Abstract
Hemophilia B occurs due to a deficiency in human blood coagulation factor IX (hFIX). Currently, no effective treatment for hemophilia B has been identified, and gene therapy has been considered the most appropriate treatment. Mesenchymal stem cells (MSCs) have homing abilities and low immunogenicity, and therefore they may be potential cell carriers for targeted drug delivery to lesional tissues. The present study constructed an adeno-associated virus integration site 1 (AAVS1)-targeted vector termed AAVS1-green fluorescent protein (GFP)-hFIX and a zinc finger nuclease (ZFN) expression vector. Nucleofection was used to co-transfect the targeting vector and the ZFN expression vector into human MSCs. The GFP-positive cells were selected using flow cytometry. Site-specific integration clones were obtained following the monoclonal culture, subsequent detections were performed using polymerase chain reaction and Southern blotting. Following the confirmation of stem cell traits of the site-specific integration MSCs, the in vivo and in vitro expression levels of hFIX were detected. The results demonstrated that the hFIX gene was successfully transfected into the AAVS1 locus in human MSCs. The clones with the site-specific integration retained stem cell traits of the MSCs. In addition, hFIX was effectively expressed in vivo and in vitro. No significant differences in expression levels were identified among the individual clones. In conclusion, the present study demonstrated that the exogenous gene hFIX was effectively expressed following site-specific targeting into the AAVS1 locus in MSCs; therefore, MSCs may be used as potential cell carriers for gene therapy of hemophilia B.
Keywords: hemophilia B, gene targeting, AAVS1, zinc finger nuclease, MSCs
Introduction
Hemophilia B is a common X-linked recessive genetic disease, with an incidence rate of 1/30,000 in males in the UK (1). Hemophilia B is primarily caused by a deficiency in human blood coagulation factor IX (hFIX). The major clinical symptoms include spontaneous or traumatic bleeding, particularly bleeding and hematomas in the muscles and joints, which may lead to disability and become life-threatening due to intracranial and joint bleeding. No effective treatment has been identified to successfully treat hemophilia B. Traditional treatment entails supplementation of plasma products with enriched hFIX. This therapy may be effective; however, the risk of viral contamination, such as human immunodeficiency virus or hepatitis B, may pose a risk (2). Furthermore, the existing therapeutics, including the products of purified and recombinant hFIX, are extremely expensive and repeated transfusions of hFIX are required due to its short half-life in vivo (3). The hFIX inhibitory antibody is generated following repeated infusions of hFIX products into the patient, thereby limiting the use of this protein replacement therapy. Hemophilia B is considered one of the genetic diseases that are most suitable for gene therapy (4). A slightly increased hFIX concentration in the plasma (>1% or 50 ng/ml) may yield a significant therapeutic effect and a hFIX concentration of up to 150% of the normal level does not lead to complications, such as thrombosis (5).
As pluripotent adult stem cells, human mesenchymal stem cells (hMSCs) are widely distributed across various tissues and organs in the body. MSCs are a potential resource for regenerative medicine due to the simplicity of isolating them with density gradient centrifugation (6–8). In addition to their homing ability on lesions and damaged tissues, and due to their low immunogenicity, MSCs are considered to be the optimal cell vectors for gene therapy (9,10). However, the efficient introduction and effective expression of an exogenous gene is critical for successful gene therapy using MSCs.
Currently, various methods have been used to modify MSCs derived from different tissues for the gene therapy of cancer and genetic diseases. However, these modifications are primarily based on random integration. The uncertainty of the insertion point may lead to various problems. Random integration may occur in the heterochromatin region, resulting in gene silencing (11), or in the coding region of endogenous genes and damage the function of these genes, thereby affecting the transcription and expression of the surrounding genes, which may result in the occurrence of malignant tumors (12). Due to the risks associated with random genetic integration, targeting of an exogenous gene into a ‘safe site’ of the genome has become a more desirable method of genetic modification. Adeno-associated virus integration site 1 (AAVS1) has been confirmed to be a safe transgenic locus and when combined with zinc finger nuclease (ZFN), an exogenous gene may be efficiently targeted into the AAVS1 locus in various cell lines (13). The present study aimed to introduce the hFIX gene into the AAVS1 locus in MSCs to investigate the feasibility of MSCs as potential cell vectors for future gene therapy of hemophilia B.
