ABSTRACT
Due to their simple architecture and control mechanism, regulatory RNA modules are attractive building blocks in synthetic biology. This is especially true for riboswitches, which are natural ligand-binding regulators of gene expression. The discovery of various tandem riboswitches inspired the design of combined RNA modules with activities not yet found in nature. Riboswitches were placed in tandem or in combination with a ribozyme or temperature-responsive RNA thermometer resulting in new functionalities. Here, we compare natural examples of tandem riboswitches with recently designed artificial RNA regulators suggesting substantial modularity of regulatory RNA elements. Challenges associated with modular RNA design are discussed.
KEYWORDS: Riboswitch, ribozyme, RNA thermometer, synthetic biology, posttranscriptional regulation
Tandem arrangement of natural riboswitches
Our view on RNA function was long reduced to the transfer of genetic information from DNA to protein level. With the discovery of ribozymes as catalytic RNAs that execute their own excision, RNA molecules received attention as mediators of gene regulation.1 Since then manifold regulatory RNAs have been identified and characterized in diverse species. A widespread class of RNA regulators is the so-called riboswitch, a natural metabolite or ion sensor. Riboswitches consist of an aptamer domain that binds small molecules with high specificity. Ligand binding to the aptamer domain is transmitted to an expression platform, which regulates expression of the downstream gene.2,3 While the aptamer domain is highly conserved for recognition of the cognate ligand, the expression platform is variable and can modulate different cellular processes like transcription termination, translation initiation or mRNA stability with transcription termination being the most prevalent mechanism of riboswitch-mediated regulation. In bacteria, riboswitches typically reside in 5’ untranslated regions (5’UTRs) of mRNAs, allowing them to react to changes of cellular metabolites before the full-length mRNA is transcribed or translated into a protein.4
In addition to individual riboswitches, several riboswitch pairs have been found in nature. The most common tandem arrangement exists in the 5′UTRs of the gcvT mRNA of diverse bacterial species like Bacillus subtilis or Vibrio cholerae.5 Two glycine sensor domains are separated by a short linker (Fig. 1A). Both aptamers can bind one molecule of glycine in the bulge of helix P3.6 Cooperative glycine binding leads to dimerization of both aptamers, in which stem P1 of the first aptamer provides the scaffold for the dimer interface. The dimerization of both aptamers stabilizes stem P1 of the second aptamer leading to structural rearrangements in the expression platform allowing transcription of glycine catabolism genes. The glycine riboswitch represents together with the adenine riboswitch one of the rare on-switches that positively regulates gene expression.5,7
