Abstract
Osteosarcoma, which is the most common type of primary bone tumor in adolescents, is characterized by complex genetic alterations and frequent resistance to conventional treatments. MicroRNAs (miRs) have emerged as fundamental regulators in gene expression through their ability to silence gene expression at post-transcriptional and translational levels. The present study investigated the role of miR-410 in the progression of osteosarcoma. The results demonstrated that the expression of miR-410 was markedly downregulated in human osteosarcoma tissues, and U2OS and MG-63 osteosarcoma cell lines. Clinicopathological significance suggested that miR-410 may be a potential biomarker for chemotherapy-resistant osteosarcoma. Furthermore, overexpression of miR-410 exhibited a limited effect on cell viability in U2OS and MG-63 cells. Target prediction algorithms (TargetScan and miRanda) indicated that autophagy related 16-like 1 (ATG16L1) was a potential target gene of miR-410. A luciferase reporter assay demonstrated that miR-410 directly decreased ATG16L1 expression by targeting its 3′-untranslated region. In addition, the results revealed that miR-410 was able to markedly inhibit autophagy. Accordingly, autophagy was activated as a protective mechanism when osteosarcoma cells were exposed to three common anticancer drugs, including rapamycin, doxorubicin and cisplatin. Furthermore, the autophagy inhibitor 3-methyladenine and miR-410 expression were able to improve the therapeutic response of the cells to chemotherapy drugs (rapamycin, doxorubicin and cisplatin), thus indicating that miR-410 enhanced chemosensitivity through autophagy inhibition in osteosarcoma cells. In conclusion, studies regarding the function of miR-410 on autophagy provided insight into the biological function of miR-410 in osteosarcoma and may offer a promising approach for the treatment of osteosarcoma.
Keywords: microRNA-410, ATG16L1, autophagy, chemoresistance, osteosarcoma
Introduction
Osteosarcoma is one of the most common malignant bone tumors that affects adolescents and children worldwide. Frequent acquisition of drug resistance is often associated with the chemotherapeutic treatment of osteosarcoma (1). Therefore, there is an urgent requirement to elucidate the underlying molecular mechanisms of chemoresistance in osteosarcoma, in order to facilitate the development of therapeutic strategies for patients.
Previous studies have demonstrated that autophagy is usually activated as a protective mechanism in tumor cells against numerous chemotherapeutics. Limited efficacy of doxorubicin (Dox), cisplatin (DDP) and methotrexate due to the induction of autophagy has been certified in osteosarcoma cells (2–4). Autophagy is a highly conserved process that entails the degradation of intracellular components to regenerate metabolites for energy and growth through the lysosomal machinery (5). However, the molecular mechanisms underlying autophagy-mediated chemotherapy resistance of osteosarcoma cells remain largely unknown.
MicroRNAs (miRNAs; miRs) are single-stranded, small noncoding RNAs ~22 nucleotides in length, which negatively regulate gene expression through base-pairing to the 3′-untranslated region (3′UTR) of target mRNA (6,7). miRNAs are involved in crucial biological processes, and the dysregulation of miRNAs is associated with tumorigenesis and the development of numerous malignancies, including osteosarcoma (8). Previous studies have investigated the role of miR-410 in several types of cancer (9–11); however, the role of miR-410 in osteosarcoma remains unknown.
The present study demonstrated that miR-410 expression was downregulated in osteosarcoma cell lines and tissues. Target prediction algorithms (TargetScan and miRanda) indicated that autophagy related 16-like 1 (ATG16L1) was a potential target gene of miR-410. ATG16L1 is a component of a large protein complex essential for autophagy, and autophagy is a tightly regulated intracellular catabolic pathway involving the lysosomal degradation of cytoplasmic organelles and proteins. Previous studies have demonstrated miRNAs mediate the regulation of proteins in the autophagy pathway, and autophagy is commonly activated as a protective mechanism when cancer cells were exposed to three commonly used anticancer drugs (12–14). In the current study, it was demonstrated that miR-410 overexpression is able to improve the therapeutic response of cells to chemotherapy [including rapamycin (Rap), Dox and DDP]. Furthermore, miR-410 could enhance the chemosensitivity of those drugs via autophagy inhibition through targeting ATG16L1 in osteosarcoma cells. The present study provided an insight into the biological function of miR-410 in osteosarcoma and offered a promising approach for future osteosarcoma treatment.
Materials and methods
Osteosarcoma tissues, cell lines and transfection
U2OS and MG-63 human osteosarcoma cell lines, the hFOB 1.19 human osteoblast cell line, and human embryonic kidney (HEK)293T cells were purchased from the Cell Resource Center of Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China). The cells were cultured in Dulbecco's modified Eagle's medium (GlutaMAX™; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (12483–020; Thermo Fisher Scientific, Inc.), 100 mg/ml streptomycin and 100 IU/ml penicillin (Thermo Fisher Scientific, Inc.) at 37°C in an atmosphere containing 5% CO2. Transfection was performed when the cells had grown to 80% confluence, using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. A total of 40 paired tumor tissues and matched non-tumor tissues were obtained from patients with osteosarcoma at Renmin Hospital of Wuhan University (Wuhan, China) between 2013 and 2015. All tissues were immediately stored in liquid nitrogen until further use. Histological diagnosis of the tumors was made and agreed upon by two senior pathologists at the Department of Pathology (Renmin Hospital of Wuhan University) based on World Health Organization criteria. The present study was approved by the Institutional Review Board and Human Ethics Committee of Renmin Hospital of Wuhan University.
