Abstract
The distribution and demographic patterns of marine organisms in the north Atlantic were largely shaped by climatic changes during the Pleistocene, when recurrent glacial maxima forced them to move south or to survive in northern peri-glacial refugia. These patterns were also influenced by biological and ecological factors intrinsic to each species, namely their dispersion ability. The ballan wrasse (Labrus bergylta), the largest labrid fish along Europe's continental margins, is a target for fisheries and aquaculture industry. The phylogeographic pattern, population structure, potential glacial refugia and recolonization routes for this species were assessed across its full distribution range, using mitochondrial and nuclear markers. The existence of a marked population structure can reflect both recolonization from three distinct glacial refugia and current and past oceanographic circulation patterns. Although isolated in present times, shared haplotypes between continental and Azores populations and historical exchange of migrants in both directions point to a common origin of L. bergylta. This situation is likely to be maintained and/or accentuated by current circulation patterns in the north Atlantic, and may lead to incipient speciation in the already distinct Azorean population. Future monitoring of this species is crucial to evaluate how this species is coping with current environmental changes.
Keywords: population structure, Labridae, cleaner fish, glacial refugia, Azorean distinctiveness, incipient speciation
1. Background
The northeastern Atlantic experienced considerable climatic changes during the Pleistocene when glacial cycles acted as a major driver shaping the structure of marine communities [1]. With the glaciation of the North Sea and the loss of the warm temperate regime along the west European continental margins [1–3], marine populations were forced to move towards southernmost regions or to survive in northern peri-glacial refugia [4]. During glacial maxima the polar front extended far south and reached the north of the Bay of Biscay [3] and possibly the western Iberian coast [2,5]. Despite the drastic changes in sea surface temperature [6], several glacial refugia have been evidenced, including areas around northern Norway, the Faeroes and Iceland, the Hürd Deep, southwest Ireland and southwest Britain, southwest Continental Europe and the Mediterranean Sea [4]. The rising temperatures and resulting regression of the polar front during interglacial periods would then allow the recolonization of northern regions [7].
These long-term environmental changes led to the emergence of distinct distribution patterns and, at the same time, to similarities between the geographical ranges of species that share similar environmental tolerance ranges [8]. Within this framework, many coastal species are thought to have experienced expansions and contractions of their geographical ranges associated with demographic changes, reflecting their biogeographic origins and environmental tolerances. As such, it could be expected that warm temperate and tropical species may have only survived in the warmer Mediterranean or along the West African coast, while cold tolerant species may have also survived in northern refugia [9]. In addition, there are multiple factors involved in the changing geographical distribution of populations and species, some of them related to other environmental variables, species-specific characters such as life-history traits and stochastic effects.
It should also be expected that distinct species coped differently with the above-referred constraints. Even closely related sympatric species with similar environmental tolerances may present distinct genetic structures and demographic history patterns (for a review see [10–12]). This should explain why we can find in the northeast Atlantic fish species showing: (i) panmictic populations without latitudinal differences in genetic diversity (e.g. Lipophrys pholis [13]); (ii) significant population structure but still with similar levels of genetic diversity throughout the entire species range (e.g. Taurulus bubalis [14]); or (iii) sharp decline of genetic diversity from southern (west European) to northern (Scandinavian) populations (e.g. Pomatoschistus microps [15]; Symphodus melops [16]). Similarly, studies have indicated that some species may have persisted in northern regions even during glacial maxima (e.g. Pholis gunnellus [17]), while other only subsisted near the North Sea [14,15] or within the Mediterranean (e.g. Chromis chromis [18]; Sprattus sprattus [19]).
The ballan wrasse, Labrus bergylta Ascanius, 1767, is the largest labrid fish in continental waters of the northeastern Atlantic [20], presently ranging from southern Norway to the west coast of Morocco and the Macaronesian archipelagos [21]. It is a sedentary, obligate protogynous hermaphrodite species whereby dominant territorial males derive from sex-changing mature females, which typically live in harems [22]. Adhesive eggs are laid in rocky substrates and larvae are released to the plankton [23].
It has attracted considerable attention due to three main reasons: (i) it is a target for commercial [22] and recreational fisheries [24], (ii) it is currently used as a cleaner species in northern Europe salmonid fish farms as an alternative to chemical treatments [25,26], and (iii) its phenotypic plasticity and life-history variation have raised questions about its taxonomic status and the possibility that one or more cryptic species could exist.
In recent years, several consensual and quite complete phylogenies have been published on both the tribe Labrini (where the genus Labrus is included) [27] and on the family Labridae [28]. At the same time, the systematic classification of the northeastern Atlantic and Mediterranean Labridae had been revised due to the recent split of some taxa and the high intraspecific polymorphism, particularly in the genera Labrus and Symphodus ([27,29] and references therein). Hanel et al. [27] reported high intraspecific genetic divergence in L. bergylta and its systematic status has raised additional interest because [30] (and references therein) showed differences in life-history traits of two main morphotypes (plain and spotted body colour patterns). Genetic differences between morphotypes were reported in [31] but only with microsatellites in one restricted region. Recently, [32] found no differences between morphotypes with genetic markers widely used in fish phylogeography.
A phylogeographic study of the ballan wrasse based on the control region (CR) of mitochondrial DNA has recently been published in [33]. Focusing in the northern distribution range of the species, around the British Isles and southern Norway, this study showed reduced levels of mitochondrial genetic diversity towards northern latitudes, and the presence of two divergent clades showing evidence of population expansion.
This study extends the previous study in [33] and our potential knowledge of the species phylogeography and genetic structure by: (i) covering the species geographical distribution and (ii) analysing a nuclear fragment (the first intron of the S7 ribosomal protein gene) in addition to the mitochondrial CR fragment. The present genetic assessment will aid conservation and management of the species by providing essential information: (i) on potential effects associated with individual translocations or the exploitation of wild cleaner fish in salmonid fish farms in the North Sea and (ii) for stock assessment studies of local fisheries in the central northeastern Atlantic. To date this is the most comprehensive study describing the genetic structure, the signatures of expansion/contraction events and identifying potential Pleistocene refugia of the ballan wrasse along its distributional range.
2. Material and methods
2.1. Sampling
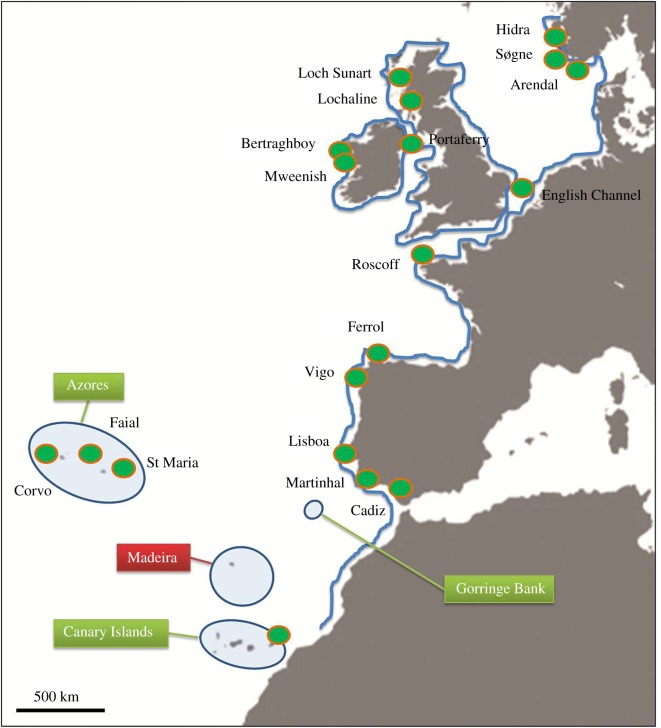
Specimens of L. bergylta were obtained from 14 locations along its distributional range in the northeastern Atlantic and North Sea (figure 1 and table 1). These sites included: Corvo, CO and Santa Maria, SM in the Atlantic archipelago of the Azores; Lisbon, LI in western Portugal; Vigo, VI and Ferrol, FE in northern Spain (Galicia); Roscoff, RO in France; Portaferry, PO in Northern Ireland; Mweenish, ME and Bertraghboy Bay, BB in western Ireland; Lochaline, LO and Loch Sunart, LS in western Scotland; Arendal, AR, Hidra, HI and Sogne, SO in southern Norway.
Figure 1.
