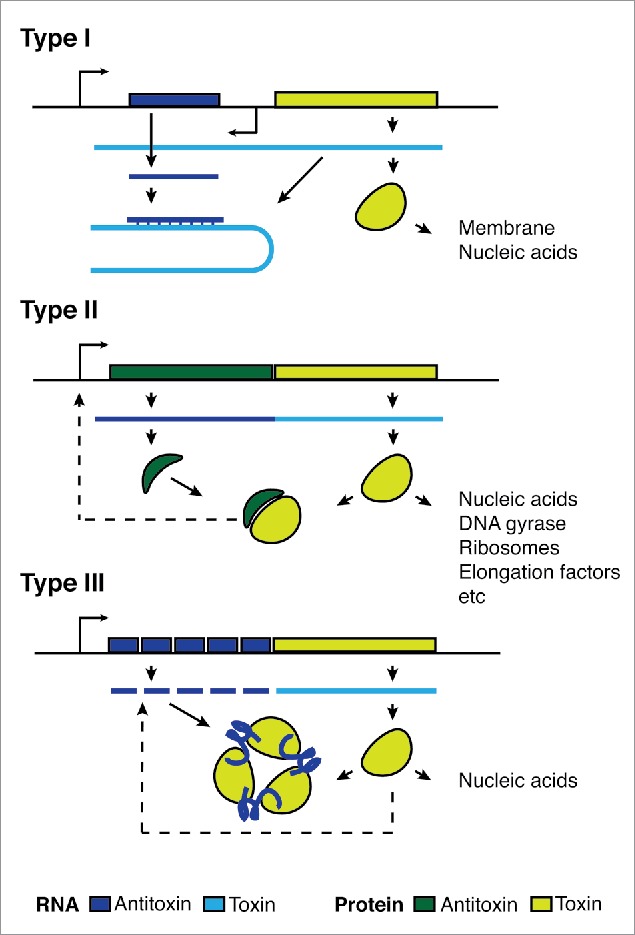
Operon organization and regulation of type I, type II and type III TA systems. Type I TA systems have an antisense RNA antitoxin that binds the toxin mRNA, leading to mRNA degradation. If the antitoxin does not bind, the toxin is translated and is free to target cellular membranes or nucleic acids.9 Type II TA systems have a protein antitoxin that directly binds the protein toxin, preventing the toxin from targeting various components of central dogma reactions. The bound toxin and antitoxin interact with their own promoter to control transcription in a process known as conditional cooperativity10 Type III TA antitoxins are RNA repeats. The cognate toxin is a nuclease that specifically cuts its own RNA repeats before the repeats directly bind and inhibit the toxin.17 All systems rely on careful titration of toxin and antitoxin in the cell. The antitoxin degrades faster in the cell, and a reduction of transcription and translation rates due to cellular stress or gene loss can free the toxin causing cell death or growth arrest.
Table 1.
Characteristics of type I TA families.
| Toxin | Discovery | Phyla:Family | Target | Regulation | References |
|---|---|---|---|---|---|
| Fst | Plasmid stability | 1:6 | Membrane damage | Cis | 42 |
| Hok | Plasmid stability | 1:4 | Membrane damage | Cis | 19 |
| Ibs | Repeats in sequence data | 1:3 | Membrane damage | Cis, | 6 |
| Ldr | Repeats in sequence data | 1:1 | Membrane damage | Cis | 33 |
| ShoB | Screening for sRNA | 1:1 | Membrane damage | Trans | 4 |
| SymE | Screening for sRNA | 5:24 | Ribonuclease | Cis | 4 |
| TisB | Screening for sRNA | 1:1 | Membrane damage | Trans. | 45 |
| TxpA | Screening for sRNA | 1:1 | Membrane damage | Cis | 3 |
| XCV2162 | Screening for sRNA | 1:11 | Predicted membrane domain | Cis | 35 |
It has been proposed that the antisense RNA-regulated type I TAs are more likely to be duplicated on chromosomes in a lineage specific manner12 and that they are less horizontally mobile than the more promiscuous type II TAs.12-16 This hypothesis is primarily driven by genomic screens finding type I TAs only in a narrow range of species while type II TAs are more widely distributed and more likely to be on mobile elements. We compare the antisense RNA and protein regulated TA systems directly, along with the more recently described type III systems, which encode an RNA antitoxin that directly binds the protein toxin,17,18 to see if they differ in their distribution across species and mobile replicons.