Figure 1.
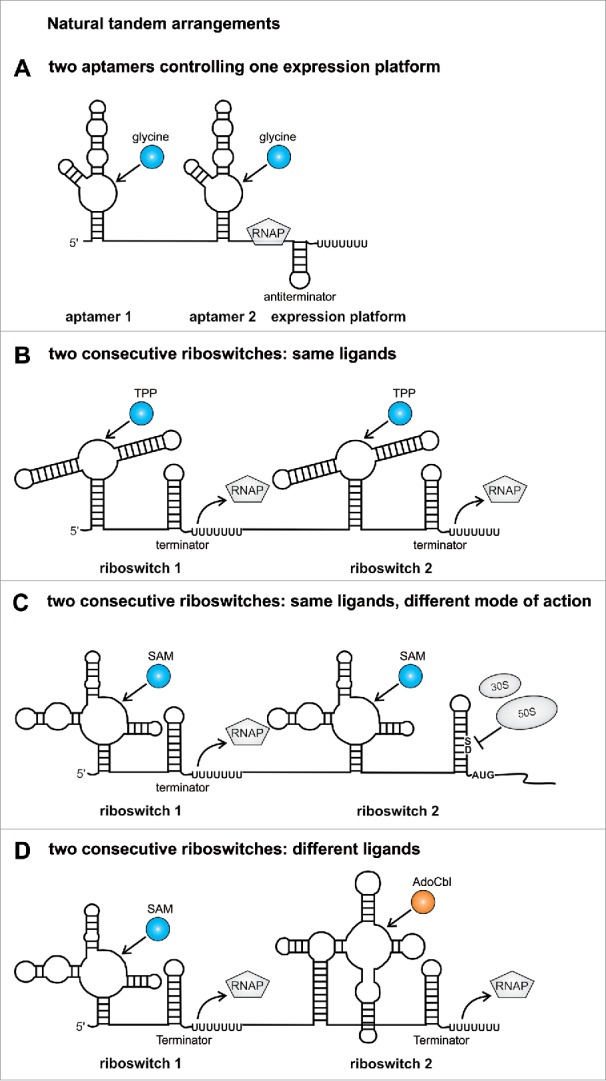
Tandem riboswitches occurring in nature. (A) In the glycine gcvT-5′UTR of B. subtilis and V. cholerae 2 glycine aptamers control a single expression platform.5 (B) Two consecutive TPP riboswitches regulate transcription termination in the tenA-5′UTR of B. anthracis.10 (C) In the bhmT-5′UTR of Pelagibacter ubique a transcriptional riboswitch is followed by a translational riboswitch both controlling gene expression in response to SAM.9 (D) The metE-5′UTR of B. clausii harbors 2 riboswitches that regulate transcription termination independently depending on 2 chemical inputs.10 Revised from reference.47 RNAP: RNA polymerase, TPP: thiamine pyrophosphate, SAM: S-adenosylmethionine, AdoCbl: adenosylcobalamin, fourU: fourU motif, SD: Shine-Dalgarno sequence, AUG: translational start codon.
Two complete riboswitches are located in the tenA-5′UTR of Bacillus anthracis. Here, both riboswitches individually control termination of transcription by sensing thiamine pyrophosphate (TPP) (Fig. 1B).8 Each riboswitch represents an off-switch that terminates transcription prematurely upon TPP binding. The serial fusion of 2 riboswitches decreases the ligand concentration needed to modulate gene expression. A riboswitch consisting of a single aptamer domain and expression platform requires an ∼80-fold change in metabolite concentration to switch gene expression from 90 to 10%. The combined activity of the tandem riboswitch reduces the required metabolite concentration from ∼80-fold to 40-fold.8
A similar arrangement of 2 riboswitches sensing the same ligand was found in the bhmT-5′UTR of the marine α-proteobacterium Pelagibacter ubique. Two consecutive riboswitches sense S-adenosylmethionine (SAM).9 However, the first riboswitch controls transcription termination, while the second riboswitch sequesters the ribosome binding site in presence of SAM (Fig. 1C). As high intracellular SAM levels would directly lead to transcription termination by the first riboswitch, it was suggested that the translational riboswitch could act as a backup system, which inhibits translation of bhmT when SAM concentrations increase after transcriptional regulation has already occurred.
