ABSTRACT
MicroRNA (miRNA) are a class of ‘18–25’ nt RNA molecules which regulate gene expression and play an important role in several biologic processes including fatty acid metabolism. Here we used S-Poly (T) and high-throughput sequencing to evaluate the expression of miRNA and mRNA during early-lactation and in the non-lactating (“dry”) period in goat mammary gland tissue. Results indicated that miR-148a, miR-17–5p, PPARGC1A and PPARA are highly expressed in the goat mammary gland in early-lactation and non-lactating periods. Utilizing a Luciferase reporter assay and Western Blot, PPARA, an important regulator of fatty acid oxidation, and PGC1a (PPARGC1A), a major regulator of fat metabolism, were demonstrated to be targets of miR-148a and miR-17–5p in goat mammary epithelial cells (GMECs). It was also revealed that miR-148a expression can regulate PPARA, and miR-17–5p represses PPARGC1A in GMECs. Furthermore, the overexpression of miR-148a and miR-17–5p promoted triacylglycerol (TAG) synthesis while the knockdown of miR-148a and miR-17–5p impaired TAG synthesis in GMEC. These findings underscore the importance of miR-148a and miR-17–5p as key components in the regulation of TAG synthesis. In addition, miR-148a cooperates with miR-17–5p to regulate fatty acid metabolism by repressing PPARGC1A and PPARA in GMECs. Further studies on the functional role of miRNAs in lipid metabolism of ruminant mammary cells seem warranted.
Keywords: miR-148a, miR-17–5p, PPARA, PPARGC1A, triacylglycerol synthesis
Introduction
Compared with cow milk, goat milk (milk of Capra hircus) contains a higher level of unsaturated fatty acids, along with total fat, vitamin, calcium, carbohydrate, and protein content.2,46 The nutritional value of goat milk, especially fat composition and content(15,44, can be further enhanced through dietary manipulation.25,27,43 As a result, there have been several research studies on the molecular mechanisms regulating milk fat synthesis; however, they have been primarily focused on analyses of single gene function (20,54,61.
MiRNA are a series of single-stranded non-coding RNA with 22 nucleotides in length. MiRNA regulate gene expression at a post-transcriptional level. Generally, miRNA bind to the 3′-UTR of target mRNA to downregulate transcription. Currently, hundreds of miRNA have been uncovered in humans, and bioinformatics predictions indicate that they could regulate the expression of approximately 30 percent of genes in the genome. Despite the vast amount of work on miRNA in ruminant species, there are few studies focused on the function and mechanisms by which miRNA synergistically regulate the process of milk fat synthesis in ruminants.
MiR-17–5p together with miR-20a, miR-20b,miR-93, miR-106a and miR-106b is a member of the miR-17 family which is derived from the miR-17–92 cluster.45 Although previous studies have shown that miR-148a and miR-17–5p also influence adipocyte differentiation37 and regeneration, the function of miR-148a and miR-17–5p in the goat mammary gland is unknown. Therefore, the main purpose of the present work was to investigate the synergistic role that miR-148a and miR-17–5p play in the control of mammary cell lipid metabolism using primary goat mammary epithelial cells (GMEC).
Results
microRNA/mRNA profiling and regulatory fat metabolism network
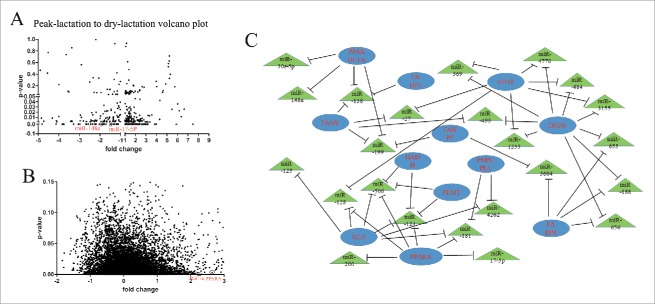
Some research reports have revealed that miRNAs are involved in the regulation of milk fat metabolism.38,39,40 To investigate the about between the miRNA regulation and this physiologic process, we profiled the miRNAs expression between early-lactation and in the non-lactating (“dry”) period with S-Poly (T) real method. The analyses included not only 436 goat miRNA and 267 sheep miRNA retrieved from miRBase, but also 793 miRNA detected in lactating mammary tissue by second-generation sequencing. In the screening, all the miRNA with p-value < 0.05 and with a 4-fold change were chosen as candidate miRNA (Fig. 1A) (Table S1).
Figure 1.
microRNA/mRNA profiling and regulatory fat metabolism network (A) Screening for miRNA associated with early-lactation and non-lactation stages. Mammary tissue used was from 3 goats at each stage of lactation. The expression of 18s rRNA was used as a normalization control. (B) Illumina de novo RNA sequencing of mRNA in the early-lactation and non-lactating samples All experiments were run in duplicate and repeated 3 times. Values are presented as means ± standard errors, *, P < 0.05; **, P < 0.01. (C) Target genes associated with the lipid transport and metabolism category of KOG in early lactation.
For the mRNA sequencing, the Illumina sequencing results showed a total of 51299 unigenes. These unigenes were compared with the transcriptome database for annotation, and a total of 12763 unigenes were annotated. There were 12,239 and 12713 unigenes detected for the samples from the early-lactation and non-lactating (“dry”) periods. The sequencing information is presented in file S2, and the length distribution of universal genes is shown in Fig. 1B and Table. S2. All sequence reads were deposited at NCBI (accession number SPR040710).
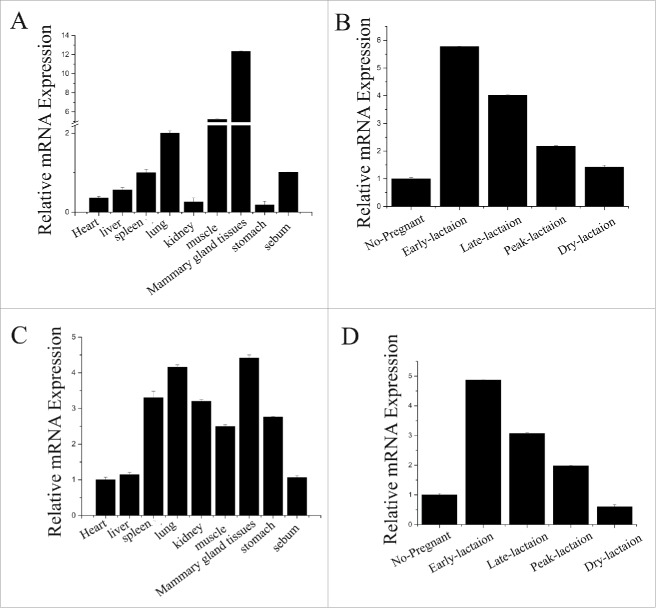
The KEGG and eukaryotic Ortholog Groups of Protein (KOG) classification were performed using the Blast 2 GO pipeline with the default parameters. The GO was used to classify the function among the universal genes. The results of the early-lactation mammary tissue revealed 12 genes with the highest expression level and closely associated with the lipid transport and metabolism KOG category. The subsequent functional analysis (Table 1, 2; Fig. 1C) provided confirmation that PPARGC1A and PPARA work in cooperation. The analysis also indicated that they are direct targets of miR-148a and miR-17–5p, which were highly expressed in lactating mammary tissue (Fig. 1C). Therefore, we focused on miR-148a and miR-17–5p for more detailed studies. For instance, we measured miR-148a and miR-17–5p expression in different tissues of dairy goats (Fig. 2A, 2C), and mammary gland tissue at different stages of lactation (Fig. 2B, 2D). Both miR-148a and miR-17–5p were primarily expressed in mammary tissue, supporting the idea that miR-148a and miR-17–5p played an important role in mechanisms associated with lactation.
Table 1.
Predicted miR-148a target genes are associated with fat metabolism.
| DO term | Number of target genes | p-value |
|---|---|---|
| Lipid synthesis | 6 | 1.8E-1 |
| Regulation of lipid metabolic process | 4 | 7.4E-1 |
| Lipid modification | 7 | 2.9E-2 |
| Lipid oxidation | 3 | 3.8E-1 |
| Lipid transporter activity | 4 | 3.6E-1 |
| Lipid transport | 5 | 7.3E-1 |
| Lipid localization | 5 | 7.8E-1 |
| Fatty acid biosynthetic process | 8 | 1.7E-2 |
| Regulation of fatty acid oxidation | 4 | 6.8E-2 |
| Fatty acid biosynthesis | 4 | 1.4E-1 |
| Regulation of fatty acid metabolic process | 4 | 2.3E-1 |
The p-value is calculated using Fisher's exact test.
Table 2.