Materials and methods
Isolation and culture of MSCs
Written informed consent was obtained from two patients on 6/13/2013 and 8/19/2013, respectively, and the present study was approved by the Ethics Committee of Central South University (Changsha, China). Iliac bone marrow was collected from the two healthy male volunteers aged 23 and 25, respectively. A histopaque-1077 density gradient was used for the separation, as previously described (14,15). The cells were cultured in MSC medium composed of Dulbecco's modified Eagle's medium (DMEM) low glucose (11885076; Thermo Fisher Scientific, Inc., Waltham, MA, USA,) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) and 10 ng/ml basic fibroblast growth factor (Thermo Fisher Scientific, Inc.).
Vector construction
AAVS-SA-2A-puro was purchased from Addgene, Inc. (Cambridge, MA, USA). The restriction enzymes were purchased from Fermentas; Thermo Fisher Scientific, Inc. The internal ribosome entry 2 (IRES2)-Ac green fluorescent protein 1 (GFP1) fragment was obtained by polymerase chain reaction (PCR) from the plasmid p-cytomegalovirus (CMV)-IRES2-AcGFP (purchased from Clontech Laboratories, Inc., Mountainview, CA, USA) and was ligated to the plasmid AAVS-SA-2A-puro (XhoI/XbaI) to obtain AAVS-AcGFP1. The plasmid pCMV-hFIX was purchased from OriGene Technologies, Inc. (Rockville, MD, USA). The CMV-hFIX expression cassette obtained by PCR was inserted into the SalI site of AAVS-AcGFP1 to obtain the plasmid vector AAVS-AcGFP1-hFIX. The construction of the ZFN expression vector was performed as previously described (16,17). The amino acid sequences in the recognition sites for the four zinc fingers of ZFN-left (L) were YNWHLQR, RSDHLTT, HNYARDC and QNSTRIG. The key amino acid sequences in the four zinc fingers of ZFN-right (R) were QSSNLAR, RTDYLVD, YNTHLTR, and QGYNLAG.
Gene transfection
MSCs were separated and amplified to passage 3. Subsequently, AAVS-AcGFP1-hFIX, ZFN-L and ZFN-R, were co-transfected using nucleofection according to the manufacturer's protocol (cat. no. VVPE-1001; Lonza, Basel, Switzerland). The quantity of the AAVS-AcGFP1-hFIX donor vector and the ZFN expression vector were 5 µg and 1 µg (0.5 µg of ZFN-L and ZFN-R each), and MSCs were seeded at a density of 3×105 cells. Subsequently, the transfected cells were cultured in DMEM containing 10% FBS for 48 h in culture incubator (37°C, 5% CO2) and the GFP-positive cells were screened 48 h post-nucleofection transfection using flow cytometry (BD FACSARIA III; BD Biosciences, Franklin Lakes, NJ, USA) and the single GFP positive cells were sorted into single wells of the 96-well plate for monoclonal culture.
Identification of the site-specific integration clones
To identify the clones with site-specific integration, gDNA of the single clones was extracted by using the PureLink™ Genomic DNA Mini kit (cat. no. K182001; Thermo Fisher Scientific, Inc.) and subsequently tested using PCR with the following primers: Forward primer, 5′-AACTACCTAGACTGGATTCGTGAC-3′; reverse primer, 5′-TGAGAAGCTGATGCAAGTTATGAG-3′. The PCR was performed with Taq DNA polymerase (Takara Biotechnology Co., Ltd., Dalian, China) with the following thermocycling parameters: 98°C for 5 min followed by 40 cycles of the following procedures: 98°C for 30 sec, 58°C for 30 sec, 72°C for 2 min, 72°C for 10 min; 4°C incubation for 1 h.
