Abstract
There is substantial research on the oncogenic role of tripartite motif containing 37 (TRIM37); however, its importance in colorectal cancer (CRC) remains to be elucidated. The present study used reverse transcription-quantitative polymerase chain reaction, immunohistochemistry and western blotting to detect the expression level of TRIM37 in CRC. The importance of TRIM37 in cell proliferation, invasion and metastasis of CRC were investigated through overexpressing or knocking-down of TRIM37 in CRC cell lines, to observe its function. The present study revealed that TRIM37 was overexpressed in human CRC tissues. High TRIM37 expression resulted in increased CRC proliferation, migration and invasion. Mechanistically, it was confirmed that TRIM37 enhanced invasion and metastasis of CRC via the epithelial-mesenchymal transition pathway. In conclusion, the present study suggested that TRIM3 may contribute to CRC and act as a potential therapeutic target for CRC treatment.
Keywords: tripartite motif containing 37, colorectal cancer, epithelial-mesenchymal transition
Introduction
Colorectal cancer (CRC) is an important contributor to cancer mortality and morbidity (1). It is estimated that ~50–60% of patients diagnosed with CRC develop colorectal metastasis, which is the primary cause of mortality (2). Several molecules that promote metastasis have been investigated and considered as potential useful tumor biomarkers, such as KRAS proto-oncogene, GTPase, vascular endothelial growth factor and v-myc avian myelocytomatosis viral oncogene homolog (3–5). The basic process of progression and metastasis of CRC remains to be elucidated (6). Therefore, it is necessary to identify novel gene targets and develop target-specific therapies for CRC.
Tripartite motif containing 37 (TRIM37) is located on chromosome 17q23, encoding a protein with an estimated molecular weight of 130 kDa (7). TRIM37 contains a really interesting new gene (RING) finger domain, a hallmark of E3 ubiquitin ligases, and a recent study determined that histone H2A was a substrate (8). TRIM37 was also revealed to be associated with polycomb repressive complex 2, and may lead to extensive changes in gene expression, including the silencing of some tumor suppressor genes (8). Previous studies revealed that TRIM37 may undergo amplification in ovarian cancer (9) and may promote transformation in breast cancer (8). Additionally, the upregulation of TRIM37 promoted the growth and migration of cancer cells of pancreatic cancer (10). TRIM37 was also identified to be an independent prognostic factor of hepatocellular carcinoma (HCC), and it may promote the migration and metastasis of HCC cells by activation of Wnt/β-catenin signaling (11). However, the expression pattern and the biological functions of TRIM37 in CRC remain to be elucidated.
Epithelial-mesenchymal transition (EMT) is a critical step of metastasis, and is considered to facilitate tumor invasion (12). In colon carcinoma, EMT occurs at the invasive front, and produces single migratory cells that lose E-cadherin expression (13). These cells are considered to be the cells that eventually enter into subsequent steps of the invasion-metastasis cascade, including intravasation, transport through the circulation, extravasation, formation of micrometastasis and, ultimately, colonization, the growth of small colonies into macroscopic metastasis (14,15). Several key proteins that regulate EMT have previously been identified, such as snail family transcriptional repressor 1, twist family bHLH transcription factor 1 and zinc finger E-box binding homeobox 1 (16–18). However, the complex molecular network and various steps of this mechanistic model require direct experimental validation (19). Therefore, in the present study, in vitro experiments were performed by inducing overexpression and interference using short hairpin RNA (shRNA) to determine that TRIM37 contributed to CRC cell migration and invasion. In addition, in vitro metastasis assays were performed in the present study, which confirmed that the overexpression of TRIM37 increased the motility of CRC cells via EMT signaling.
Materials and methods
Cell culture
SW480 and SW620 CRC cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin, incubated at 37°C in a humidified incubator in an atmosphere of 5% CO2. The cell culture media and supplements were from Gibco; Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
Tissue samples
A total of 30 CRC tissues and paired non-cancerous tissues were obtained from the Department of General Surgery, Huashan Hospital (Shanghai, China). Written informed consent was obtained from all patients. The protocol for the current study was approved by the Human Research Ethics Committee of Huashan Hospital. None of the patients with cancer received any treatment prior to surgery, such as radiation therapy, chemotherapy or immunotherapy. CRC tissues and paired non-cancerous tissues were stored at −80°C until required for further processing.