Reagents and antibodies
Reagents used were as follows: 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; M5655; Sigma-Aldrich, St. Louis, MO, USA) and Hoechst 3342 (B2261; Sigma-Aldrich), both of which were dissolved in phosphate-buffered saline (PBS). The following antibodies were obtained: Anti-microtubule-associated protein 1A/1B-light chain 3 (LC3)I/II (1:1,000; ab58610; Abcam, Cambridge, MA, USA), anti-caspase 3 (1:1,000; ab2171; Abcam), anti-ATG161L (1:1,000; 8089S; Cell Signaling Technology, Inc., Danvers, MA, USA) and anti-β-actin (1:2,000; ab6276; Abcam). 3-methyladenine (3-MA; M9281; Sigma-Aldrich); Rap (37094; Sigma-Aldrich); Dox (D1515; Sigma-Aldrich). DDP (Chengdu Organic chemicals Co., Ltd., Wuhou, China). All drugs were dissolved in dimethyl sulfoxide and were stored at −20°C until the cells were treated with the drugs for 48 h (Rap, 20 nM; Dox, 0.1 µg/ml; DDP, 10 µM).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Tissues were homogenized prior to RNA extraction. Total RNA was extracted from the cells and tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and was reverse transcribed using Revert Aid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. qPCR was performed using the Maxima SYBR Green/Fluorescein qPCR Master Mix (Thermo Fisher Scientific, Inc.) on the Real-Time PCR Detection system (iQ5; Bio-Rad Laboratories, Inc., Hercules, CA, USA). The cDNA templates were diluted to 1:10 and 20 µl per reaction (SYBR Green Mix 10 µl, forward primer 2 µl, reverse primer 2 µl, RNase Free dH2O 4.5 µl) was analyzed. The PCR conditions were as follows: DNA denaturation at 94°C for 4 min, followed by 40 cycles of amplification at 94°C for 40 sec, 52°C for 40 sec and 72°C for 40 sec for data collection. The Bulge-Loop™ hsa-miR-410 qRT-PCR primer set (miRQ0002171-1-2) was purchased from Guangzhou RiboBio Co., Ltd (Guangzhou, China). The qPCR experiments were run three times on separate days, each with independent biological samples (n=4 per group); within each experiment run, relative expression values were normalized to the standard deviations from the mean. The relative expression levels of miR-410 were normalized to those of the internal control gene U6 using the 2-ΔΔCq cycle quantification method (15).
Cell transfection
The sequences of miR-410 were retrieved from the miRNA database miRBase (www.mirbase.org) (AAUAUAACACAGAUGGCCUGU). miR-410 (miR-410) and negative control (NC) mimics were purchased from Guangzhou RiboBio Co., Ltd. and were transfected into the U2OS and MG-63 cells using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) or 24 h or 48 h in different experiments. qPCR assays were used to detect the expression levels of miR-410 subsequent to each transfection.
Cell proliferation and apoptosis assays
Cells were seeded in 96-well culture plates at 30% confluence 1 day prior to transfection. Post-transfection with miR-410 or NC the cells were evaluated using an MTT assay. The MTT assay was used to determine relative cell growth, according to the manufacturer's protocol. Subsequently, medium was replaced with 0.1 ml dimethyl sulfoxide (Sigma-Aldrich) and the 96-well culture plates were agitated at room temperature for 1 min. The absorbance was subsequently measured at 490 nm. In addition, a Hoechst assay was used to detect cell apoptosis. Cells were seeded in 6-well culture plates at 40% confluence 1 day prior to transfection. After 24 h, the cells were washed with PBS and were fixed in 4% paraformaldehyde for 15 min. Subsequently, Hoechst 33342 diluted in PBS (final concentration, 5%) was added to each well for 15 min, the cells were washed twice with PBS (10 min/wash), the blue-stained nuclei were observed and the apoptotic rate was calculated under fluorescence microscopy (Olympus Corporation, Hamburg, Germany).
Luciferase reporter assay
The 3′UTR of the human ATG16L1 gene was predicted to interact with miR-410. A 360 bp-fragment of the ATG16L1 3′-UTR, which contained a putative binding site of miR-410, was subcloned into the 3′-UTR region of the pMIR-REPORT firefly luciferase construct (Ambion; Thermo Fisher Scientific, Inc.). The empty vector and the PCR products were digested with SacI and XbaI, and subjected to T4 DNA ligation (Promega Corporation, Madison, WI, USA) at 16°C overnight. The ligation product was then transformed. Single colonies were picked and the construct was confirmed by sequencing, yielding pMIR-REPORT ATG16L1. Mutations within the potential miR-410 binding sites were generated by nucleotide replacement of the wild-type sequence to inhibit miR-410 binding (Guangzhou RiboBio Co., Ltd.). HEK293T cells were seeded into 96-well plates (40% cell density). After a 24 h incubation, the cells were co-transfected with 0.4 mg of the reporter construct or 0.2 mg of the control vector, and miR-410 or NC. Luciferase values were subsequently determined using the Dual-Luciferase Reporter Assay system (Promega Corporation). All transfection experiments were performed in triplicate and repeated at least three times.
Detection of green fluorescent protein (GFP)-LC3 autophagic dots
pcDNA3.1-GFP-LC3 vectors were provided as a gift by Professor Shi Wu (Tsinghua University, Beijing, China). U2OS and MG-63 cells, which stably expressed high levels of GFP-LC3 protein, were established by pcDNA3.1-GFP-LC3 transfection and were selected using G418 (10131027; Thermo Fisher Scientific, Inc.). The U2OS-GFP-LC3 and MG-63-GFP-LC3 stably expressing cells were detected and confirmed by fluorescence microscopy, and used for the following experiments. Post-transfection with miR-410 or NC, the percentage of GFP-LC3 punctate-positive cells was quantified and analyzed using automated image acquisition (DMIRE2 Fluorescence Microscope; Leica Microsystems, Wetzlar, Germany) with a threshold of ≥5 dots/cell. Data are presented as the mean ± standard deviation and are representative of three independent experiments.