Map of sampling locations for Labrus bergylta.
Table 1.
Diversity measures for the collecting sites and population groups of Labrus bergylta for CR and S7: number of sequences (N), number of haplotypes (Nh), private haplotypes (Ph), haplotype diversity (h), nucleotide diversity (π) and mean number of pairwise differences (PD).
| CR |
S7 |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| location | label | coordinates | N | Nh | Ph (%) | h | π | PD | N | Nh | Ph (%) | h | π | PD |
| Arendal | AR | 58°25′ N, 08°45′ E | 17 | 7 | 57 | 0.721 | 0.018 | 6.015 | — | — | — | — | — | — |
| Hidra | HI | 58°13′ N, 06°31′ E | 49 | 12 | 42 | 0.605 | 0.020 | 6.624 | 44 | 10 | 10 | 0.839 | 0.003 | 1.398 |
| Sogne | SO | 58°04′ N, 07°48′ E | 50 | 10 | 40 | 0.481 | 0.009 | 3.109 | 46 | 11 | 9 | 0.847 | 0.003 | 1.523 |
| Norway | 116 | 19 | 74 | 0.569 | 0.015 | 5.091 | ||||||||
| Loch Sunart | LS | 56°40′ N, 05°56′ W | 24 | 15 | 27 | 0.909 | 0.028 | 9.398 | 20 | 9 | 0 | 0.879 | 0.003 | 1.658 |
| Lochaline | LO | 56°31′ N, 05°46′ W | 26 | 17 | 24 | 0.938 | 0.025 | 8.335 | 40 | 10 | 20 | 0.869 | 0.003 | 1.613 |
| Portaferry | PO | 54°23′ N, 05°33′ W | 14 | 13 | 62 | 0.989 | 0.027 | 9.044 | 24 | 6 | 17 | 0.786 | 0.003 | 1.406 |
| Bertraghboy | BB | 53°19′ N, 09°51′ W | 44 | 22 | 64 | 0.970 | 0.029 | 9.616 | — | — | — | — | — | — |
| Mweenish | ME | 53°17′ N, 09°49′ W | 72 | 39 | 54 | 0.964 | 0.026 | 8.533 | 44 | 12 | 17 | 0.849 | 0.003 | 1.571 |
| Roscoff | RO | 48°43′ N, 03°58′ W | 27 | 22 | 68 | 0.977 | 0.023 | 7.547 | 46 | 12 | 17 | 0.850 | 0.003 | 1.543 |
| Ferrol | FE | 43°27′ N, 08°17′ W | 20 | 14 | 36 | 0.953 | 0.009 | 2.858 | 36 | 7 | 0 | 0.841 | 0.002 | 1.367 |
| Vigo | VI | 42°13′ N, 08°46′ W | 21 | 17 | 47 | 0.976 | 0.017 | 5.814 | 26 | 11 | 18 | 0.901 | 0.003 | 1.692 |
| Lisbon | LI | 38°42′ N, 09°24′ W | 34 | 22 | 59 | 0.955 | 0.019 | 6.241 | 68 | 14 | 21 | 0.810 | 0.002 | 1.302 |
| Europe | 282 | 120 | 96 | 0.964 | 0.024 | 7.857 | 394 | 27 | 96 | 0.856 | 0.003 | 1.533 | ||
| Santa Maria | SM | 36°57′ N, 25°06′ W | 15 | 13 | 92 | 0.971 | 0.013 | 4.362 | 24 | 5 | 20 | 0.696 | 0.002 | 0.880 |
| Corvo | CO | 39°41′ N, 31°05′ W | 15 | 12 | 92 | 0.971 | 0.016 | 5.343 | 26 | 5 | 40 | 1.062 | 0.002 | 1.062 |
| Azores | 30 | 24 | 100 | 0.982 | 0.015 | 4.880 | 50 | 7 | 86 | 0.696 | 0.002 | 0.971 | ||
| all | 428 | 158 | — | 0.946 | 0.037 | 12.198 | 444 | 33 | — | 0.862 | 0.003 | 1.583 | ||
All sampling and handling of fish were performed by experienced personnel in accordance with relevant legislation in each country (see the Ethics section for detailed information). A field campaign to Madeira in 2014 provided no fish samples or even sightings despite this species being listed to the region [34]. In all cases, a small piece of fin tissue was clipped and preserved in 96% ethanol.
2.2. DNA extraction, amplification and sequencing
Total genomic DNA was extracted with the REDExtract-N-Amp Kit (Sigma-Aldrich) following the manufacturer's instructions. The mitochondrial CR and the first intron of the nuclear S7 ribosomal protein gene (S7) were amplified using L-pro1 and H-DL1 [35], and S7RPEX1F and S7RPEX2R [36], respectively (see electronic supplementary material, appendix I for details).
Sequences were edited with Codon Code Aligner (Codon Code Corporation) and aligned with Clustal X 2.1 [37]. Whenever possible, both strands of the same specimen were recovered for S7 following the approach of [38]. Sequences obtained were deposited in GenBank (accession nos. KU751889-KU752182). Additional sequences available in GenBank were obtained from [32,33] (electronic supplementary material, table S1 provides the original reference for each individual sequence).
2.3. DNA analyses
The appropriate model of sequence evolution for each fragment was determined using the jModeltest program [39,40], applying the Akaike information criterion [41]. Haplotype networks were built with the software TCS 1.21 [42] using the parsimony method in [43]. For these analyses, some additional sequences from Faial, FA (Azores); Canary Islands, CI; Martinhal, MA and Cádiz, CA (south Iberia); and English Channel, EC were added in order to extend the sampling area (electronic supplementary material, table S1).
The ARLEQUIN software package v. 3.5 [44] was used to estimate genetic diversity within each sample, to evaluate potential population differentiation and to perform neutrality tests. The same software was used to perform analyses of molecular variance (AMOVA) [45], to compute pairwise FST estimates and corrected pairwise differences between populations. In the case of the S7 gene fragment, the analyses were also run in ARLEQUIN after allowing the program to reconstruct the haplotypes using the ELB algorithm [46]. The number of migrants was calculated for each population pair using the same software. The correlation between geographical distance and FST was computed with the Mantel test [47,48] (also in ARLEQUIN with 10 000 permutations; geographical distances measured along the shore line).
The spatial analysis of molecular variance (SAMOVA 1.0) [49] was used to identify groups of sampling locations that are geographically and genetically homogeneous and maximally differentiated from each other. The most likely number of groups was identified by running SAMOVA with 2 to 13 groups and choosing the partition scheme with the highest FCT value. The sequences of the locations included in each of the groups that maximized FCT were pooled together. Mismatch analysis [50,51], Fu's Fs [52] and Tajima's D [53] tests were performed in ARLEQUIN to test for possible bottlenecks and population expansion in each group. For the mismatch analyses parameters τ, θ0, θ1 and M and their confidence intervals were obtained by a parametric bootstrap approach using 10 000 replicates. Effective population sizes were calculated before (N0) and after (N1) a sudden expansion event, the effective population size prior to a spatial expansion event (N), the time since the expansion (t, in years), and the migration rate (m), using the equations τ = 2μτ and θ = 2Nμ.
A Markov chain Monte Carlo (MCMC) approach taking into account phylogenetic relationships among haplotypes as implemented in LAMARC 2.1.9 [54] was used to estimate effective population size (Nef), the exponential growth parameter (g) and the migration rates among adjacent groups of populations, using 10 runs of 12 short chains of 1000 steps and five long chains of 50 000 steps, with a burn-in of 10 000 steps. In order to compute estimates of effective population size, their changes with time and the age of populations, the following mutation rates were used: 5% for CR [55] and 0.23% for S7 [56]. MtDNA CR mutation rates in fish are widely variable (e.g. 2.2–4.5%/MY between lineages for East African cichlids [57]; 15–20%/MY for Indo-Pacific sardines [58]). In the absence of a clock calibration for the CR of L. bergylta, we address the uncertainty by tentatively assuming a within lineage mutation rate of 5%/MY, which is within the range of values found for other fish species
Past population demography of L. bergylta was inferred using the linear Bayesian skyline plot (BSP) model [59] as implemented in BEAST v. 1.7 [60], employing the Bayesian MCMC coalescent method and a strict clock. The Bayesian distribution was generated using results from five independent runs of 150 million MCMC steps obtaining effective samples sizes (ESS) of parameter estimates of over 200, with a burn-in of 10%. The time to most recent common ancestor (tMRCA) and the median and corresponding credibility intervals of the BSP were depicted using Tracer v. 1.6 [61].
