Abstract
Hypertrophic scars (HS) area fibroproliferative disorder of the skin, which causes aesthetic and functional impairment. However, the molecular pathogenesis of this disease remains largely unknown and currently no efficient treatment exists. MicroRNAs (miRNAs) are involved in a variety of pathophysiological processes, however the role of miRNAs in HS development remains unclear. To investigate the miRNA expression signature of HS, microarray analysis was performed and 152 miRNAs were observed to be differentially expressed in HS tissue compared with normal skin tissues. Of the miRNAs identified, miRNA-21 (miR-21) was significantly increased in HS tissues and hypertrophic scar fibroblasts (HSFBs) as determined by reverse transcription-quantitative polymerase chain reaction analysis. It was also observed that, when miR-21 in HSFBs was blocked through use of an antagomir, the phenotype of fibrotic fibroblasts in vitro was reversed, as demonstrated by growth inhibition, induction of apoptosis and suppressed expression of fibrosis-associated genes collagen type I α 1 chain (COL1A1), COL1A2 and fibronectin. Furthermore, miR-21 antagomir administration significantly reduced the severity of HS formation and decreased collagen deposition in a rabbit ear HS model. The total scar area and scar elevation index were calculated and were demonstrated to be significantly decreased in the treatment group compared with control rabbits. These results indicated that the miR-21 antagomir has a therapeutic effect on HS and suggests that targeting miRNAs may be a successful and novel therapeutic strategy in the treatment of fibrotic diseases that are difficult to treat with existing methods.
Keywords: microRNA-21, hypertrophic scar, fibrosis, antagomir, fibroblasts
Introduction
Hypertrophic scar (HS) is a fibroproliferative disorder of the skin characterized by aberrant wound healing and pathological, excessive dermal fibrosis (1). Skin damage, as occurs with burns, activates fibroblasts and causes them to produce extracellular matrix proteins, including collagen I and III, and growth factors, including transforming growth factor-β and insulin-like growth factor I. Excessive extracellular matrix formation combined with inadequate remodeling of scar tissue results in an aesthetically and functionally unsatisfactory, painful, pruritic scar that may impair skin functions. Current advances in molecular biology and genetics have led to novel non-surgical preventive and therapeutic options, however none of these treatments are efficient (2). Thus, it is important to further elucidate the pathophysiology of HS and to identify potential targets for the development of an effective strategy for HS prevention and treatment.
MicroRNAs (miRNAs) are small noncoding RNAs of 19–25 nucleotides. miRNAs regulate gene expression at the post-transcriptional level by degradation or translational repression of a target mRNA via binding 3′untranslated regions (3′-UTR) of target genes. It has been estimated that up to 90% of human genes are regulated by miRNAs and >3,700 miRNAs have been identified in human cells (3,4). miRNAs are involved in the regulation of the majority, if not all, molecular and cellular processes, and aberrant expression of miRNAs is associated with the initiation and progression of numerous pathological disorders, including cancer, cardiovascular disease and multiple fibrotic diseases (5,6). However, the full miRNA expression signature of HS has not been previously identified and the functions of miRNAs in the pathology of HS remain poorly understood. An in-depth understanding of the specific miRNAs that are involved in the pathogenesis of HS may provide important novel targets for the prevention and treatment of HS.
The present study aimed to identify potential therapeutic targets for the clinical treatment of HS. Specifically, the current study determined miRNA expression associated with HS by miRNA microarray analysis. Of the miRNAs differentially expressed between HS and control tissues, miRNA-21 (miR-21) was significantly upregulated in vitro and in vivo. Therefore, the present study focused on investigating the function of miR-21 in HS and its potential as a target for the prevention and treatment of HS.
