Abstract
Triple-negative breast cancer (TNBC) is a heterogeneous disease characterized by an aggressive phenotype and reduced survival. The aim of the present study was to investigate the molecular mechanisms involved in the carcinogenesis of TNBC and to identify novel target molecules for therapy. The differentially expressed genes (DEGs) in TNBC and normal adjacent tissue were assessed by analyzing the GSE41970 microarray data using Qlucore Omics Explorer, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes. Pathway enrichment analyses for DEGs were performed using the Database for Annotation, Visualization and Integrated Discovery online resource. A protein-protein interaction (PPI) network was constructed using Search Tool for the Retrieval of Interacting Genes, and subnetworks were analyzed by ClusterONE. The PPI network and subnetworks were visualized using Cytoscape software. A total of 121 DEGs were obtained, of which 101 were upregulated and 20 were downregulated. The upregulated DEGs were significantly enriched in 14 pathways and 83 GO biological processes, while the downregulated DEGs were significantly enriched in 18 GO biological processes. The PPI network with 118 nodes and 1,264 edges was constructed and three subnetworks were extracted from the entire network. The significant hub DEGs with high degrees were identified, including TP53, glyceraldehyde-3-phosphate dehydrogenase, cyclin D1, HRAS and proliferating cell nuclear antigen, which were predominantly enriched in the cell cycle pathway and pathways in cancer. A number of critical genes and pathways were revealed to be associated with TNBC. The present study may provide an improved understanding of the pathogenesis of TNBC and contribute to the development of therapeutic targets for TNBC.
Keywords: disease, enrichment analysis, protein-protein interaction network, hub genes, therapeutic targets
Introduction
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer with a poor prognosis and high mortality, which is diagnosed more frequently in young and premenopausal women (1,2). Due to the absence of estrogen receptors, progesterone receptor and the human epidermal growth factor receptor-2 in TNBC tumor types, patients with TNBC do not respond to hormone- or trastuzumab-based therapies, leaving cytotoxic chemotherapy as the current therapy (3). Therefore, the identification of critical genes involved in TNBC carcinogenesis may provide a strategy for molecular target therapy of TNBC (4).
Microarray technology, which may be used to detect the global gene expression, has provided an alternative method for the molecular classification of different types of cancer, as well as the exploration of potential prognostic biomarkers and therapeutic targets. Based on microarray analysis, breast cancer has been divided into distinctive subtypes according to different gene expression patterns (5–11). Furthermore, Rakha et al (4) performed a microarray analysis on a relatively large set of 1,944 cases of invasive breast cancer that contained TNBC tissues, as well as information on tumor size, lymph node involvement and androgen receptor expression levels, and determined these three parameters as the most useful prognosticators of TNBC. Therefore, a systematic analysis of gene expression patterns in TNBC may aid researchers to comprehend the development and the treatment of this disease and to identify novel therapeutic targets.
In the present study, biological microarray analysis was used to analyze the gene expression profile of TNBC and to screen the differentially expressed genes (DEGs) between TNBC and adjacent normal tissues. Furthermore, altered biological pathways in TNBC were identified using bioinformatics tools. Additionally, a protein-protein interaction (PPI) network of DEGs was constructed in order to identify the crucial genes involved in the process of TNBC. The present study aimed to improve the understanding of the underlying pathological mechanism and facilitate the discovery of potential novel therapeutic targets for TNBC.
Materials and methods
Gene expression data
The GSE41970 gene expression profiles of TNBC were downloaded from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) of the National Center for Biotechnology Information (Bethesda, MD, USA) based on the GPL16299 platform data (NanoString nCounter mRNA Human Cancer Reference Kit; NanoString, Inc., Ticino, Bellinzona, Switzerland) (12). In total, 200 specimens, including 160 primary TNBC specimens and 40 normal samples, were used in the present study.
Data processing and differential analysis
Qlucore Omics Explorer (QOE) software (version 3; Qlucore AB, Lund, Sweden) was used to analyze the data from the microarray (13,14). Intensity values of each probe-set were log2 transformed, and a gene-specific t-test was subsequently performed between TNBC samples and matched normal samples. P<0.01 and [log2(fold change)>2] were regarded as the cut-off criteria to screen out DEGs. To generate an overview of the gene expression profile, hierarchical clustering was also performed by QOE.