3. Results
The CR dataset consisted of a total of 333 bp fragment after alignment (433 sequences) and yielded 160 distinct haplotypes with 93 polymorphic sites (94 transitions, 12 transversions and six indels). For the S7, a fragment of 537 bp was analysed, with 39 haplotypes obtained for the 478 sequences (corresponding to 239 individuals). Differences between haplotypes included 12 transitions, six transversions and no indels, in 18 polymorphic sites.
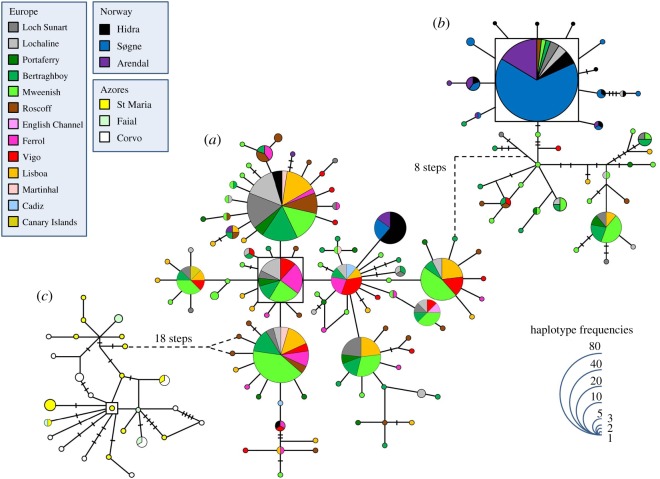
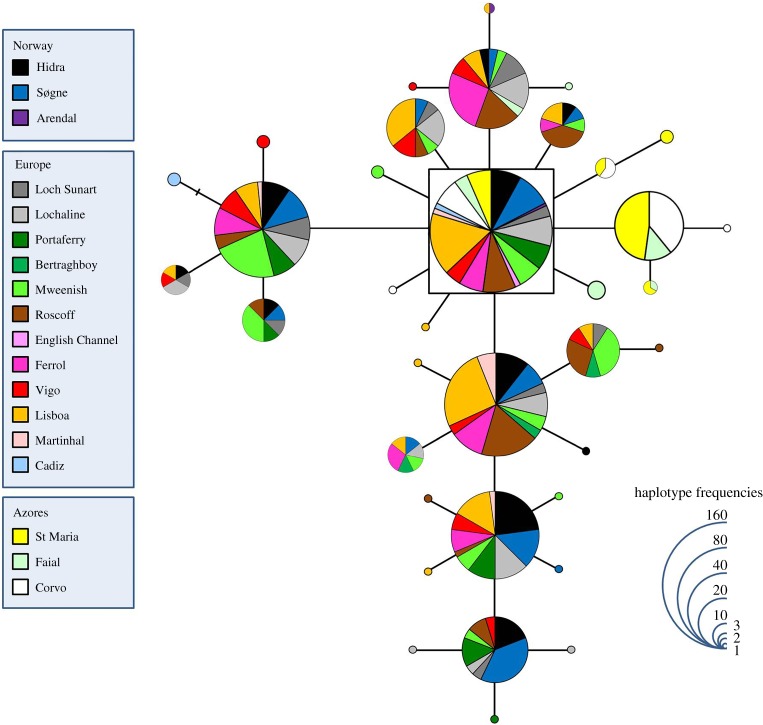
The haplotype network for the CR of L. bergylta revealed three distinct groups (figure 2). The first group (sub-network (a) in figure 2) included most sequences from the Atlantic European shores and some Norwegian samples, showing star-like patterns with haplotypes shared between several sampling locations. The inferred ancestral haplotype included 17 individuals from Spain, Ireland and Scotland (outgroup weight 0.096). The second sub-network grouped ((b) in figure 2) together the majority of the Norwegian and the remaining Atlantic European sequences. The ancestral haplotype inferred for this sub-network included 82 fish from Norway, Ireland, Scotland and France (outgroup weight 0.238). Interestingly, the only sample caught in the Canary Islands shared its haplotype with fish from Ireland, Scotland, northern Spain and western Portugal. The third sub-network ((c) in figure 2) includes only fish from the Azores (outgroup weight 0.109). The haplotype network obtained for the S7 of the ballan wrasse is less deep and diverse, and the geographical pattern obtained is not as obvious (figure 3). The inferred ancestral, dominant and in the centre of several star-like patterns, includes 138 fish from Norway, Ireland, Scotland, the English Channel, France, Spain and Portugal (mainland and Azores) (outgroup weight 0.104).
Figure 2.
(a–c) Haplotype network for the CR of Labrus bergylta. The haplotype with the highest out group probability is displayed as a square, other haplotypes as circles. The area of the circles is proportional to each haplotype frequency. Colours refer to the region in which haplotypes were found. In the case where haplotypes are shared among regions, shading is proportional to the frequency of the haplotype in each region.
Figure 3.
Haplotype network for the S7 of Labrus bergylta. The haplotype with the highest out group probability is displayed as a square, other haplotypes as circles. The area of the circles is proportional to each haplotype frequency. Colours refer to the region in which haplotypes were found. In the case where haplotypes are shared among regions, shading is proportional to the frequency of the haplotype in each region.
Table 1 showed the genetic diversity indices per sampling site. For the CR, haplotype diversity indices were higher for the Atlantic locations, comparing to Norwegian ones. Azorean locations presented a much higher percentage of private haplotypes when compared with all other sites. Concerning the S7, the genetic indices showed homogeneity along sites.
AMOVA analyses revealed genetic structure along the distribution area of L. bergylta for both genetic markers (CR: FST = 0.462, p < 0.001; S7: FST = 0.107, p < 0.001). Genetic differentiation and gene flow between sampling locations are shown in table 2 and corrected pairwise differences in electronic supplementary material, table S2. For the CR, significant FST and corrected pairwise differences were found between locations from Norway, Azores and the rest of European sites, confirming the existence of three populations. Significant differences were also found between FE and some other adjacent locations; however, migration among these locations was consistently higher than the threshold limit of 1. The pattern of differentiation was not as straightforward for the S7. Significant FST and corrected pairwise differences were found between Azorean and all other sites. These sites revealed no evident pattern, with significant differences being found between some of them. Considering the number of migrants (Nm), isolation was only found between the Azores and the remaining locations.
Table 2.
Gene flow among collecting sites of Labrus bergylta represented by FST (below diagonal) and Nm (number of migrants; above diagonal). Significant values of probability p are shown in italics.