Materials and methods
Patient specimens and primary cell culture
Hypertrophic scar fibroblasts (HSFBs) were established as primary cell lines from HS tissues obtained from patients recovering from severe burns. A total 22 patients (13 male and 9 female; age range, 17–42) were recruited in the Department of Plastic and Aesthetic Surgery, Southwest Hospital (Chongqing, China) between 2014 and 2015. The scars were all in a proliferative phase and there was no local infection or ulceration present. None of the cases had been previously treated with glucocorticosteroids or radiotherapy. NS specimens were obtained from donor sites during scar resection. The experimental protocols were approved by the ethics committee of the Third Military Medical University (Chongqing, China) and written informed consent was obtained from each patient and donor. NS and HS tissues were cut into a size of ~1×1×0.5 cm and incubated in a solution of collagenase type I (0.5 mg/ml) and trypsin (0.2 mg/ml) at 37°C for 6 h. Primary fibroblasts were cultured in Dulbecco's modified Eagle medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). HSFBs from each donor were used at passages 3–6 in the experiments. When required, the chemically synthesized miR-21 antagomir (5′-UAGCUUAUCAGACUGAUGUUGA-3′) and antagomir control 5′-UCACAACCUCCUAGAAAGAGUAGA-3′; Guangzhou RiboBio Co., Ltd., (Guangzhou, China) were diluted to final concentrations of 20 nM in cell culture media to disturb miR-21 expression. Cells were incubated with miRNA antagomir for 6 h at 37°C prior to changing the medium. Following a further 48 h incubation, the cells were used for further experiments.
Microarray analysis
miRNAs were isolated from HS and corresponding normal skin (NS) tissues using the mirVana miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc.). Total RNA was quantified using the NanoDrop 2000 (Thermo Fisher Scientific, Inc.) and RNA integrity was assessed using the Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA, USA). Each group consisted of three samples and miRNA expression profiling was performed by miRNA microarray analysis using a chip containing 2158 mature miRNAs (Human miRNA Microarray Slide, Design ID: 046064; Agilent Technologies, Inc.). Proprietary ‘spike-in’ controls were used at each step of the process. Feature Extraction software (version no. 10.7.1.1; Agilent Technologies, Inc.) was used to analyze array images to obtain raw data. GeneSpring GX software (version no. 12.5; Agilent Technologies, Inc.) was employed to perform basic analysis of the raw data. Using this software, the raw data was normalized with the quantile algorithm. Hierarchical clustering was performed to highlight distinguishable miRNA expression patterns within samples.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) for collagen type I α1chain (COL1A1), collagen type I α 2 chain (COL1A2), fibronectin (Fn) and miR-21 analysis. RT-qPCR was performed to detect miRNA expression according to the manufacturer's protocol using the TaqMan® microRNA assay kit (Applied Biosystems; Foster City, CA). Reactions were performed using Stratagene Mx3000P instrument (Agilent Technologies, Inc.) in triplicate. A total of ten pairs of HS/NS samples obtained from the patients, and all the samples obtained from the hypertrophic scar model of rabbit ears were used in the qPCR. Relative miRNA expression was calculated using the 2-ΔΔCq method (7) and levels were normalized to the expression of U6 small nuclear RNAs. The sequences of the primers were as follows: Forward, 5′-TCTAGACATGTTCAGCTTTGTGGAC-3′, and reverse, 5′-TCTGTACGCAGGTGATTGGTG-3′ for human COL1A1; forward, 5′-GAGGGCAACAGCAGGTTCACTTA-3′ and reverse, 5′-TCAGCACCACCGATGTCCA-3′ for human COL1A2; forward, 5′-ACCCAGTCCCGAAGGCACTA-3′ and reverse, 5′-CCTGTACATCTAAAGGCATGAAGCA-3′ for human fibronectin; forward, 5′-GCCCGCTAGCTTATCAGACTGATG-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′ for miR-21; and forward, 5′-GCGCGTCGTGAAGCGTTC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′ for U6.
In situ hybridization (ISH)
ISH was performed as previously described (8). To determine the location of miR-21, an antisense locked nucleic acid (LNA)-modified oligonucleotide probe was used (Exiqon A/S, Vedbaek, Denmark). LNA/DNA oligos contained locked nucleic acids at eight consecutive centrally located bases and had the following sequence: LNA-miR-21, 5′-TCAACATCAGTCTGATAAGCTA-3′.