Reverse transcription-quantitative polymerase reaction (RT-qPCR)
Total RNA from 30 CRC cancer and adjacent non-cancerous mucosa were extracted according to the manufacturer's protocol using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was reverse-transcribed into cDNA by PrimeScript® 1st Strand cDNA Synthesis kit (Takara Biotechnology Company, Ltd., Dalian, China) and used as a template with SYBR-Green PCR kit (Takara Biotechnology Company, Ltd.). The following primers were used for TRIM37: Forward 5′-TCAGCTGTATTAGGCGCTGG-3′ and reverse 5′-ACTTCTTCTGCCCAACGACA-3′. β-actin mRNA was amplified using a sense primer, 5′-CTGGGACGACATGGAGAAAA-3′ and an antisense primer 5′-AAGGAAGGCTGGAAGAGTGC-3′. The PCR conditions were as follows: Initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 10 sec, annealing at 58°C for 15 sec, and elongation at 72°C for 1 min. The relative mRNA level 2∆∆Cq of TRIM37 were obtained by normalizing to β-actin (20).
Immunohistochemical staining
Clinical CRC tissues and paired non-cancerous tissues (n=30) were fixed in formalin, embedded in paraffin, and cut into 5-µm-thick consecutive sections. Slides were dewaxed, rehydrated, and the epitope was subsequently retrieved. After blocking the enzymatic activity of endogenous peroxidase, slides were immunolabeled using TRIM37 primary antibody (1:200; cat. no. sc-49548, Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Subsequently, the slides were washed with PBS and incubated with secondary antibody (1:1,000; cat. no. GK500705; Dako; Aligent Technologies, Santa Clara, CA, USA) and 3,3′-diaminobenzidine using the Dako EnVision Detection System (Dako; Aligent Technologies). The slides were then counterstained with hematoxylin.
Protein extraction and western blotting
A cocktail of proteinase inhibitors and phosphatase inhibitors were added to the radioimmunoprecipitation assay lysis buffer (Thermo Fisher Scientific, Inc.), which was used to extract total protein from tumor cells according to the manufacturer's protocol. Equivalent quantities of protein (30 µg) from each sample were electrophoresed on a 10% SDS-PAGE gel, and then transferred to 0.2 mm polyvinylidene fluoride membranes (Merck Millipore, Darmstadt, Germany). Primary antibodies for TRIM37 (1:500; cat. no. sc-49548, Santa Cruz Biotechnology, Inc.), GAPDH (1:1,000; cat. no. sc-293335, Santa Cruz Biotechnology, Inc.), E-cadherin (1:500; cat. no. ab76055, Abcam, Cambridge, UK), N-cadherin (1:500; cat. no. ab12221, Abcam) and vimentin (1;500; cat. no. ab72547, Abcam) were incubated overnight at 4°C. Following incubation with secondary goat anti-mouse/rabbit-horseradish peroxidase antibody (1:5,000; Santa Cruz Biotechnology, USA), the proteins were detected using an enhanced chemiluminescence kit (Pierce; Thermo Fisher Scientific, Inc.), and then exposed on an X-ray film visualizer (Thermo Fisher Scientific, Inc.).
TRIM37 knockdown and overexpression
Commercially available TRIM37 shRNA and overexpression constructs were obtained from Genechem Co. Ltd. (Shanghai, China) and used to knock down TRIM37 in SW620 cells (TRIM37-KD) and to overexpress TRIM37 in SW480 cells (TRIM37-OV). Scrambled sequences were used as a control. Cell transfection was performed using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
Cell proliferation and colony formation assays
CRC cell proliferation was measured with Cell Counting Kit 8 (CCK8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Each group of cells was seeded in 96-well plates at a density of 1,500 cells/well and cultured in a humidified 5% CO2 atmosphere at 37°C for 24, 48, 72, and 96 h. CCK8 solution was added to the wells and cultured for 2 h, and cell viability was determined by measuring the absorbance at 570 nm using Gen5 microplate reader (Bio-Rad Laboratories, Hercules, CA, USA). A cell growth curve was generated by using the growth data, which was expressed as the mean ± standard deviation. In order to evaluate colony formation ability, equal quantities of control cells and experimental cells were seeded at a density of 1,000 cells/well in 12-well plates and cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco; Thermo Fishers Scientific, Inc.) supplemented with 10% FBS. Cells were cultured for 14 days, and subsequently they were fixed and stained with 0.5% crystal violet. Colonies were then counted in 10 randomly selected fields and images captured using used Nikon Digital ECLIPSE C1 system (Nikon Corporation, Tokyo, Japan). The tests were independently performed in triplicates.