Interestingly, one of the first TA systems to be identified was Hok-Sok, a plasmid-borne type I system discovered through its ability to stabilize mobile elements19 After transcription, the stable Hok mRNA is slowly processed at the 3′ end into a translatable isoform.20 This process is attenuated by the highly expressed RNA Sok, which forms a duplex with Hok mRNA leading to subsequent degradation of both transcripts. Should transcription of the TA operon cease, the less stable antitoxin RNA rapidly degrades, allowing unprocessed Hok mRNA in the cytoplasm to mature and be translated into a toxic protein that destabilizes cellular membranes.19,21 This causes the cell to die or stop replicating upon gene loss, an effect known as post-segregational killing (PSK). Genes that are able to induce PSK, which include some restriction-modification systems, abortive-infection systems, and bacteriocins,15,22,23 are prevalent on mobile elements. Plasmids containing PSKs are advantaged over non-PSK plasmids22,24-26 under conditions of horizontal gene transfer (HGT), and individual TA loci on plasmids are more likely to maintain functional toxins than chromosomal loci.16,27
While TA systems were discovered for their effects on plasmids, TAs of all types are also abundant on bacterial chromosomes. The role of TA systems on chromosomes is still uncertain,11,27 with theories ranging from them being important components in cellular function to being genomic parasites that persist due to difficulties in displacing them. Proposed cellular functions for the various types of TA systems are mostly stress related, including bacteriostasis, programmed cell death and persister cell formation.2,28,29 Other functions are related to their ability to cause PSK: stabilizing genomic regions,30-32 neutralizing PSK from plasmid borne TAs,16 and acting in abortive infection of bacteriophages.33,34 Some functions have been well characterized for specific loci, such as the ability of type I TA TisB-IstR to increase resistance to antibiotics in E. coli,29 but many others have not, with individual knockouts showing little change in phenotype.11
Within each type of TA system are multiple families of related toxins. Many type I TA families have only been found on chromosomes to date12 while most type II families are found on both plasmids and chromosomes.14 We analyzed the distribution of type I, type II and type III TA systems across bacterial species and mobile replicons using iterative searches with profile hidden Markov models (HMMs), a powerful approach for identifying remote homologues. Nine type I TA toxin families were included in the analysis (Table 1). The family XCV2162 (also known as Plasmid_toxin), has only been described computationally35 but was included due to its reported distribution, which is consistent with HGT. Of the 11 type II TA toxin families investigated, most are part of large, well-described families except for the recently described families GinA and GinB.14 Three type III TA families described so far36 were also included.
Phage, plasmid, and bacterial chromosome sequence data were downloaded from the EMBL nucleotide archive (http://www.ebi.ac.uk/ena/, October 2014). These were translated in 6 frames to derive all possible amino acid sequences from the genomes including the short ORFs that are a characteristic of type I toxins (and make them difficult to detect). This database was analyzed with profile HMMs, derived from the known amino acid sequences for each toxin family as downloaded from Pfam and GenBank. The HMMs for the more recently described families GinA and GinB14 and CptN and TenpN36 were derived from loci reported in the literature.
Consistent with previous claims, most RNA-antisense regulated (type I) TA families exhibited a narrow, phyla-specific distribution and were more rare on plasmids than either type II or type III systems, where the toxin is regulated through direct binding of the antitoxin. Interestingly, though, this pattern was less consistent on more recently discovered systems. Reasons for these differences, including toxin function and gene regulation, ability to exhibit PSK, and biases in the databases and discovery process, are discussed here.