Gene ontology (GO) and pathway enrichment analysis
To determine the biological pathways altered in TNBC, GO biological process terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed on the DEGs using Database for Annotation, Visualization and Integration Discovery (version 6.7; DAVID; http://david.abcc.ncifcrf.gov) (15). P<0.01 and false discovery rate (FDR) <0.01 were set as the cut-off value.
Construction and analysis of PPI network
It is possible to utilize PPI research to study DEG protein functions at the molecular level, and cellular regulatory mechanisms can be interpreted by elucidation of genome-wide protein interactions (16). The PPI network was constructed for the DEGs using information provided by the Search Tool for the Retrieval of Interacting Genes, version 10 (STRING; http://string-db.org) (17), and was subsequently visualized using Cytoscape version 3.2.1 (http://cytoscape.org) (18). The interactions of protein pairs with a combined score >0.5 were retained in the network and the hub genes were screened according to the degree score (the number of neighbors). The subnetworks were then analyzed by Clustering with Overlapping Neighborhood Expansion (ClusterONE; http://www.paccanarolab.org/clusterone). GO functional and KEGG pathway enrichment analyses of the most significant subnetworks were performed with a threshold of P<0.01 and FDR<0.01.
Results
Identification of DEGs in TNBC
Using QOE, 121 DEGs were identified between 160 TNBC and 40 normal tissues, including 101 upregulated genes and 20 downregulated genes. The 10 most significantly up- or downregulated genes are listed in Table I.
Table I.
Top 10 most significantly up- or downregulated differentially expressed genes in triple-negative breast cancer compared with normal tissue.
| Gene symbol | P-value | Fold-change |
|---|---|---|
| Upregulated genes | ||
| BIRC5 | 6.50×10−36 | 48.63 |
| MYBL2 | 4.32×10−34 | 57.33 |
| TOP2A | 3.49×10−31 | 41.91 |
| CDC2 | 4.07×10−29 | 32.75 |
| MMP9 | 2.09×10−23 | 21.35 |
| CHEK1 | 8.68×10−23 | 13.37 |
| SPP1 | 2.21×10−22 | 40.41 |
| TYMS | 4.26×10−22 | 18.21 |
| E2F1 | 4.38×10−21 | 11.18 |
| PCNA | 1.27×10−20 | 6.87 |
| Downregulated genes | ||
| IGFBP6 | 2.19×10−14 | 0.12 |
| ESR1 | 1.09×10−13 | 0.09 |
| DLC1 | 1.27×10−12 | 0.14 |
| EGR1 | 6.87×10−11 | 0.20 |
| IGF1 | 1.72×10−10 | 0.27 |
| TGFBR3 | 3.37×10−10 | 0.16 |
| PPARG | 3.84×10−10 | 0.23 |
| NGFR | 3.93×10−10 | 0.12 |
| CD34 | 6.67×10−9 | 0.27 |
| FOS | 5.83×10−8 | 0.30 |
Clustering of DEGs
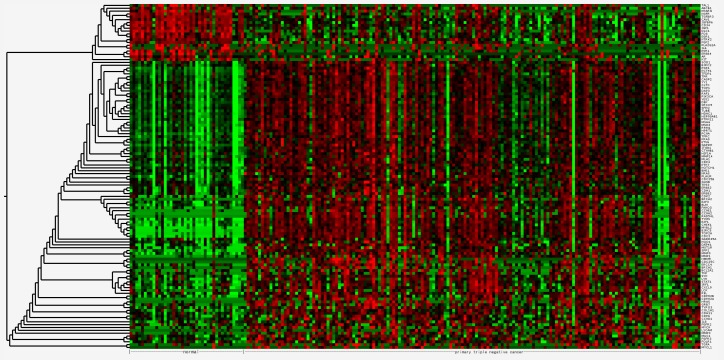
The 121 DEGs in TNBC compared with normal tissues were selected for hierarchical clustering analysis. As presented in Fig. 1, the DEGs were divided into two major groups, separating 101 upregulated genes from 20 downregulated genes.
Figure 1.
Cluster analysis of differentially expressed genes. Red highlights genes with high expression, green shows cells with low expression and black indicates no significant change in the expression level between the mean and sample.
GO of biological process enrichment analysis of up- and downregulated DEGs
The upregulated DEGs were significantly enriched in 83 biological processes and the top five significantly enriched processes were mainly associated with the regulation of cell proliferation, cell cycle and cell apoptosis (Table II). The downregulated DEGs were significantly enriched in 18 biological processes, among which the most significant were mainly involved in the regulation of cell proliferation, macromolecule metabolic process and cell migration (Table II).