Cell proliferation and apoptosis assay
Fibroblasts were transfected with the miR-21 antagomir or antagomir control (Guangzhou RiboBio Co., Ltd.) according to the manufacturer's protocols. Fibroblasts were initially seeded at 50–60% confluence and treated for 6 h at 37°C with miR-21 antagomir or antagomir control at a final concentration of 20 nM. The transfected cells were then prepared as a single-cell suspension. A total of 1×104 cells were seeded into 96-well plates and cultured for 5 days. At designated time points, 20 µl 0.5% MTT was added to each well and incubated for 4 h prior to measuring the absorbance of the plates at 570 nm using a microplate reader (Model 680; Bio-Rad Laboratories, Inc., Hercules, CA, USA). The Annexin V-FITC Apoptosis Detection kit (eBioscience, Inc., San Diego, CA, USA) was used to analyze apoptosis. Apoptotic cells were then analyzed by a FACSAria flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) using CellQuest software version 5.1 (BD Biosciences).
Collagen synthesis analysis by H3-proline incorporation assay
Cells at the logarithmic phase (5×104/ml) were incubated in 96-well plates for 48 h at 37°C, with 200 µl of medium in each well., A 100 µl mixture of H3-proline (5 µCi/ml; Yahui Co., Beijing, China), vitamin C (25 µg/ml) and β-aminopropionitrile (50 µg/ml; Yahui Co.) was added and cells were then cultured for another 10 h. The supernatants were then discarded, and cells were digested with trypsin for 2 min, washed and collected onto a glass fiber filter (0.5×104/well). Following precipitation and fixation with 10% trichloroacetic acid for 20 min at −4°C and absolute ethanol, the filter was dried at 70°Cin an oven and placed in a scintillation bottle containing 5 ml of scintillation fluid. A Beckman LS6500 scintillation counter (Beckman Coulter Inc., Brea, CA, USA) was used to count γ-rays. The results were expressed as counts per min.
Hypertrophic scar model of rabbit ear and drug administration
Adult male New Zealand white rabbits (n=8) weighing between 2.2 and 2.5 kg (age, 5–7 months) were used in this study. All animals were obtained from the Laboratory Animal Center of the Third Military Medical University (Chongqing, China), and housed alone at 15–25°C, with a 12-h light/dark cycle and access to food and water provided ad libitum. The HS model was produced following the standardized protocol as previously described (9). The animals were anaesthetized with intramuscular ketamine (40 mg/kg) and xylazine (4 mg/kg). Cleaning of the ears was conducted with iodine and no local anesthesia was used. Four wounds, reaching down to bare cartilage on the ventral surface of each ear, were created by means of a 7 mm punch biopsy. Experiments were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals of Third Military Medicine University, and the experimental protocols used in the present study were approved by the Animal Care Committee of TMMU.
The rabbits were randomly assigned to either the intervention group or the control group (n=4 in each group). Chemically synthesized antagomir or antagomir control (RiboBio Co., Ltd.) were used to inhibit miR-21 expression. Experimental wounds were injected, under the new epithelium, with 100 µl solution containing 10 nM miR-21 antagomir or antagomir control. The injections were carried out on days 15 and 25 after wounding. On day 65 post-wounding, rabbits in each group were anaesthetized with intramuscular ketamine (40 mg/kg) and xylazine (4 mg/kg) and then scars were harvested. One half of each scar was fixed in 4% paraformaldehyde for 24 h, embedded in paraffin, cut in 4-mm sections, and stained with hematoxylin and eosin. The thickness of the epidermis in uninjured skin and of the epidermis of the scar was measured at ×400 magnification under optical microscope (Olympus Corporation, Tokyo, Japan). The other half was frozen for RT-qPCR analysis, which was performed according to the aforementioned description. The degree of dermal hypertrophy of each scar was expressed as the scar elevation index (SEI). This index is the ratio of the area of newly formed dermis of the scar to the area of surrounding normal dermis. An SEI value greater than 1 was considered to indicate a hypertrophic dermis.