A more complicated arrangement was described for Bacillus clausii, where the metE-5′UTR harbors 2 transcriptional riboswitches, which are affected by different ligands.10 The first riboswitch reduces transcription upon SAM binding, while the second riboswitch recognizes coenzyme B12 (AdoCbl) (Fig. 1D). Both riboswitches partially inhibit transcription of metE encoding an enzyme that converts homocysteine to methionine. As a second enzyme (MetH) was found that catalyzes the same reaction more efficiently, it was suggested that SAM and AdoCbl repress expression of the less efficient MetE enzyme if sufficient amounts of MetH are present in the cell.11,12
Synthetic RNA regulators: From single to tandem arrangements
The ever growing number of natural riboswitches has raised considerable interest in synthetic biology and biotechnology,13,14 in particular when the first tandem riboswitches were discovered. A major objective of synthetic biology is the engineering of RNAs with novel functionalities, which can be achieved through recombination of already existing RNA elements or by de novo design.15
Aptamer-driven riboswitches are attractive building blocks in synthetic biology because RNA modules with specificity for any ligand of choice can be generated by the SELEX technology (systematic evolution of ligands by exponential enrichment),16,17 SELEX enriches aptamers by an in vitro selection process that is based on iterative cycles of incubation of an RNA pool with the favored ligand and removal of unbound RNAs by washing steps. Using this method, several aptamers had been engineered de novo long before the first natural riboswitches were discovered.18 Among these synthetic aptamers is the highly selective theophylline aptamer, which discriminates between the structurally related compounds theophylline and caffeine just by the presence of an H and a CH3 group, respectively.19 The theophylline aptamer served as backbone for several designer riboswitches with unprecedented functionalities. For instance, a theophylline riboswitch regulates translation by a helix slippage mechanism.20 The riboswitch is positioned close to the ribosomal binding site, thereby hindering ribosome access. Ligand binding shifts the riboswitch by one nucleotide into a position that allows ribosome binding and translation initiation. In another approach, the theophylline aptamer was used to engineer transcriptional on-switches de novo.21 The theophylline aptamer was separated from a sequence with partial complementarity to the aptamer by a spacer region ranging from 6 to 20 nucleotides in length. In the absence of theophylline, a terminator composed of the aptamer 3’-part, the spacer region and a poly(U) tail inhibited transcription, while the binding-competent structure allowed read-through (Fig. 2A). The theophylline aptamer was also used to construct a series of ligand-inducible riboswitches that control translation in various Gram-negative and Gram-positive bacteria.22 In every species at least one of the 5 tested riboswitches repressed translation in the absence of theophylline and provided a clear increase in gene expression (at least 25-fold) when the ligand was present.
Figure 2.
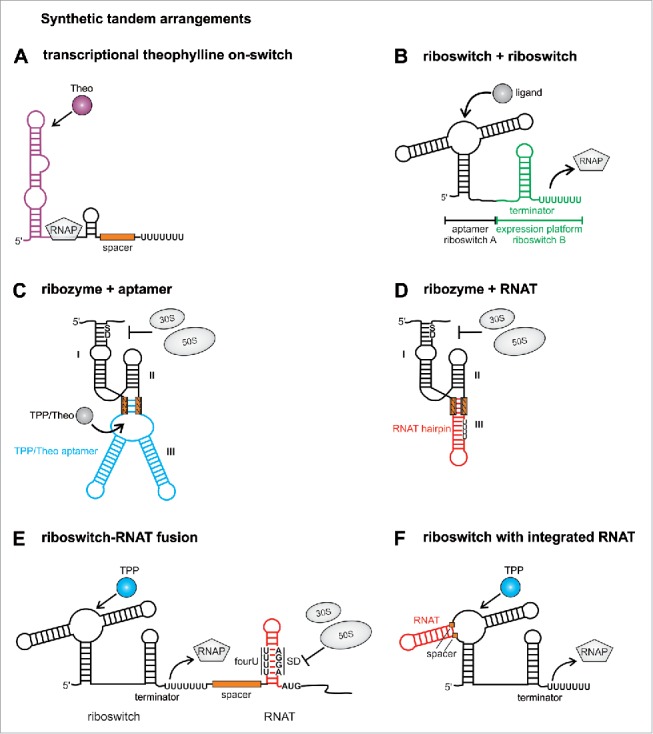
Synthetic tandem riboswitches. (A) The theophylline aptamer domain was coupled to an expression platform resulting in the first synthetic transcriptional on switch. (B) An RNA chimera consisting of aptamers and expression platforms from different riboswitches was designed without the need of a communication module. (C) An aptazyme was constructed by fusing the hammerhead ribozyme to the TPP or theophylline aptamer, respectively. (D) The temperature responsive fourU RNAT hairpin was substituted against hairpin III of the hammerhead ribozyme leading to temperature dependent mRNA cleavage. (E) Tandem arrangement of riboswitch and RNAT leads to transcriptional and translational control by the two inputs ligand binding and temperature sensing. (F) An RNAT is integrated into a riboswitch aptamer resulting in a temperature controlled riboswitch. RNAP: RNA polymerase, TPP: thiamine pyrophosphate, Theo: theophylline, fourU: fourU motif, SD: Shine Dalgarno sequence, AUG: translational start codon. Positions of inserted spacers or communication modules are labeled with orange boxes.