Table II.
Top five significantly enriched biology processes for differentially expressed genes.
| GO ID | Term | Gene counts | P-value | FDR |
|---|---|---|---|---|
| Upregulated | ||||
| GO:0042127 | Regulation of cell proliferation | 35 | 2.22×10−18 | 3.76×10−15 |
| GO:0051726 | Regulation of cell cycle | 24 | 8.20×10−17 | 1.89×10−13 |
| GO:0008284 | Positive regulation of cell proliferation | 26 | 8.35×10−17 | 1.89×10−13 |
| GO:0042981 | Regulation of apoptosis | 31 | 2.15×10−14 | 3.63×10−11 |
| GO:0043067 | Regulation of programmed cell death | 31 | 2.79×10−14 | 4.72×10−11 |
| Downregulated | ||||
| GO:0042127 | Regulation of cell proliferation | 12 | 1.19×10−9 | 1.88×10−6 |
| GO:0010604 | Positive regulation of macromolecule metabolic process | 11 | 5.40×10−8 | 8.57×10−5 |
| GO:0030334 | Regulation of cell migration | 7 | 8.25×10−8 | 1.31×10−4 |
| GO:0040012 | Regulation of locomotion | 7 | 1.76×10−7 | 2.79×10−4 |
| GO:0051270 | Regulation of cell motion | 7 | 1.81×10−7 | 2.88×10−4 |
GO, gene ontology; Gene counts, number of genes; FDR, false discovery rate.
KEGG pathway enrichment analysis of up- and downregulated DEGs
The upregulated DEGs were significantly enriched in 14 pathways, which mainly involved the cell cycle and pathways associated with cancer, including the p53 signaling pathway (Table III). In the downregulated DEGs, four pathways were identified, however they were without statistical significance (P>0.01 or FDR>0.01).
Table III.
Significantly enriched pathways for differentially expressed genes.
| KEGG ID | Term | Gene counts | P-value | FDR |
|---|---|---|---|---|
| hsa05200 | Pathways in cancer | 37 | 3.77×10−24 | 4.19×10−21 |
| hsa05219 | Bladder cancer | 17 | 8.90×10−20 | 9.87×10−17 |
| hsa05223 | Non-small cell lung cancer | 14 | 3.04×10−13 | 3.38×10−10 |
| hsa04110 | Cell cycle | 18 | 1.03×10−12 | 1.14×10−9 |
| hsa05215 | Prostate cancer | 16 | 1.05×10−12 | 1.16×10−9 |
| hsa05212 | Pancreatic cancer | 14 | 1.59×10−11 | 1.76×10−8 |
| hsa05220 | Chronic myeloid leukemia | 14 | 2.74×10−11 | 3.04×10−8 |
| hsa05214 | Glioma | 13 | 5.32×10−11 | 5.91×10−8 |
| hsa05218 | Melanoma | 13 | 2.33×10−10 | 2.59×10−7 |
| hsa05213 | Endometrial cancer | 11 | 2.22×10−9 | 2.46×10−6 |
| hsa05222 | Small cell lung cancer | 12 | 2.30×10−8 | 2.56×10−5 |
| hsa05216 | Thyroid cancer | 8 | 1.25×10−7 | 1.39×10−4 |
| hsa05210 | Colorectal cancer | 10 | 2.74×10−6 | 3.04×10−3 |
| hsa04115 | p53 signaling pathway | 9 | 5.03×10−6 | 5.58×10−3 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; Gene counts, number of genes; FDR, false discovery rate.