Western blotting
Protein concentrations were quantified using the DC Protein Assay kit (500–0121; Bio-Rad Laboratories, Inc.) following cell protein extraction in radioimmunoprecipitation assay buffer (EMD Millipore, Billerica, MD, USA). The proteins (20 µg) were then subjected to 10% sodium dodecyl sulfate-polyacrylimide gel electrophoresis and were transferred onto nitrocellulose membranes. The membranes were blocked with 5% milk for 1 h at room temperature. The membranes were then incubated with the aforementioned primary rabbit antibodies at 4°C for 18 h. The membranes were then incubated with horseradish peroxidase-conjugated rabbit IgG secondary antibodies (1:3,000; cat. no. 7074; Cell Signaling Technology, Inc.) at room temperature for 60 min. Immunoreactivities were visualized using enhanced chemiluminescence reagents (WBKLS0500; EMD Millipore). The relative expression levels of the proteins of interest were obtained by normalizing their band densities to that of β-actin.
Immunofluorescence
Cells were plated on coverslips, and were fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.1% Triton X-100 for 15 min, and blocked with 5% bovine serum albumin (Sigma-Aldrich) for 1 h at room temperature post-treatment. After being rinsed three times with Tris-buffered saline containing 1% Tween-20, the cells were incubated with anti-ATG16L1 overnight at 4°C, followed by an incubation with conjugated secondary antibody for 2 h at 25°C, with 0.1 M 4′6-diamidino-2-phenylindole in PBS staining as the control. Images were acquired using the Leica DMIRE2 fluorescence microscope.
Statistical analysis
All statistical analyses were performed using SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA). All experiments were repeated three times and data are presented as the mean ± standard deviation. Student's t-test was used to determine significant differences between results. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-410 is downregulated in osteosarcoma tissues and cell lines
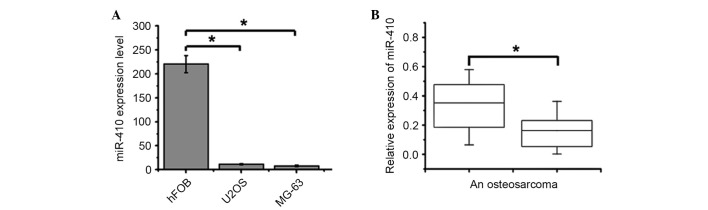
qPCR was used to detect the expression levels of miR-410 in U2OS and MG-63 human osteosarcoma cell lines. The expression levels of miR-410 were significantly downregulated in U2OS and MG-63 cells compared with in the hFOB 1.19 human osteoblast cell line (Fig. 1A). In addition, the expression levels of miR-410 were significantly downregulated in 34 out of 40 osteosarcoma tissues compared with in the matched normal non-tumor tissues (Fig. 1B). Further investigations indicated that miR-410 expression was not associated with age, gender or clinical stage; however, it had a close association with response to chemotherapy. Osteosarcoma tissues with high miR-410 expression exhibited a good response to chemotherapy (Table I; Chi-square test). Collectively, these results suggest that miR-410 may exert an antitumor function and be associated with chemoresistance in osteosarcoma.
Figure 1.
miR-410 expression is downregulated in osteosarcoma. (A) Expression levels of miR-410 in hFOB 1.19 cells and U2OS and MG-63 osteosarcoma cell lines were measured by RT-qPCR after a 48 h culture. Data are presented as the mean ± standard deviation of triplicate experiments (*P<0.05). (B) A total of 40 pairs of osteosarcoma tissues and matched adjacent normal non-tumor tissues were collected from patients and were assessed by RT-qPCR analysis (*P<0.05). Values were standardized to U6 expression. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; AN, adjacent normal non-tumor tissue; miR-410, microRNA-410.
Table I.
Association between clinicopathological features and miR-410 expression levels.
| miR-410 expression | ||||
|---|---|---|---|---|
| Factor | Cases (n) | High (6) | Low (34) | P-value |
| Age (years) | NS | |||
| ≥25 | 15 | 2 | 13 | |
| <25 | 25 | 4 | 21 | |
| Gender | NS | |||
| Male | 24 | 3 | 21 | |
| Female | 16 | 3 | 13 | |
| Tumor size (cm) | NS | |||
| ≥8 | 19 | 4 | 15 | |
| <8 | 21 | 2 | 19 | |
| Serum level of lactate dehydrogenase | NS | |||
| Elevated | 26 | 4 | 22 | |
| Normal | 14 | 2 | 12 | |
| Serum level of alkaline dehydrogenase | NS | |||
| Elevated | 28 | 5 | 23 | |
| Normal | 12 | 1 | 11 | |
| Clinical stage | NS | |||
| IIA | 23 | 3 | 20 | |
| IIB/III | 17 | 3 | 14 | |
| Response to chemotherapy | <0.05 | |||
| Good | 14 | 6 | 9 | |
| Poor | 26 | 0 | 26 | |
miR-410, microRNA-410; NS, not significant.
miR-410 exerts a limited influence on the proliferation of osteosarcoma cells
To determine whether miR-410 was able to affect the proliferation of osteosarcoma cells, proliferation assays were conducted in U2OS and MG-63 cells transfected with miR-410 or NC mimics. As shown in Fig. 2 overexpression of miR-410 had almost no affect on the viability of U2OS and MG-63 cells. Therefore, further studies were conducted to examine whether other mechanisms associated with antitumor function were affected by miR-410.
Figure 2.