Type I families occur across a narrow phylogenetic range and are less likely to be on mobile elements than type II and type III families
Despite the number of new, unannotated loci found in the family-based searches, all type I toxins except for SymE were found in only one phylum (Fig. 2 and Fig. S1). Some toxins were especially narrow in their distribution: toxins Ldr, ShoB, Txp and TisB were found on no mobile elements and in less than 5% of species within that phylum (Fig. 2) and those species were all in the same family, either Enterobacteriaceae within Proteobacteria or Bacillaceae within Firmicutes (Table 1).
Figure 2.
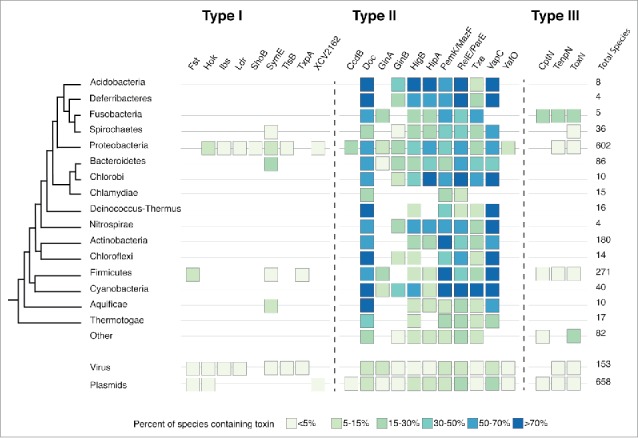
Percent of species within each phyla or replicon that contain a loci from a given type I, type II and type III TA toxin family. HMMs for each TA family were derived from known amino acid sequences and used to search a database of phage, plasmid, and bacterial chromosome sequences subjected to 6-frame translations to derive all possible amino acid sequences from that sequence. This includes short ORFs that are typical of type I toxins. For each phyla or replicon, the percent of total species in the database (left of figure) that contain at least one locus of that toxin is reported (boxes).
Most type II families were found across phyla (Fig. 2 and Fig. S1). The families Doc, MazF/PemK, RelE/ParE and VapC37 were found in all bacterial phyla analyzed, as well as in viruses and plasmids. These TA toxins were prevalent within phyla as well, found on over 70% of species within the phyla Acidobacteria and Cyanobacteria. The 2 type II toxin families CcdB and YafO were found in only one phylum. Compared to single-phylum type I toxins, these type II toxins were observed in a higher percentage of species, across more taxonomic families within the phylum as well as on mobile elements.
Type III families were intermediate between type I and type II TAs. All 3 families were found in both Gram-negative and Gram-positive phyla and on plasmids,36 though none were on greater than 30% of the species in the database of translated genomes. Different factors may account for chromosomal-only and narrowly distributed type I families, as discussed below.
Presence on mobile elements and ability to exhibit PSK as a factor in TA family distribution
None of the most narrowly distributed type I toxin families were found on elements that could be identified as being mobile. Generally, we see a correlation between presence on mobile elements and a larger taxonomic range (though the low number of sequenced mobile element prevents showing a strong association). The 3 type I families found on mobile elements, Fst, Hok, XCV2162, were found in more taxonomic families (6, 4, and 11, respectively) within their phylum (Table 1) though even mobile type I TAs are narrowly distributed compared with type II and III TAs, with most of these families found on mobile elements.
Chromosome-only TAs may not be widely distributed on mobile elements simply because they are unable to mediate phenotypes beneficial to them. PSK genes confer an advantage to horizontally mobile elements24,25 and with few exceptions TA loci on plasmids are able to mediate PSK.28,38 Most chromosome-specific type I systems have not been tested for their ability to confer a PSK phenotype, except Ldr-RdlD, which did not.33,39 However, because chromosome- and plasmid-borne homologues may be under different selective pressures and have different phenotypes, chromosomal homologues of plasmid-borne TAs often do not exhibit PSK themselves.16,39,40 This makes it difficult to determine if inability to confer a PSK phenotype is an inherent feature of these chromosome-only TA families, or simply that the particular gene pairs analyzed do not show it.