Southern blot analysis
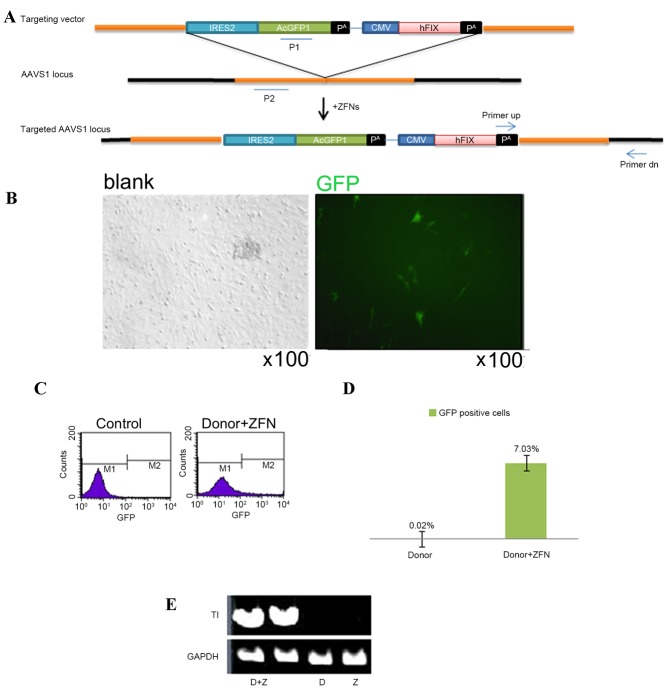
A total of 5 µg gDNA was digested with AhdI overnight and transferred onto a positively charged nylon membrane following 0.8% agarose electrophoresis. The probes were labeled with digoxigenin-deoxyuridine triphosphate by using the PCR DIG Probe Synthesis kit (cat. no. 11636090910; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany USA) and were hybridized to the gDNA fragments on the nylon membrane at 42°C overnight. The hybridization signals were detected using CDP-Star chemiluminescent substrate (cat. no. C0712-100ML; Sigma-Aldrich; Merck Millipore). The probes are presented in Fig. 1A. P1 was a 6,443–6,885 base pair fragments of the protein phosphatase 1 regulatory subunit 12C (PPP1R12C) gene and probe P2 was the amplification product with the AAVS-AcGFP1-hFIX template, using the primers 5′-ACCGGCACCGATTTCAAGGAGGAT-3′ and 5′-CAGGGGGAGGTGTGGGAGGTTTTT-3′.
Figure 1.
Targeting of MSCs by AAVS-AcGFP1-hFIX. (A) Targeting vector AAVS-AcGFP1-hFIX was composed of the 5′ and 3′ homologous arms, the promoterless GFP expression cassette IRES2-AcGFP1 and the hFIX expression cassette CMV-hFIX-pA under the control of the CMV promoter. The recognition site for ZFN was located at the AAVS1 locus of the first intron of the PPP1R12C gene. ‘primer up’ and ‘primer dn’ were the primers used to validate the site-specific integration clones, which were located at the pA and outside the 3′ end of the 3′ homologous arm, respectively. P1 and P2 were the probes used for the Southern blot to identify the site-specific integration clones. (B) ZFN expression in transfected MSCs. (C and D) Efficiency of GFP expression, as determined using by flow cytometry. (E) Polymerase chain reaction detection for the site-specific integration events. IRES1, internal ribosome entry 2; GFP, green fluorescent protein; CMV, cytomegalovirus; hFIX, human blood coagulation factor IX; AAVS1, adeno-associated virus integration site 1; ZFN, zinc finger nuclease; MSCs, mesenchymal stem cells; D, donor; Z, ZFN; TI, targeted integration.
Determination of pluripotency
MSCs were differentiated into osteoblasts, adipocytes, and chondrocytes to determine their pluripotency using the StemPro® Osteogenesis, Chondrogenesis and Adipogenesis Differentiation kits (Thermo Fisher Scientific, Inc., cat nos. A1007201, A1007101 and A1007001), respectively, according to the manufacturer's protocol. The differentiated osteoblasts, adipocytes, and chondrocytes were stained with Alizarin red, Oil Red O and Alcian blue, respectively. Images were captured using a Leica microscope (Leica DM6000 B) and analyzed using LAS AF software version 1.6.0 (Leica Microsystems GmbH, Wetzlar, Germany).
Flow cytometry analysis
Cultured MSCs at passage 5 were detached using 0.25% trypsin/EDTA and centrifuged at 400 × g for 5 min. The supernatant was discarded and the cell pellet was re-suspended in D-PBS containing 1% fetal bovine serum to the concentration of 105 cells/ml. Cells were then incubated with 10 µl of phycoerythrin-conjugated antibodies anti-CD34 (1:50, cat. no. MA1-19645; Thermo Fisher Scientific, Inc.), anti-CD44 (1:50, cat. no. MHCD4404; Thermo Fisher Scientific, Inc.), anti-CD73 (1:50, cat. no. FAB5795P; R&D Systems, Inc., Minneapolis, MN, USA), anti-CD90 (1:50, cat. no. A15794; Thermo Fisher Scientific, Inc.), anti-CD105 (1:50, cat. no. MA1-80944; Thermo Fisher Scientific, Inc.) at 25°C for 45 min. Then cells centrifuged at 300 × g for 15 min and washed with flow cytometry washing buffer (PBS with 0.5–1% bovine serum albumin; Sigma-Aldrich; Merck Millipore), the cells analyzed by flow cytometry analyzer (BD FACSARIA III, BD Biosciences).
ELISA
MSCs were cultured for 24 h, subsequently the supernatant was harvested, and the total number of cells was counted. ELISA was used to detect the hFIX protein quantity according to the manufacturer's protocol (cat. no. LS-F10414; LifeSpan BioSciences, Inc., Seattle, WA, USA). Three independent experiments were performed. The standard curve was prepared using hFIX samples of established concentrations, the concentration of the sample to be tested was evaluated accordingly.