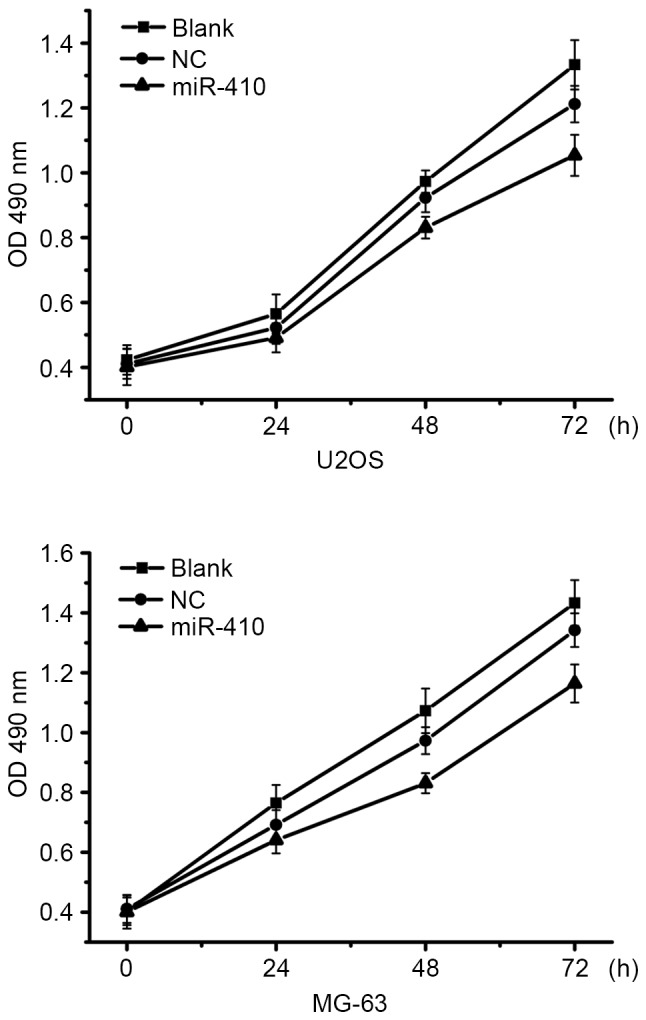
miR-410 has limited influence on osteosarcoma cell proliferation. MTT assays were performed to detect the cell viability of U2OS and MG-63 cells post-transfection with miR-410 or NC mimics for 48 h. Data are presented as the mean ± standard deviation. Blank control, untransfected group; miR-410, microRNA-410; NC, negative control; OD, optical density.
miR-410 directly targets ATG161L in osteosarcoma cell lines
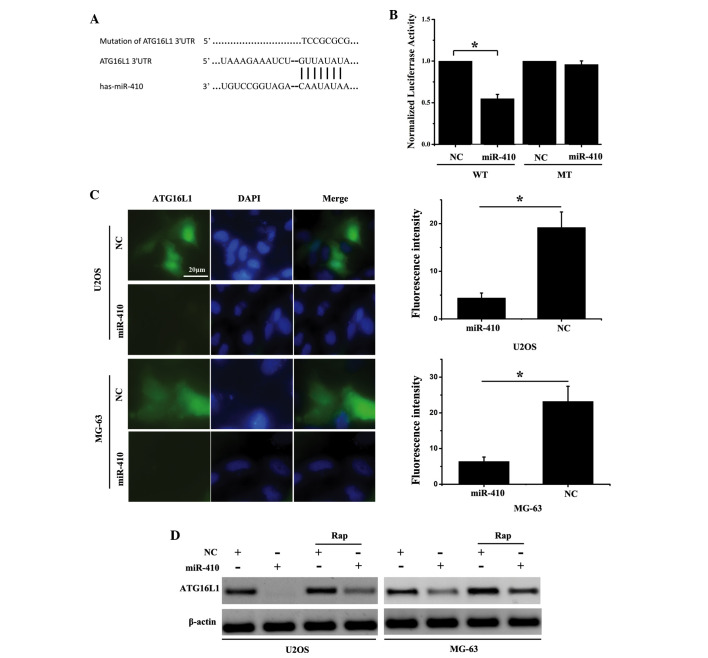
Target prediction algorithms [TargetScan (http://www.targetscan.org/vert_71/) and miRanda (http://www.microrna.org/microrna/home.do)] were used to predict the targets of miR-410 in osteosarcoma cells. ATG16L1 was predicted to be a target gene of miR-410, and putative miR-410 binding sites were predicted to be present in the 3′UTR of ATG16L1 mRNA. ATG16L1 is a component of a large protein complex, which is essential for autophagosome formation. Autophagy is generally considered to be a pro-survival mechanism that preserves cell viability in response to cancer therapy (16). To validate the prediction that ATG16L1 is a direct target of miR-410, luciferase reporter vectors containing wild-type (WT) or mutant ATG16L1 3′UTR were generated (Fig. 3A). The luciferase assay indicated that miR-410 inhibited luciferase activity in the ATG16L1 WT 3′UTR group; however, the inhibitory effects were abolished when the ATG16L1 3′UTR binding sites were mutated (Fig. 3B). Furthermore, an immunofluorescence assay demonstrated that overexpression of miR-410 suppressed ATG16L1 expression in U2OS and MG-63 cells (Fig. 3C). Concordantly, western blot analysis revealed that miR-410 was able to suppress ATG16L1 protein expression levels. Notably, ATG16L1 expression levels were stimulated by Rap (an autophagy activator), whereas miR-410 overexpression effectively decreased ATG16L1 expression (Fig. 3D). These results indicate that miR-410 may directly target ATG16L1 in osteosarcoma cells.