PPI network analysis
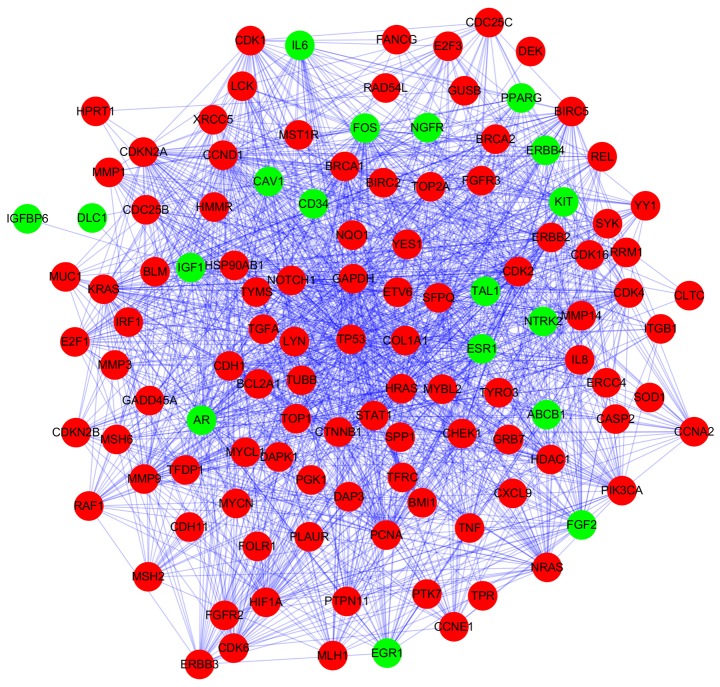
Based on STRING database analysis, a total of 1,264 protein pairs with the combined score >0.5 were identified. The PPI network including 118 nodes and 1,264 edges was constructed (Fig. 2) and the connectivity degree of each node was calculated. The top five nodes TP53 (degrees, 86), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; degrees, 62), cyclin D1 (CCND1; degrees, 58), HRAS (degrees, 58) and proliferating cell nuclear antigen (PCNA; degrees, 52) with degrees >50 were screened as hub proteins in the PPI network.
Figure 2.
Protein-protein interaction network of differentially expressed genes. The red nodes represent upregulated genes and the green nodes represent downregulated genes. The lines represent the interaction between proteins.
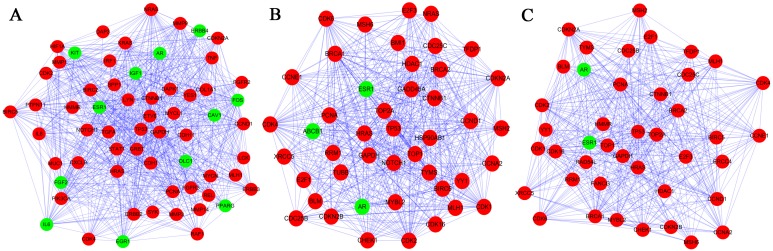
Three subnetworks (1,2 and 3) with P<0.05 were established using the ClusterONE plugin (Fig. 3). The hub proteins TP53, GAPDH, CCND1, HRAS and PCNA were demonstrated to be involved in all of these three subnetworks. Subnetwork 1 was mainly associated with regulation of cell proliferation and cell communication, while the significant pathways were mainly associated with pathways in cancer (Tables IV and V). By contrast, the other subnetworks were mainly associated with the cell cycle (Table IV). Additionally, the significant pathways correlated with subnetwork 2 and 3 were the cell cycle, the p53 signaling pathway and pathways in cancer (Table V).
Figure 3.
Three subnetworks in the protein-protein interaction network of differentially expressed genes. (A) Subnetwork 1; (B) Subnetwork 2; and (C) Subnetwork 3 were determined. The red nodes represent upregulated genes and the green nodes represent downregulated genes. The lines represent the interaction relationship between proteins.
Table IV.
Top five significantly enriched biology processes for differentially expressed genes in three subnetworks.
| GO ID | Term | Gene counts | P-value | FDR |
|---|---|---|---|---|
| Subnetwork 1 | ||||
| GO:0042127 | Regulation of cell proliferation | 30 | 3.15×10−20 | 5.31×10−17 |
| GO:0010647 | Positive regulation of cell communication | 21 | 7.11×10−18 | 1.20×10−14 |
| GO:0009967 | Positive regulation of signal transduction | 20 | 1.89×10−17 | 3.19×10−14 |
| GO:0008284 | Positive regulation of cell proliferation | 22 | 3.59×10−17 | 6.06×10−14 |
| GO:0009719 | Response to endogenous stimulus | 21 | 4.01×10−16 | 7.44×10−13 |
| Subnetwork 2 | ||||
| GO:0022402 | Cell cycle process | 24 | 1.05×10−20 | 1.65×10−17 |
| GO:0007049 | Cell cycle | 26 | 3.03×10−20 | 4.79×10−17 |
| GO:0051726 | Regulation of cell cycle | 19 | 3.49×10−18 | 5.51×10−15 |
| GO:0022403 | Cell cycle phase | 20 | 7.57×10−18 | 1.20×10−14 |
| GO:0051329 | Interphase of mitotic cell cycle | 13 | 3.33×10−16 | 5.22×10−13 |
| Subnetwork 3 | ||||
| GO:0022402 | Cell cycle process | 23 | 1.66×10−20 | 2.59×10−17 |
| GO:0007049 | Cell cycle | 25 | 2.88×10−20 | 4.51×10−17 |
| GO:0051726 | Regulation of cell cycle | 19 | 4.22×10−19 | 6.61×10−16 |
| GO:0022403 | Cell cycle phase | 19 | 2.27×10−17 | 3.55×10−14 |
| GO:0006259 | DNA metabolic process | 20 | 3.35×10−17 | 5.25×10−14 |
GO, gene ontology; Gene counts, number of genes; FDR, false discovery rate.