Detection of in vivo anti-hFIX antibody
All animal experiments were approved by the Ethics Committee of Xiangya Hospital, Central South University (Changsha, China). Female Kunming mice aged 4-weeks-old were purchased from the Experimental Animal Center of Hunan Province (Changsa, China). Mice were housed three per cage, in a room maintained at 23±2°C and 60±10% humidity, in a 12/12 h light/dark cycle. Food and water were supplied ad libitum. A total of 106 MSCs with site-specific integration of the hFIX gene were injected through the tail vein of the 6 mice that weighed 50–60 g, Tail vein blood (~150 ml) was collected following anesthesia using ketamine/xylazine at a dosage of 100 mg/kg (Sigma-Aldrich; Merck Millipore) at 14 days following MSC injection. The blood plasma was obtained by centrifugation (3,000 × g, 10 min, 4°C) and then stored at −20°C. The anti-hFIX antibody in the mouse plasma was detected with the modified Bethesda method, as previously described (4). Sacrifice of mice was performed using cervical dislocation.
Statistical analysis
Results of mean ± standard deviation were analyzed using a one-way analysis of variance, the mean of each MSC site-specific clone was compared to that of the control MSC respectively using Dunnett's multiple comparison test. The hFIX expression level of each MSC site-specific integration clone was compared with the wild type MSC control group. In vivo anti-hFIX antibody level of each mouse injected with an MSC clone was compared with that of control mice without injection. P<0.05 was considered to indicate a statistically significant difference.
Results
Construction of the targeting system
The AAVS-AcGFP1- hFIX vector targeting AAVS1 was constructed, which contained the promoterless GFP expression cassette. Following integration at the AAVS1 locus, the expression was initiated using the PPP1R12C gene promoter. In addition, this vector also contained a hFIX expression cassette with the CMV promoter (Fig. 1A).
Gene targeting MSCs
The AAVS1-AcGFP1-hFIX donor vector and the ZFN expression vector were nucleofected into the MSCs. 48 h post-transfection, GFP expression was observed under a microscope (Fig. 1B). Flow cytometry demonstrated that the GFP-positive cells accounted for 7.03% of the total cells (Fig. 1C and D). In the MSC control group, which was transfected with the donor vector but not with the ZFN vector, the proportion of GFP-positive cells was only 0.02% (Fig. 1C and D). The PCR analysis revealed an expected target band at 1.6 kb in the co-transfection group, which was not evident in the blank control group and the donor transfection group (Fig. 1E). These findings indicated that the hFIX expression cassette was successfully integrated into the AAVS1 locus.
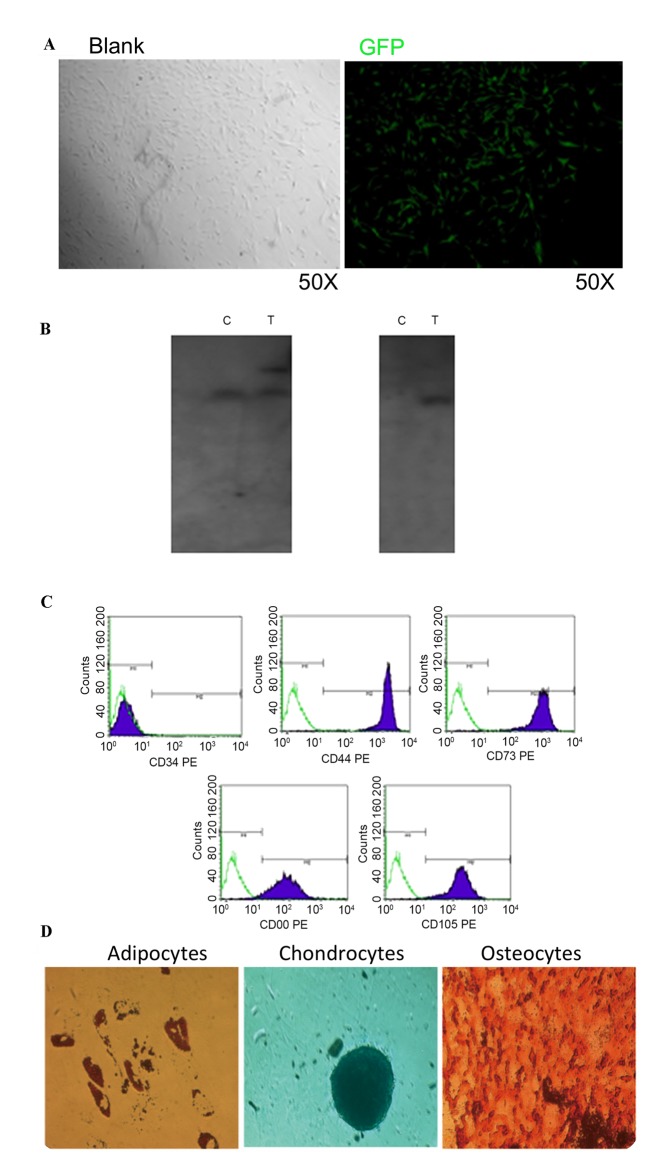
The GFP-positive MSCs were isolated by flow cytometry. The GFP-positive MSCs were isolated from 3.0×106 transfected cells for monoclonal culture (Fig. 2A). The clone with the target band of 18 kb was detected and defined as the site-specific integration clone using Southern blotting (Fig. 2B).