Predicted miR-17a-5p target genes are associated with fat metabolism.
| DO term | Number of target genes | p-value |
|---|---|---|
| Lipid metabolism | 11 | 3.0E-1 |
| Lipid transport | 4 | 6.6E-1 |
| Lipid localization | 9 | 7.2E-1 |
| Cholesterol metabolism | 6 | 2.7E-2 |
| Cholesterol homeostasis | 3 | 6.7E-1 |
| Cholesterol transport | 4 | 3.8E-1 |
| Lipid homeostasis | 3 | 8.1E-1 |
| Protein-lipid complex | 3 | 5.4E-1 |
| Lipid degradation | 7 | 3.2E-1 |
| Lipid catabolic process | 10 | 7.0E-1 |
| Fatty acid transport | 3 | 5.0E-1 |
The p-value is calculated using Fisher's exact test.
Figure 2.
The expression level of miR-148a and miR-17–5p (A) miR-148a expression in various tissues of milk goats The miR-148a expression level is detected in heart, liver, spleen, lung, kidney, muscle, mammary gland, stomach and sebum. The expression of 18s rRNA is used as a normalization control. (B) miR-148a expression in different lactation period of mammary gland. The miR-148a expression level is detected in no-pregnancy, early-lactation (15 d after parturition) and peak-lactation (60 d after parturition), late-lactation (150 d after parturition) and non- lactation. The expression of 18s r RNA is used as a normalization control. (C) miR-17–5p expression in various tissues of milk goats The miR-17–5p expression level is detected in heart, liver, spleen, lung, kidney, muscle, mammary gland, stomach and sebum. The expression of 18s rRNA is used as a normalization control. (D) miR-17–5p expression in different lactation period of mammary gland The miR-17–5p expression level is detected in no-pregnancy, early-lactation (15 d after parturition) and peak-lactation (60 d after parturition), late-lactation (150 d after parturition) and non-lactation. The expression of 18s rRNA is used as a normalization control.
miR-148a via PPARGC1A and miR-17–5p via PPARA synergistically regulate TAG
MiR-148a and miR-17–5p synergistically regulate TAG synthesis in GMEC
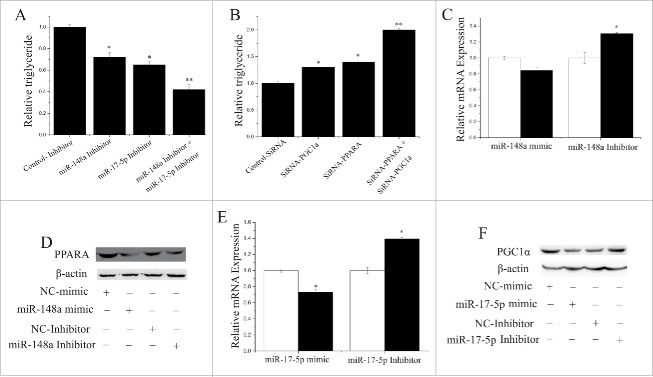
siRNA technology and TAG concentration were used to determine the potential functional relationship between miR-148a and miR-17–5p. In Fig. 3A, compared with the negative control the expression of miR-148a and miR-17–5p decreased (P < 0.05) when the inhibitors were transfected into GMEC. In contrast, the TAG level decreased significantly (P < 0.05) when miR-148a (30 nM) as well as miR-17–5p (30 nM) were inhibited (Fig. 3A).
Figure 3.
Relationship between miR- 148a, miR-17–5p, PPARGC1A and PPARA (A) TAG concentrations in cells transfected with NC-Inhibitor (60 nM), miR-148a-Inhibitor (60 nM), miR-17–5p-Inhibitor (60 nM), miR-148a-Inhibitor (30 nM) and miR-17–5p-Inhibitor (30 nM); TAG concentrations are compared with the control (n = 6). (B) TAG concentrations in cells transfected with NC-Inhibitor (60 nM), SiRNA-PPARGC1A (60 nM), SiRNA-PPARA (60 nM), SiRNA-PPARGC1A (30 nM) and SiRNA-PPARA (30 nM); TAG concentrations are compared with the control (n = 6). (C) PPARA expression quantified by RT-qPCR (n = 6) in GMECs transfected with miR-148a mimic or inhibitor for 48 h. White bars: negative control; black bars: miR-148a mimic or inhibitor. (D) Western blot analysis of PPARA expression in the miR-148a mimic and NC treatment experiments. The effect of miR-148a mimics and Inhibitor on PPARA protein expression in GMECs was evaluated by Western blot analysis. Total protein were harvested after 48 h post-transfection, respectively. (E) PPARGC1A expression quantified by RT-qPCR (n = 6) in GMECs transfected with miR-17–5p mimic or inhibitor for 48 h. White bars: negative control; black bars:miR-17–5p mimic or inhibitor. (F) Western blot analysis of PPARGC1A expression in the miR-17–5p mimic and NC treatment experiments. The effect of miR-17–5p mimics and Inhibitor on PPARGC1A protein expression in GMECs was evaluated by Western blot analysis. Total protein were harvested after 48 h post-transfection.
Compared with the negative control (Fig. 3B), the TAG level increased (P < 0.05) in GMEC cultured with siRNA-PPARGC1A (60 nM) and siRNA-PPARA (60 nM). We also examined the mRNA level PPARGC1A and PPARA in GMEC while miR-148a and miR-17–5p were inhibited or activated. Interestingly, compared with the negative control (Fig. 3C, 3D), the expression of PPARGC1A decreased (P < 0.05) in GMEC cultured with a miR-17–5p mimic. In contrast, the expression of PPARA decreased significantly (P < 0.05) by overexpression of miR-148a (Fig. 3E, 3F).
MiR-148a targets PPARGC1A, and miR-17–5p targets PPARA in GMEC
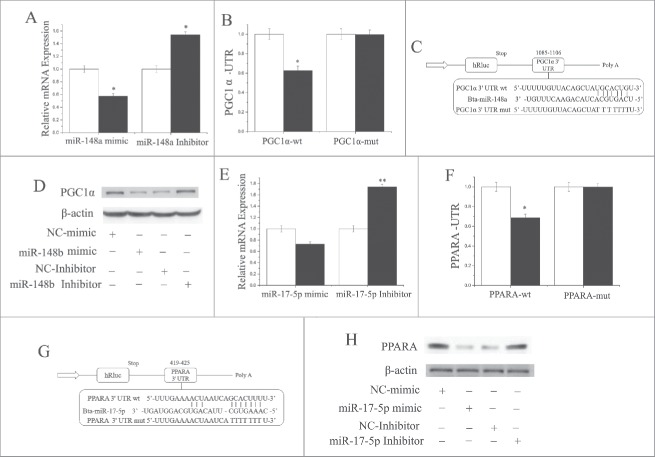
Based on the 3′-UTR complementary prediction with Target Scan 6.2 and miRNA functional analysis by DAVID (https://david.ncifcrf.gov/summary), it is evident that many genes are potential targets of miR-148a (Fig. S1;Table. S1,S2). We chose PPARGC1A for functional validation because it is known to be an important regulator of multiple metabolic processes. Furthermore, at least in non-ruminants, PPARGC1A is an important PPARA co-activator in tissues that undergo extensive oxidative metabolism. The results indicated that PPARGC1A was downregulated (P < 0.05) by overexpression of miR-148a, and upregulated (P < 0.05) by inhibition of miR-148a (Fig. 4A). Furthermore, as depicted in Fig. 4C, PPARGC1A has a binding site for miR-148a in the 3′-UTR.
Figure 4.
MiR-148a targets PPARGC1A, and miR-17–5p targets PPARA (A) PPARGC1A expression level is quantified by RT-qPCR (n = 6) GMECs are transfected with miR-148a mimic or inhibitor for 48h, and. White bars: negative control; black bars: miR-148a mimic or inhibitor. (B and C) Target site of miR-148a in the PPARGC1A 3′-UTR and the construction of the luciferase (Luc) expression vector fused with the PPARGC1A 3′-UTR. WT: Luc reporter vector with the WT PPARGC1A 3′-UTR (1085 to 1106); MU: Luc reporter vector with the mutation at the miR-148a site in PPARGC1A 3′-UTR. (D) Western blot analysis of PPARGC1A expression in the miR-148a mimic and NC treatment experiments. The effect of miR-148a mimics and Inhibitor on PPARGC1A protein expression in GMECs was evaluated by western blot analysis. Total protein was harvested after 48 h post-transfection, respectively. (E) PPARA expression quantified by RT-qPCR (n = 6) in GMECs transfected with miR-17–5p mimic or inhibitor for 48 h. White bars: negative control; black bars: miR-17–5p mimic or inhibitor. (F and G) Target site of miR-17–5p in PPARA3′-UTR and the construction of the luciferase (Luc) expression vector fused with the PPARA3′-UTR. WT: Luc reporter vector with the WT PPARA 3′-UTR (419 to 425); MU: Luc reporter vector with the mutation at miR-17–5p site in PPARA 3′-UTR. (H) Western blot analysis of PPARA expression in the miR-17–5p mimic and NC treatment experiments. The effect of miR-17–5p mimics and Inhibitor on PPARA protein expression in GMECs was evaluated by Western blot analysis. Total protein was harvested after 48 h post-transfection. All experiments were duplicated and repeated 3 times. Values are presented as means ± standard errors, *, P < 0.05; **, P < 0.01.