A prerequisite for the design of more complex RNA-based devices is that the used RNA elements retain their functions in a foreign genetic context. Several studies demonstrated that regulatory RNA elements from different sources can be combined in a modular fashion. Coupling of aptamer domain and expression platforms from different riboswitches demonstrated their high variability.23 Six aptamer domains, including 2 synthetic aptamers (theophylline and tetracycline) were fused to the expression platforms of the metE, yitJ or lysC riboswitches from B. subtilis (Fig. 2B). Each aptamer was able to direct the chosen expression platform in in vitro transcription studies as well as in an in vivo reporter gene system in Escherichia coli. The main criteria for the selection was that aptamer domain and expression platform did not overlap in their sequence requirements in the riboswitch P1 helix.23 Thus, chimeric riboswitches could be engineered by a simple combination of already existing building blocks suggesting a profound modularity of regulatory RNA elements.
A general drawback of RNA-based systems is that they often suffer from high background activity and a low dynamic range.23,24 To reduce background activity and increase the dynamic range, riboswitches can be toggled in series as demonstrated for synthetic theophylline or tetracycline riboswitches.25,26 A serial arrangement of 2 or 3 transcriptional theophylline riboswitches drastically reduced read-trough by the RNA polymerase in E. coli, while addition of a second or third copy of the tetracycline aptamer improved the strength of regulation in a GFP-based reporter gene system in yeast.25,26 To control gene expression in response to more than one signal, multi-input systems can be constructed by the assembly of RNA modules specific for the desired input pathways.
A number of novel functionalities have been generated by such combinatorial approaches. For instance, catalytic RNA cleavage of a ribozyme was coupled to ligand sensing of an aptamer domain leading to a so-called aptazyme, in which ligand binding triggers mRNA cleavage.27,28 The synthetic theophylline aptamer or the natural TPP aptamer were used to replace hairpin III of a hammerhead ribozyme from Schistosoma mansonii. A communication module derived from 3 to 6 randomized nucleotides was necessary for a functional connection of both elements (Fig. 2C). Aptazymes that cleaved the mRNA in response to the cognate ligands theophylline and TPP were designed. The theophylline aptazyme cleaved the mRNA in presence of the ligand thereby liberating the SD sequence and initiating translation of the mRNA. Interestingly, in a screen for aptazymes based on the TPP aptamer both positive and negative regulators were found.27 A similar design consisting of the theophylline RNA aptamer and a hammerhead ribozyme was reported to be functional in yeast29 demonstrating that at least theophylline aptazymes are applicable in different kingdoms of life and retain their functionality in various genetic contexts.
A follow-up study demonstrated that ribozyme cleavage can also be triggered by a temperature shift. The hammerhead ribozyme was fused to a temperature-responsive RNA hairpin, called RNA thermometer (RNAT).30 RNATs form temperature-sensitive RNA structures31 allowing translation of the mRNA only at elevated temperatures. At low temperatures a stable hairpin is formed in the 5′UTR, which occludes the Shine-Dalgarno (SD) sequence thereby inhibiting binding of the 30S ribosomal subunit. With increasing temperatures, the RNAT hairpin unfolds partially and releases the SD sequence leading to translation initiation. Replacement of hairpin III of the hammerhead ribozyme by an RNAT resulted in an outcome opposite to natural RNATs. While typical RNATs induce translation at high temperatures, the synthetic thermozyme cleaves the mRNA at low temperatures (when the overall structure is intact) thereby liberating the SD sequence. In contrast, high temperatures unfold the RNAT, inhibit the cleavage reaction and prevent translation (Fig. 2D).