Figure 3.
miR-410 directly targets ATG161L in osteosarcoma cell lines. (A) miR-410/ATG16L1 alignment by miRanda analysis, and schematic diagram of the pMIR-ATG16L1/pMIR- ATG16L1mut paired sequences for miR-410. (B) Normalized luciferase activity of pMIR-ATG16L1/pMIR-ATG16L1mu reporter in human embryonic kidney 293T cells transfected with miR-410 or NC mimics. *P<0.05. (C) Immunofluorescent detection of ATG16L1 expression in U2OS and MG-63 cells transfected with miR-410 or NC for 48 h. (D) Western blotting was conducted to detect the effects of miR-410 on ATG16L1 expression post-transfection with miR-410 or NC for 48 h. Rapamycin (20 nM), which is an activator of autophagy, increased ATG16L1 expression. Data are presented as the mean ± standard deviation; *P<0.05. ATG161L, autophagy related 16-like 1; miR-410, microRNA-410; NC, negative control; WT, wild-type; mut/MT, mutant; Rap, rapamycin; UTR, untranslated region; DAPI, 4′,6-diamidino-2-phenylindole.
miR-410 inhibits autophagy in osteosarcoma cell lines
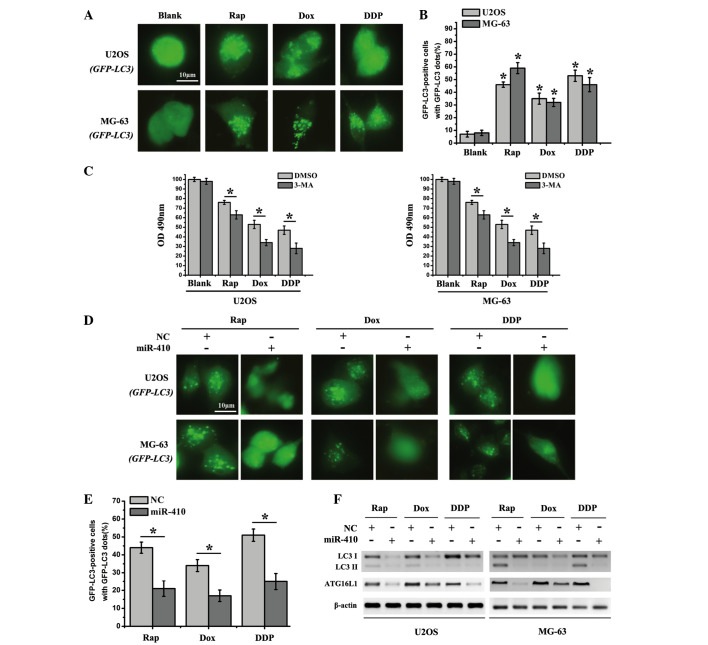
In relation to tumorigenesis, the role of autophagy is complex and depends on the environmental cues that cells are exposed to. Accordingly, intrinsic resistance to Dox and DDP through autophagic activity has been certified in osteosarcoma cells (17). Therefore, since miR-410 directly targeted ATG16L1 in osteosarcoma cancer cells, it was hypothesized that miR-410 may be able to reverse chemoresistance via autophagy inhibition. In order to verify this hypothesis, three common chemotherapy drugs, Rap, Dox and DDP were used to treat the cells in subsequent experiments. Initially, cell strains that stably expressed GFP-LC3 were generated (U2OS-GFP-LC3 and MG-63-GFP-LC3) by transfection of U2OS and MG-63 cells with GFP-LC3 (data not shown). U2OS-GFP-LC3 and MG-63-GFP-LC3 stably expressed high levels of the GFP-LC3 protein. The cytosolic form of LC3 (LC3-I) is conjugated to phosphatidylethanolamine to form the LC3-phosphatidylethanolamine conjugate (LC3-II), which is recruited to autophagosomal membranes and reflects autophagic activity. The GFP-LC3 expressing cells were subsequently exposed to the drugs, and autophagosomes (fluorescent dots) were accumulated due to the GFP-LC3 translocation to the structural components of the double-membrane autophagosome. As shown in Fig. 4A and B, the percentage of GFP-LC3 puncta-positive cells was significantly increased in the drug treatment groups. Furthermore, 3-MA significantly enhanced the sensitivity of cells to chemotherapy drugs in the drug treatment groups (Fig. 4C). In order to determine the effects of miR-410 on autophagy inhibition, U2OS and MG-63 cells, which were transfected with miR-410 or NC, were treated with Rap, Dox and DDP. The number of fluorescent dots and GFP-LC3 puncta-positive cells was significantly reduced in the cells transfected with miR-410 compared with in the NC group (Fig. 4D and E). These results suggest that miR-410 may function as a potential autophagy inhibitor in U2OS and MG-63 cells. In addition, western blotting was employed to detect the expression levels of ATG16L1 and the autophagy marker LC3I/LC3II in the drug treatment groups. The expression levels of ATG16L1 and LC3I/LC3II were markedly decreased in cells transfected with miR-410 (Fig. 4F). Taken together, miR-410 may act as a potential autophagy inhibitor in osteosarcoma cell lines.