Table V.
Top five significantly enriched pathways for differentially expressed genes in three subnetworks.
| KEGG ID | Term | Gene counts | P-value | FDR |
|---|---|---|---|---|
| Subnetwork 1 | ||||
| hsa05200 | Pathways in cancer | 32 | 3.04×10−25 | 3.30×10−22 |
| hsa05219 | Bladder cancer | 15 | 4.66×10−19 | 5.05×10−16 |
| hsa05215 | Prostate cancer | 14 | 1.23×10−12 | 1.34×10−9 |
| hsa05213 | Endometrial cancer | 11 | 3.80×10−11 | 4.12×10−8 |
| hsa05218 | Melanoma | 12 | 4.29×10−11 | 4.65×10−8 |
| Subnetwork 2 | ||||
| hsa04110 | Cell cycle | 19 | 3.60×10−20 | 3.72×10−17 |
| hsa05200 | Pathways in cancer | 21 | 5.11×10−15 | 5.27×10−12 |
| hsa05215 | Prostate cancer | 11 | 3.82×10−10 | 3.94×10−7 |
| hsa04115 | p53 signaling pathway | 10 | 7.34×10−10 | 7.57×10−7 |
| hsa05220 | Chronic myeloid leukemia | 10 | 1.81×10−9 | 1.86×10−6 |
| Subnetwork 3 | ||||
| hsa04110 | Cell cycle | 18 | 2.32×10−19 | 2.37×10−16 |
| hsa05200 | Pathways in cancer | 19 | 2.03×10−13 | 2.07×10−10 |
| hsa04115 | p53 signaling pathway | 9 | 9.25×10−9 | 9.43×10−6 |
| hsa05220 | Chronic myeloid leukemia | 9 | 2.04×10−8 | 2.08×10−5 |
| hsa05223 | Non-small cell lung cancer | 8 | 4.40×10−8 | 4.49×10−5 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; Gene counts, number of genes; FDR, false discovery rate.
Discussion
TNBC is one of the most deadly breast cancer subtypes due to the lack of an effective treatment. The improvement of the diagnosis and therapeutic methods of this disease relies on the discovery of novel potential molecular markers. In the present study, a total of 121 DEGs between TNBC and normal tissues were identified, consisting of 101 upregulated and 20 downregulated genes. With 121 gene signatures mapped from the STRING database, a giant component PPI network was established with 118 nodes and 1,264 edges. After applying the ClusterONE clustering algorithm, three significant subnetworks with highly connected nodes were obtained. The top five ranked genes as hub nodes with maximum degrees were identified, including TP53, GAPDH, CCND1, HRAS and PCNA. In addition, it was revealed that these hub nodes existed in all of the three subnetworks, which contributed to the biological processes of cell growth and the cell cycle and connected the cell cycle, p53 signaling pathway and pathways in cancer.
TP53, also termed p53, encodes a tumor suppressor protein that controls the cell cycle arrest, apoptosis, senescence, DNA repair and changes in metabolism (19). Mutations in this gene are associated with a variety of human cancer types, including TNBC, lung cancer and high-grade serous ovarian tumor types (20). p53 is mutated and overexpressed in ~25–30% of human breast cancer (21), and had an increased incidence in TNBC (22,23). Nishimura et al (24) also found that high proliferation rate and frequent p53 overexpression occurred in TNBC. Furthermore, p53 is also expressed as smaller isoforms, some of which inhibit wild-type p53. Avery-Kiejda et al (25) reported that Δ40p53, one of the p53 isoforms, was significantly upregulated in tumor tissue when compared with the normal breast and was closely associated with TNBC. Mutation and isoforms of p53 may provide an alternate explanation for the malfunction of p53 pathway and deregulated p53 signaling in TNBC. The present study demonstrated that the p53 gene was elevated in the TNBC samples and that p53 was a hub protein with a degree score of 86 in the established PPI network. Therefore, the p53 gene may be a key regulator in TNBC development.