To ascertain that miR-148a directly targets this site, we synthesized a 3′-UTR segment of PPARGC1A including the miR-148a target site, and cloned it into the psi-CHECK2 vector to construct a 3′-UTR report plasmid. The luciferase assay indicated that overexpression of miR-148a decreased (P < 0.05) the relative luciferase activity of the reporter containing the wild-type 3′-UTR rather than the one with mutations in the seed sequences (Figs. 4B and C). Furthermore, the protein expression level of PPARGC1A after miR-148a mimic treatment was consistent with the mRNA expression data (Fig. 4D). These findings illustrated that miR-148a directly interacts with the target site of the PPARGC1A mRNA, and negatively regulated its expression, which partly explains the function of miR-148a during lactation.
Our results also revealed that PPARA was downregulated (P < 0.05) by overexpression of miR-17–5p (Fig. 4E). Furthermore, as illustrated in Fig. 3G, PPARA has a binding site for miR-17–5p in the 3′-UTR. To ascertain that miR-17–5p could directly target this site, we synthesized a 3′-UTR segment of PPARA including the miR-17–5p target site, and cloned it into the psi-CHECK2 vector to construct a 3′-UTR reporter plasmid. Luciferase assay indicated that overexpression of miR-17–5p decreased (P < 0.05) the relative luciferase activity of the reporter with a wild-type 3′-UTR rather than the one with mutations in the seed sequences (Fig. 4F and Fig. 4G). Furthermore, the protein expression level of PPARA after miR-17–5p mimic treatment was consistent with mRNA expression data (Fig. 4H). These findings illustrate that miR-17–5p directly interacts with the target site of the PPARA mRNA, and negatively regulates its expression, which partly explains the function of miR-17–5p during lactation.
Functional evaluation of miR-148a, miR-17–5p, PPARGC1A and PPARA in GMEC
Expression of miR-148a and miR-17–5p increased the level of TAG and milk fat droplet accumulation in GMEC
The expression of miR-148a was 31 times higher (P < 0.05) in the GMEC cultured with the miR-148a mimic than the negative control, but expression decreased (P < 0.05) more than 99% in the miR-148a-inhibited group (Fig. S2). The miR-17–5p expression was 85 times higher (P < 0.05) in the miR-17–5p mimic-transfected GMEC than the negative control, but expression decreased more than 99% in the miR-17–5p-inhibited group (Fig. S2).
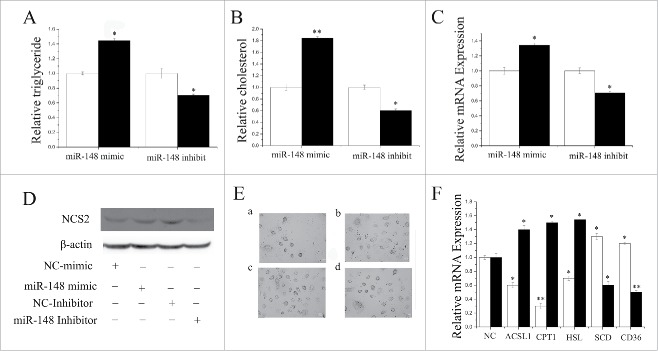
Milk fat exists as lipid droplets composed almost exclusively of TAG.18,13 Hence, we sought to detect the lipid droplet and cellular TAG content in GMEC with overexpression or inhibition of miR-148a. Compared with the negative control, the TAG content increased (P < 0.05) by 1.4-fold in miR-148a-mimic transfected cells (Fig. 5A). In contrast, TAG content decreased significantly (P < 0.05) when miR-148a was inhibited (Fig. 5A). Compared with the negative control, the cholesterol content increased (P < 0.05) by 1.8-fold in miR-148a-mimic transfected cells (Fig. 5B). Furthermore, β-casein and lipid droplet formation increased (P < 0.05) in the miR-148a mimics (Figs. 5C-E). Our findings revealed that miR-148a plays a crucial role in milk TAG synthesis, and could promotes milk fat secretion in goat mammary cells.
Figure 5.
Functional evaluation of miR-148a (A) TAG concentrations in cells transfect with miR-148a mimic (60 nM) or inhibitor (60 nM); TAG concentrations were compared with that of control (n = 6). White bars: negative control; black bars:miR-148a mimic or inhibitor. (B) Cholesterol concentrations in cells transfected with miR-148a mimic (60 nM) or inhibitor (60 nM); cholesterol concentrations were compared with that of control (n = 6). White bars: negative control; black bars: miR-148a mimic or inhibitor. (C) mRNA expression of β-Casein quantified by RT-qPCR (n = 6) in GMECs transfected with miR-148a mimic or inhibitor for 48 h. White bars: negative control; black bars: miR-148a mimic or inhibitor. (D) Western blot analysis of β-Casein expression in the miR-148a mimic or inhibitor treatment experiments. The effect of miR-148a mimic or inhibitor for 48 h on β-Casein protein expression in GMECs was evaluated by Western blot analysis. Total protein was harvested for 48 after post-treatment. (E) Changes in the lipid content of GMECs transfected with miR-148a mimic or inhibitor for 48 h. Cells were stained with oil red O. After examination microscopically, the oil red was extracted with 400 µl of isopropanol and its absorbance determined at 510 nm. The relative fat droplet content was normalized to control transfected cells. a: NC mimic treatment, b: miR-148a mimic treatment, c: NC inhibitor treatment, d: miR-148a inhibitor treatment (F) Expression of fat metabolism related genes GMECs are transfected with miR-148a mimic or inhibitor for 48h, and the mRNA expression of ACSL1, CPT1, HSL, SCD and CD36 are quantified by RT-qPCR (n = 6). White bars: miR-148a mimic; black bars: miR-148a inhibitor. All experiments were duplicated and repeated 3 times. Values are presented as means ± standard errors, *, P < 0.05; **, P < 0.01.
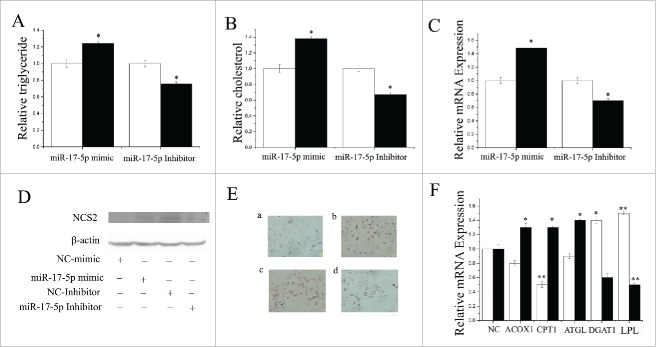
We also evaluated the function of miR-17–5p using a series of methods mentioned in the preceding sections. Compared with the negative control, the TAG content increased (P < 0.05) by 1.2-fold in miR-17–5p-mimic transfected cells (Fig. 6A). In contrast, TAG content decreased significantly (P < 0.05) when miR-17–5p was inhibited (Fig. 6A). Compared with the negative control, the cholesterol content increased (P < 0.05) by 1.3-fold in miR-17–5p-mimic transfected cells, but decreased significantly (P < 0.05) when miR-17–5p was inhibited (Fig. 6B). The expression of milk fat metabolic marker genes (β-casein) increased significantly when miR-17–5p was overexpressed (Figs. 6C, D). Additionally, we used oil red O staining to determine that the overexpression of miR-17–5p promoted fat droplet formation (Fig. 6E). Our findings revealed that miR-17–5p plays a crucial role in milk TAG synthesis, and could promote milk fat secretion in goat mammary gland.
Figure 6.