Another design strategy used tetracycline aptamers to mediate pre-mRNA splicing in yeast.32 The efficiency of regulation could be improved when the construct with the highest regulation factor was combined with a second tetracycline aptamer that controlled translation initiation. This led to an increase of reporter gene activity from 16-fold for the single aptamer controlling splicing to 32-fold for the tandem arrangement regulating splicing and translation.
Motivated by these successful mix-and-match strategies, we engineered several novel RNA regulators based on riboswitch and RNAT activity.33 In a first strategy we designed a tandem arrangement of riboswitch and RNAT (Fig. 2E). Two transcriptional off-switches binding TPP34,35 or lysine36-38 and one on-switch recognizing theophylline were placed in front of the fourU RNAT from Salmonella enterica39 resulting in a 2-input system controlled on the transcriptional and translational level by ligand binding and temperature sensing, respectively. Only the proper combination of elevated temperature and presence or absence of a ligand, depending on whether an on- or off-switch was used, led to efficient expression of the downstream gene. Contrary to known natural tandem arrangements, the riboswitch-RNAT fusions respond to a chemical and a physical cue. They control gene expression on 2 layers and are able to reduce background activity as compared with the individual control elements.33
In a second approach, riboswitches were rendered susceptible to temperature by the substitution of an internal hairpin of the riboswitch aptamer domain by an RNAT (Fig. 2F). The tenA TPP riboswitch34,35 and the lysC lysine riboswitch36-38 from B. subtilis were used as scaffold for the design of these thermoswitches. By the exchange of one hairpin in the riboswitch aptamer domain against the RNAT, the riboswitch acquired temperature sensing ability. Ligand binding to the aptamer is only possible at low temperatures when the RNAT hairpin is in a stable double-stranded conformation and maintains the aptamer structure. A temperature upshift unfolds the RNAT hairpin thereby deforming the ligand-binding pocket and impeding the conformational switch to the off-state.33 Thus, the conformation of the RNAT hairpin controls the regulatory outcome of the riboswitch in a temperature-dependent manner resulting in a thermosensitive riboswitch.
Challenges of synthetic RNA design
Over the last years, numerous new RNA regulators have been engineered by combination of naturally occurring RNA elements or via de novo design using in vitro selection or computationally designed RNA sequences. A common challenge of RNA design remains that the regulatory outcome of the engineered RNAs are difficult to predict and the functionality of every newly designed regulator has to be tested in vivo, preferably directly in the organism of choice. Occasionally, RNA modules can be combined directly without further optimization, whereas other engineered RNA regulators need extensive improvement to be functional. A study by Ceres and coworkers demonstrated that riboswitch aptamer domains, including natural and synthetic aptamers, can be mixed and matched with expression platforms derived from 3 riboswitches.23 The chosen expression platform was controlled by the cognate ligand of each aptamer and no spacer or communication modules were necessary for cooperation of both domains (Fig. 2B). In contrast, the aptazymes consisting of the hammerhead ribozyme and the TPP or theophylline aptamers and the thermozyme comprising the hammerhead ribozyme coupled with an RNAT needed comprehensive follow-up work. Hundreds of variants had to be screened in vivo to find a few functional constructs that triggered ribozyme activity in response to the cognate ligand or in response to temperature.27,28,30 Short sequences of 3 to 6 randomized nucleotides were used to connect the selected RNA modules and to facilitate communication between mRNA cleavage and ligand binding or temperature sensing (Fig. 2C). Several screening strategies like colorimetric assays,40 flow cytometry41 or motility assays42 have been developed to facilitate a fast and efficient survey of new regulators.