Wound healing assay
Cells (5×105) were cultured in 6-well plates until they reached 90% confluence. A wound was generated by scraping the plate with a 200 µl tip. Images of the wounded monolayer were captured, and cell migration was assessed by quantifying the gap sizes at 3 fields between 0 and 48 h and ImageJ (version 1.48; imagej.nih.gov/ij/) was used to compare the difference between groups.
Matrigel invasion assay
The invasion assay was performed using a specialized Chemicon invasion chamber (Merck Millipore). The inserts contained an 8 µm pore-size polycarbonate membrane with a precoated thin layer of ECMatrix basement membrane matrix. Cells were seeded at a density of 1×104 cells/well, starved in serum-free medium for 24 h and plated on the top chambers. The bottom chambers were filled with medium containing 10% FBS. Cells which invaded the lower surface of the membrane and the ECMatrix had migrated through the polycarbonate membrane, were fixed in 4% paraformaldehyde and stained with crystal violet. Next, these cells were counted and 10 fields were randomly selected and captured at a magnification of ×200 by phase-contrast microscope (Nikon Digital ECLIPSE C1 system, Nikon Corporation).
Statistical analysis
The differences between two experimental groups were analyzed by using a two-tailed Student's t-test. P<0.05 was considered indicate a statistically significant difference. All analyses were performed using SPSS version 21.0 (IBM SPSS, Armonk, NY, USA). Data are presented as the mean ± standard error.
Results
TRIM37 levels increase in CRC tissues
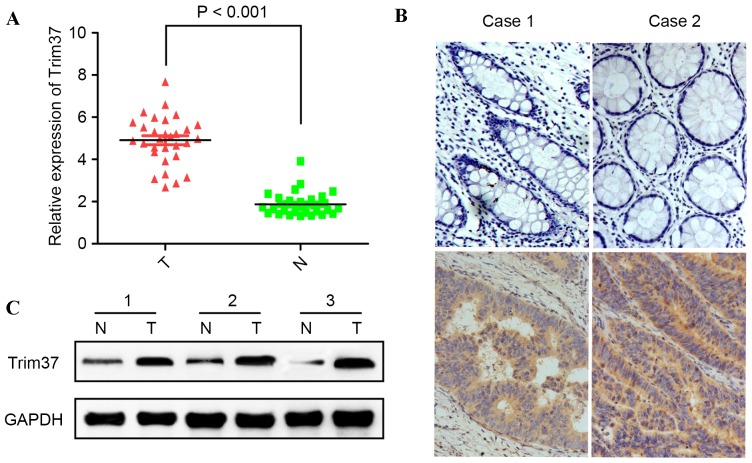
In order to determine the expression of TRIM37 in CRC, TRIM37 expression levels in 30 CRC tissues and paired adjacent normal tissues were quantified using RT-qPCR. It was determined that TRIM37 was significantly upregulated in CRC tissues compared with the matched adjacent normal tissues (P<0.001; Fig. 1A). Next, TRIM37 protein expression was evaluated in primary colorectal cancerous tissue and paired normal colorectal mucosa using western blotting and immunohistochemistry. It was revealed that TRIM37 was highly expressed in cancerous tissue compared with non-cancerous mucosa (Fig. 1B and C). These observations suggested that TRIM37 may have an oncogenic function in CRC.
Figure 1.
mRNA and protein expression levels of TRIM37 are increased in CRC cells relative to normal cells. (A) The mRNA expression level of TRIM37 in 30 paired CRC samples and adjacent normal tissues was detected by reverse transcription-quantitative polymerase chain reaction. The TRIM37 expression was normalized to β-actin. *P<0.001. (B) Representative immunohistochemical images of TRIM37 in matched CRC tissues (lower panel) and adjacent non-cancerous tissues (upper panel). Magnification, ×200. (C) Western blot analysis was used to examine the protein expression level of TRIM37 in CRC samples and paired adjacent normal tissues. CRC, colorectal cancer; TRIM37, tripartite motif containing 37; T, CRC tissues; N, non-cancerous adjacent tissues.