Figure 2.
Screening for a single clone with site-specific integration and the identification of its stem cell traits. (A) A single GFP-positive cell selected by flow cytometry was cultured to be a cell clone. (B) Southern blot identification of the site-specific integration event. (C) Expression analysis of the cell surface markers. The PE-conjugated antibodies (CD34, CD45, CD73, CD90 and CD105) were incubated with MSCs and then detected with flow cytometry. (D) Differentiation of the MSCs with site-specific integration into adipocytes, osteoblasts and chondrocytes was investigated by staining with Oil Red O, Alizarin red and Alcian blue, respectively. GFP, green fluorescent protein; MSCs, mesenchymal stem cells; C, wild-type MSC control group; T, MSCs with site-specific integration; P1/2, probe 1/2; PE, phycoerythrin; CD, cluster of differentiation.
Characterization of the MSC clone with site-specific integration
To examine stem cell traits in the MSCs with site-specific integration, the phenotype and pluripotency of the MSCs were investigated. It was determined that the GFP-positive MSCs with site-specific integration maintained the typical expression pattern of the MSC surface markers, including cluster of differentiation (CD) 34-, CD44+, CD73+, CD90+ and CD105+ (Fig. 2C). In addition, these cells retained pluripotency of differentiation into osteoblasts, adipocytes and chondrocytes (Fig. 2D).
Expression of hFIX in the clones with site-specific integration
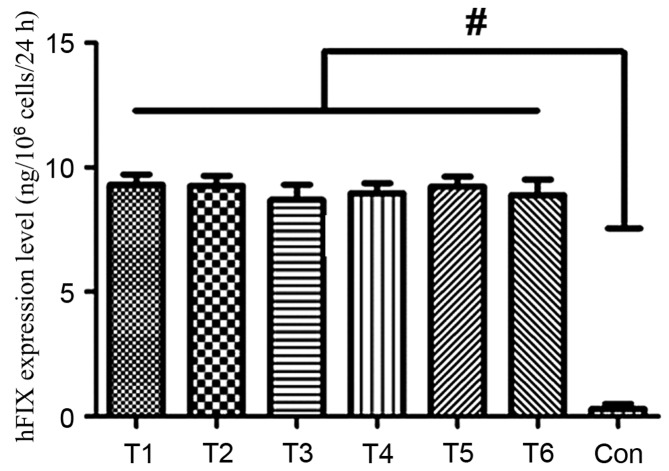
To determine whether hFIX may be effectively expressed from the AAVS1 locus of the MSCs, the cell culture supernatants from 6 modified cells with site-specific integration were harvested, and the hFIX expression levels were determined using an ELISA. The hFIX expression levels of the six representative cells were 9.3 ng/106 cells/24 h, 9.26 ng/106 cells/24 h, 8.7 ng/106 cells/24 h, 8.97 ng/106 cells/24 h, 9.23 ng/106 cells/24 h and 8.9 ng/106 cells/24 h. The expression levels of hFIX in the individual clone groups were significantly greater compared with the control group (P<0.05; Fig. 3).
Figure 3.
Detection of the in vitro expression of hFIX in the MSCs. A total of 6 MSC clones with site-specific integration were randomly selected to harvest the cell culture supernatant and the corresponding hFIX expression levels were detected by ELISA. The detection for each clone was repeated three times. #P<0.05 vs. Con. hFIX, human blood coagulation factor IX; MSCs, mesenchymal stem cells; T1-6, clone IDs of the individual MSCs clones with site-specific integration; Con, unmodified MSC control.