In some cases 2 individual RNA components can be fused to a functional regulator with only minor adjustments. For instance, linkers of a defined length can be used to reduce background activity as shown for the transcriptional theophylline on-switches.21 Most of the designed theophylline riboswitches displayed low on/off ratios but background activity could be strongly reduced when a 19 nt spacer was inserted between the terminator and the ribosomal binding site of the downstream reporter gene (Fig. 2A). Moreover, the dynamic range could be improved by toggling 2 or 3 of the engineered riboswitches in series thereby increasing the on/off ratio from 3-fold to 10-fold or 23-fold.25 Spacer sequences can also be applied to overcome folding problems when 2 RNAs are connected. In our study, a linker of 20 nt was inserted between the theophylline-sensing riboswitch and the consecutive fourU RNAT to guarantee correct folding of each individual element (Fig. 2E).33 While the 2 off-switches used in our study could be combined with the RNAT without further improvement, the thermoswitch based on the TPP riboswitch needed insertion of single nucleotides as spacers to ensure that riboswitch activity depended on temperature (Fig. 2F). In contrast, the thermoswitch adapted from a lysine riboswitch was functional at the first attempt although the structure of the lysine aptamer is more complex than the TPP aptamer structure.33
The examples presented in this article demonstrate that regulatory RNA elements can be mixed and matched in various combinations but often require adjustments to be functional in the desired fashion. It is not at all clear why some RNAs are more suitable than others. The ongoing discovery of new riboswitches,43,44 ribozymes45 and RNA thermometers,46 opens new playgrounds for the artificial arrangement of sensory and regulatory RNA modules.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgement
We are grateful to Lisa-Marie Bittner for critical reading of the manuscript.
Funding
This work was supported by a grant from the German Research Foundation (NA 240/10–1) to F.N. and by a fellowship from the Studienstiftung des Deutschen Volkes to J.R.
References
- 1.Cech TR, Zaug AJ, Grabowski PJ. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell 1981; 27:487-96; PMID:6101203; http://dx.doi.org/ 10.1016/0092-8674(81)90390-1 [DOI] [PubMed] [Google Scholar]
- 2.Peselis A, Serganov A. Themes and variations in riboswitch structure and function. Biochim Biophys Acta 2014; 1839:908-18; PMID:4643838; http://dx.doi.org/ 10.1016/j.bbagrm.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serganov A, Nudler E. A decade of riboswitches. Cell 2013; 152:17-24; PMID:4215550; http://dx.doi.org/ 10.1016/j.cell.2012.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell 2011; 43:867-79; PMID:4140403; http://dx.doi.org/ 10.1016/j.molcel.2011.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 2004; 306:275-9; PMID:15472076; http://dx.doi.org/ 10.1126/science.1100829 [DOI] [PubMed] [Google Scholar]
- 6.Ruff KM, Strobel SA. Ligand binding by the tandem glycine riboswitch depends on aptamer dimerization but not double ligand occupancy. RNA 2014; 20:1775-88; PMID:4201829; http://dx.doi.org/ 10.1261/rna.047266.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol 2004; 11:29-35; PMID:14718920; http://dx.doi.org/ 10.1038/nsmb710 [DOI] [PubMed] [Google Scholar]
- 8.Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA 2007; 13:573-82; PMID:1831863; http://dx.doi.org/ 10.1261/rna.407707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poiata E, Meyer MM, Ames TD, Breaker RR. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA 2009; 15:2046-56; PMID:2764483; http://dx.doi.org/ 10.1261/rna.1824209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science 2006; 314:300-4; PMID:17038623; http://dx.doi.org/ 10.1126/science.1130716 [DOI] [PubMed] [Google Scholar]