Knockdown of TRIM37 expression reduces the proliferation and migration of CRC cells
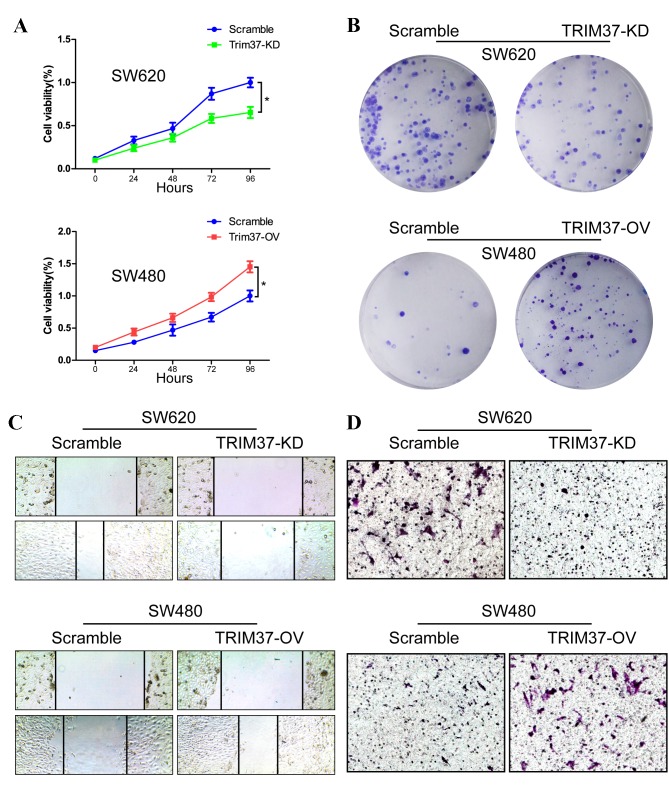
In order to investigate the function of TRIM37, its expression was downregulated in SW620 cells (the TRIM37-KD group). Reduced expression of TRIM37 in SW620 cells significantly inhibited their growth rate compared with the scramble cell group (P<0.05; Fig. 2A). Colony formation assays also revealed reduction in the number of colonies in the TRIM37-KD group compared with the scramble group (Fig. 2B). To determine the effect of TRIM37-KD on migration, cells were wounded by scratching, and then maintained at 37°C for an additional 48 h. Downregulation of TRIM37 attenuated the flattening and spreading of SW620 cells (Fig. 2C). These observations were confirmed by invasion assays, as the invasion ability of cells in the TRIM37-KD group was reduced (Fig. 2D).
Figure 2.
TRIM37 promotes the proliferation, migration and invasion abilities in CRC cells. (A) TRIM37-KD in SW620 cells inhibited cell proliferation, whereas TRIM37-OV in SW480 cells increased proliferation. *P<0.05. (B) Crystal violet assay was used to analyze the effects of TRIM37 on the growth of CRC cells. (C) Representative images of the wound healing assay of TRIM37-KD and TRIM37-OV cells with control. Magnification, ×200. (D) Representative images of the Transwell assays of TRIM37-KD and TRIM37-OV cells with control. Magnification, ×200. Data are representative of three independent experiments. CRC, colorectal cancer; TRIM37, tripartite motif containing 37; TRIM 37-KD, TRIM37 knockdown; TRIM37-OV, TRIM37 overexpression.
Overexpression of TRIM37 promotes the growth and migration of CRC cells
The effects of TRIM37 overexpression (TRIM37-OV group) on the viability of the CRC cells was also investigated. CCK8 and colony formation assays revealed that TRIM37-OV cells had significantly increased cell viability (P<0.05; Fig. 2A and B). The levels of migration and invasion in the TRIM37-OV SW480 cells were higher compared with the control cells in the wound healing and Matrigel chamber assay (Fig. 2C and D). Therefore, overexpression of TRIM37 increased the growth, migratory and invasive properties of CRC cells.