Functional evaluation of miR-17–5p (A) TAG levels in cells transfect with miR-17–5p mimic (60nM) or inhibitor (60nM); TAG levels are compared with that of control (n = 6). White bars: negative control; black bars:miR-17–5p mimic or inhibitor. (B) Cholesterol levels in cells transfect with miR-17–5p mimic (60nM) or inhibitor (60nM); cholesterol levels are compared with that of control (n = 6). White bars: negative control; black bars:miR-17–5p mimic or inhibitor. (C) GMECs are transfected with miR-17–5p mimic or inhibitor for 48h, and the mRNA expression of β-Caseinis quantified by RT-qPCR (n = 6). White bars: negative control; black bars: miR-17–5p mimic or inhibitor. (D) Western blot analysis of β-Casein expression in the miR-17–5p mimic or inhibitor treatment experiments The effect of miR-17–5p mimic or inhibitor for 48h on β-Casein protein expression is evaluated by western blot analysis in GMECs. Total proteins are harvested for 48h post-treatment, respectively. (E) Changes in the lipid contents of GMECs transfect with miR-17–5p mimic or inhibitor for 48h. Cells were stained by oil red. After examined microscopically, the oil red o is extracted with 400 µl of isopropanol and its absorbance is determined at 510 nm. The relative fat droplet contents are normalized to control transfected cells. a: NC mimic treatment, b: miR-17–5p mimic treatment, c: NC inhibitor treatment, d: miR-17–5p inhibitor treatment (F) Expression of fat metabolism related genes GMECs are transfected with miR-17–5p mimic or inhibitor for 48h, and the mRNA expression of ACOX1, CPT1, ATGL, DGAT1 and LPL are quantified by RT-qPCR (n = 6). White bars:miR-17–5p mimic; black bars:miR-17–5p inhibitor. All experiments are duplicated and repeated for 3 times. Values are presented as means ± standard errors, *, P < 0.05; **, P < 0.01.
Several genes work in a coordinated fashion to control mammary lipid and protein metabolism in ruminants.6,7 To study the role of miR-148a and miR-17–5p in the regulation of lipid metabolism gene expression, we detected the mRNA level of various genes. Fatty acids outside the cell could be hydrolyzed by LPL (Lipoprotein lipase) and then transported into cells by CD36 (CD 36 molcule (thrombospondin receptor).65 SCD (Stearoyl-CoA desaturase (delta-9-desaturase)) is relate to triglyceride synthesis.41 The results showed that overexpression of miR-148a upregulated (P < 0.05) the mRNA expression of SCD and CD36 (Fig. 5F, Table S3). The results showed that overexpression of miR-148a upregulated (P < 0.05) the mRNA expression of SCD and CD36 (Fig. 5F, Table S3). In contrast, miR-148a inhibitor elicited a marked downregulation (P < 0.05) of a series of genes related to fat metabolism including ACSL1, CPT1 and HSL (Fig. 5F, Table S3), indicating that miR-148a plays an important role in their regulation.
The results also indicated that ectopic overexpression of miR-17–5p strongly upregulated (P < 0.05) the mRNA expression of LPL and DGAT1 (Fig. 6F, Table S3). In contrast, cells transfected with the miR-17–5p inhibitor displayed marked downregulation (P < 0.05) of several fat metabolism-related genes including ACOX1, CPT1 and ATGL (Fig. 6F, Table S3), indicating that miR-17–5p plays an important role in regulation of these genes.
Expression of PPARGC1A and PPARA increases TAG levels and milk fat droplet accumulation in GMEC
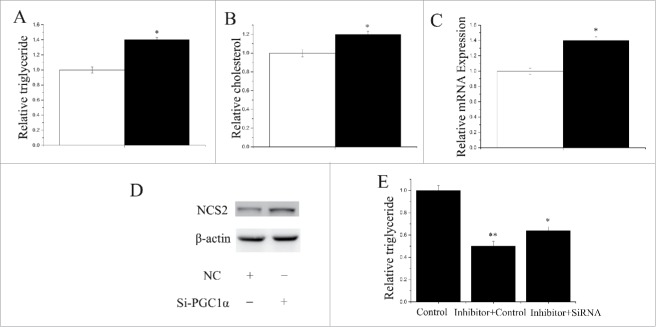
Both PPARGC1A siRNA and PPARA siRNA were used to explore their function in GMEC obtained from individual lactating goats. Compared with the negative control, the level of PPARGC1A decreased (P < 0.05) by 75% in GMEC transfected with the PPARGC1A siRNA (Fig. S2). Similarly, compared with the negative control, the PPARA level decreased (P < 0.05) by 72% in the GMEC transfected with PPARA siRNA (Fig. S2). As depicted in Fig. 7A, compared with the negative control, the TAG content increased (P < 0.05) by 1.38-fold in GMEC transfected with PPARGC1A siRNA. Compared with the negative control, the cholesterol content increased (P < 0.05) by 1.2-fold in GMEC transfected with PPARGC1A siRNA (Fig. 7B). In addition, we uncovered that PPARGC1A promoted the expression of milk fat metabolic marker genes including miRNA and protein (Fig. 7C,D). Our data indicated that PPARGC1A influences milk TAG synthesis and could promote aspects of lipid metabolism in GMEC.
Figure 7.
Functional evaluation of PPARGC1A (A) TAG levels in cells transfect with Si-NC (60nM) or SiRNA-PPARGC1A (60nM); TAG levels are compared with that of control (n = 6). White bars: negative control; black bars: SiRNA-PPARGC1A. (B) Cholesterol levels in cells transfect with Si-NC (60nM) or SiRNA-PPARGC1A (60nM); cholesterol levels are compared with that of control (n = 6). White bars: negative control; black bars: SiRNA-PPARGC1A. (C) GMECs are transfected with Si-NC (60nM) or SiRNA-PPARGC1A (60nM) for 48h, and the mRNA expression of β-Casein is quantified by RT-qPCR (n = 6). White bars: negative control; black bars: SiRNA-PPARGC1A. (D) Western blot analysis of β-Casein expression in the Si-NC (60nM) or SiRNA-PPARGC1A (60nM) treatment experiments The effect of Si-NC (60nM) or SiRNA-PPARGC1A(60nM) for 48h on β-Casein protein expression is evaluated by Western blot analysis in GMECs. Total proteins are harvested for 48h post-treatment, respectively. (E) TAG levels in cells transfect with Control inhibitor (50nM) + Control siRNA (50nM), Inhibitor-miR-148a (50nM) + Control siRNA (50nM) and Inhibitor-miR-148a (50nM) + siRNA-PPARGC1A(50nM); TAG levels are compared with that of control (n = 6).
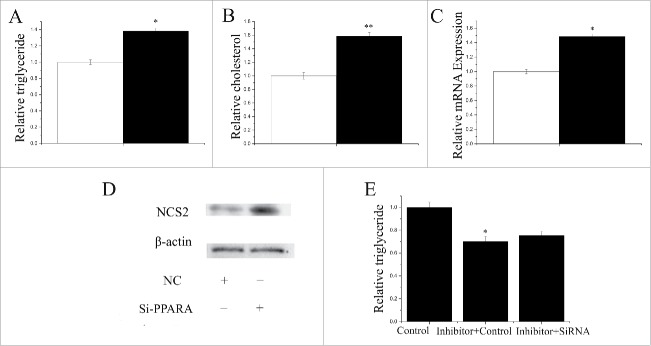
The TAG content increased (P < 0.05) by 1.35-fold in GMEC transfected with PPARA siRNA (Fig. 8A). Compared with the negative control, the cholesterol content increased (P < 0.05) by 1.6-fold in GMEC transfected with PPARA siRNA (Fig. 8B). We also observed that PPARA promoted the expression of milk fat metabolic marker genes including miRNA and protein (Fig. 8C,D). Our data indicated that PPARA influences milk fat synthesis by increasing TAG synthesis, and regulating the expression of lipid metabolism-related genes in GMEC.
Figure 8.
Functional evaluation of PPARA (A) TAG levels in cells transfect with Si-NC (60nM) or SiRNA-PPARA (60nM); TAG levels are compared with that of control (n = 6). White bars: negative control; black bars: SiRNA-PPARA. (B) Cholesterol levels in cells transfect with Si-NC (60nM) or SiRNA-PPARA (60nM); cholesterol levels are compared with that of control (n = 6). White bars: negative control; black bars: SiRNA-PPARA. (C) GMECs are transfected with Si-NC (60nM) or SiRNA-PPARA (60nM) for 48h, and the mRNA expression of β-Casein is quantified by RT-qPCR (n = 6). White bars: negative control; black bars: SiRNA-PPARA. (D) Western blot analysis of β-Casein expression in the Si-NC (60nM) or SiRNA-PPARA (60nM) treatment experiments The effect of Si-NC (60nM) or SiRNA-PPARA (60nM) for 48h on β-Casein protein expression is evaluated by Western blot analysis in GMECs. Total proteins are harvested for 48h post-treatment, respectively. (E) TAG levels in cells transfect with Control inhibitor(50nM) + Control siRNA (50nM), Inhibitor-miR-17–5p (50nM) + Control siRNA (50nM) and Inhibitor-miR-17–5p (50nM)+ SiRNA-PPARA (50nM); TAG levels are compared with that of control (n = 6).
siRNA-PPARGC1A and siRNA-PPARA rescue partly abolishes the decrease in TAG level induced by inhibition of miR-148a and miR-17–5p
We used a “rescue” experiment to demonstrate that miR-148a and miR-17–5p exert their function via PPARGC1A and PPARA. The siRNA-PPARGC1A rescue increased (P < 0.05) TAG in GMEC (Fig. 7E) in response to ectopic expression of the inhibitor-miR-148a. The decrease in TAG was partly alleviated by the siRNA-PPARGC1A rescue (TAG assay, P < 0.05, Fig. 7E). Interestingly, the increase in TAG was partly alleviated by siRNA-PPARA rescue (TAG assay, P < 0.05, Fig. 8E). The effects of inhibitor-miR-17–5p were also partly diminished by siRNA-PPARA rescue.