Statistical analysis
All values are presented as the mean ± standard deviation of three independent experiments. Statistical analysis was performed using analysis of variance, Chi-squared test or paired Student's t-test using SPSS17.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
miRNA expression profiling of HS
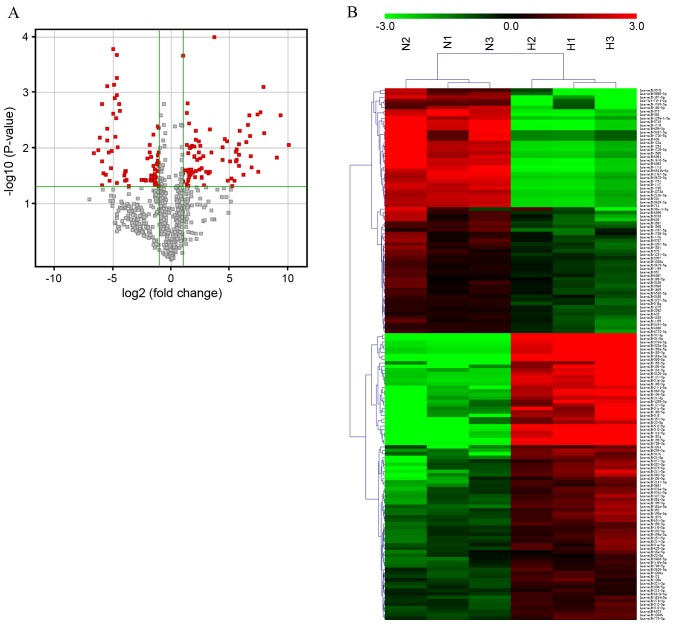
To determine whether miRNAs are involved in the development of HS, miRNA expression profiling was performed on HS and matched NS tissues (n=3). A total of 2158 miRNAs were screened using an Agilent Human miRNA Bioarray (Human miRNA Microarray Slide, Design ID 046064; Agilent Technologies, Inc.). The expression of certain miRNA was significantly different between the two groups (Fig. 1A). The raw data was normalized with the quantile algorithm. In any conditions, the probes flagged as ‘Detected’ in at least 100% of samples were selected for further data analysis. Differentially expressed miRNAs were identified through fold change (FC) and the calculation of P-values was performed using Student's t-test. The threshold set for up and downregulated genes was an FC ≥2 and P<0.05. Target genes of the miRNAs identified as differentially expressed were predicted using the following databases: Targetscan (www.targetscan.org), microRNAorg (http://www.microrna.org) and the PITA algorithm (https://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html).
Figure 1.
miRNA expression signature profiling in HS. (A) Volcano plot presenting differentially expressed miRNAs between HS and paired NS tissue. miRNA microarray expression profiling from three paired HS and NS tissues was performed. Differentially expressed miRNAs were identified by fold change and a P-value calculated using Student's t-test. The threshold set to identify up and downregulated genes was a fold change ≥2 and P<0.05. Red dots indicate points-of-interest that exhibit large-magnitude fold-changes (x-axis; log2 of the fold change) and high statistical significance (y-axis; -log10 of the P-value). (B) Hierarchical clustering showing differentially expressed miRNAs from HS samples compared with paired NS tissues. Each row represents one miRNA and each column represents one tissue sample. The relative miRNA expression is depicted according to the color scale. Red indicates upregulation and green indicates downregulation. N1-3 represents NS tissue samples, whereas H1-3 represents HS tissue samples. The differentially expressed miRNAs were clearly separated into clusters. miRNA, microRNA; hsa-miR, human microRNA; HS, hypertrophic scar; NS, normal skin.
Analysis identified a total of 152 aberrantly expressed miRNAs in HS, including 82 miRNAs that were upregulated and 70 miRNAs that were downregulated. Hierarchical clustering involves unsupervised sample clustering of candidate genes, obtained by appropriate analysis and filtering, to identify novel clusters that changed in a similar direction in the development of HS. Differentially expressed miRNAs were used to generate a heat map, and HS and NS clusters were clearly visible (Fig. 1B). Table I lists the 40 miRNAs with the greatest differences in expression between the HS and NS groups. Of the dysregulated miRNAs identified, miR-31-5p was the most upregulated and miR-3713 was the most downregulated miRNA in HS compared with NS tissue, with FC values of 1090.30 and −98.72, respectively. Dysregulated miRNAs were grouped based on their absolute FC values as follows: 12 miRNAs with an FC >100, 57 miRNAs between 10 and 100, 14 miRNAs between 5 and 10, and 69 miRNAs between 2 and 5. Given that miRNAs are important in numerous pathophysiological processes, the data suggest that these specific miRNAs may be implicated in the development of HS.