In vivo expression of anti-hFIX
A total of 4 MSC clones were transplanted into normal Kunming mice. After 14 days, serum expression of the anti-hFIX antibody was detected. The average anti-hFIX expression levels were 5.3, 7.05, 3.05 and 6.15 BU/ml, which were significantly different from the control mouse without MSC injection (Fig. 4), indicating the expression of hFIX in vivo.
Figure 4.
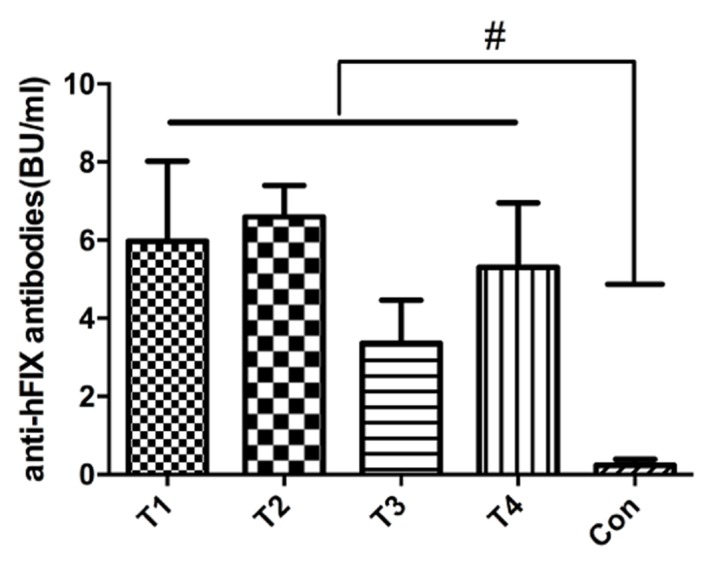
Detection of the in vivo expression of anti-hFIX in the MSCs. A total of 4 MSC clones with site-specific integration were randomly selected for transplantation into normal Kunming mice and the corresponding expression levels of the anti-hFIX antibody was detected. #P<0.05 vs. Con. hFIX, human blood coagulation factor IX; MSCs, mesenchymal stem cells; T1-T4, clone IDs of the individual MSCs clones with site-specific integration; Con, unmodified MSC control.
Discussion
MSCs may be potential cell vectors for gene therapy. The development of gene targeting technology has promoted the generation of methods for site-specific modification in MSCs. The present study used ZFN to introduce hFIX into a safe and effective docking site for exogenous genes in human MSCs, the AAVS1 locus, with effective in vitro and in vivo expression. The characteristics of MSCs make them useful for potential hemophilia gene therapy, as blood coagulation factor FVIII (18,19) and coagulation factor hFIX (20,21) may be highly expressed in MSCs. Following transplantation, MSCs may proliferate in various soft and perivascular tissues in the body (22), which facilitate the release of coagulation factors into the peripheral blood. Furthermore, MSCs may exhibit a targeting ability, which improves targeted gene therapy for damaged tissues.
Various methods for the genetic modification of MSCs have been reported (23–25). However, these methods primarily used random integration of therapeutic genes into the genome. Random integration with an exogenous expression cassette introduces the risk of insertion mutations. Furthermore, the exogenous gene may be silenced, leading to large differences in the expression of the genetically modified cells obtained (11,26). It has previously been reported that this gene silencing may be mediated by methylation-dependent and methylation-independent mechanisms (27). The present study used targeted insertion of the hFIX gene into the AAVS1 locus for expression. No gene silencing was identified in the isolated clones, which may be associated with the natural insulator in the AAVS1 locus that protects a gene from being silenced (28). A previous study demonstrated that an exogenous gene integrated into the AAVS1 locus in human and mouse embryonic stem cells may be effectively expressed (29). The present study confirmed that an exogenous gene integrated into the AAVS1 locus in MSCs may be efficiently expressed, which was important for the full application of MSCs in future gene therapy. It of note that our previous study did not identify a difference in the expression levels of tissue plasminogen activator among the different clones following integration into the AAVS1 locus (17). Similarly, the present study also demonstrated no significant difference in hFIX expression levels among individual clones of MSCs, which may contribute to the reproducible and stable efficacy of future gene therapy.