Yet, while mobility can increase access of a gene to new environments depending on the vector host range, it is not a guarantee that the gene will be maintained in those new environments. PSK would only be an advantage to a mobile element in cellular backgrounds where the phenotype was expressed. PSK requires that the toxin have a relevant target, and that a close stoichiometry of toxin and antitoxin is maintained. In cellular backgrounds where the PSK phenotype is neutralized, the genes would no longer be selected.25 Families of TA that exhibit PSK in a wide range of cellular backgrounds, may, then, be more likely to be maintained on mobile elements across hosts. It would be interesting to test if the type I TAs known to exhibit PSK but which remain phyla-specific, such Hok and Fst, were more limited in their ability to exhibit PSK across phyla.
Toxin target and regulation of toxins as factors in TA family distribution
Both toxin target14 and toxin gene regulation15 have been suggested as factors in the distribution of more widespread TA systems, but it is unclear if either can explain the narrow distribution of type I systems. It was proposed that type II toxins are successful across a wide array of species because they affect highly conserved targets14 Most type II toxins affect central dogma reactions, inhibiting DNA gyrase, ribosomes, and elongation factors or acting as nucleases.1,11 Type III toxins are all nucleases. Type I toxins are predominately predicted membrane disrupting proteins (Table 1)41; though the exact mechanisms of toxicity are not always known, the membrane is not not necessarily a phyla-specific target, as discussed with Fst below.
Crucially, having a target does not ensure toxicity if the regulon does not produce sufficient quantities of the protein. The Gram-positive, membrane disrupting type I toxin Fst was able to be toxic in E. coli when a translation regulating stem-loop in the RNA was disrupted, increasing translation of the protein.42 This suggests that regulation, rather than toxin-target was the limiting factor. Similarly, small changes in gene regulation were believed to be why Bacillus subtilis could be used to amplify clones of only some of the Fst loci from various bacterial species: others appeared to cause cell death when moved into the new cellular background.43 Furthermore, the various phenotypes associated with TA systems, PSK, abortive infection and TA-mediated stress response, all require that the toxin is inactive but can be released upon specific stimuli, often involving very close regulation of toxin and antitoxin in the cell.44
Not having a conserved toxin target or requiring specific host proteins for function, and changes in operon regulation when entering new hosts are all plausible reasons that a given TA would not be maintained across cell types. And this may be true for the families of TAs surveyed here, which maintain a narrow distribution. Yet there is no reason, a priori, that the defining aspects of type I TAs (RNA-RNA interactions) should limit their host range. We have only just begun to explore the space of possible type I TAs, with new systems being discovered, and the narrowness we seek to explain now may not continue to be a feature of all type I TAs. An example is SymE, an outlier among the type I TAs in that it is distributed across both Gram negative and Gram positive phyla (Fig. 2).
Biases in databases and discovery of type I TA families may account for apparent distribution of type I TAs
For all screens such as this, it can be difficult to make sweeping statements on the distribution of any gene across many phyla and mobile elements due to the selection of sequenced genomes and how well bioinformatics tools find different genetic elements within them. Type I TA systems in particular have historically been more difficult to detect in silico, with sequence-diverse RNA antitoxins and small toxins (under 60 amino acids), potentially reducing the number of toxin families we know of.
Many type II families reported here are actually super-families of many described toxins. Aggregating these can increase their apparent distribution. CcdB has a narrow distribution in this screen, but is often combined with MazF and related families, making the super-family relatively widely distributed. Type I toxins Ldr and Fst are considered to be related,12,14 and combining them together would also result in a broad family. As we see in this screen already, more recently described systems (SymE for type I, Gin C and D for type II) are more likely to differ in their distribution. The apparent narrowness of type I TA systems, then, may change as we develop new methods for detecting them and describe new systems that aggregate into larger families.