Table I.
Top 40 up and downregulated miRs identified by microarray analysis.
| Upregulated miRs | ||
|---|---|---|
| miR name | FC (abs) | P-value |
| Upregulated | ||
| hsa-miR-31-5p | 1090.30 | 0.009 |
| hsa-miR-503-5p | 654.82 | 0.003 |
| hsa-miR-31-3p | 532.56 | 0.015 |
| hsa-miR-542-5p | 248.59 | 0.005 |
| hsa-miR-181a-3p | 244.34 | 0.001 |
| hsa-miR-542-3p | 202.18 | 0.002 |
| hsa-miR-483-3p | 177.35 | 0.026 |
| hsa-miR-493-5p | 165.60 | 0.003 |
| hsa-miR-450a-5p | 118.70 | 0.022 |
| hsa-miR-323a-3p | 112.27 | 0.014 |
| hsa-miR-487a | 108.29 | 0.006 |
| hsa-miR-758-3p | 104.53 | 0.003 |
| hsa-miR-411-5p | 96.54 | 0.004 |
| hsa-miR-376b-3p | 87.80 | 0.008 |
| hsa-miR-154-3p | 72.48 | 0.030 |
| hsa-miR-369-5p | 57.18 | 0.005 |
| hsa-miR-409-5p | 56.06 | 0.009 |
| hsa-miR-34a-3p | 55.91 | 0.013 |
| hsa-miR-3120-3p | 55.88 | 0.021 |
| hsa-miR-24-1-5p | 48.98 | 0.016 |
| hsa-miR-136-3p | 48.82 | 0.011 |
| hsa-miR-485-3p | 46.73 | 0.018 |
| hsa-miR-424-3p | 45.83 | 0.012 |
| hsa-miR-22-5p | 42.09 | 0.038 |
| hsa-miR-455-5p | 37.82 | 0.049 |
| hsa-miR-543 | 35.33 | 0.036 |
| hsa-miR-431-3p | 32.87 | 0.017 |
| hsa-miR-34c-5p | 29.35 | 0.039 |
| hsa-miR-1285-3p | 27.86 | 0.008 |
| hsa-miR-21-3p | 24.41 | 0.007 |
| hsa-miR-485-5p | 21.43 | 0.016 |
| hsa-miR-424-5p | 21.37 | 0.010 |
| hsa-miR-214-5p | 12.94 | 0.000 |
| hsa-miR-382-5p | 10.87 | 0.012 |
| hsa-miR-299-3p | 10.15 | 0.005 |
| hsa-miR-3176 | 9.80 | 0.027 |
| hsa-miR-21-5p | 9.31 | 0.029 |
| hsa-miR-379-5p | 8.03 | 0.027 |
| hsa-miR-1261 | 7.14 | 0.047 |
| hsa-miR-337-5p | 6.57 | 0.014 |
| Downregulated | ||
| hsa-miR-3713 | 98.72 | 0.012 |
| hsa-miR-659-3p | 77.69 | 0.011 |
| hsa-miR-4748 | 69.06 | 0.007 |
| hsa-miR-4441 | 61.18 | 0.002 |
| hsa-miR-5585-3p | 60.28 | 0.047 |
| hsa-miR-383 | 60.08 | 0.017 |
| hsa-miR-375 | 55.28 | 0.029 |
| hsa-miR-187-5p | 49.09 | 0.030 |
| hsa-miR-6511b-5p | 45.33 | 0.001 |
| hsa-miR-6083 | 45.16 | 0.007 |
| hsa-miR-4640-5p | 44.61 | 0.005 |
| hsa-miR-4725-3p | 40.22 | 0.012 |
| hsa-miR-125b-1-3p | 40.07 | 0.038 |
| hsa-miR-422a | 35.22 | 0.023 |
| hsa-miR-4251 | 34.83 | 0.011 |