GAPDH, originally identified as a glycolytic enzyme is considered a housekeeping gene. It is frequently used as an internal standard for gene expression in RNA or protein experiments. However, the abnormal expression of GAPDH has been confirmed to have a close association with various types of cancer (26), as it serves an important role in carcinogenesis and cell death, as well as energy metabolism (27). Notably, increased levels of GAPDH were observed in most types of human cancer and were often correlated with reduced survival (28,29). Results of the present study indicated that GAPDH is predicated as a pivotal gene associated with TNBC and the cell cycle, and may be a potential biomarker for detection and prevention of TNBC.
CCND1, a member of the cyclin-dependent kinase regulator family, is required for the activation of CDK4 and CDK6, and is recognized as a critical modulator of the cell cycle and a positive regulator of cell proliferation (30). Aberrant amplification and overexpression of CCND1 are a driving force in 13–20% of human breast cancer, and are associated with poor disease outcome (31). However, Mylona et al (32) suggested that CCND1 overexpression may serve as a marker for prolonged survival of patients with TNBC. Although the exact role of CCND1 in TNBC remains controversial, according to the findings of the present study, CCND1 overexpression was detected in TNBC tissues, and it acted as a hub gene in the PPI network of TNBC. These data implied that CCND1 is of clinical importance for TNBC.
HRAS, a member of the ras superfamily of genes, encodes for a 21-kDa protein (p21), which takes part in the signal transduction pathways that control proliferation and apoptosis, and regulate the cell cycle. Ras genes are involved in a wide variety of human tumor types and there is a known positive correlation between HRAS activation and breast cancer (33). In addition, previous studies (34,35) have shown that HRAS can induce invasion and migration of the breast cancer cell line, MCF10A. From the present study, HRAS may serve a significant role in the pathogenesis of TNBC and, given its functional significance in various types of cancer, it may also be a potential therapeutic target in the treatment of TNBC.
PCNA is well known as a coordinator of essential cellular processes for cell growth, death, and maintenance. Accumulated evidence suggests that an enhanced level of PCNA often correlates with carcinogenesis and that PCNA levels can be used as a prognostic marker in certain cases (36–39). Yu et al (40) demonstrated that when PCNA activity was inhibited by a peptide, it suppressed the growth of TNBC cells. Given the critical function of PCNA in cancer growth, it is possible that the targeting of PCNA may be a viable therapeutic method for TNBC.
Pathways in cancer and pathways involving the cell cycle were demonstrated to be highly enriched in the DEGs for the PPI network and the subnetworks. All these pathways have been implicated in the development of breast cancer. Notably, the most significantly enriched pathways were pathways in cancer, which refers to some classical cancer pathways, including p53 signaling, Wnt signaling, and Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling pathways. Certain previous studies (41–43) suggested that these critical signaling nodes, including p53, β-catenin and JAK/STAT3, can be used as therapeutic targets for the treatment of TNBC. In addition, cancer has been viewed as a cell cycle disease (44). Substantial evidence has indicated that the hub genes, P53, CCND1, HRAS and PCNA, are related to the pathways of the cell cycle, which is in accordance with the functions of networks identified in the present study.
In conclusion, 121 genes were revealed to be differentially expressed between TNBC and normal tissues by integrated analysis. Among them were TP53, GAPDH, CCND1, HRAS and PCNA, and these may be involved in TNBC progression via the cell cycle pathways and pathways in cancer. These findings may improve the understanding of the pathogenesis of TNBC and the development of targeted treatments for TNBC. Further experiments are required to confirm the findings of this work and the hypotheses put forward.
Acknowledgements
The current research was supported by the Institute of Genetic Engineering at the Southern Medical University (Guangzhou, China) and the present study was funded by the National Natural Science Foundation of China (grant no. 39880032), the Guangdong Foundation for Leading Talented Scientists (grant no. C1030925) and the China Postdoctoral Science Foundation Funded Project (grant no. 2016M602487). The authors would like to acknowledge the excellent technical assistance of Dr Kaiyuan Ji (School of Basic Medicine, Southern Medical University, China) and Dr Zijun Qiao (Institutes of Biomedical Sciences, Fudan University, China) during the present study.