| CR | AR | HI | SO | LS | LO | PO | BB | ME | RO | FE | VI | LI | SM | CO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AR | inf | 60.345 | 0.633 | 0.595 | 0.619 | 0.727 | 0.573 | 0.440 | 0.205 | 0.354 | 0.348 | 0.130 | 0.145 | |
| HI | −0.026 | 9.562 | 0.699 | 0.684 | 0.701 | 0.799 | 0.654 | 0.509 | 0.305 | 0.447 | 0.422 | 0.154 | 0.164 | |
| SO | 0.008 | 0.050 | 0.304 | 0.293 | 0.370 | 0.387 | 0.254 | 0.229 | 0.117 | 0.183 | 0.195 | 0.078 | 0.085 | |
| LS | 0.442 | 0.417 | 0.622 | inf | inf | inf | inf | inf | 5.787 | 38.305 | 31.941 | 0.195 | 0.212 | |
| LO | 0.457 | 0.422 | 0.630 | −0.032 | inf | inf | inf | inf | 6.311 | 184.129 | 32.578 | 0.180 | 0.196 | |
| PO | 0.466 | 0.433 | 0.663 | −0.037 | −0.039 | inf | inf | inf | 5.300 | inf | 98.994 | 0.174 | 0.193 | |
| BB | 0.408 | 0.385 | 0.563 | −0.020 | −0.018 | −0.036 | inf | 32.978 | 5.172 | 29.067 | 15.283 | 0.219 | 0.234 | |
| ME | 0.447 | 0.416 | 0.575 | −0.013 | −0.014 | −0.033 | −0.011 | 46.576 | 6.718 | 73.917 | 25.630 | 0.203 | 0.214 | |
| RO | 0.532 | 0.496 | 0.686 | −0.005 | −0.003 | −0.011 | 0.015 | 0.011 | 25.520 | inf | inf | 0.166 | 0.180 | |
| FE | 0.709 | 0.621 | 0.810 | 0.080 | 0.073 | 0.086 | 0.088 | 0.069 | 0.019 | 26.002 | 505.969 | 0.081 | 0.094 | |
| VI | 0.586 | 0.528 | 0.732 | 0.013 | 0.003 | −0.006 | 0.017 | 0.007 | −0.005 | 0.019 | inf | 0.125 | 0.140 | |
| LI | 0.590 | 0.543 | 0.719 | 0.015 | 0.015 | 0.005 | 0.032 | 0.019 | −0.010 | 0.001 | −0.013 | 0.139 | 0.150 | |
| SM | 0.793 | 0.764 | 0.865 | 0.720 | 0.735 | 0.742 | 0.695 | 0.712 | 0.751 | 0.860 | 0.799 | 0.783 | 44.69298 | |
| CO | 0.775 | 0.753 | 0.855 | 0.702 | 0.718 | 0.721 | 0.682 | 0.701 | 0.735 | 0.842 | 0.781 | 0.769 | 0.011 |
| S7 | HI | SO | LS | LO | PO | ME | RO | FE | VI | LI | SM | CO | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HI | inf | 6.893 | 38.323 | inf | 24.919 | 6.286 | 8.191 | 10.416 | 6.540 | 0.773 | 0.834 | |||
| SO | −0.014 | 8.104 | 30.246 | inf | 36.058 | 6.526 | 7.881 | 9.747 | 5.638 | 0.832 | 0.905 | |||
| LS | 0.068 | 0.058 | inf | 9.797 | 1653.034 | 11.143 | inf | inf | 10.972 | 1.315 | 1.506 | |||
| LO | 0.013 | 0.016 | −0.009 | 22.087 | 312.661 | 12.842 | 53.152 | inf | 23.044 | 1.133 | 1.284 | |||
| PO | −0.003 | −0.015 | 0.049 | 0.022 | inf | 3.958 | 5.402 | 13.078 | 3.684 | 0.744 | 0.781 | |||
| ME | 0.020 | 0.014 | 0.000 | 0.002 | −0.005 | 7.616 | 12.118 | 355.708 | 7.606 | 1.109 | 1.257 | |||
| RO | 0.074 | 0.071 | 0.043 | 0.037 | 0.112 | 0.062 | 153.494 | 8.173 | 152.166 | 1.311 | 1.540 | |||
| FE | 0.058 | 0.060 | −0.002 | 0.009 | 0.085 | 0.040 | 0.003 | 69.923 | 80.944 | 1.219 | 1.415 | |||
| VI | 0.046 | 0.049 | −0.037 | −0.014 | 0.037 | 0.001 | 0.058 | 0.007 | 10.130 | 1.178 | 1.327 | |||
| LI | 0.071 | 0.081 | 0.044 | 0.021 | 0.120 | 0.062 | 0.003 | 0.006 | 0.047 | 1.261 | 1.510 | |||
| SM | 0.393 | 0.375 | 0.275 | 0.306 | 0.402 | 0.311 | 0.276 | 0.291 | 0.298 | 0.284 | inf | |||
| CO | 0.375 | 0.356 | 0.249 | 0.280 | 0.390 | 0.285 | 0.245 | 0.261 | 0.274 | 0.249 | −0.007 |
SAMOVA results for the CR dataset yielded a maximized FCT (0.623, p = 0.009) for two groups, the two locations from the Azores (CO and SM) versus the 12 locations belonging to other European shores (AR, HI, SO, LS, LO, PO, BB, ME, RO, FE, VI and LI). The three gene pools structured test (Azores versus Norway versus all remaining sites in Continental Europe) was also significant (FCT = 0.610, p < 0.001). Concerning the S7, the SAMOVA analysis revealed a maximized FCT (0.268, p = 0.016) for two gene pools: Azores versus all remaining sites.
Isolation by distance was found for both the CR and the S7 of the ballan wrasse, as the Mantel test yielded significant correlations between genetic and geographical distances (r = 0.738, p < 0.001 and r = 0.573, p < 0.001, respectively). Taking together the results from the FST, Nm and SAMOVA, the following groups were considered for further demographic and phylogeographic analyses: three populations for the CR—Azores (CO and SM) versus Atlantic (LS, LO, PO, BB, ME, RO, FE, VI and LI) versus Norway (AR, HI and SO); and two populations for the S7—Azores (CO and SM) versus Continental Europe (HI, SO, LS, LO, PO, ME, RO, FE, VI and LI). These two schemes ensured highly significant differences (p < 0.001) among groups of populations: CR (FST Azores-Atlantic = 0.718, FST Azores-Norway = 0.801 and FST Atlantic-Norway = 0.519) and S7 (FST Azores-Continental Europe = 0.718).
Neutrality tests yielded negative values for both markers (table 3) with the exception of the Azorean population for the S7. However, significant values suggesting demographic expansion were only present for the FS of Azores and Atlantic (CR) and Continental Europe (S7).
Table 3.
Demographic parameters of Labrus bergylta based on CR and S7. Significant values of probability p are shown with an asterisk. Neutrality tests: Fs (Fu's), D (Tajima's). Mismatch distributions: t (time in years, 95% CI in parenthesis), N0 (female effective population size before the expansion), N1 (female effective population size after the expansion), SSD (sum of square deviation) and Hri (Harpending's Raggedness index). Estimates of population parameters with LAMARC: θ (theta), Nf (female effective population size), G (growth rate) and N1% (age of population, accessed as the age at which Nf drops below 1%). Estimates of tMRCA (time to most recent common ancestor) with BEAST. n.a., not applicable. nc, no convergence.
| CR |
S7 |
||||
|---|---|---|---|---|---|
| Azores | Atlantic | Norway | Azores | Continental Europe | |
| neutrality tests | |||||
| FS | −17.880* | −24.339* | −0.939 | −2.083 | −17.224* |
| D | −1.284 | −1.068 | −0.306 | 0.199 | −0.368 |
| mismatch distribution | |||||
| demographic expansion | |||||
| t (95% CI) (ky) | 154 (114–185) | 46 (16–261) | — | — | — |
| N0 | 106 | 130 378 | — | — | — |
| N1 | 3 002 972 973 | 3 002 972 973 | — | — | — |
| SSD | 0.008 | 0.017 | 0.406* | 0.012 | 0.011* |
| Hri | 0.029 | 0.011 | 0.112 | 0.125* | 0.102* |
| spatial expansion | |||||
| t (95% CI) (ky) | 154 (97–180) | 471 (28–898) | 519 (10–868) | — | — |
| N | 22 | 260 213 | 27 244 | — | — |
| Nm | 3 002 972 973 | 4848 | 12 426 | — | — |
| SSD | 0.008 | 0.026 | 0.027 | 0.012* | 0.011* |
| Hri | 0.029 | 0.011 | 0.112 | 0.125* | 0.102* |
| LAMARC | |||||
| θ (95% CI) | 0.196 (0.069–0.749) | 0.068 (0.033–0.181) | 0.003 (0.001–0.009) | 0.005 (0.001–0.036) | 0.006 (0.002–0.041) |
| Nf (95% CI) | 1 960 888 (692 093–7 498 888) | 678 830 (332 300–1,812,773) | 30 758 (11 270–93 085) | 1,082,120 (260 181–7,854,130) | 1 369 457 (398 533–8,842,717) |
| G (95% CI) | 361 (149–750) | 87 (−11–306) | −154 (−757–314) | 1230 (−1323–8023) | 1041 (−549–6,940) |
| N1% (95% CI) (ky) | 255 (123–619) | 1060 (300–n.a.) | n.a. (293–n.a.) | 1628 (250–n.a.) | 1923 (288–n.a.) |
| BEAST | |||||
| tMRCA (95% CI) (ky) | 220 (49–2230) | nc | 548 (113–4616) | 0 (0–0) | nc |
The analyses of the mismatch distributions for the CR of L. bergylta were compatible with the models of sudden and spatial expansion for the Azores and all remaining Atlantic populations, except for the sudden demographic expansion in the case of Norway (table 3). Visual inspection of the mismatch graphics reveals clear differences among the three populations: the Azorean population presents a narrower width in comparison with the other two populations, revealing a strong peak at five differences; the Atlantic population revealed a stronger peak at four differences, while Norway presented a peak at 0 differences (electronic supplementary material, figure S1). The average estimated time for the demographic expansion yielded 154 thousand years (ky) for the Azores and 46 ky for the Atlantic. Considering the average spatial expansion, it was estimated to have occurred in the Late Pleistocene for the Azores (154 kya) and in the Middle Pleistocene for the Atlantic (471 kya) and Norway (519 kya). The S7 dataset, on the other hand, did not conform to neither the demographic nor the spatial model, for both populations.