Ultimately, methods that go beyond sequence-based features of known TA systems are more likely to yield families with novel characteristics. Many type I TAs were first described on chromosomes (Table 1). Many of the best-studied and most widely distributed type II systems were discovered due to a phenotype, usually the ability to stabilize plasmids in monocultures. It is not surprising, then, that these families were later found to be widely distributed on mobile elements. The narrowly distributed type II families YafO, GinC and GinD were discovered bioinformatically14 due to their association with known antitoxins (guilt by association) rather than sequence features of the toxin. They exhibited patterns of distribution similar to many type I families (Fig. 2). Another approach which may yield novel types of TA systems is that of Sberro et al.45 This group studied genomes that had been randomly fragmented and inserted into E. coli plasmids for sequencing. They identified genes that were only present on fragments (and thus could be amplified in E. coli) when an adjacent ORF was present, implying a toxin and antitoxin function. These were filtered for genes that appeared as homologues across species, suggesting HGT, and in regions of the genome associated with phage defense to find novel TA systems.45
Conclusion
We found that type I toxin families are less often found on known mobile elements or distributed across large taxonomic ranges when compared with type II and type III families despite increases in sequencing data and our ability to detect them. Though TA systems have common characteristics, including organization into an operon, antitoxin-mediated regulation of toxin transcription (type II, type III) or translation (type I), and high lability of the antitoxin relative to the toxin.17,19,43,46,47 it has frequently been suggested that type I TAs are more lineage specific than type II TA systems. However, the broader phylogenetic distribution of type I toxin SymE and type III TA systems seen here would suggest one of the defining features of type I systems, the presence of an RNA antitoxin, does not account for the difference.
The factors that select for maintenance of horizontally acquired genes vary. Genes on both chromosomes and plasmids can be selected by within-host forces.26 All TA systems consist of a tightly regulated antitoxin and toxin capable of stopping bacterial growth-some systems even have reversible effects, able to be turned off when conditions change. These make for versatile modules with the potential to fill a variety of functions, from plasmid competition (eg, PSK) to cellular stress response to phage-plasmid competition (eg abortive infection). The function of a given locus will depend in part on its history, though some families may have features that make them more able to fill certain roles. Aspects of the toxin target and gene regulation have been proposed as reasons that type II and type III TAs are successful in HGT, and these may also affect the ability of some type I TAs to stably establish in new species.
This work only surveys the distribution of known type I TA systems, which remain narrowly distributed across methodologies. But it is interesting to consider whether this narrowness is only a feature of the families found so far. New families of TAs are being described at a rapid pace,13,48-50 some of which have alternative distributions. We see this already, with type I TA toxins SymE and type II toxins like YafO, GinB and GinC that are narrowly distributed. The potential type I TA XCV2162 is plasmid-borne and has an erratic phylogenetic distribution consistent with HGT.35 As we find more families, they may aggregate into the superfamilies of type II TAs and gain a broader distribution. It may be that the patterns of lineage dependence so far attributed to type I TAs as a group will turn out to be a feature of specific families within all types of TAs, and we simply found the narrowly-distributed families of type I TAs and the broadly distributed central-dogma targeting type II TAs first.
Supplementary Material
Disclosure of potential conflicts of interest
The authors report no potential conflict of interest in the publication of this material.
Funding
This work was supported by the Marsden Fund under Grant M1138, Rutherford Discovery Fellowship and Biomolecular Interaction Center.