Figure 4.
miR-410 effectively inhibits autophagy in osteosarcoma cell lines. (A) U2OS-GFP-LC3/MG-63-GFP-LC3 cells which stably express GFP-LC3 were used to detect autophagy after the cells were treated with various agents for 48 h (Rap, 20 nM; Dox, 0.1 µg/ml; DDP, 10 µM). (B) Percentage of GFP-LC3 punctate-positive cells was quantified and analyzed by automated image acquisition using a threshold of ≥5 dots/cell. (C) Cell viability was detected after the cells were treated with various agents for 48 h with the presence or absence of the autophagy inhibitor 3-MA. (D) U2OS-GFP-LC3/MG-63-GFP-LC3 cells which stably express GFP-LC3 were used to detect autophagy after the cells were treated with Rap (20 nM), Dox (0.1 µg/ml) or DDP (10 µM), and were transfected with miR-410 or NC mimics for 48 h. (E) Percentage of GFP-LC3 punctate-positive cells was quantified and analyzed by automated image acquisition using a threshold of ≥5 dots/cell (U2OS-GFP-LC3). (F) Western blot analysis of LC3I/II and ATG16L1 expression from the lysates of U2OS and MG-63 cells transfected with NC or miR-410 for 48 h. β-actin was used as an internal control. Data are presented as the mean ± standard deviation and are representative of three independent experiments (*P<0.05). ATG161L, autophagy related 16-like 1; miR-410, microRNA-410; NC, negative control; Rap, rapamycin; Dox, doxorubicin; DDP, cisplatin; GFP, green fluorescent protein; LC3I/II, microtubule-associated protein 1A/1B-light chain 3; OD, optical density; 3-MA, 3-methyladenine; DMSO, dimethyl sulfoxide.
miR-410 enhances chemosensitivity of cells to Rap, Dox and DDP via autophagy inhibition
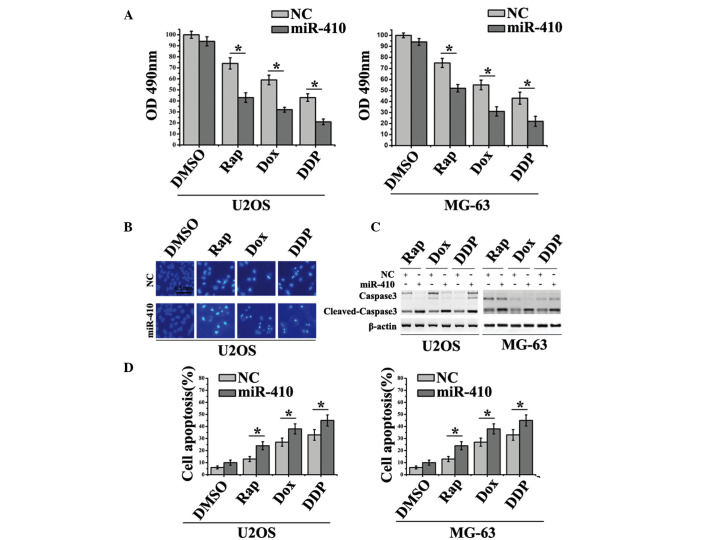
To further determine the influence of miR-410-induced autophagy inhibition on chemoresistance in osteosarcoma cell lines, an MTT assay was used to detect cell viability in U2OS and MG-63 cells. As shown in Fig. 5A, miR-410 sensitized U2OS and MG-63 cells to Rap, Dox and DDP, thus suggesting that the presence of miR-410 could improve the therapeutic response to those agents. Subsequently, cell apoptosis was detected by Hoechst assay. Cells treated with miR-410 and chemotherapy agents exhibited enhanced cell apoptosis compared with the cells treated with chemotherapy agents alone (Fig. 5B). Notably, Rap-, Dox- and DDP-treated groups all activated the cleavage of caspase 3 (early molecular marker of apoptosis), whereas miR-410 enhanced the apoptotic-inducing ability of these agents (Fig. 5C). The cell apoptotic rate of the aforementioned groups is presented in Fig. 5D. These results suggest that miR-410 may reverse the resistance of U2OS and MG-63 cells to Rap, Dox and DDP through autophagy inhibition. In conclusion, miR-410 regulates ATG16L1 expression and enhances chemosensitivity of cells via autophagy inhibition in osteosarcoma (Fig. 6).
Figure 5.
miR-410 enhances chemosensitivity of Rap (20 nM), Dox (0.1 µg/ml) and DDP (10 µM) through autophagy inhibition. (A) MTT assays were used to detect cell viability of U2OS and MG-63 cells treated with the various chemotherapeutic agents post-transfection with miR-410 or NC mimics for 48 h . (B) Hoechst assays were performed to detect apoptosis of U2OS and MG-63 cells treated with various chemotherapeutic agents post-transfection with miR-410 or NC for 48 h. (C) Western blot analysis of cleaved caspase 3 expression from U2OS and MG-63 lysates after treatment with various agents for 48 h. β-actin was used as the internal control. (D) Cell apoptotic rate was determined in the various groups. Data are presented as the mean ± standard deviation (*P<0.05). ATG161L, autophagy related 16-like 1; miR-410, microRNA-410; NC, negative control; Rap, rapamycin; Dox, doxorubicin; DDP, cisplatin; OD, optical density.
Figure 6.
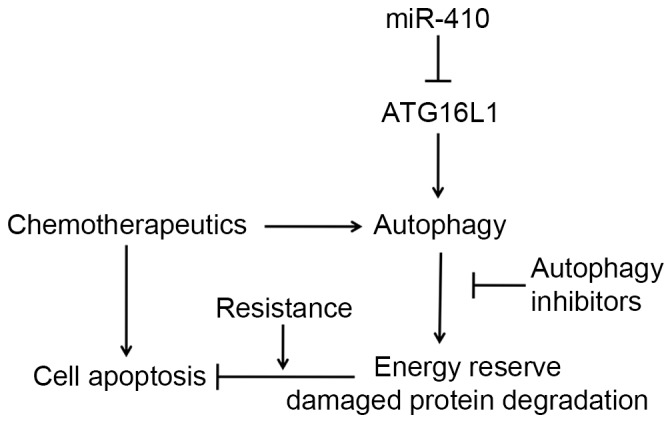
miR-410 regulates autophagy-related gene ATG16L1 expression and enhances chemosensitivity of cells via autophagy inhibition. miR-410, microRNA-410; ATG161L, autophagy related 16-like 1.