References
- 1.Dawson SJ, Provenzano E, Caldas C. Triple negative breast cancers: Clinical and prognostic implications. Eur J Cancer. 2009;45:S27–S40. doi: 10.1016/S0959-8049(09)70013-9. (Suppl 1) [DOI] [PubMed] [Google Scholar]
- 2.Reis-Filho JS, Tutt AN. Triple negative tumours: A critical review. Histopathology. 2008;52:108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- 3.Anders CK, Carey LA. Biology, metastatic patterns and treatment of patients with triple-negative breast cancer. Clin Breast Cancer. 2009;9:S73–S81. doi: 10.3816/CBC.2009.s.008. (Suppl 2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. doi: 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- 5.Kapp AV, Jeffrey SS, Langerød A, Børresen-Dale AL, Han W, Noh DY, Bukholm IR, Nicolau M, Brown PO, Tibshirani R. Discovery and validation of breast cancer subtypes. BMC Genomics. 2006;7:231. doi: 10.1186/1471-2164-7-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Rehim Abd DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer. 2005;116:340–350. doi: 10.1002/ijc.21004. [DOI] [PubMed] [Google Scholar]
- 9.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 12.Cascione L, Gasparini P, Lovat F, Carasi S, Pulvirenti A, Ferro A, Alder H, He G, Vecchione A, Croce CM, et al. Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PLoS One. 2013;8:e55910. doi: 10.1371/journal.pone.0055910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma H, Morey R, O'Neil RC, He Y, Daughtry B, Schultz MD, Hariharan M, Nery JR, Castanon R, Sabatini K, et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177–183. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Killian JK, Kim SY, Miettinen M, Smith C, Merino M, Tsokos M, Quezado M, Smith WI, Jr, Jahromi MS, Xekouki P, et al. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discov. 2013;3:648–657. doi: 10.1158/2159-8290.CD-13-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman BT, da Huang W, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Knowledgebase: A gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freed-Pastor WA, Prives C. Mutant p53: One name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tennis M, Krishnan S, Bonner M, Ambrosone CB, Vena JE, Moysich K, Swede H, McCann S, Hall P, Shields PG, Freudenheim JL. p53 Mutation analysis in breast tumors by a DNA microarray method. Cancer Epidemiol Biomarkers Prev. 2006;15:80–85. doi: 10.1158/1055-9965.EPI-05-0444. [DOI] [PubMed] [Google Scholar]
- 22.Hanby AM. Aspects of molecular phenotype and its correlations with breast cancer behaviour and taxonomy. Br J Cancer. 2005;92:613–617. doi: 10.1038/sj.bjc.6602421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XR, Liu M, Zhang YJ, Wang JD, Zheng YQ, Li J, Ma B, Song X. CK5/6, EGFR, Ki-67, cyclin D1, and nm23-H1 protein expressions as predictors of pathological complete response to neoadjuvant chemotherapy in triple-negative breast cancer patients. Med Oncol. 2011;28:S129–S134. doi: 10.1007/s12032-010-9742-6. (Suppl 1) [DOI] [PubMed] [Google Scholar]
- 24.Nishimura R, Arima N. Is triple negative a prognostic factor in breast cancer? Breast Cancer. 2008;15:303–308. doi: 10.1007/s12282-008-0042-3. [DOI] [PubMed] [Google Scholar]
- 25.Avery-Kiejda KA, Morten B, Wong-Brown MW, Mathe A, Scott RJ. The relative mRNA expression of p53 isoforms in breast cancer is associated with clinical features and outcome. Carcinogenesis. 2014;35:586–596. doi: 10.1093/carcin/bgt411. [DOI] [PubMed] [Google Scholar]
- 26.Caradec J, Sirab N, Revaud D, Keumeugni C, Loric S. Is GAPDH a relevant housekeeping gene for normalisation in colorectal cancer experiments? Br J Cancer. 2010;103:1475–1476. doi: 10.1038/sj.bjc.6605851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colell A, Green DR, Ricci JE. Novel roles for GAPDH in cell death and carcinogenesis. Cell Death Differ. 2009;16:1573–1581. doi: 10.1038/cdd.2009.137. [DOI] [PubMed] [Google Scholar]
- 28.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Guo C, Liu S, Sun MZ. Novel insight into the role of GAPDH playing in tumor. Clin Transl Oncol. 2013;15:167–172. doi: 10.1007/s12094-012-0924-x. [DOI] [PubMed] [Google Scholar]