For the CR dataset of the ballan wrasse, higher female effective population size was found for the population of Azores, and the Norwegian group presented the lowest value for this parameter. The same pattern was found for the growth rates, with the Scandinavian population presenting a negative growth rate. For the S7, the yielded values were similar for the two analysed groups (table 3). BSP runs failed to converge for the CR of the Atlantic group and for the S7 of Continental Europe. Estimates of tMRCA were consistent with estimates from previous analyses.
For the CR of L. bergylta, it was not possible to estimate migration rates between the Azorean group and the rest, as the connectivity assumption of LAMARC was not met. Migration rate was higher from Norway to the Atlantic (20.564) than in the opposite direction (0.014). Considering the S7 dataset, the migration rate was higher from the Azores into the European group (390.521) versus 106.857 in the opposite direction.
4. Discussion
The results presented in this study highlight four main features concerning the phylogeography of the ballan wrasse. First, the northward decrease in genetic diversity along the species' distributional range. Second, the existence of genetic structure along the sampled area. Third, the distinctiveness of the Azorean population. Finally, the demographic and spatial expansion of most of the species' populations. Taken together, these findings agree with and expand those found in [33] for the North Sea.
4.1. Genetic diversity and population structure
In general, the pattern of haplotype diversity found for the ballan wrasse follows that reported for other fishes [15,16], with higher indices in the central north Atlantic and lower in the northern locations (CR dataset). The Azorean locations clearly stood out, with higher percentages of private haplotypes. This fact supports the distinctiveness of the population from this isolated archipelago in the central northern Atlantic. Similar results were described for other fish species like Coryphoblennius galerita [62] and L. pholis [63], calling attention to the genetic distinctiveness and importance of the Azorean fish populations.
Are these differences related to life-history traits such as the pelagic larval duration (PLD)? The correlation between the PLD and FST estimates has been found to be very strong in recent modelling approaches [64]. However, there is an ongoing debate on this topic, as other studies (e.g. [65]) refute the conventional use of PLD as a good predictor for the magnitude of gene flow. Several studies reported the absence of genetic structure for species with relatively long PLD, such as the rocky intertidal L. pholis [13] or the highly migratory Thunnus thynnus [66]. Although somehow expected, this is not a consensual pattern for species with such a life-history trait. In fact, population structure has also been recorded for a variety of fishes [14,62]. This also seems to be the case of the ballan wrasse, which has a relatively long PLD of 37–49 days [67] but revealed genetic structure along the sampled area, especially when considering the CR dataset. The locations from Norway, Azores and the rest of European sites were considered to be significantly different. Although the results from the slow evolving S7 gene do not show such a clear cut, Azores keeps its singularity, being isolated (with no migration) from the remaining European locations (average Nm of 0.160 between Azorean locations and the remaining ones). Similar findings related to the peripheral position of Azorean haplotypes were also reported for several other species [62,68].
Are these differences related to ocean current patterns in the northeastern Atlantic? The North Atlantic circulation pattern can at least partially explain the isolation of the Azorean population as it prevents gene flow between European and Azorean locations. This isolation is mainly related to the Azores recent Miocene origin (1–8 My [69–71]), its distance from the closest continental shore (approx. 1300 km from mainland Europe), and the fact that predominant sea surface currents cross the North Atlantic in an eastward direction. Main current patterns may show regional variations that can be detected at a seasonal scale [72], yearly scale from decades to thousand years [73], or may occasionally generate considerable mesoscale variability (e.g. according to [74] strong eddies promote northwestward transport from Africa or Madeira towards the Azores).
4.2. Complexity and depth of the genealogies
All the analyses reveal expansion events and population ages clearly older than the Last Glacial Maximum (LGM, 21–18 kya) [75] with lineages coalescing in Late and Middle Pleistocene. Similar findings were reported for other fish in northeastern Atlantic (e.g. [62]). The close relative S. melops, on the other hand, showed a different pattern with exclusive haplotypes for the Scandinavian population and an expansion event much younger than the LGM [16].
Migration rate was higher from Norway to the Atlantic than in the opposite direction (CR), unlike what was found for other species (e.g. [14,16]). Considering the S7 dataset, the migration rate was higher from the Azores into the European group compared with the opposite direction. This pattern is uncommon and needs further evaluation. Nevertheless, this historical migration was previously advanced in [76] in a model of blenniid speciation in the northeastern Atlantic. According to this author, fish from European mainland could occasionally be transported to the Atlantic islands and survive glacial maxima, potentially evolving to become incipient new species. The Azores would thus act as potential speciation centre, exporting them back to European shores by predominant currents (e.g. Parablennius ruber [77]).
4.3. Glacial refugia
For L. bergylta, the present results suggest three distinct refugia during glacial maxima: North Sea, western and southern Iberian coasts and Azores. Considering the fact that L. bergylta is able to breed at high latitudes in southern Norway [21], it is very likely that it was able to survive in European shores during the last glaciation. During the LGM, the North Sea surface was almost entirely frozen [2]. However, ballan wrasse populations may have survived in the vicinity of the North Sea and in more southern refugia. In fact, L. bergylta could have used unglaciated parts of these shores as glacial refugia, as previously suggested for other north Atlantic fish species (e.g. [14,16]). Our results agree with this hypothesis, namely the reported deep networks and haplotypes shared between northern and southern populations along the northeastern Atlantic.
Western and southern Iberian shores could have kept their populations and may have functioned as glacial refugia for this species. According to several authors, during the LGM the temperatures could have been kept within the thermal limits tolerated by L. bergylta (e.g. [2,5]). While analysing the post-glacial recolonization events in the marine coastal environment, one must also consider the close dependence between dispersal patterns and the ecology, life-history traits and behaviour of the various organisms [7]. The relatively long PLD reported for L. bergylta is probably related to the low degree of genetic differentiation found in this study along European coasts. A long PLD might also be associated with the species' ability to long-distance dispersal in a single or a few generations. This would explain the existence of shared haplotypes between southern and northern locations for both markers, thus diluting the source effect of their glacial refugia.
Another potential refugium where L. bergylta may have survived Pleistocene glaciations is the Azores. Water temperatures in this Macaronesian archipelago would have been suitable for this species during the LGM, as the drop in sea surface temperature was moderate [78]. The distinct nature of Azorean haplotypes and the absence of gene flow between Azorean and Continental European locations support this hypothesis. The existence of an Azorean glacial refugium has been reported in the literature for several fish species (e.g. Raja clavata [79], C. galerita [62]).
4.4. Final remarks on species cohesiveness, phenotype plasticity and implications to management
The ballan wrasse is the target of local fisheries across its entire distribution range and is a resource of paramount importance for local fisheries in northern Spain (e.g. [22,24]). It is currently being used as a cleaner species in northern Europe salmonid fish farms (e.g. [25]). Its absence from the archipelago of Madeira (the species was reported in [34]), if confirmed in the next few years, suggests that L. bergylta may be under important selective pressures. This is particularly relevant in its southern distribution limit. In the future, under a warming climate prospective, this could imply a northward regression of this species along its southern limit. Therefore, a thorough monitoring programme should be conducted both in Madeira (where it seems to be currently absent) and also along the Portuguese continental shore.
It is important that the results of this paper are taken into account together with the recent attention given to the wide phenotypic plasticity [80] and the systematic status of the ballan wrasse [22,27–32]. Although this study did not control for morphotypes, and this is still an ongoing subject of research, results obtained in [32] using the same genetic markers point to the conclusion that our results are not affected by it.