Discussion
Osteosarcoma is the most common type of primary bone tumor in children and adolescents, which accounts for 8% of the incidence rate and is a leading cause of cancer-associated mortality among young adults (18). Resistance to chemotherapeutic agents remains a major clinical obstacle to effective therapy; however, the mechanisms underlying osteosarcoma chemoresistance remain largely unknown.
Previous studies have confirmed that autophagy, a chief mechanism for bulk degradation of superfluous or aberrant cytoplasmic components, functions as a protective mechanism that degrades and enables reuse of abnormal proteins and organelles as energy sources to promote cancer cell survival in response to cancer treatment (5,19,20). Mammalian target of rapamycin (mTOR) has generally been regarded as a negative regulator of autophagy, and autophagy-related genes are regulated by proteins upstream of mTOR signaling, including phosphatase and tensin homolog, phosphoinositide-dependent kinase-1, Akt and tuberous sclerosis 1/2 (20). In addition, upregulated autophagy has been identified in various cancer cells following therapeutic stress. Therefore, suppressing cancer cell autophagy is emerging as a novel approach to enhance the efficiency of chemotherapy in cancer treatment (21–23).
miRNAs have emerged as fundamental regulators of gene expression, and are able to silence gene expression at the post-transcriptional and translational levels (8). Dysregulation of miRNAs has been demonstrated to be associated with the tumorigenesis and progression of various types of tumor, including osteosarcoma (24). Downregulation of miR-410 has been detected in several malignant tumors and overexpression of miR-410 inhibits tumor growth and promotes cell apoptosis (11,25,26). In the present study, the role of miR-410 in the progression of osteosarcoma was investigated. The results demonstrated that miR-410 expression was markedly downregulated in human osteosarcoma tissues, and U2OS and MG-63 osteosarcoma cell lines; however, the overexpression of miR-410 exhibited a limited effect on the viability of U2OS and MG-63 cells. Furthermore, ATG16L1 was identified as a direct target of miR-410, and overexpression of miR-410 inhibited the expression of ATG16L1 at the mRNA and protein level in osteosarcoma cell lines. It was also demonstrated that miR-410 was able to markedly inhibit autophagy. Therefore, miR-410 may have the potential to reverse chemoresistance via autophagy inhibition.
The regulatory mechanisms between miRNAs and autophagy remain largely unknown. Chen et al reported that increased miR-155 expression levels upregulated autophagy in osteosarcoma cells, and ameliorated Dox-induced decreases in cell viability (27). Li et al demonstrated that overexpression of miR-22 targeted the 3′UTR of high mobility group box 1 (HMGB1) and inhibited HMGB1-induced autophagy, which contributed to chemotherapy resistance in osteosarcoma in vitro (28). The present study revealed that autophagy was activated after cells were treated with Rap, Dox and DDP. Conversely, the presence of autophagy inhibitor 3-MA or transfection with miR-410 was able to effectively reduce autophagic activation. Further investigations indicated that, compared with the Rap-, Dox- and DDP-treated groups, cell viability was significantly decreased and apoptosis was significantly increased in the drug-treated groups transfected with miR-410.
Taken together, the results of the present study provided novel evidence suggesting that ATG16L1 is a direct target of miR-410. Overexpression of miR-410 significantly sensitized osteosarcoma cells to Rap, Dox and DDP. Understanding the miR-410-mediated tumor suppressor pathways and the potential ability to suppress the autophagic process in human osteosarcoma may provide invaluable therapeutic targets for the treatment of osteosarcoma.
The present study demonstrated that miR-410 expression was downregulated in osteosarcoma cell lines and tissues; however, miR-410 overexpression exhibited limited effects on the viability of U2OS and MG-63 cells. Furthermore, target prediction algorithms identified ATG16L1 as a potential target gene of miR-410, and luciferase reporter assays indicated that miR-410 directly suppressed ATG16L1 expression by targeting its 3′UTR. Furthermore, miR-410 was shown to effectively inhibit autophagy. Accordingly, autophagy was activated as a protective mechanism when osteosarcoma cells were exposed to anticancer drugs, including Rap, Dox and DDP (29,30). The present study demonstrated that the autophagy inhibitor 3-MA and miR-410 expression were able to improve the therapeutic response of cells to chemotherapy drugs (Rap, Dox and DDP), thus indicating that miR-410 may reverse chemoresistance via autophagy inhibition in osteosarcoma cells. The studies conducted regarding the function of miR-410 on autophagy provide insight into the biological function of miR-410 in osteosarcoma, and offer a promising approach for osteosarcoma treatment.
Acknowledgements
The authors would like to thank Professor Shi Wu for providing kindly technological support assistance.