Discussion
Some research has illustrated that miRNA play an important role in mammary development and lactation.10,47,66 With increaesd miRNA research, researchers are trying to combine the data from different approaches (genomics, proteomics, and miRNA genomics) to obtain more effective data for practical application.57,60 Although there are many methods used to analyze mRNA and miRNA for lipid metabolism55, there has been no miRNA-mRNA regulation analysis performed in milk goats. The main purpose of this article is to examine the role of mRNA and miRNA in the regulation of milk fat. Through the comparison of miRNA-mRNA between groups, this study provides insight into the regulation mechanism of lipid metabolism. In this study, we selected 3 goats for each period, early-lactation and non-lactation (“dry”). This is a small sample number was small. However, the primary objective of this study was to identify miRNA-mRNAs differential expression to see if there is a regulatory relationship with milk fat metabolism. After selecting our potential pathways of target miRNA-mRNA, we will need a series of experiments to validate this miRNA-mRNA, including assessing function and regulatory relationships. Therefore, the number of selected goats is quite small in this study, but the extensive sequencing data provides reliability.
To understand the exact molecular mechanism and function of miR-148a and miR-17–5p, we performed some functional experiments, including the detection of lipid contents and fat droplet formation, under conditions of miR-148a and miR-17–5p overexpression and inhibition. Interestingly, the triglyceride content and lipid accumulation were increased by miR-148a and miR-17–5p, suggesting that the functions of miR-148a and miR-17–5p are related to milk fat secretion and metabolism. In support of this finding, fat metabolism related gene expression when miR-148a and miR-17–5p were overexpressed or inhibited indicated that miR-148a and miR-17–5p may be involved in lipid metabolism signaling pathways. Many genes have been reported that are involved in lipid metabolism signaling pathways. For example, ACSL (acyl-CoA synthetase), CPT (carnitine palmitoyltransferase), ACOX1 (acyl-CoA oxidase 1) were reported to play crucial roles in β-oxidation53, and HSL (hormone-sensitive lipase) and ATGL (Adipose triglyceride lipase) are very important in lipolysis.67 Fatty acids outside the cell could be hydrolyzed by LPL (Lipoprotein lipase) and then transported into cells by CD36 (CD 36 molcule (thrombospondin receptor).65 SCD (Stearoyl-CoA desaturase (delta-9-desaturase)) and DGAT1 (Diacylglycerol acyltransferase 1) are related to triglyceride synthesis.41 The experimental results supported our hypothesis that miR-148a and miR-17–5p regulate fat metabolism in GMECs. On the other hand, the β-casein gene serves as a kind of marker gene for breast fat metabolism. In this study, we effectively monitored breast fat metabolism and indirectly explained triglyceride synthesis based on the expression level of β-casein. For this reason, we examined the protein and mRNA levels of β-casein in different experimental treatments.
Although the specific biologic functions of some miRNA have been reported, information concerning the function of miRNA in GMEC is limited. MiR-148a was highly-expressed in hMSCs-Ad during adipogenesis.42 Thus, the role of miR-148a in TAG synthesis in GMEC was further clarified in the present study. Because miR-148a has different functions in different tissues, we speculate that miR-148a can regulate TAG metabolism in ruminant mammary cells. When mimic and inhibitor of miR-148a were simultaneously transfected in GMEC, we detected a much stronger response than individually. Our data also indicated that miR-17–5p could regulate TAG synthesis. Interestingly, miR-148a could regulate PPARA, which is a target gene of miR-17–5p; and miR-17–5p could regulate PPARGC1A, which is target gene of miR-148a. PPARA is a member of the family of nuclear hormone receptors that functions as a ligand-activated transcription factor with a signature type II zinc finger DNA binding. It controls the expression of specific genes involved in fatty acid utilization.1,8 CPT1A expression is controlled by a complex of transcription factors including PPARGC1A and PPARA.8
In conclusion, our findings indicated that miR-148a could target and repress PPARGC1A, which in turn could regulate PPARA (i.e. miR-17–5p targets PPARA) to regulate milk TAG synthesis. (Fig. 9). The results have uncovered an important role miR-148a and miR-17–5p in TAG synthesis in GMEC. The miR-148a and miR-17–5p seem to synergistically regulate lipid catabolism via the control of PPARGC1A and PPARA in GMEC. In the long-term, these findings might be helpful in developing practical means to improve the quality of ruminant milk.
Figure 9.
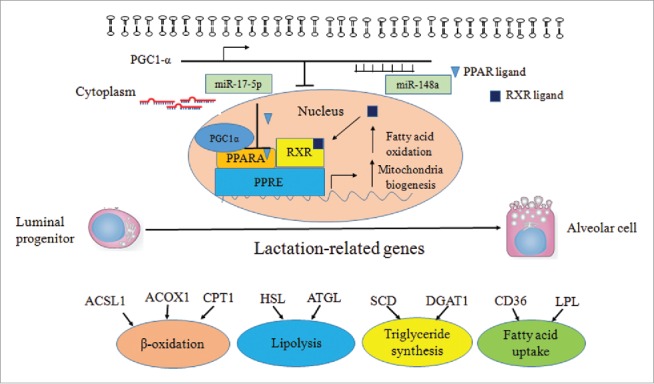
Diagram summarizing our findings: miR-148a and miR-17–5p synergistically regulating fat oxidation via PPARGC1A and PPARA in goat
Material and methods
Ethical statement
All experimental materials and protocols were approved by the Animal Care and Use Committee of Northwest A&F University. The methods were performed in accordance with the approved guidelines.
Animals and samples of RNA extraction
The elite herd of Xinong Saanen dairy goats used in this research were from the experimental farm at Northwest Agricultural and Forestry University of China. Three 3-year-old goats of similar weight at each of the following stages of lactation were used: non-pregnant, early-lactation (15 d after parturition), peak-lactation (60 d after parturition), late-lactation (150 d after parturition), and non-lactation (“dry” period). Spleen, stomach, heart, liver, sebum, mammary gland tissue, muscle, lung and kidney were collected after slaughter. Three biologic samples per tissue were snap-frozen in liquid nitrogen as soon as possible. Using Trizol reagent (Invitrogen, USA), total RNA was extracted in accordance with instructions. The quality and quantity of RNA were detected by a ND-1000 spectrophotometer (NanoDrop, USA), and the RNA was stored in −80°C before experiment.
Annotation, classification and abundance analysis of gene expression
Annotation and classification: The sequencing data provided unique sequences and the corresponding copy number of each unique sequence. The resulting sequences were compared with the mRNA database. The process allows only a single base mismatch, and the gene expression is the sum of the sequencing frequencies. The selection of different genes was performed using values of P < 0.05 and FDR < 0.05. Clustering, enrichment of GO, and analysis of KEGG enrichment were also performed.
Gene expression abundance analysis: Based on the data obtained from the sequencing, the dairy goat mammary transcriptome data obtained by 52 were used as reference sequences. Using the software bowtie 0.12.8, gene expression was measured based on the RPKM (Reads Per Kilobase of exon model per Million mapped reads) value (S2).
Cell culture and transfection
The GMEC were cultured in DMEM/F12 medium (Invitrogen Corp., USA) containing 10% FBS, 10 ng/ml EGF-1 (epidermal growth factor 1, Gibco), 5 mg/ml insulin and 0.25 mmol/L hydrocortisone in 37°C in a humidified atmosphere with 5% CO2. The GMEC were cultured and fractionated in accordance with previous research.51 To induce lactogenesis, GMEC were cultured in a lactogenic medium for 48 h before initial experiments.(31,48 Cells were transfected with either the miR-148a, miR-17–5p-mimic (60 nM) or inhibitor (60 nM) (Invitrogen, USA) using Lipofectamine™ RNAMAX (Invitrogen, USA) according to manufacturer's instructions. Cells were harvested after 48 h of transfection. The sequences of mimic, inhibitor and siRNA were listed in S1 and S3.