5. Conclusion
On the one hand, this study points towards the common origin of the historical population of L. bergylta with shared haplotypes between the Azores and mainland and exchanges of migrants in both directions (S7 dataset). On the other hand, the mitochondrial results reveal a complete isolation of the Azorean archipelago. The present-day pattern of oceanographic currents in the northeastern Atlantic is likely to maintain and/or accentuate the isolation and distinctiveness of this insular population of the ballan wrasse. This situation can possibly lead to a case of incipient speciation, although endemic fish species are rare in this archipelago (see [8]). This trend for the genetic isolation of the Azores can be occasionally reversed by severe or atypical weather events (e.g. [68]).
The relatively long PLD of the ballan wrasse and the absence of geographical barriers could lead us to infer unconstrained gene flow along European shores. However, the patterns found for this species reveal a population substructure, which can reflect both a recolonization from two distinct glacial refugia and the circulation pattern along the continental coast.
Commercial exploitation, together with the ongoing taxonomic discussion on distinct morphotypes, suggests that special attention should be given to this species in the near future. The genetic structure and phylogeographic pattern of L. bergylta still need further enlightenment and it would be interesting to complement this study with microsatellites and/or genomic tools, comparing specimens from both the plain and the spotted morphotypes along their entire distribution area.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful to Pedro Duarte Coelho and Fernando Roneberg for their help with fieldwork. We would like to thank the help of Patricia Carvalho during fieldwork and Manuel Biscoito, Ricardo Araújo and Mafalda Freitas for their support in the campaign to Madeira. We are also grateful to Catarina Craveiro and Filipe Tadeu for their help with laboratory work. We thank Station Biologique de Roscoff (France—ASSEMBLE project) for providing samples from Roscoff. We are grateful to José António González Pérez for providing the sample from the Canaries. We dedicate this paper to the loving memory of Professor Richard Fitzgerald whose tragic loss we suffered during the course of this work.
Ethics
All sampling and handling of fish were performed by experienced personnel (scientists in Portugal, France, Ireland, UK and Norway and by commercial fishers in Spain) in accordance with relevant legislation in each country: Norway (Dyrevelferdsloven 112 (law of animal welfare: http://www.lovdata.no; accessed July 2016)); UK (Animal Welfare Act 2006 (http://www.legislation.gov.uk; accessed June 2016)).
In Portugal, including the Azores, samples were collected by spearfishing carried out by authorized divers. In Spain, Ireland, UK and Norway samples were collected by trammel-nets, pots, and/or fish traps. Spear fishing in Portugal was carried out by divers with a permit (no. 246/2000). There is no protection status or fishing restrictions for this species in any of the countries where the samples were collected or by European law. It is commercially harvested and used as a ‘cleaner’ fish in the salmonid aquaculture industry. For those reasons, no special permits are required for sampling the species either for research or commercial purposes.
In the Canary Islands this species is classified as ‘especie de interes para los ecosistemas canarios’ (Boletín Oficial de Canarias 112, 15200, LEY 4/2010, 4 June, Catálogo Canario de Especies Protegidas) which lists the species that are not threatened but are under close observation. One single sample was provided from the collection of Prof. Jose Antonio González Pérez (Universidad de Las Palmas de Gran Canaria).
Data accessibility
DNA sequences are deposited in GenBank (accession nos. KU751889-KU752182).
Authors' contributions
F.A. collected field samples, carried out molecular laboratory work, data analysis and helped draft the manuscript. S.M.F. carried out molecular laboratory work, sequence alignment, data analysis and helped draft the manuscript. C.S.L. carried out molecular laboratory work, sequence alignment and helped draft the manuscript. R.F., L.M., D.V.-R., F.S.-R., P.A. and T.M. collected field samples and revised critically the manuscript. S.B. carried out molecular laboratory work, participated in data analysis and revised critically the manuscript. J.I.R. coordinated the study and helped draft the manuscript. All authors gave final approval for publication.
Competing interests
The authors declare no competing interests.
Funding
This study was funded by the Eco-Ethology Research Unit Strategic Plan (PEst-OE/MAR/UI0331/2011)—Fundação para a Ciência e a Tecnologia—FCT (partially FEDER funded), now included in MARE (UID/MAR/04292/2013). F.A. (SFRH/BPD/63170/2009) and S.M.F. (SFRH/BPD/84923/2012) were supported by FCT grants. C.S.L. was supported by MARE-ISPA/BI/004/2015. D.V.-R. was supported by a Marie Curie Intra European Fellowship within the 7th European Community Framework Programme (grant no. 625852). Sampling in the Azores was supported by CLIPE (PRAXIS/3/3.2/EMG/1957/95) and in continental Iberia by the European Science Foundation's MarinERA project ‘Marine phylogeographic structuring during climate change: the signature of leading and rear edge of range shifting populations'.
References
- 1.Briggs JC. 1995. Global biogeography. Developments in palaeontology and sratigraphy. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 2.Climap Project Members. 1984. The last interglacial ocean. Quat. Res. 21, 123–224. (doi:10.1016/0033-5894(84)90098-x) [Google Scholar]
- 3.Hayes A, Kucera M, Kallel N, Sbaffi L, Rohling EJ. 2005. Glacial Mediterranean sea surface temperatures based on the planktonic foraminiferal assemblages. Quat. Sci. Rev. 24, 999–1016. (doi:10.1016/j.quascirev.2004.02.018) [Google Scholar]
- 4.Maggs CA, et al. 2008. Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology 89, S108–S122. (doi:10.1890/08-0257.1) [DOI] [PubMed] [Google Scholar]
- 5.Alveirinho-Dias J, Rodrigues A, Magalhãees F. 1997. Evolução da linha de costa, em Portugal, desde o último máximo glaciárico até à actulalidade: Síntese dos conhecimentos. Estud. Quat. 1, 53–66. [Google Scholar]
- 6.Lambeck K, Esat TM, Potter E-K. 2002. Links between climate and sea levels for the past three million years. Nature 419, 199–206. (doi:10.1038/nature01089) [DOI] [PubMed] [Google Scholar]
- 7.Wares JP, Cunningham CW. 2001. Phylogeography and historical ecology of the North Atlantic intertidal. Evolution 55, 2455–2469. (doi:10.1111/j.0014-3820.2001.tb00760.x) [DOI] [PubMed] [Google Scholar]
- 8.Almada VC, et al. 2013. Complex origins of the Lusitania biogeographic province and northeastern Atlantic fishes. Front. Biogeogr. 5, 14493. [Google Scholar]
- 9.Briggs JC, Bowen BW. 2012. A realignment of marine biogeographic provinces with particular reference to fish distributions. J. Biogeogr. 39, 12–30. (doi:10.1111/j.1365-2699.2011.02613.x) [Google Scholar]
- 10.Patarnello T, Volckaert FAM, Castilho R. 2007. Pillars of Hercules: is the Atlantic-Mediterranean transition a phylogeographical break? Mol. Ecol. 16, 4426–4444. (doi:10.1111/j.1365-294X.2007.03477.x) [DOI] [PubMed] [Google Scholar]
- 11.Kettle AJ, Morales-Muñiz A, Roselló-Izquierdo E, Heinrich D, Vollestad A. 2011. Refugia of marine fish in the northeast Atlantic during the last glacial maximum: concordant assessment from archaeozoology and paleotemperature reconstructions. Clim. Past 7, 181–201. (doi:10.5194/cp-7-181-2011) [Google Scholar]