TRIM37 promotes the migration of CRC cells by EMT
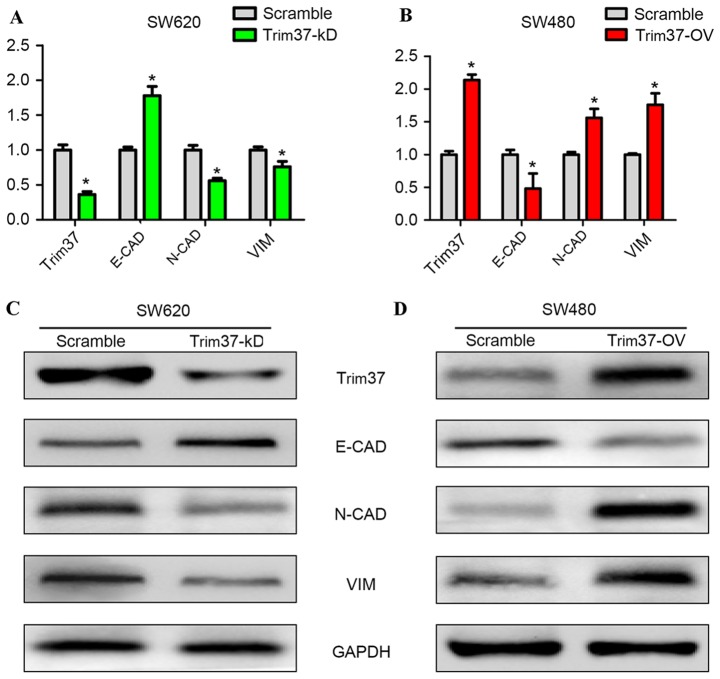
The underlying molecular mechanisms through which TRIM37 promoted the migration and invasion of CRC cells were investigated in the present study. EMT is considered to be the critical mechanism contributing to cancer metastasis (16). To determine the effect of altered TRIM37 expression on the induction of EMT in CRC cells, RT-qPCR and western blot analyses were performed in order to determine the changes in mRNA and protein expression levels of EMT markers. It was determined that, in the TRIM37-KD group, vimentin and N-cadherin mRNA levels were significantly reduced compared with the scramble group (P<0.05; Fig. 3A). However, the E-cadherin mRNA expression level was significantly increased in the TRIM37-KD group compared with the scramble group (P<0.05; Fig. 3A). In contrast with the TRIM37-OV group, vimentin and N-cadherin mRNA expression levels were significantly increased compared with the scramble group (P<0.05; Fig. 3B). The E-cadherin mRNA expression level was significantly reduced in the TRIM37-OV group compared with the scramble group (P<0.05; Fig. 3B). The identical patterns were evident for protein expression levels of the EMT markers in the TRIM37-KD (Fig. 3C) and TRIM37-OV (Fig. 3D) groups. These results clearly indicated that altered TRIM37 expression levels affected the epithelial and mesenchymal phenotype of the CRC cells.
Figure 3.
TRIM37 promotes the invasion of colorectal cancer cells via EMT. (A) Reverse transcription-quantitative polymerase chain reaction was performed to determine the levels of several components that were involved in EMT. In the TRIM37-KD SW620 cells, N-CAD and VIM were downregulated, whereas E-CAD was upregulated. *P<0.05. (B) In the TRIM37-OV SW480 cells, N-CAD and VIM were upregulated, whereas E-CAD was downregulated. *P<0.05. (C) Western blot analysis demonstrated that, in TRIM37-KD SW620 cells, the epithelial marker E-CAD was upregulated, whereas N-CAD and VIM, the mesenchymal markers, were downregulated. (D) In TRIM37-OV SW480 cells, E-CAD was downregulated, whereas N-CAD and VIM were upregulated. TRIM37, tripartite motif containing 37; TRIM 37-KD, TRIM37 knockdown; TRIM37-OV, TRIM37 overexpression; E-CAD, E-cadherin; N-CAD, N-cadherin; VIM, vimentin.
Discussion
The primary cause of mortality in patients with CRC is metastasis (21). The TRIMs family has been identified to be involved in the progression, transformation, autophagy and metastasis of cancer (22–26). Previous studies have demonstrated altered expression levels of TRIM37 in breast and ovarian cancer, which confirmed its oncogenic function (8,9). The present study used RT-qPCR, immunostaining and western blotting to determine that the increased expression of TRIM37 occurred at the transcriptional and post-transcriptional levels in CRC, suggesting that it may contribute to the progression of malignancies.
Previous studies revealed that TRIM37 may interact with β-catenin and promote growth and metastasis in pancreatic and hepatocellular carcinoma, suggesting that there may be co-operation between TRIM37 and the β-catenin/transcription factor complex in the regulation of cancer progression (10,11). Additionally, the present study revealed that TRIM37 positively regulated the proliferation and migration of CRC cells. The TRIM37-OV cells were more effective in wound repair, and their invasive ability was improved, whereas the TRIM37-KD cells had reduced wound healing and invasive capacity, which supported the notion that TRIM37 is an oncogene that contributes to the progression of malignancies.