Staining of oil red O
Oil red O staining was used as mentioned in the previous research with modifications.66 In brief, GMEC were transfected with miR-148a, miR-17–5p mimic or inhibitor, were cleaned several times in PBS (phosphate buffer solution), and were fixed in 10% paraformaldehyde for 30 minutes. Afterwards, cells were stained using 10% oil red O in isopropanol for 10 minutes, were cleaned by PBS, and were detected microscopically.
Assay of cellular TAG content
The GMEC were transfected with either miR-148a, miR-17–5p mimic or inhibitor. Cells were obtained with lysis buffer (1% Triton X-100, pH 7.4, 150 mmol/NaCl, 50mmol/l Tris-HCL) after 48 h of incubation. TAG was measured using a commercial kit following the manufacturer's instructions (Loogen, China) on an XD 811G Biochemistry Analyzer (Odin Science &Technology Company, Shanghai China). The values acquired were normalized to the content of total protein with the BCA protein assay kit (Thermo crop, Prod#23227).
Assay of cellular cholesterol content
The GMEC were transfected with either miR-148a, miR-17–5p mimic or inhibitor and siRNA (20ng). Cells were obtained with lysis buffer (1% Triton X-100, pH 7.4, 150 mmol/NaCl, 50 mmol/l Tris-HCL) after 48 h of incubation. Cholesterol was measured using a serum cholesterol kit on the basis of the instructions (Loogen, China) on an XD 811G Biochemistry Analyzer (Odin Science &Technology Company, Shanghai China). The values obtained were normalized to the content of total protein.
RT-qPCR and Western blot
RT-qPCR for miRNA, the mature miRNA expression level was determined using the S-Poly(T) assay.32 The S1 primer was used as specific reverse primer, respectively. Briefly, reverse transcription was performed as follows: a 10 µl reaction including 0.2 µg total RNA, 2.5 µl of 4× reaction buffer, 1 µl of polyA/RT enzyme mix, 1µl of 0.5 µM RT primer. The reaction was performed at 37°C for 30 minutes, followed by 42°C for 30 minutes, then 75°C for 5 minute. The RT products were amplified and detected using a universal TaqMan® probe, a 20 µl PCR reaction containing 0.3 µl of RT products, 4 µl of 5×qPCR probe Mix, 0.5 unit of Go TaqMan® Hot Start Polymerase (Promega, USA), 0.2 mM universal TaqMan® probe, and 0.5µM forward primer and universal reverse primer, respectively. The PCR reaction was performed at 95°C for 3min, followed by 40 cycles of 95°C for 10s and 60°C for 30s. The 18S rRNA was used as internal control.
RT-qPCR for mRNA, 0.5 µg of total RNA was synthesized into cDNA using the Prime Script® RT Reagent Kit (Perfect Real time, Takara, Japan). The RT-qPCR assays were performed according to instructions (SYBR Premix TaqMan®, Perfect Real Time, Takara, Japan) with the specific primers reported previously.9,31 The expression was normalized to UXT. The sequence of primers was listed in S3. All real-time reactions, including controls with no templates, were performed on a Bio-Rad CFX96 real-time PCR detection system (Bio-Rad, USA) in triplicate. Relative expression was calculated using 2−△△Ct.38–40,51
As to western blot analysis, cells were obtained and lysed in RIPA buffer (Solarbio, China). Proteins extracted from cells were separated by SDS-PAGE, transferred to nitrocellulose membrane (Millipore, USA) and probed with the primary monoclonal rabbit anti- PGC-1α (Cell Signaling Technology Kit #2178), Polyclonal anti-rabbit PPARA15540–1-AP, China), rabbit anti-β-Casein (Proteintech Group, 51067–2-AP, China) and monoclonal mouse anti-β-action (Proteintech Group, 66009–1-IG, China), respectively. Polyclonal goat anti-rabbit HRP-conjugated IgG (Tiangen, China) was used as secondary antibody. All antibodies were used on the basis of the instructions. Signals were detected by the chemiluminescent ECL Western blot system (Pierce, USA).
Luciferase reporter assay
To generate a reporter construct luciferase assays, a segment containing a miRNA target site in the 3′-UTR of PPARGC1A and PPARA was inserted into the psiCHECK-2 vector (Promega, USA) between the NotⅠand Xho sites immediately downstream of the Renilla luciferase gene. The wild-type segment and mutant-type sequence were established by PCR overlap technology. The sequence of primers is listed in S5. All constructs were verified by sequencing. The GMEC were seeded in 384-well plates at a density of 50,000 cells per well one day before transfection. A total of 0.33 g of each reporter construct was transiently transfected using the X-treme GENE HP DNA Transfection Reagent (Roche, Switzerland) on the basis of the protocol. Cells were then transfected with either miR-148a, miR-17–5p mimic or inhibitor using Lipofectamine™ RNAMAX after a 6 h recovery period in medium. At 48 h post-transfection, firefly and Renilla luciferase activities were measured with the Dual-Glo luciferase assay system following manufacturer's instructions (Promega, USA).
Statistical analysis
Statistical analysis was performed using the software package SPSS (SPSS 19.0). Research data are presented as mean ± SE (standard error) of 3 independent experiments. Differences between the groups were detected at *p < 0.05, **p < 0.01.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was jointly supported by the National Natural Science Foundation of China (No. 31372281), the Special Fund for Agro-scientific Research in the Public Interest (201103038), and the Transgenic New Species Breeding Program of China (2014ZX08009–051B).
References
- 1.Alaynick WA. Nuclear receptors, mitochondria and lipid metabolism. Mitochondrion 2008; 8(4):329-37. PMID:18375192; http://dx.doi.org/ 10.1016/j.mito.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopoulos AK, Katsafadou AI, Pierros V, Kontopodis E, Fthenakis GC, Arsenos G, Karkabounas S, Tzora A, Skoufos I, Tsangaris GT. Milk of Greek sheep and goat breeds; characterization by means of proteomics. J Proteomics 2016; 147:76-84. PMID:27102495; http://dx.doi.org/ 10.1016/j.jprot.2016.04.008 [DOI] [PubMed] [Google Scholar]
- 3.Avery-Kiejda KA, Braye SG, Mathe A, Forbes JF, Scott RJ. Decreased expression of key tumour suppressor microRNAs is associated with lymph node metastases in triple negative breast cancer. BMC Cancer 2014; 14:51. PMID:24479446; http://dx.doi.org/ 10.1186/1471-2407-14-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avril-Sassen S, Goldstein LD, Stingl J, Blenkiron C, Le Quesne J, Spiteri I, Karagavriilidou K, Watson CJ, Tavare S, Miska EA, et al.. Characterisation of microRNA expression in post-natal mouse mammary gland development. BMC Genomics 2009; 10:548. PMID:19930549; http://dx.doi.org/ 10.1186/1471-2164-10-548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao H, Kommadath A, Sun X, Meng Y, Arantes AS, Plastow GS, Guan LL, Stothard P. Expansion of ruminant-specific microRNAs shapes target gene expression divergence between ruminant and non-ruminant species. BMC Genomics 2013; 14:609. PMID:24020371; http://dx.doi.org/ 10.1186/1471-2164-14-609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bionaz M, Loor JJ. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics 2008; 9(1):366. PMID:18671863; http://dx.doi.org/ 10.1186/1471-2164-9-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bionaz M, Loor JJ. Gene networks driving bovine mammary protein synthesis during the lactation cycle. Bioinform Biol Insights 2011; 83-92; PMID:2169807315363638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnefont JP, Djouadi F, Prip-Buus C, Gobin S, Munnich A, Bastin J. 2004. Carnitine palmitoyltransferases 1 and 2: biochemical, molecular and medical aspects. Mol Aspects Med 25(5-6):495-520. PMID:15363638; http://dx.doi.org/ 10.1016/j.mam.2004.06.004 [DOI] [PubMed] [Google Scholar]
- 9.Bonnet M, Bernard L, Bes S, Leroux C. Selection of reference genes for quantitative real-time PCR normalisation in adipose tissue, muscle, liver and mammary gland from ruminants. Animal 2013; 7(08):1344-53. PMID:23552195; http://dx.doi.org/ 10.1017/S1751731113000475 [DOI] [PubMed] [Google Scholar]