References
- 1.Van Melderen L, De Bast MS. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genetics 2009; 5(3); PMID:19325885; http://dx.doi.org/15864262 10.1371/journal/pgen.1000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol 2005; 3(5):371-82; PMID:15864262; http://dx.doi.org/ 10.1038/nrmicro1147 [DOI] [PubMed] [Google Scholar]
- 3.Silvaggi JM, Perkins JB, Losick R. Small untranslated RNA antitoxin in Bacillus subtilis. J Bacteriol 2005; 187(19):6641-50; PMID:16166525; http://dx.doi.org/ 10.1128/JB.187.19.6641-6650.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5′-and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res 2005; 33(3):1040-50; PMID:15718303; http://dx.doi.org/ 10.1093/nar/gki256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darfeuille F, Unoson C, Vogel Jr, Wagner EGH. An antisense RNA inhibits translation by competing with standby ribosomes. Mol Cell 2007; 26(3):381-92; PMID:17499044; http://dx.doi.org/ 10.1016/j.molcel.2007.04.003 [DOI] [PubMed] [Google Scholar]
- 6.Kawano M, Aravind L, Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol Microbiol 2007; 64(3):738-54; PMID:17462020; http://dx.doi.org/ 10.1111/j.1365-2958.2007.05688.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fozo EM, Kawano M, Fontaine F, Kaya Y, Mendieta KS, Jones KL, Ocampo A, Rudd KE, Storz G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol Microbiol 2008; 70(5):1076-93; PMID:18710431; http://dx.doi.org/ 10.1111/j.1365-2958.2008.06394.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol Rev 2008; 72(4):579-89; PMID:19052321; http://dx.doi.org/ 10.1128/MMBR.00025-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerdes K, Nielsen A, Thorsted P, Wagner EGH. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable Hok, SrnB and PndA effector messenger RNAs. J Mol Biol 1992; 226(3):637-49; PMID:1380562; http://dx.doi.org/ 10.1016/0022-2836(92)90621-P [DOI] [PubMed] [Google Scholar]
- 10.Cataudella I, Trusina A, Sneppen K, Gerdes K, Mitarai N. Conditional cooperativity in toxin–antitoxin regulation prevents random toxin activation and promotes fast translational recovery. Nucleic acids Res 2012; 40(14):6424-34; PMID:22495927; http://dx.doi.org/21041110 10.1093/nar/gks297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Melderen L. Toxin-antitoxin systems: why so many, what for? Curr Opin Microbiol 2010; 13(6):781-5; PMID:21041110; http://dx.doi.org/ 10.1016/j.mib.2010.10.006 [DOI] [PubMed] [Google Scholar]
- 12.Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res 2010; 38(11):3743-59; PMID:20156992; http://dx.doi.org/ 10.1093/nar/gkq054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biology Direct 2009; 4(1):19; PMID:19493340; http://dx.doi.org/ 10.1186/1745-6150-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leplae R, Geeraerts D, Hallez R, Guglielmini J, Drèze P, Van Melderen L. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res 2011; 39(13):5513-25; PMID:21422074; http://dx.doi.org/ 10.1093/nar/gkr131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mruk I, Kobayashi I. To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res 2014; 42(1):70-86; PMID:23945938; http://dx.doi.org/ 10.1093/nar/gkt711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mine N, Guglielmini J, Wilbaux M, Van Melderen L. The decay of the chromosomally encoded ccdO157 toxin-antitoxin system in the Escherichia coli species. Genetics 2009; 181(4):1557-66; PMID:19189956; http://dx.doi.org/ 10.1534/genetics.108.095190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GPC. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci 2009; 106(3):894-9; http://dx.doi.org/ 10.1073/pnas.0808832106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goeders N, Chai R, Chen B, Day A, Salmond GP. Structure, evolution, and functions of bacterial type III toxin-antitoxin systems. Toxins 2016; 8(10):282; PMID:27690100; http://dx.doi.org/3517851 10.3390/toxins8100282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdes K, Rasmussen PB, Sr Molin. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A 1986; 83(10):3116-20; PMID:3517851; http://dx.doi.org/ 10.1073/pnas.83.10.3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franch T, Gultyaev AP, Gerdes K. Programmed cell death by hok/sok of plasmid R1: processing at the hok mRNA 3′-end triggers structural rearrangements that allow translation and antisense RNA binding. J Mol Biol 1997; 273(1):38-51; PMID:9367744; http://dx.doi.org/ 10.1006/jmbi.1997.1294 [DOI] [PubMed] [Google Scholar]