- 11.González JC, Peariso K, Penner-Hahn JE, Matthews RG. Cobalamin-independent methionine synthase from Escherichia coli: a zinc metalloenzyme. Biochemistry 1996; 35:12228-34; PMID:8823155; http://dx.doi.org/ 10.1021/bi9615452 [DOI] [PubMed] [Google Scholar]
- 12.Pejchal R, Ludwig ML. Cobalamin-independent methionine synthase (MetE): a face-to-face double barrel that evolved by gene duplication. PLoS Biol 2005; 3:e31; PMID:539065; http://dx.doi.org/ 10.1371/journal.pbio.0030031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weigand JE, Suess B. Aptamers and riboswitches: perspectives in biotechnology. Appl Microbiol Biotechnol 2009; 85:229-36; PMID:19756582; http://dx.doi.org/ 10.1007/s00253-009-2194-2 [DOI] [PubMed] [Google Scholar]
- 14.Wittmann A, Suess B. Engineered riboswitches: Expanding researchers' toolbox with synthetic RNA regulators. FEBS Lett 2012; 586:2076-83; PMID:22710175; http://dx.doi.org/ 10.1016/j.febslet.2012.02.038 [DOI] [PubMed] [Google Scholar]
- 15.Liang JC, Bloom RJ, Smolke CD. Engineering biological systems with synthetic RNA molecules. Mol Cell 2011; 43:915-26; PMID:3176441; http://dx.doi.org/ 10.1016/j.molcel.2011.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990; 249:505-10; PMID:2200121 ; http://dx.doi.org/ 10.1126/science.2200121 ; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 17.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature 1990; 346:818-22; PMID:1697402; http://dx.doi.org/ 10.1038/346818a0 [DOI] [PubMed] [Google Scholar]
- 18.Vazquez-Anderson J, Contreras LM. Regulatory RNAs: charming gene management styles for synthetic biology applications. RNA Biol 2013; 10:1778-97; PMID:3917981; http://dx.doi.org/ 10.4161/rna.27102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science 1994; 263:1425-9; PMID:7510417 http://dx.doi.org/ 10.1126/science.7510417 [DOI] [PubMed] [Google Scholar]
- 20.Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res 2004; 32:1610-4; PMID:390306; http://dx.doi.org/ 10.1093/nar/gkh321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wachsmuth M, Findeiss S, Weissheimer N, Stadler PF, Mörl M. De novo design of a synthetic riboswitch that regulates transcription termination. Nucleic Acids Res 2013; 41:2541-51; PMID:3575828; http://dx.doi.org/ 10.1093/nar/gks1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Topp S, Reynoso CM, Seeliger JC, Goldlust IS, Desai SK, Murat D, Shen A, Puri AW, Komeili A, Bertozzi CR, et al. Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl Environ Microbiol 2010; 76:7881-4; ; http://dx.doi.org/ 10.1128/AEM.01537-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceres P, Garst AD, Marcano-Velázquez JG, Batey RT. Modularity of select riboswitch expression platforms enables facile engineering of novel genetic regulatory devices. ACS Synth Biol 2013; 2:463-72; PMID:3742664; http://dx.doi.org/ 10.1021/sb4000096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chappell J, Watters KE, Takahashi MK, Lucks JB. A renaissance in RNA synthetic biology: new mechanisms, applications and tools for the future. Curr Opin Chem Biol 2015; 28:47-56; PMID:26093826; http://dx.doi.org/ 10.1016/j.cbpa.2015.05.018 [DOI] [PubMed] [Google Scholar]
- 25.Wachsmuth M, Domin G, Lorenz R, Serfling R, Findeiss S, Stadler PF, Mörl M. Design criteria for synthetic riboswitches acting on transcription. RNA Biol 2015; 12:221-31; PMID:25826571; http://dx.doi.org/ 10.1080/15476286.2015.1017235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kötter P, Weigand JE, Meyer B, Entian KD, Suess B. A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res 2009; 37:e120; ; http://dx.doi.org/ 10.1093/nar/gkp578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wieland M, Benz A, Klauser B, Hartig JS. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew Chem Int Ed Engl 2009; 48:2715-8; PMID:19156802; http://dx.doi.org/ 10.1002/anie.200805311 [DOI] [PubMed] [Google Scholar]
- 28.Wieland M, Hartig JS. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew Chem Int Ed Engl 2008; 47:2604-7; PMID:18270990; http://dx.doi.org/ 10.1002/anie.200703700 [DOI] [PubMed] [Google Scholar]