The development of an appropriate enrichment method for homologous recombination is also important for gene therapy. The use of promoter trapping has been identified as an effective enrichment method for homologous recombination. For targeting vectors with promoter trapping, the selectable marker gene lacks a promoter; following integration by homologous recombination into the genome, the promoter for the targeted gene may be used to increase the expression level of the selectable gene. In the present study, the PPP1R12C gene promoter was used to promote the expression of GFP gene without a promoter before integrating into the MSC genome, and the MSCs with site-specific modifications were screened by flow cytometry. A previous study regarding gene targeting primarily screened genes using drugs, such as neomycin and puromycin; however, this screening process may require 2–4 weeks (17), whereas the enrichment method used in the present study only required 24 h to obtain the modified MSCs. A previous study has highlighted that during the in vitro MSC culturing process, stem cell traits may be reduced with increased culture time (30). Therefore, gene therapy and cell treatment using MSCs should aim to minimize the in vitro culture time. As the anti-h hFIX was detected in vivo, it indicates that MSCs obtained by screening in the present study may successfully mediate the in vivo expression of hFIX, which was important for the rapid screening of gene-modified MSCs.
Acknowledgements
The present study was supported by a grant from the National Natural Science Foundation of China (grant no. 81401631).
References
- 1.Giannelli F, Choo KH, Rees DJ, Boyd Y, Rizza CR, Brownlee GG. Gene deletions in patients with haemophilia B and anti-factor IX antibodies. Nature. 1983;303:181–182. doi: 10.1038/303181a0. [DOI] [PubMed] [Google Scholar]
- 2.Beltrami EM, Williams IT, Shapiro CN, Chamberland ME. Risk and management of blood-borne infections in health care workers. Clin Microbiol Rev. 2000;13:385–407. doi: 10.1128/CMR.13.3.385-407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters RT, Low SC, Kamphaus GD, Dumont JA, Amari JV, Lu Q, Zarbis-Papastoitsis G, Reidy TJ, Merricks EP, Nichols TC, Bitonti AJ. Prolonged activity of factor IX as a monomeric Fc fusion protein. Blood. 2010;115:2057–2064. doi: 10.1182/blood-2009-08-239665. [DOI] [PubMed] [Google Scholar]
- 4.Kung SH, Hagstrom JN, Cass D, Tai SJ, Lin HF, Stafford DW, High KA. Human factor IX corrects the bleeding diathesis of mice with hemophilia B. Blood. 1998;91:784–790. [PubMed] [Google Scholar]
- 5.VandenDriessche T, Collen D, Chuah MK. Viral vector-mediated gene therapy for hemophilia. Curr Gene Ther. 2001;1:301–315. doi: 10.2174/1566523013348508. [DOI] [PubMed] [Google Scholar]
- 6.Bittner RE, Schöfer C, Weipoltshammer K, Ivanova S, Streubel B, Hauser E, Freilinger M, Höger H, Elbe-Bürger A, Wachtler F. Recruitment of bone-marrow-derived cells by skeletal and cardiac muscle in adult dystrophic mdx mice. Anat Embryol (Berl) 1999;199:391–396. doi: 10.1007/s004290050237. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira E, Potier E, Logeart-Avramoglou D, Salomskaite-Davalgiene S, Mir LM, Petite H. Optimization of a gene electrotransfer method for mesenchymal stem cell transfection. Gene ther. 2008;15:537–544. doi: 10.1038/gt.2008.9. [DOI] [PubMed] [Google Scholar]
- 8.Baksh D, Song L, Tuan RS. Adult mesenchymal stem cells: Characterization, differentiation, and application in cell and gene therapy. J Cell Mol Med. 2004;8:301–316. doi: 10.1111/j.1582-4934.2004.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benabdallah BF, Allard E, Yao S, Friedman G, Gregory PD, Eliopoulos N, Fradette J, Spees JL, Haddad E, Holmes MC, Beauséjour CM. Targeted gene addition to human mesenchymal stromal cells as a cell-based plasma-soluble protein delivery platform. Cytotherapy. 2010;12:394–399. doi: 10.3109/14653240903583803. [DOI] [PubMed] [Google Scholar]
- 10.Kucerova L, Altanerova V, Matuskova M, Tyciakova S, Altaner C. Adipose tissue-derived human mesenchymal stem cells mediated prodrug cancer gene therapy. Cancer Res. 2007;67:6304–6313. doi: 10.1158/0008-5472.CAN-06-4024. [DOI] [PubMed] [Google Scholar]
- 11.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 12.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. Science. Vol. 302. New York, N.Y.: 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1; pp. pp415–419. [DOI] [PubMed] [Google Scholar]