- 21.Pecota DC, Wood TK. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J Bacteriol 1996; 178(7):2044-50; PMID:8606182; http://dx.doi.org/ 10.1128/jb.178.7.2044-2050.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science 1995; 267(5199):897; PMID:7846533; http://dx.doi.org/ 10.1126/science.7846533 [DOI] [PubMed] [Google Scholar]
- 23.Inglis RF, Bayramoglu B, Gillor O, Ackermann M. The role of bacteriocins as selfish genetic elements. Biology letters 2013; 9(3):20121173; PMID:23616642; http://dx.doi.org/ 10.1098/rsbl.2012.1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper TF, Heinemann JA. Selection for plasmid post-segregational killing depends on multiple infection: evidence for the selection of more virulent parasites through parasite-level competition. Proc Biol Sci 2005; 272(1561):403; PMID:15734695; http://dx.doi.org/ 10.1098/rspb.2004.2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper TF, Heinemann JA. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. Proc Natl Acad Sci U S A 2000; 97(23):12643; PMID:11058151; http://dx.doi.org/ 10.1073/pnas.220077897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper TF, Paixão T, Heinemann JA. Within-host competition selects for plasmid-encoded toxin-antitoxin systems. Proc Biol Sci 2010; 277(1697):3149; PMID:20504809; http://dx.doi.org/ 10.1098/rspb.2010.0831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsilibaris V, Maenhaut-Michel G, Mine N, Van Melderen L. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J Bacteriol 2007; 189(17):6101; PMID:17513477; http://dx.doi.org/ 10.1128/JB.00527-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christensen-Dalsgaard M, Gerdes K. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol Microbiol 2006; 62(2):397-411; PMID:17020579; http://dx.doi.org/ 10.1111/j.1365-2958.2006.05385.x [DOI] [PubMed] [Google Scholar]
- 29.Dörr T, Lewis K, Vulić M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genetics 2009; 5(12):e1000760; PMID:20011100; http://dx.doi.org/ 10.1371/journal.pgen.1000760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowe-Magnus DA, Mazel D. Integrons: natural tools for bacterial genome evolution. Curr Opin Microbiol 2001; 4(5):565-9; PMID:11587934; http://dx.doi.org/ 10.1016/S1369-5274(00)00252-6 [DOI] [PubMed] [Google Scholar]
- 31.Szekeres S, Dauti M, Wilde C, Mazel D, Rowe-Magnus DA. Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol Microbiol 2007; 63(6):1588-605; PMID:17367382; http://dx.doi.org/ 10.1111/j.1365-2958.2007.05613.x [DOI] [PubMed] [Google Scholar]
- 32.Brantl S. Bacterial type I toxin-antitoxin systems. RNA biology 2012; 9(12):1488-90; PMID:23324552; http://dx.doi.org/ 10.4161/rna.23045 [DOI] [PubMed] [Google Scholar]
- 33.Kawano M, Oshima T, Kasai H, Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol Microbiol 2002; 45(2):333-49; PMID:12123448; http://dx.doi.org/ 10.1046/j.1365-2958.2002.03042.x [DOI] [PubMed] [Google Scholar]
- 34.Blower TR, Evans TJ, Przybilski R, Fineran PC, Salmond GPC. Viral evasion of a bacterial suicide system by RNA-based molecular mimicry enables infectious altruism. PLoS Genetics 2012; 8(10):e1003023; PMID:23109916; http://dx.doi.org/ 10.1371/journal.pgen.1003023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.S Findeiß, Schmidtke C, Stadler PF, Bonas U. A novel family of plasmid-transferred anti-sense ncRNAs. RNA biology 2010; 7(2):120-4; PMID:20220307; http://dx.doi.org/ 10.4161/rna.7.2.11184 [DOI] [PubMed] [Google Scholar]