- 12.Francisco SM, Robalo JI, Levy A, Almada VC. 2014. In search of phylogeographic patterns in the northeastern Atlantic and adjacent seas. In Evolutionary biology, genome, speciation, coevolution and origin of life (ed. Pontarotti P.), pp. 323–338. New York, NY: Springer. [Google Scholar]
- 13.Francisco SM, Faria C, Lengkeek W, Vieira MN, Velasco EM, Almada VC. 2011. Phylogeography of the shanny Lipophrys pholis (Pisces: Blenniidae) in the NE Atlantic records signs of major expansion event older than the last glaciation. J. Exp. Mar. Biol. Ecol. 403, 14–20. (doi:10.1016/j.jembe.2011.03.020) [Google Scholar]
- 14.Almada V, Almada F, Francisco S, Castilho R, Robalo JI. 2012. Unexpected high genetic diversity at the extreme northern geographic limit of Taurulus bubalis (Euphrasen, 1786). PLoS ONE 7, e44404 (doi:10.1371/journal.pone.0044404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gysels ES, Helllemans B, Pampoulie C, Volckaert FAM. 2004. Phylogeography of the common goby, Pomatoschistus microps, with particular emphasis on the colonization of the Mediterranean and the North Sea. Mol. Ecol. 13, 403–417. (doi:10.1046/j.1365-294X.2003.02087.x) [DOI] [PubMed] [Google Scholar]
- 16.Robalo JI, Castilho R, Francisco SM, Almada F, Knutsen H, Jorde PE, Pereira AM, Almada VC. 2012. Northern refugia and recent expansion in the North Sea: the case of the wrasse Symphodus melops (Linnaeus, 1758). Ecol. Evol. 2, 153–164. (doi:10.1002/ece3.77) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickerson MJ, Cunningham CW. 2006. Nearshore fish (Pholis gunnellus) persists across the North Atlantic through multiple glacial episodes. Mol. Ecol. 15, 4095–4107. (doi:10.1111/j.1365-294X.2006.03085.x) [DOI] [PubMed] [Google Scholar]
- 18.Domingues VS, Bucciarelli G, Almada VC, Bernardi G. 2005. Historical colonization and demography of the Mediterranean damselfish, Chromis chromis. Mol. Ecol. 14, 4051–4063. (doi:10.1111/j.1365-294X.2005.02723.x) [DOI] [PubMed] [Google Scholar]
- 19.Debes PV, Zachos FE, Hanel R. 2008. Mitochondrial phylogeography of the European sprat (Sprattus sprattus L., Clupeidae) reveals isolated climatically vulnerable populations in the Mediterranean Sea and range expansion in the northeast Atlantic. Mol. Ecol. 17, 3873–3888. (doi:10.1111/j.1365-294X.2008.03872.x) [DOI] [PubMed] [Google Scholar]
- 20.Bañon R, Villegas-Ríos D, Serrano A, Mucientes G, Arronte JC. 2010. Marine fishes from Galicia (NW Spain): an updated checklist. Zootaxa 2667, 1–27. (doi:10.15468/hiiv7z) [Google Scholar]
- 21.Quignard JP, Pras A. 1986. Labridae. In The fishes of the northeastern Atlantic and the Mediterranean, vol. 2 (eds Whitehead PJP, Bauchot M-L, Hureau J-C, Nielsen J, Tortonese E), pp. 919–942. Paris, France: UNESCO. [Google Scholar]
- 22.Villegas-Ríos D, Alonso-Fernández A, Domínguez-Petit R, Saborido-Rey F. 2013. Intraspecific variability in reproductive patterns in the temperate hermaphrodite Labrus bergylta. Mar. Fresh. Res. 64, 1156–1168. (doi:10.1071/MF12362) [Google Scholar]
- 23.D'Arcy J, Dunaevskaya E, Treasurer JW, Ottesen O, Maguire J, Zhuravleva N, Karlsen A, Rebours C, FitzGerald RD. 2012. Embryonic development in ballan wrasse Labrus bergylta. J. Fish Biol. 81, 1101–1110. (doi:10.1111/j.1095-8649.2012.03337.x) [DOI] [PubMed] [Google Scholar]
- 24.Villegas-Ríos D, Alós J, March D, Palmer M, Mucientes G, Saborido-Rey F. 2013. Home range and diel behavior of the ballan wrasse, Labrus bergylta, determined by acoustic telemetry. J. Sea Res. 80, 61–71. (doi:10.1016/j.seares.2013.02.009) [Google Scholar]
- 25.Skiftesvik AB, Bjelland RM, Durif CM, Johansen IS, Browman HI. 2013. Delousing of Atlantic salmon (Salmo salar) by cultured vs. wild ballan wrasse (Labrus bergylta). Aquaculture 402, 113–118. (doi:10.1016/j.aquaculture.2013.03.032) [Google Scholar]
- 26.Skiftesvik AB, et al. 2014. Wrasse (Labridae) as cleaner fish in salmonid aquaculture—the Hardangerfjord as a case study. Mar. Biol. Res. 10, 289–300. (doi:10.1080/17451000.2013.810760) [Google Scholar]
- 27.Hanel R, Westneat MW, Sturmbauer C. 2002. Phylogenetic relationships, evolution of broodcare behavior, and geographic speciation in the wrasse tribe Labrini. J. Mol. Evol. 55, 776–789. (doi:10.1007/s00239-002-2373-6) [DOI] [PubMed] [Google Scholar]
- 28.Baliga VB, Law CJ. 2016. Cleaners among wrasses: phylogenetics and evolutionary patterns of cleaning behavior within Labridae. Mol. Phylogenet. Evol. 94, 424–435. (doi:10.1016/j.ympev.2015.09.006) [DOI] [PubMed] [Google Scholar]
- 29.Almada VC, Almada F, Henriques M, Santos RS, Brito A. 2002. On the phylogenetic affinities of Centrolabrus trutta and Centrolabrus caeruleus (Perciforms: Labridae). Molecular and meristic evidence. Arquipélago Life Mar. Sci. 19, 85–92. [Google Scholar]
- 30.Villegas-Ríos D, Alonso-Fernández A, Domínguez-Petit R, Saborido-Rey F. 2014. Energy allocation and reproductive investment in a temperate protogynous hermaphrodite, the ballan wrasse Labrus bergylta. J. Sea Res. 86, 76–85. (doi:10.1016/j.seares.2013.11.010) [Google Scholar]
- 31.Quintela M, Danielsen EA, Lopez L, Barreiro R, Svasand T, Knutsen H, Skiftesvik AB, Glover KA. 2016. Is the ballan wrasse, Labrus bergylta, two species? Genetic analysis reveals within-species divergence associated with plain and spotted morphotype frequencies. Integr. Zool. 11, 162–172. (doi:10.1111/1749-4877.12186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almada F, Casas L, Francisco SM, Villegas-Ríos D, Saborido-Rey F, Irigoien X, Robalo JI. 2016. On the absence of genetic differentiation between morphotypes of the ballan wrasse Labrus bergylta (Labridae). Mar. Biol. 163, 86 (doi:10.1007/s00227-016-2860-8) [Google Scholar]
- 33.D'Arcy J, Mirimin L, FitzGerald R. 2013. Phylogeographic structure of a protogynous hermaphrodite species, the ballan wrasse Labrus bergylta, in Ireland, Scotland, and Norway, using mitochondrial DNA sequence data. ICES J. Mar. Sci. 70, 685–693. (doi:10.1093/icesjms/fst018) [Google Scholar]
- 34.Wirtz P, Fricke R, Biscoito MJ. 2008. The coastal fishes of Madeira Island—new records and an annotated check-list. Zootaxa 1715, 1–26. [DOI] [PubMed] [Google Scholar]
- 35.Ostellari L, Bargelloni L, Penzo E, Patarnello P, Patarnello T. 1996. Optimization of single-strand conformation polymorphism and sequence analysis of the mitochondrial control region in Pagellus bogaraveo (Sparidae, Teleostei): rationalized tools in fish population biology. Anim. Genet. 27, 423–427. (doi:10.1111/j.1365-2052.1996.tb00510.x) [DOI] [PubMed] [Google Scholar]
- 36.Chow S, Hazama K. 1998. Universal PCR primers for S7 ribosomal protein gene introns in fish. Mol. Ecol. 7, 1247–1263. (doi:10.1007/s40071-016-0122-5) [PubMed] [Google Scholar]
- 37.Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. (doi:10.1093/bioinformatics/btm404) [DOI] [PubMed] [Google Scholar]
- 38.Sousa-Santos C, Robalo IJ, Collares-Pereira MJ, Almada VC. 2005. Heterozygous indels as useful tools in the reconstruction of DNA sequences and in the assessment of ploidy level and genomic constitution of hybrid organisms. DNA Seq. 16, 462–467. (doi:10.1080/10425170500356065) [DOI] [PubMed] [Google Scholar]
- 39.Guindon S, Gascuel O. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. (doi:10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 40.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. (doi:10.1093/molbev/msn083) [DOI] [PubMed] [Google Scholar]
- 41.Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York, NY: Oxford University Press. [Google Scholar]