- 13.Lombardo A, Cesana D, Genovese P, Di Stefano B, Provasi E, Colombo DF, Neri M, Magnani Z, Cantore A, Lo Riso P, et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- 14.Bartsch E, Park AL, Kingdom JC, Ray JG. Risk threshold for starting low dose aspirin in pregnancy to prevent preeclampsia: An opportunity at a low cost. PloS one. 2015;10:e0116296. doi: 10.1371/journal.pone.0116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedenstein AJ, Chailakhjan RK, Lalykina KS. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Tissue Kinet. 1970;3:393–403. doi: 10.1111/j.1365-2184.1970.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 16.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li SJ, Shi RZ, Bai YP, Hong D, Yang W, Wang X, Mo L, Zhang GG. Targeted introduction of tissue plasminogen activator (TPA) at the AAVS1 locus in mesenchymal stem cells (MSCs) and its stable and effective expression. Biochem Biophys Res Commun. 2013;437:74–78. doi: 10.1016/j.bbrc.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 18.Doering CB. Retroviral modification of mesenchymal stem cells for gene therapy of hemophilia. Methods Mol Biol. 2008;433:203–212. doi: 10.1007/978-1-59745-237-3_12. [DOI] [PubMed] [Google Scholar]
- 19.Van Damme A, Thorrez L, Ma L, Vandenburgh H, Eyckmans J, Dell'Accio F, De Bari C, Luyten F, Lillicrap D, Collen D, et al. Efficient lentiviral transduction and improved engraftment of human bone marrow mesenchymal cells. Stem Cells. 2006;24:896–907. doi: 10.1634/stemcells.2003-0106. [DOI] [PubMed] [Google Scholar]
- 20.Coutu DL, Cuerquis J, El Ayoubi R, Forner KA, Roy R, François M, Griffith M, Lillicrap D, Yousefi AM, Blostein MD, Galipeau J. Hierarchical scaffold design for mesenchymal stem cell-based gene therapy of hemophilia B. Biomaterials. 2011;32:295–305. doi: 10.1016/j.biomaterials.2010.08.094. [DOI] [PubMed] [Google Scholar]
- 21.Sayyar B, Dodd M, Wen J, Ma S, Marquez-Curtis L, Janowska-Wieczorek A, Hortelano G. Encapsulation of factor IX-engineered mesenchymal stem cells in fibrinogen-alginate microcapsules enhances their viability and transgene secretion. J Tissue Eng. 2012;3:2041731412462018. doi: 10.1177/2041731412462018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chamberlain J, Yamagami T, Colletti E, Theise ND, Desai J, Frias A, Pixley J, Zanjani ED, Porada CD, Almeida-Porada G. Efficient generation of human hepatocytes by the intrahepatic delivery of clonal human mesenchymal stem cells in fetal sheep. Hepatology. 2007;46:1935–1945. doi: 10.1002/hep.21899. [DOI] [PubMed] [Google Scholar]
- 23.Shi D, Chen G, Lv L, Li L, Wei D, Gu P, Gao J, Miao Y, Hu W. The effect of lentivirus-mediated TH and GDNF genetic engineering mesenchymal stem cells on Parkinson's disease rat model. Neurol Sci. 2011;32:41–51. doi: 10.1007/s10072-010-0385-3. [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Xu X, Li Z, Lendlein A, Ma N. Genetic engineering of mesenchymal stem cells by non-viral gene delivery. Clin Hemorheol Microcirc. 2014;58:19–48. doi: 10.3233/CH-141883. [DOI] [PubMed] [Google Scholar]
- 25.Lee K, Majumdar MK, Buyaner D, Hendricks JK, Pittenger MF, Mosca JD. Human mesenchymal stem cells maintain transgene expression during expansion and differentiation. Mol Ther. 2001;3:857–866. doi: 10.1006/mthe.2001.0327. [DOI] [PubMed] [Google Scholar]
- 26.Yao S, Sukonnik T, Kean T, Bharadwaj RR, Pasceri P, Ellis J. Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol Ther. 2004;10:27–36. doi: 10.1016/j.ymthe.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Liew CG, Draper JS, Walsh J, Moore H, Andrews PW. Transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25:1521–1528. doi: 10.1634/stemcells.2006-0634. [DOI] [PubMed] [Google Scholar]
- 28.Ogata T, Kozuka T, Kanda T. Identification of an insulator in AAVS1, a preferred region for integration of adeno-associated virus DNA. J Virol. 2003;77:9000–9007. doi: 10.1128/JVI.77.16.9000-9007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith JR, Maguire S, Davis LA, Alexander M, Yang F, Chandran S, ffrench-Constant C, Pedersen RA. Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells. 2008;26:496–504. doi: 10.1634/stemcells.2007-0039. [DOI] [PubMed] [Google Scholar]
- 30.Sethe S, Scutt A, Stolzing A. Aging of mesenchymal stem cells. Ageing Res Rev. 2006;5:91–116. doi: 10.1016/j.arr.2005.10.001. [DOI] [PubMed] [Google Scholar]