- 29.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science 2008; 322:456-60; PMID:2805114; http://dx.doi.org/ 10.1126/science.1160311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saragliadis A, Krajewski SS, Rehm C, Narberhaus F, Hartig JS. Thermozymes: Synthetic RNA thermometers based on ribozyme activity. RNA Biol 2013; 10:1010-6; PMID:4111729; http://dx.doi.org/ 10.4161/rna.24482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krajewski SS, Narberhaus F. Temperature-driven differential gene expression by RNA thermosensors. Biochim Biophys Acta 2014; 1839:978-88; PMID:24657524; http://dx.doi.org/ 10.1016/j.bbagrm.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 32.Weigand JE, Suess B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res 2007; 35:4179-85; ; http://dx.doi.org/ 10.1093/nar/gkm425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roßmanith J, Narberhaus F. Exploring the modular nature of riboswitches and RNA thermometers. Nucleic Acids Res 2016; 44:5410-23; PMID:27060146 http://dx.doi.org/ 10.1093/nar/gkw232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol 2005; 12:1325-35; PMID:16356850; http://dx.doi.org/ 10.1016/j.chembiol.2005.10.007 [DOI] [PubMed] [Google Scholar]
- 35.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 2002; 111:747-56; PMID:12464185; http://dx.doi.org/ 10.1016/S0092-8674(02)01134-0 [DOI] [PubMed] [Google Scholar]
- 36.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev 2003; 17:2688-97; PMID:280618; http://dx.doi.org/ 10.1101/gad.1140003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grundy FJ, Lehman SC, Henkin TM. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci U S A 2003; 100:12057-62; PMID:218712; http://dx.doi.org/ 10.1073/pnas.2133705100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch? Nucleic Acids Res 2003; 31:6748-57; PMID:290268; http://dx.doi.org/ 10.1093/nar/gkg900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. FourU: a novel type of RNA thermometer in Salmonella. Mol Microbiol 2007; 65:413-24; PMID:17630972 http://dx.doi.org/ 10.1111/j.1365-2958.2007.05794.x [DOI] [PubMed] [Google Scholar]
- 40.Lynch SA, Desai SK, Sajja HK, Gallivan JP. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem Biol 2007; 14:173-84; PMID:1858662; http://dx.doi.org/ 10.1016/j.chembiol.2006.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lynch SA, Gallivan JP. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Res 2009; 37:184-92; PMID:2615613; http://dx.doi.org/ 10.1093/nar/gkn924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Topp S, Gallivan JP. Random walks to synthetic riboswitches–a high-throughput selection based on cell motility. Chembiochem 2008; 9:210-3; PMID:18098254 http://dx.doi.org/ 10.1002/cbic.200700546 [DOI] [PubMed] [Google Scholar]
- 43.Dar D, Shamir M, Mellin JR, Koutero M, Stern-Ginossar N, Cossart P, Sorek R. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 2016; 352:aad9822; PMID:27120414; http://dx.doi.org/ 10.1126/science.aad9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sommer MO, Suess B. RIBOSWITCHES. (Meta-)genome mining for new ribo-regulators. Science 2016; 352:144-5; PMID:27124438; http://dx.doi.org/ 10.1126/science.aaf6189 [DOI] [PubMed] [Google Scholar]
- 45.Weinberg Z, Kim PB, Chen TH, Li S, Harris KA, Lunse CE, Breaker RR. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol 2015; 11:606-10; PMID:26167874; http://dx.doi.org/ 10.1038/nchembio.1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Righetti F, Nuss AM, Twittenhoff C, Beele S, Urban K, Will S, Bernhart SH, Stadler PF, Dersch P, Narberhaus F. Temperature-responsive in vitro RNA structurome of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A 2016; 113:7237-42; PMID:27298343; http://dx.doi.org/ 10.1073/pnas.1523004113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol 2012; 4:a003566; PMID:3281570; http://dx.doi.org/ 10.1101/cshperspect.a003566 [DOI] [PMC free article] [PubMed] [Google Scholar]