- 30.Hadzisejdić I, Mustać E, Jonjić N, Petković M, Grahovac B. Nuclear EGFR in ductal invasive breast cancer: Correlation with cyclin-D1 and prognosis. Mod Pathol. 2010;23:392–403. doi: 10.1038/modpathol.2009.166. [DOI] [PubMed] [Google Scholar]
- 31.Courjal F, Cuny M, Simony-Lafontaine J, Louason G, Speiser P, Zeillinger R, Rodriguez C, Theillet C. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: Definition of phenotypic groups. Cancer Res. 1997;57:4360–4367. [PubMed] [Google Scholar]
- 32.Mylona E, Tzelepis K, Theohari I, Giannopoulou I, Papadimitriou C, Nakopoulou L. Cyclin D1 in invasive breast carcinoma: Favourable prognostic significance in unselected patients and within subgroups with an aggressive phenotype. Histopathology. 2013;62:472–480. doi: 10.1111/his.12013. [DOI] [PubMed] [Google Scholar]
- 33.Watson DM, Elton RA, Jack WJ, Dixon JM, Chetty U, Miller WR. The H-ras oncogene product p21 and prognosis in human breast cancer. Breast Cancer Res Treat. 1991;17:161–169. doi: 10.1007/BF01806365. [DOI] [PubMed] [Google Scholar]
- 34.Moon A, Kim MS, Kim TG, Kim SH, Kim HE, Chen YQ, Kim HR. H-ras, but not N-ras, induces an invasive phenotype in human breast epithelial cells: A role for MMP-2 in the H-ras-induced invasive phenotype. Int J Cancer. 2000;85:176–181. doi: 10.1002/(SICI)1097-0215(20000115)85:2%3C176::AID-IJC5%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 35.Yong HY, Hwang JS, Son H, Park HI, Oh ES, Kim HH, Kim DK, Choi WS, Lee BJ, Kim HR, Moon A. Identification of H-Ras-specific motif for the activation of invasive signaling program in human breast epithelial cells. Neoplasia. 2011;13:98–107. doi: 10.1593/neo.101088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malkas LH, Herbert BS, Abdel-Aziz W, Dobrolecki LE, Liu Y, Agarwal B, Hoelz D, Badve S, Schnaper L, Arnold RJ, et al. A cancer-associated PCNA expressed in breast cancer has implications as a potential biomarker. Proc Natl Acad Sci USA. 2006;103:19472–19477. doi: 10.1073/pnas.0604614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bukholm IR, Bukholm G, Holm R, Nesland JM. Association between histology grade, expression of HsMCM2, and cyclin A in human invasive breast carcinomas. J Clin Pathol. 2003;56:368–373. doi: 10.1136/jcp.56.5.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JS, Kim HS, Jung JJ, Kim YB, Park CS, Lee MC. Correlation between angiogenesis, apoptosis and cell proliferation in invasive ductal carcinoma of the breast and their relation to tumor behavior. Anal Quant Cytol Histol. 2001;23:161–168. [PubMed] [Google Scholar]
- 39.Aziz SA, Pervez S, Khan SM, Kayani N, Nasir MI. Prognostic value of proliferating cell nuclear antigen (PCNA) in infiltrating ductal carcinoma breast. J Coll Physicians Surg Pak. 2005;15:225–229. [PubMed] [Google Scholar]
- 40.Yu YL, Chou RH, Liang JH, Chang WJ, Su KJ, Tseng YJ, Huang WC, Wang SC, Hung MC. Targeting the EGFR/PCNA signaling suppresses tumor growth of triple-negative breast cancer cells with cell-penetrating PCNA peptides. PLoS One. 2013;8:e61362. doi: 10.1371/journal.pone.0061362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner N, Moretti E, Siclari O, Migliaccio I, Santarpia L, D'Incalci M, Piccolo S, Veronesi A, Zambelli A, Del Sal G, Di Leo A. Targeting triple negative breast cancer: Is p53 the answer? Cancer Treat Rev. 2013;39:541–550. doi: 10.1016/j.ctrv.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Dey N, Barwick BG, Moreno CS, Ordanic-Kodani M, Chen Z, Oprea-Ilies G, Tang W, Catzavelos C, Kerstann KF, Sledge GW, Jr, et al. Wnt signaling in triple negative breast cancer is associated with metastasis. BMC Cancer. 2013;13:537. doi: 10.1186/1471-2407-13-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poage GM, Hartman ZC, Brown PH. Revealing targeted therapeutic opportunities in triple-negative breast cancers: A new strategy. Cell Cycle. 2013;12:2705–2706. doi: 10.4161/cc.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]