- 10.Bu DP, Nan XM, Wang F, Loor JJ, Wang JQ. Identification and characterization of microRNA sequences from bovine mammary epithelial cells. J Dairy Sci 2015; 98(3):1696-705. PMID:25622872; http://dx.doi.org/ 10.3168/jds.2014-8217 [DOI] [PubMed] [Google Scholar]
- 11.Calvano Filho CM, Calvano-Mendes DC, Carvalho KC, Maciel GA, Ricci MD, Torres AP, Filassi JR, Baracat EC. Triple-negative and luminal A breast tumors: differential expression of miR-18a-5p, miR-17-5p, and miR-20a-5p. Tumour Biol 2014; 35(8):7733-41. PMID:24810926; http://dx.doi.org/ 10.1007/s13277-014-2025-7 [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Luo J, Ma L, Wang H, Cao W, Xu H, Zhu J, Sun Y, Li J, Yao D, et al.. MiR130b-Regulation of PPARgamma Coactivator- 1alpha Suppresses Fat Metabolism in Goat Mammary Epithelial Cells. PloS one 2015; 10(11):e0142809. PMID:26579707; http://dx.doi.org/ 10.1371/journal.pone.0142809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Qiu H, Ma L, Luo J, Sun S, Kang K, Gou D, Loor JJ. miR-30e-5p and miR-15a Synergistically Regulate Fatty Acid Metabolism in Goat Mammary Epithelial Cells via LRP6 and YAP1. Int J Mol Sci 2016a; 17(11).; PMID:2785432927616141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z, Shi H, Sun S, Xu H, Cao D, Luo J. MicroRNA-181b suppresses TAG via target IRS2 and regulating multiple genes in the Hippo pathway. Exp Cell Res 2016b; 348(1):66-74. PMID:27616141.; http://dx.doi.org/ 10.1016/j.yexcr.2016.09.004 [DOI] [PubMed] [Google Scholar]
- 15.Chilliard Y, Ferlay A, Rouel J, Lamberet G. A review of nutritional and physiological factors affecting goat milk lipid synthesis and lipolysis. J Dairy Sci 2003; 86(5):1751-70. PMID:12778586; http://dx.doi.org/ 10.3168/jds.S0022-0302(03)73761-8 [DOI] [PubMed] [Google Scholar]
- 16.Danza K, Silvestris N, Simone G, Signorile M, Saragoni L, Brunetti O, Monti M, Mazzotta A, De Summa S, Mangia A, et al.. Role of miR-27a, miR-181a and miR-20b in gastric cancer hypoxia-induced chemoresistance. Cancer Biol Ther 2016; 17(4):400-6; PMID:2679399215531879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature 2004; 432(7014):231-5. PMID:15531879; http://dx.doi.org/ 10.1038/nature03049 [DOI] [PubMed] [Google Scholar]
- 18.Destaillats F, Angers P. Base-catalyzed derivatization methodology for FA analysis. Application to Milk fat and Celery Seed Lipid TAG. Lipids 2002; 37(5):527-32.; PMID:1205659719366863 [DOI] [PubMed] [Google Scholar]
- 19.Estall JL, Kahn M, Cooper MP, Fisher FM, Wu MK, Laznik D, Qu L, Cohen DE, Shulman GI, Spiegelman BM. Sensitivity of lipid metabolism and insulin signaling to genetic alterations in hepatic peroxisome proliferator-activated receptor-gamma coactivator-1alpha expression. Diabetes 2009; 58(7):1499-508. PMID:19366863; http://dx.doi.org/ 10.2337/db08-1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Ann Rev Biochem 2010; 79:351-79. PMID:20533884; http://dx.doi.org/ 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- 21.Fu J, Tang W, Du P, Wang G, Chen W, Li J, Zhu Y, Gao J, Cui L. Identifying microRNA-mRNA regulatory network in colorectal cancer by a combination of expression profile and bioinformatics analysis. BMC Systems Biol 2012; 6:68. PMID:22703586; http://dx.doi.org/ 10.1186/1752-0509-6-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci S, Irkkan C, Morrison C, Croce CM, Shapiro CL, et al.. microRNA expression profiling identifies a 4 microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget 2014; 5(5):1174-1184. PMID:24632568; http://dx.doi.org/ 10.18632/oncotarget.1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004; 432(7014):235-40. PMID:15531877; http://dx.doi.org/ 10.1038/nature03120 [DOI] [PubMed] [Google Scholar]
- 24.Gu Z, Eleswarapu S, Jiang H. Identification and characterization of microRNAs from the bovine adipose tissue and mammary gland. FEBS Lett 2007; 581(5):981-8. PMID:17306260; http://dx.doi.org/ 10.1016/j.febslet.2007.01.081 [DOI] [PubMed] [Google Scholar]
- 25.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. 2004. The Drosha-DGCR8 complex in primary microRNA processing. Gen Dev 18(24):3016-27.; PMID:1557458917018837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocrine Rev 2006; 27(7):728-35. PMID:17018837; http://dx.doi.org/ 10.1210/er.2006-0037 [DOI] [PubMed] [Google Scholar]
- 27.Hinrichs J. Mediterranean milk and milk products. Eur J Nutr 2004; 43 Suppl 1:I/12-17.; PMID:1505249424925028 [DOI] [PubMed] [Google Scholar]
- 28.Humphries B, Wang Z, Oom AL, Fisher T, Tan D, Cui Y, Jiang Y, Yang C. MicroRNA-200b targets protein kinase Calpha and suppresses triple-negative breast cancer metastasis. Carcinogenesis 2014; 35(10):2254-63. PMID:24925028; http://dx.doi.org/ 10.1093/carcin/bgu133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jabed A, Wagner S, McCracken J, Wells DN, Laible G. Targeted microRNA expression in dairy cattle directs production of beta-lactoglobulin-free, high-casein milk. Proc Natl Acad Sci U S A 2012; 109(42):16811-6. PMID:23027958; http://dx.doi.org/ 10.1073/pnas.1210057109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji Z, Wang G, Xie Z, Zhang C, Wang J. Identification and characterization of microRNA in the dairy goat (Capra hircus) mammary gland by Solexa deep-sequencing technology. Mol Biol Rep 2012; 39(10):9361-71. PMID:22763736; http://dx.doi.org/ 10.1007/s11033-012-1779-5 [DOI] [PubMed] [Google Scholar]
- 31.Kadegowda AKG, Bionaz M, Piperova LS, Erdman RA, Loor JJ. Peroxisome proliferator-activated receptor-γ activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents. J Dairy Sci 2009; 92(9):4276-89. PMID:19700688; http://dx.doi.org/ 10.3168/jds.2008-1932 [DOI] [PubMed] [Google Scholar]
- 32.Kang K, Zhang X, Liu H, Wang Z, Zhong J, Huang Z, Peng X, Zeng Y, Wang Y, Yang Y, et al.. A novel real-time PCR assay of microRNAs using S-Poly(T), a specific oligo(dT) reverse transcription primer with excellent sensitivity and specificity. PloS one 2012; 7(11):e48536. PMID:23152780; http://dx.doi.org/ 10.1371/journal.pone.0048536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang K, Zhong J, Jiang L, Liu G, Gou CY, Wu Q, Wang Y, Luo J, Gou D. Identification of microRNA-Like RNAs in the filamentous fungus Trichoderma reesei by solexa sequencing. PloS one 2013; 8(10):e76288. PMID:24098464; http://dx.doi.org/ 10.1371/journal.pone.0076288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee CH, Olson P, Evans RM. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology 2003a; 144(6):2201-07. PMID:12746275; http://dx.doi.org/ 10.1210/en.2003-0288 [DOI] [PubMed] [Google Scholar]
- 35.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al.. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003b; 425(6956):415-9. PMID:14508493; http://dx.doi.org/ 10.1038/nature01957 [DOI] [PubMed] [Google Scholar]
- 36.Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, et al.. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol 2005; 3(4):e101. PMID:15760270; http://dx.doi.org/ 10.1371/journal.pbio.0030101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Li T, Wang S, Wei J, Fan J, Li J, Han Q, Liao L, Shao C, Zhao RC. miR-17-5p and miR-106a are involved in the balance between osteogenic and adipogenic differentiation of adipose-derived mesenchymal stem cells. Stem Cell Res 2013; 10(3):313-24. PMID:23399447; http://dx.doi.org/ 10.1016/j.scr.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 38.Lin X, Luo J, Zhang L, Wang W, Gou D. MiR-103 controls milk fat accumulation in goat (Capra hircus) mammary gland during lactation. PloS one 2013a; 8(11):e79258. PMID:24244462; http://dx.doi.org/ 10.1371/journal.pone.0079258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin X, Luo J, Zhang L, Zhu J. MicroRNAs synergistically regulate milk fat synthesis in mammary gland epithelial cells of dairy goats. Gene Expression 2013b; 16(1):1-13. PMID:24397207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin XZ, Luo J, Zhang LP, Wang W, Shi HB, Zhu JJ. MiR-27a suppresses triglyceride accumulation and affects gene mRNA expression associated with fat metabolism in dairy goat mammary gland epithelial cells. Gene 2013c; 521(1):15-23. PMID:23537996; http://dx.doi.org/ 10.1016/j.gene.2013.03.050 [DOI] [PubMed] [Google Scholar]