- 42.Clement M, Posada D, Crandall KA. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1660. (doi:10.1046/j.1365-294x.2000.01020.x) [DOI] [PubMed] [Google Scholar]
- 43.Templeton AR, Crandall KA, Sing CF. 1992. A cladistic-analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA-sequence data. III. Cladogram estimation. Genetics 132, 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Excoffier L, Lischer H. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. (doi:10.1111/j.1755-0998.2010.02847.x) [DOI] [PubMed] [Google Scholar]
- 45.Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Excoffier L, Laval G, Balding D. 2003. Gametic phase estimation over large genomic regions using an adaptive window approach. Hum. Genome 1, 7–19. (doi:10.1186/1479-7364-1-1-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220. [PubMed] [Google Scholar]
- 48.Smouse PE, Long JC, Sokal RR. 1986. Multiple regression and correlation extensions of the Mantel Test of matrix correspondence. Syst. Zool. 35, 627–632. (doi:10.2307/2413122) [Google Scholar]
- 49.Dupanloup I, Schneider S, Excoffier L. 2002. A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 11, 2571–2581. (doi:10.1046/j.1365-294X.2002.01650.x) [DOI] [PubMed] [Google Scholar]
- 50.Rogers AR, Harpending HC. 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569. [DOI] [PubMed] [Google Scholar]
- 51.Rogers AR. 1995. Genetic evidence for a Pleistocene population explosion. Evolution 49, 608–615. (doi:10.2307/2410314) [DOI] [PubMed] [Google Scholar]
- 52.Fu YX. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tajima F. 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105, 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuhner MK. 2006. LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics 22, 768–770. (doi:10.1093/bioinformatics/btk051) [DOI] [PubMed] [Google Scholar]
- 55.Bowen BW, Muss A, Rocha LA, Grant WS. 2006. Shallow mtDNA coalescence in Atlantic pygmy angelfishes (genus Centropyge) indicates a recent invasion from the Indian Ocean. J. Hered. 97, 1–12. (doi:10.1093/jhered/esj006) [DOI] [PubMed] [Google Scholar]
- 56.Bernardi G, Lape J. 2005. Tempo and mode of speciation in the Baja California disjunct fish species Anisotremus davidsonii. Mol. Ecol. 14, 4085–4096. (doi:10.1111/j.1365-294X.2005.02729.x) [DOI] [PubMed] [Google Scholar]
- 57.Sato A, Takezaki N, Tichy H, Figueroa F, Mayer WE, Klein J. 2003. Origin and speciation of haplochromine fishes in East African Crater Lakes investigated by the analysis of their mtDNA, Mhc genes, and SINEs. Mol. Biol. Evol. 20, 1448–1462. (doi:10.1093/molbev/msg151) [DOI] [PubMed] [Google Scholar]
- 58.Bowen BW, Grant WS. 1997. Phylogeography of the sardines (Sardinops spp.): assessing biogeographic models and population histories in temperate upwelling zones. Evolution 51, 1601–1610. (doi:10.2307/2411212) [DOI] [PubMed] [Google Scholar]
- 59.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192. (doi:10.1093/molbev/msi103) [DOI] [PubMed] [Google Scholar]
- 60.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. (doi:10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1.6. See http://beast.bio.ed.ac.uk/Tracer.
- 62.Francisco SM, Almada VC, Faria C, Velasco EM, Robalo JI. 2014. Phylogeographic pattern and glacial refugia of a rocky shore species with limited dispersal capability: the case of Montagu's blenny (Coryphoblennius galerita, Blenniidae). Mar. Biol. 161, 2509–2520. (doi:10.1007/s00227-014-2523-6) [Google Scholar]
- 63.Stefanni S, Domingues V, Bouton N, Santos RS, Almada F, Almada V. 2006. Phylogeny of the shanny, Lipophrys pholis, from the NE Atlantic using mitochondrial DNA markers. Mol. Phylogenet. Evol. 39, 282–287. (doi:10.1016/j.ympev.2005.07.001) [DOI] [PubMed] [Google Scholar]
- 64.Faurby S, Barber PH. 2012. Theoretical limits to the correlation between pelagic larval duration and population genetic structure. Mol. Ecol. 21, 3419–3432. (doi:10.1111/j.1365-294X.2012.05609.x) [DOI] [PubMed] [Google Scholar]
- 65.Weersing K, Toonen RJ. 2009. Population genetics, larval dispersal, and connectivity in marine systems. Mar. Ecol. Prog. Ser. 393, 1–12. (doi:10.3354/meps08287) [Google Scholar]
- 66.Bremer JRA, Viñas J, Mejuto J, Ely B, Pla C. 2005. Comparative phylogeography of Atlantic blue fin tuna and swordfish: the combined effects of vicariance, secondary contact, introgression, and population expansion on the regional phylogenies of two highly migratory pelagic fishes. Mol. Phylogenet. Evol. 36, 169–187. (doi:10.1016/j.ympev.2004.12.011) [DOI] [PubMed] [Google Scholar]
- 67.Ottesen OH, Dunaevskaya E, D'Arcy J. 2012. Development of Labrus bergylta (Ascanius 1767) larvae from hatching to metamorphosis. J. Aquac. Res. Development 3, 127 (doi:10.4172/2155-9546.1000127) [Google Scholar]
- 68.Stefanni S, et al. 2015. Establishment of a coastal fish in the Azores: recent colonization or sudden expansion of an ancient relict population? Heredity 115, 527–537. (doi:10.1038/hdy.2015.55) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Féraud G, Kaneoka I, Allègre CJ. 1980. K/Ar ages and stress pattern in the Azores: geodynamic implications. Earth Planet. Sci. Lett. 46, 275–286. (doi:10.1016/0012-821X(80)90013-8) [Google Scholar]
- 70.Azevedo JMM, Ferreira MP, Martins JA. 1991. The emergent volcanism of Flores Island, Azores (Portugal). Arquipélago Life Earth Sci. 9, 37–46. [Google Scholar]
- 71.Serralheiro A, Madeira J. 1993. Stratigraphy and geochronology of Santa Maria island (Azores). Açoreana 7, 575–592. [Google Scholar]
- 72.Gould WJ. 1985. Physical oceanography of the Azores Front. Prog. Oceanogr. 14, 167–190. (doi:10.1016/0079-6611(85)90010-2) [Google Scholar]
- 73.Trouet V, Scourse JD, Raible CC. 2012. North Atlantic storminess and Atlantic meridional overturning circulation during the last millennium: reconciling contradictory proxy records of NAO variability. Glob. Planet. Change 84, 48–55. (doi:10.1016/j.gloplacha.2011.10.003) [Google Scholar]
- 74.Santos RS, Hawkins S, Monteiro LR, Alves M, Isidro EJ. 1995. Marine research, resources and conservation in the Azores. Aquatic conservation. Mar. Freshwater Ecosyst. 5, 311–354. (doi:10.1002/aqc.3270050406) [Google Scholar]
- 75.Shakun JD, Carlson AE. 2010. A global perspective on Last Glacial Maximum to Holocene climate change. Quat. Sci. Rev. 29, 1801–1816. (doi:10.1016/j.quascirev.2010.03.016) [Google Scholar]
- 76.Zander CD. 1973. Evolution of Blennioidei in the Mediterranean Sea. Rev. Trav. Inst. Pêches Marit. 37, 215–221. [Google Scholar]
- 77.Almada VC, Domingues VS, Monteiro NM, Almada F, Santos RS. 2007. Molecular data confirm the validity of the Portuguese blenny (Parablennius ruber, Valenciennes, 1836) and its presence in Western Europe. J. Fish Biol. 70, 248–254. (doi:10.1111/j.1095-8649.2007.01397.x) [Google Scholar]
- 78.Crowley TJ. 1981. Temperature and circulation changes in the eastern north Atlantic during the last 150,000 years: evidence from the planktonic foraminiferal record. Mar. Micropaleontol. 6, 97–129. (doi:10.1016/0377-8398(81)90001-3) [Google Scholar]
- 79.Chevolot M, Hoarau G, Rinjsdorp AD, Stam WT, Olsen JL. 2006. Phylogeography and population structure of thornback rays (Raja clavata L., Rajidae). Mol. Ecol. 15, 3693–3705. (doi:10.1111/j.1365-294X.2006.03043.x) [DOI] [PubMed] [Google Scholar]
- 80.Phillips AC, Carleton KL, Marshall NJ. 2015. Multiple genetic mechanisms contribute to visual sensitivity variation in the Labridae. Mol. Biol. Evol. 33, 201–215. (doi:10.1093/molbev/msv213) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences are deposited in GenBank (accession nos. KU751889-KU752182).