- 36.Blower TR, Short FL, Rao F, Mizuguchi K, Pei XY, Fineran PC, Luisi BF, Salmond GPC. Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res 2012; 40(13):6158-73; PMID:22434880; http://dx.doi.org/ 10.1093/nar/gks231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenzie JL, Robson J, Berney M, Smith TC, Ruthe A, Gardner PP, Arcus VL, Cook GM. A VapBC toxin-antitoxin module is a posttranscriptional regulator of metabolic flux in mycobacteria. J Bacteriol 2012; 194(9):2189-204; PMID:22366418; http://dx.doi.org/ 10.1128/JB.06790-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milunovic B, Morton RA, Finan TM. Cell growth inhibition upon deletion of 4 toxin-antitoxin loci from the megaplasmids of Sinorhizobium meliloti. J Bacteriol 2014; 196(4):811-24; PMID:24317400; http://dx.doi.org/ 10.1128/JB.01104-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawano M. Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E. coli. RNA biology 2012; 9(12):1520-7; PMID:23131729; http://dx.doi.org/ 10.4161/rna.22757 [DOI] [PubMed] [Google Scholar]
- 40.Pedersen K, Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol 1999; 32(5):1090-102; PMID:10361310; http://dx.doi.org/ 10.1046/j.1365-2958.1999.01431.x [DOI] [PubMed] [Google Scholar]
- 41.Brielle R, Pinel-Marie M-L, Felden B. Linking bacterial type I toxins with their actions. Curr Opin Microbiol 2016; 30:114-21; PMID:26874964; http://dx.doi.org/18641135 10.1016/j.mib.2016.01.009 [DOI] [PubMed] [Google Scholar]
- 42.Shokeen S, Patel S, Greenfield TJ, Brinkman C, Weaver KE. Translational regulation by an intramolecular stem-loop is required for intermolecular RNA regulation of the par addiction module. J Bacteriol 2008; 190(18):6076-83; PMID:18641135; http://dx.doi.org/ 10.1128/JB.00660-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weaver KE, Reddy SG, Brinkman CL, Patel S, Bayles KW, Endres JL. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology 2009; 155(9):2930-40; PMID:19542006; http://dx.doi.org/ 10.1099/mic.0.030932-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coray DS, Kurenbach B, Heinemann JA. Exploring the parameters of post segregation killing using heterologous expression of secreted toxin barnase and antitoxin barstar in an E. coli case study. Microbiology 2016; PMID:27902436; http://dx.doi.org/ 10.1099/mic.0.000395 [DOI] [PubMed] [Google Scholar]
- 45.Sberro H, Leavitt A, Kiro R, Koh E, Peleg Y, Qimron U, Sorek R. Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning. Mol Cell 2013; 50(1):136-48; PMID:23478446; http://dx.doi.org/ 10.1016/j.molcel.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sat B, Hazan R, Fisher T, Khaner H, Glaser G, Engelberg-Kulka H. Programmed cell death in escherichia coli: some antibiotics can trigger mazEF lethality. J Bacteriol 2001; 183(6):2041-5; PMID:11222603; http://dx.doi.org/ 10.1128/JB.183.6.2041-2045.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christensen SK, Pedersen K, Hansen FG, Gerdes K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol 2003; 332(4):809-19; PMID:12972253; http://dx.doi.org/ 10.1016/S0022-2836(03)00922-7 [DOI] [PubMed] [Google Scholar]
- 48.Wen J, Fozo EM. sRNA antitoxins: more than one way to repress a toxin. Toxins 2014; 6(8):2310-35; PMID:25093388; http://dx.doi.org/ 10.3390/toxins6082310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masuda H, Tan Q, Awano N, KÄ Wu, Inouye M. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol Microbiol 2012; 84(5):979-89; PMID:22515815; http://dx.doi.org/ 10.1111/j.1365-2958.2012.08068.x [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Lord DM, Cheng H-Y, Osbourne DO, Hong SH, Sanchez-Torres V, Quiroga C, Zheng K, Herrmann T, Peti W. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol 2012; 8(10):855-61; PMID:22941047; http://dx.doi.org/ 10.1038/nchembio.1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