- 41.Listenberger LL, Han X, Lewis SE, Cases S, Farese RV Jr., Ory DS, Schaffer JE. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci U S A 2003; 100(6):3077-82. PMID:12629214; http://dx.doi.org/ 10.1073/pnas.0630588100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, et al.. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A 2008; 105(36):13556-61. PMID:18768788.; http://dx.doi.org/ 10.1073/pnas.0803055105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luna P, Bach A, Juarez M, de la Fuente MA. Effect of a diet enriched in whole linseed and sunflower oil on goat milk fatty acid composition and conjugated linoleic acid isomer profile. J Dairy Sci 2008; 91(1):20-28. PMID:18096921; http://dx.doi.org/ 10.3168/jds.2007-0447 [DOI] [PubMed] [Google Scholar]
- 44.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A 2010; 107(27):12228-32. PMID:20566875; http://dx.doi.org/ 10.1073/pnas.1005191107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell 2008; 133(2):217-222. PMID:18423194; http://dx.doi.org/ 10.1016/j.cell.2008.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreno-Fernandez J, Diaz-Castro J, Alferez MJ, Nestares T, Ochoa JJ, Sanchez-Alcover A, Lopez-Aliaga I. Fermented goat milk consumption improves melatonin levels and influences positively the antioxidant status during nutritional ferropenic anemia recovery. Food & function 2016; 7(2):834-42.; PMID:2666204125580536 [DOI] [PubMed] [Google Scholar]
- 47.Peng J, Zhao JS, Shen YF, Mao HG, Xu NY. MicroRNA expression profiling of lactating mammary gland in divergent phenotype swine breeds. Int J Mol Sci 2015; 16(1):1448-65. PMID:25580536; http://dx.doi.org/ 10.3390/ijms16011448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peterson DG, Matitashvili EA, Bauman DE. The Inhibitory Effect of trans-10, cis-12 CLA on Lipid Synthesis in Bovine Mammary Epithelial Cells Involves Reduced Proteolytic Activation of the Transcription Factor SREBP-1. J Nutrition 2004; 134(10):2523-7. PMID:15465741. [DOI] [PubMed] [Google Scholar]
- 49.Sharma SB, Lin CC, Farrugia MK, McLaughlin SL, Ellis EJ, Brundage KM, Salkeni MA, Ruppert JM. MicroRNAs 206 and 21 cooperate to promote RAS-extracellular signal-regulated kinase signaling by suppressing the translation of RASA1 and SPRED1. Mol Cell Biol 2014; 34(22):4143-64. PMID:25202123; http://dx.doi.org/ 10.1128/MCB.00480-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi C, Zhang M, Tong M, Yang L, Pang L, Chen L, Xu G, Chi X, Hong Q, Ni Y, et al.. miR-148a is Associated with Obesity and Modulates Adipocyte Differentiation of Mesenchymal Stem Cells through Wnt Signaling. Scientific Rep 2015a; 5:9930. PMID:26001136; http://dx.doi.org/ 10.1038/srep09930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi H, Luo J, Zhu J, Li J, Sun Y, Lin X, Zhang L, Yao D, Shi H. PPAR gamma Regulates Genes Involved in Triacylglycerol Synthesis and Secretion in Mammary Gland Epithelial Cells of Dairy Goats. PPAR Res 2013; 2013:310948. PMID:23710163; http://dx.doi.org/ 10.1155/2013/310948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi H, Zhu J, Luo J, Cao W, Shi H, Yao D, Li J, Sun Y, Xu H, Yu K, Loor JJ. Genes regulating lipid and protein metabolism are highly expressed in mammary gland of lactating dairy goats. Functional & integrative genomics 2015b; 15(3):309-21.; PMID:2543370819256553 [DOI] [PubMed] [Google Scholar]
- 53.Shimoda H, Tanaka J, Kikuchi M, Fukuda T, Ito H, Hatano T, Yoshida T. Effect of polyphenol-rich extract from walnut on diet-induced hypertriglyceridemia in mice via enhancement of fatty acid oxidation in the liver. J Agricultural Food Chem 2009; 57(5):1786-92. PMID:19256553; http://dx.doi.org/ 10.1021/jf803441c [DOI] [PubMed] [Google Scholar]
- 54.Shirasaki T, Honda M, Shimakami T, Horii R, Yamashita T, Sakai Y, Sakai A, Okada H, Watanabe R, Murakami S, et al.. MicroRNA-27a regulates lipid metabolism and inhibits hepatitis C virus replication in human hepatoma cells. J Virol 2013; 87(9):5270-86. PMID:23449803; http://dx.doi.org/ 10.1128/JVI.03022-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tabe Y, Hatanaka Y, Nakashiro M, Sekihara K, Yamamoto S, Matsushita H, Kazuno S, Fujimura T, Ikegami T, Nakanaga K, et al.. Integrative genomic and proteomic analyses identifies glycerol-3-phosphate acyltransferase as a target of low-dose ionizing radiation in EBV infected-B cells. Int J Radiat Biol 2016; 92(1):24-34. PMID:26809544; http://dx.doi.org/ 10.3109/09553002.2015.1106021 [DOI] [PubMed] [Google Scholar]
- 56.van Iterson M, Bervoets S, de Meijer EJ, Buermans HP, t Hoen PA, Menezes RX, Boer JM. Integrated analysis of microRNA and mRNA expression: adding biological significance to microRNA target predictions. Nucleic Acids Res 2013; 41(15):e146. PMID:23771142; http://dx.doi.org/ 10.1093/nar/gkt525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vincent M, Oved K, Morag A, Pasmanik-Chor M, Oron-Karni V, Shomron N, Gurwitz D. Genome-wide transcriptomic variations of human lymphoblastoid cell lines: insights from pairwise gene-expression correlations. Pharmacogenomics 2012; 13(16):1893-904. PMID:23215882; http://dx.doi.org/ 10.2217/pgs.12.179 [DOI] [PubMed] [Google Scholar]
- 58.Wang J, Tsouko E, Jonsson P, Bergh J, Hartman J, Aydogdu E, Williams C. miR-206 inhibits cell migration through direct targeting of the actin-binding protein coronin 1C in triple-negative breast cancer. Mol Oncol 2014; 8(8):1690-702. PMID:25074552; http://dx.doi.org/ 10.1016/j.molonc.2014.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warrington JM, Kim JJ, Stahel P, Cieslar SR, Moorehead RA, Coomber BL, Corredig M, Cant JP. Selenized milk casein in the diet of BALB/c nude mice reduces growth of intramammary MCF-7 tumors. BMC cancer 2013; 13:492. PMID:24152862; http://dx.doi.org/ 10.1186/1471-2407-13-492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilbert ML, Yeo GW. Genome-wide approaches in the study of microRNA biology. Wiley Interdiscip Rev Syst Biol Med 2011; 3(5):491-512. PMID:21197653; http://dx.doi.org/ 10.1002/wsbm.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab 2007; 91(3):209-17. PMID:17521938; http://dx.doi.org/ 10.1016/j.ymgme.2007.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu J, Liao M, Zhu H, Kang K, Mu H, Song W, Niu Z, He X, Bai C, Li G, et al.. CD49f-positive testicular cells in Saanen dairy goat were identified as spermatogonia-like cells by miRNA profiling analysis. J Cell Biochem 2014; 115(10):1712-23. PMID:24817091; http://dx.doi.org/ 10.1002/jcb.24835 [DOI] [PubMed] [Google Scholar]
- 63.Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, Seale P, Fernando P, van Ijcken W, Grosveld F, et al.. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab 2013; 17(2):210-24. PMID:23395168; http://dx.doi.org/ 10.1016/j.cmet.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu S, Reddy JK. Transcription coactivators for peroxisome proliferator-activated receptors. Biochimica et biophysica acta 2007; 1771(8):936-51. PMID:17306620; http://dx.doi.org/ 10.1016/j.bbalip.2007.01.008 [DOI] [PubMed] [Google Scholar]
- 65.Zhou J, Febbraio M, Wada T, Zhai Y, Kuruba R, He J, Lee JH, Khadem S, Ren S, Li S, et al.. Hepatic fatty acid transporter Cd36 is a common target of LXR, PXR, and PPARgamma in promoting steatosis. Gastroenterology 2008; 134(2):556-67. PMID:18242221; http://dx.doi.org/ 10.1053/j.gastro.2007.11.037 [DOI] [PubMed] [Google Scholar]
- 66.Zhu J, Sun Y, Luo J, Wu M, Li J, Cao Y. Specificity protein 1 regulates gene expression related to fatty acid metabolism in goat mammary epithelial cells. Int J Mol Sci 2015; 16(1):1806-20. PMID:25594872; http://dx.doi.org/ 10.3390/ijms16011806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, et al.. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 2004; 306(5700):1383-6. PMID:15550674; http://dx.doi.org/ 10.1126/science.1100747 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.