References
- 1.Tsai YC, Wu CT, Hong RL. Response of refractory osteosarcoma to thalidomide and celecoxib. Lancet Oncol. 2005;6:997–999. doi: 10.1016/S1470-2045(05)70468-X. [DOI] [PubMed] [Google Scholar]
- 2.Zhang W, Li Q, Song C, Lao L. Knockdown of autophagy-related protein 6, Beclin-1, decreases cell growth, invasion, and metastasis and has a positive effect on chemotherapy-induced cytotoxicity in osteosarcoma cells. Tumour Biol. 2015;36:2531–2539. doi: 10.1007/s13277-014-2868-y. [DOI] [PubMed] [Google Scholar]
- 3.Xie Z, Xie Y, Xu Y, Zhou H, Xu W, Dong Q. Bafilomycin A1 inhibits autophagy and induces apoptosis in MG63 osteosarcoma cells. Mol Med Rep. 2014;10:1103–1107. doi: 10.3892/mmr.2014.2281. [DOI] [PubMed] [Google Scholar]
- 4.Kim HJ, Lee SG, Kim YJ, Park JE, Lee KY, Yoo YH, Kim JM. Cytoprotective role of autophagy during paclitaxel-induced apoptosis in Saos-2 osteosarcoma cells. Int J Oncol. 2013;42:1985–1992. doi: 10.3892/ijo.2013.1884. [DOI] [PubMed] [Google Scholar]
- 5.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1415. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Fu J, Jiang M, Zhang X, Cheng L, Xu X, Fan Z, Zhang J, Ye Q, Song H. MiR-410 is overexpressed in liver and colorectal tumors and enhances tumor cell growth by silencing FHL1 via a direct/indirect mechanism. PLoS One. 2014;9:e108708. doi: 10.1371/journal.pone.0108708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labialle S, Marty V, Bortolin-Cavaillé ML, Hoareau-Osman M, Pradère JP, Valet P, Martin PG, Cavaillé J. The miR-379/miR-410 cluster at the imprinted Dlk1-Dio3 domain controls neonatal metabolic adaptation. EMBO J. 2014;33:2216–2230. doi: 10.15252/embj.201387038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Zhang J, Feng Y, Li R, Sun X, Du W, Piao X, Wang H, Yang D, Sun Y, et al. MiR-410 regulates MET to influence the proliferation and invasion of glioma. Int J Biochem Cell Biol. 2012;44:1711–1717. doi: 10.1016/j.biocel.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Ni J, Liu K, Yu Y, Xie M, Kang R, Vernon P, Cao L, Tang D. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res. 2012;72:230–238. doi: 10.1158/0008-5472.CAN-11-2001. [DOI] [PubMed] [Google Scholar]
- 13.Lambert LA, Qiao N, Hunt KK, Lambert DH, Mills GB, Meijer L, Keyomarsi K. Autophagy: A novel mechanism of synergistic cytotoxicity between doxorubicin and roscovitine in a sarcoma model. Cancer Res. 2008;68:7966–7974. doi: 10.1158/0008-5472.CAN-08-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Z, Tao L, Shen C, Liu B, Yang Z, Tao H. Silencing of Barkor/ATG14 sensitizes osteosarcoma cells to cisplatin-induced apoptosis. Int J Mol Med. 2014;33:271–276. doi: 10.3892/ijmm.2013.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Cadwell K, Patel KK, Komatsu M, Virgin HW, IV, Stappenbeck TS. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250–252. doi: 10.4161/auto.5.2.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao D, Yuan H, Yi F, Meng C, Zhu Q. Autophagy prevents doxorubicin-induced apoptosis in osteosarcoma. Mol Med Rep. 2014;9:1975–1981. doi: 10.3892/mmr.2014.2055. [DOI] [PubMed] [Google Scholar]
- 18.Mu Y, Zhang H, Che L, Li K. Clinical significance of microRNA-183/Ezrin axis in judging the prognosis of patients with osteosarcoma. Med Oncol. 2014;31:821. doi: 10.1007/s12032-013-0821-3. [DOI] [PubMed] [Google Scholar]
- 19.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao F, Wang G, Wei W, Tu Y, Tong H, Sun S. An autophagy inhibitor enhances the inhibition of cell proliferation induced by a proteasome inhibitor in MCF-7 cells. Mol Med Rep. 2012;5:84–88. doi: 10.3892/mmr.2011.590. [DOI] [PubMed] [Google Scholar]
- 22.Yang C, Han LO. Knockdown of Beclin 1 inhibits vitamin K3-induced autophagy, but promotes apoptosis of human hepatoma SMMC-7721 cells. Mol Med Rep. 2010;3:801–807. doi: 10.3892/mmr.2010.347. [DOI] [PubMed] [Google Scholar]
- 23.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Yan YG, Wang C, Zhang SJ, Yu XH, Wang WJ. MicroRNAs in osteosarcoma. Clin Chim Acta. 2015;444:9–17. doi: 10.1016/j.cca.2015.01.025. [DOI] [PubMed] [Google Scholar]
- 25.Shen J, Niu W, Zhou M, Zhang H, Ma J, Wang L, Zhang H. MicroRNA-410 suppresses migration and invasion by targeting MDM2 in gastric cancer. PLoS One. 2014;9:e104510. doi: 10.1371/journal.pone.0104510. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Chen N, Wang J, Hu Y, Cui B, Li W, Xu G, Liu L, Liu S. MicroRNA-410 reduces the expression of vascular endothelial growth factor and inhibits oxygen-induced retinal neovascularization. PLoS One. 2014;9:e95665. doi: 10.1371/journal.pone.0095665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Jiang K, Jiang H, Wei P. miR-155 mediates drug resistance in osteosarcoma cells via inducing autophagy. Exp Ther Med. 2014;8:527–532. doi: 10.3892/etm.2014.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Wang S, Chen Y, Liu G, Yang X. miR-22 targets the 3′ UTR of HMGB1 and inhibits the HMGB1-associated autophagy in osteosarcoma cells during chemotherapy. Tumour Biol. 2014;35:6021–6028. doi: 10.1007/s13277-014-1797-0. [DOI] [PubMed] [Google Scholar]
- 29.Shen C, Wang W, Tao L, Liu B, Yang Z, Tao H. Chloroquine blocks the autophagic process in cisplatin-resistant osteosarcoma cells by regulating the expression of p62/SQSTM1. Int J Mol Med. 2013;32:448–456. doi: 10.3892/ijmm.2013.1399. [DOI] [PubMed] [Google Scholar]
- 30.Wu W, Li W, Zhou Y, Zhang C. Inhibition of beclin1 affects the chemotherapeutic sensitivity of osteosarcoma. Int J Clin Exp Pathol. 2014;7:7114–7122. [PMC free article] [PubMed] [Google Scholar]