The present study determined that TRIM37 promoted cell migration and invasion by inducing EMT. Previous studies have determined that the expression levels of epithelial cell markers, such as E-cadherin and cytokeratins, were reduced, whereas upregulation of mesenchymal cell markers, including vimentin and N-cadherin, was observed (27,28). The present study revealed that overexpression of TRIM37 may be accompanied by an increased mesenchymal phenotype and reduced expression of E-cadherin, whereas, by contrast, the knockdown of TRIM37 may lead to an increase in E-cadherin and a decrease in the levels of mesenchymal markers. When cells undergo EMT, they express anchorage-independent growth and obtain the capacity to travel throughout the body, which may lead to metastasis (29–31). The present study may be improved by increasing the clinical sample size to determine the expression of TRIM3. Future studies should focus on discussing the correlation between the expression of TRIM37 and the survival of patients with CRC.
In conclusion, the findings of the present study have supported the notion that TRIM37 upregulation leads to increased vimentin and N-cadherin expression levels, and a reduced expression of E-cadherin. This has provided novel insights into the function of TRIM37 in CRC. Additionally, when TRIM37 was overexpressed in CRC cells, they were more aggressive, with higher tumorigenicity and increased migratory and invasive capability. As TRIM37 has been revealed to contribute to EMT, inhibition of EMT may involve the downregulation of TRIM37 expression. Therefore, TRIM37 may be an effective therapeutic target in the treatment of the progression of CRC.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21314. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 3.Fu B, Yachida S, Morgan R, Zhong Y, Montgomery EA, Iacobuzio-Donahue CA. Clinicopathologic and genetic characterization of traditional serrated adenomas of the colon. Am J Clin Pathol. 2012;138:356–366. doi: 10.1309/AJCPVT7LC4CRPZSK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertucci F, Salas S, Eysteries S, Nasser V, Finetti P, Ginestier C, Charafe-Jauffret E, Loriod B, Bachelart L, Montfort J, et al. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene. 2004;23:1377–1391. doi: 10.1038/sj.onc.1207262. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Jatkoe T, Zhang Y, Mutch MG, Talantov D, Jiang J, McLeod HL, Atkins D. Gene expression profiles and molecular markers to predict recurrence of Dukes' B colon cancer. J Clin Oncol. 2004;22:1564–1571. doi: 10.1200/JCO.2004.08.186. [DOI] [PubMed] [Google Scholar]
- 6.Tejpar S, Bertagnolli M, Bosman F, Lenz HJ, Garraway L, Waldman F, Warren R, Bild A, Collins-Brennan D, Hahn H, et al. Prognostic and predictive biomarkers in resected colon cancer: Current status and future perspectives for integrating genomics into biomarker discovery. The oncologist. 2010;15:390–404. doi: 10.1634/theoncologist.2009-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kallijarvi J, Avela K, Lipsanen-Nyman M, Ulmanen I, Lehesjoki AE. The TRIM37 gene encodes a peroxisomal RING-B-box-coiled-coil protein: Classification of mulibrey nanism as a new peroxisomal disorder. Am J Hum Genet. 2002;70:1215–1228. doi: 10.1086/340256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhatnagar S, Gazin C, Chamberlain L, Ou J, Zhu X, Tushir JS, Virbasius CM, Lin L, Zhu LJ, Wajapeyee N, Green MR. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature. 2014;516:116–120. doi: 10.1038/nature13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlberg S, Lipsanen-Nyman M, Lassus H, Kallijärvi J, Lehesjoki AE, Butzow R. Gynecological tumors in Mulibrey nanism and role for RING finger protein TRIM37 in the pathogenesis of ovarian fibrothecomas. Mod Pathol. 2009;22:570–578. doi: 10.1038/modpathol.2009.13. [DOI] [PubMed] [Google Scholar]
- 10.Jiang J, Tian S, Yu C, Chen M, Sun C. TRIM37 promoted the growth and migration of the pancreatic cancer cells. Tumour Biol. 2016;37:2629–2634. doi: 10.1007/s13277-015-4078-7. [DOI] [PubMed] [Google Scholar]
- 11.Jiang J, Yu C, Chen M, Tian S, Sun C. Over-expression of TRIM37 promotes cell migration and metastasis in hepatocellular carcinoma by activating Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2015;464:1120–1127. doi: 10.1016/j.bbrc.2015.07.089. [DOI] [PubMed] [Google Scholar]
- 12.Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: A central regulator of cancer progression. Trends in cell biology. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao H, Xu E, Liu H, Wan L, Lai M. Epithelial-mesenchymal transition in colorectal cancer metastasis: A system review. Pathol Res Pract. 2015;211:557–569. doi: 10.1016/j.prp.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Pasquier J, Abu-Kaoud N, Al Thani H, Rafii A. Epithelial to mesenchymal transition in a clinical perspective. J Oncol. 2015;2015:792182. doi: 10.1155/2015/792182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, Weinberg RA. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan Q, Qiu MT, Zhu Z, Zhou JH, Chen L, Zhou Y, Gu W, Wang LH, Li ZN, Xu Y, et al. Twist induces epithelial-mesenchymal transition in cervical carcinogenesis by regulating the TGF-β/Smad3 signaling pathway. Oncol Rep. 2015;34:1787–1794. doi: 10.3892/or.2015.4143. [DOI] [PubMed] [Google Scholar]
- 18.Zhang P, Sun Y, Ma L. ZEB1: At the crossroads of epithelial-mesenchymal transition, metastasis, and therapy resistance. Cell Cycle. 2015;14:481–487. doi: 10.1080/15384101.2015.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zoni E, van der Pluijm G, Gray PC, Kruithof-de Julio M. Epithelial plasticity in cancer: Unmasking a MicroRNA network for TGF-β-, Notch-, and Wnt-Mediated EMT. J Oncol. 2015;2015:198967. doi: 10.1155/2015/198967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Rubin MA. Insights into the mechanism of organ-specific cancer metastasis. Cancer Discov. 2014;4:1262–1264. doi: 10.1158/2159-8290.CD-14-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, Silvestri G, Münch J, Kirchhoff F, Simonsen A, et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev Cell. 2014;30:394–409. doi: 10.1016/j.devcel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandell MA, Kimura T, Jain A, Johansen T, Deretic V. TRIM proteins regulate autophagy: TRIM5 is a selective autophagy receptor mediating HIV-1 restriction. Autophagy. 2014;10:2387–2388. doi: 10.4161/15548627.2014.984278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee OH, Lee J, Lee KH, Woo YM, Kang JH, Yoon HG, Bae SK, Songyang Z, Oh SH, Choi Y. Role of the focal adhesion protein TRIM15 in colon cancer development. Biochim Biophys Acta. 2015;1853:409–421. doi: 10.1016/j.bbamcr.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Qiu F, Xiong JP, Deng J, Xiang XJ. TRIM29 functions as an oncogene in gastric cancer and is regulated by miR-185. Int J Clin Exp Pathol. 2015;8:5053–5061. [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, He D, Yang L, Wen B, Dai J, Zhang Q, Kang J, He W, Ding Q, He D. TRIM26 functions as a novel tumor suppressor of hepatocellular carcinoma and its downregulation contributes to worse prognosis. Biochem Biophys Res Commun. 2015;463:458–465. doi: 10.1016/j.bbrc.2015.05.117. [DOI] [PubMed] [Google Scholar]
- 27.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wicki A, Lehembre F, Wick N, Hantusch B, Kerjaschki D, Christofori G. Tumor invasion in the absence of epithelial-mesenchymal transition: Podoplanin-mediated remodeling of the actin cytoskeleton. Cancer cell. 2006;9:261–272. doi: 10.1016/j.ccr.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 30.Godde NJ, Galea RC, Elsum IA, Humbert PO. Cell polarity in motion: Redefining mammary tissue organization through EMT and cell polarity transitions. J Mammary Gland Biol Neoplasia. 2010;15:149–168. doi: 10.1007/s10911-010-9180-2. [DOI] [PubMed] [Google Scholar]
- 31.Baum B, Settleman J, Quinlan MP. Transitions between epithelial and mesenchymal states in development and disease. Semin Cell Dev Biol. 2008;19:294–308. doi: 10.1016/j.semcdb.2008.02.001. [DOI] [PubMed] [Google Scholar]