| hsa-miR-4785 | 32.05 | 0.000 |
| hsa-miR-2467-3p | 31.71 | 0.002 |
| hsa-miR-4754 | 29.63 | 0.001 |
| hsa-miR-4447 | 28.50 | 0.001 |
| hsa-miR-6081 | 27.06 | 0.006 |
| hsa-miR-3675-3p | 26.78 | 0.003 |
| hsa-miR-711 | 25.55 | 0.000 |
| hsa-miR-23c | 25.25 | 0.001 |
| hsa-miR-3620-3p | 25.08 | 0.001 |
| hsa-miR-3945 | 24.74 | 0.043 |
| hsa-miR-4508 | 24.33 | 0.010 |
| hsa-miR-2116-3p | 21.99 | 0.002 |
| hsa-miR-1273d | 20.75 | 0.002 |
| hsa-miR-636 | 16.16 | 0.031 |
| hsa-let-7f-1-3p | 15.26 | 0.038 |
| hsa-miR-4726-5p | 15.17 | 0.027 |
| hsa-miR-3187-3p | 15.14 | 0.026 |
| hsa-miR-486-5p | 14.49 | 0.037 |
| hsa-miR-4769-3p | 12.68 | 0.048 |
| hsa-miR-630 | 5.74 | 0.038 |
| hsa-miR-5703 | 4.69 | 0.038 |
| hsa-miR-6090 | 4.47 | 0.039 |
| hsa-miR-1587 | 4.13 | 0.015 |
| hsa-miR-30c-1-3p | 3.67 | 0.009 |
| hsa-miR-4707-5p | 3.54 | 0.012 |
P<0.05 was considered to indicate a statistically significant difference. miR, microRNA; hsa-miR, human microRNA; FC, fold change.
miR-21 expression is upregulated in HS tissues and HSFBs
Numerous differentially expressed miRNAs, which may be associated with HS development, were identified by the preliminary expression profiling study (Table I). Of those identified, miR-21, also termed miR-21-5p, was upregulated in HS tissue compared with NS tissue and was selected for further analysis, because a previous study had investigated the function of phosphatase and tensin homolog (PTEN) in hypertrophic scar (10) and miR-21 was demonstrated to directly target PTEN (11). Additionally, existing evidence suggested that miR-21 promotes fibrosis in other diseases (12,13). An miR-21 antagomir may have the potential to be used therapeutically in HS, which, to the best of our knowledge, has not been previously investigated. Therefore, the present study selected miR-21 for investigation as a therapeutic target in HS.
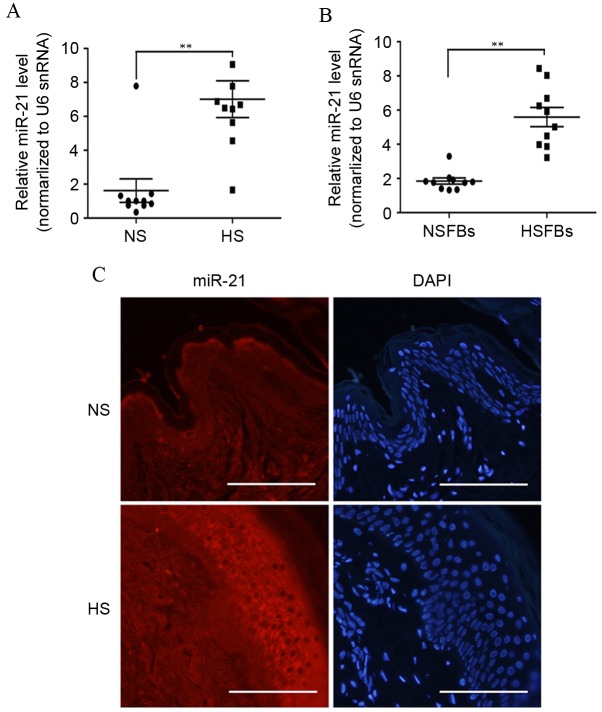
In order to further validate the functions of miR-21in the pathogenesis and progression of HS, RT-qPCR analysis of miR-21 expression in HS and NS tissue samples was performed in more patients. As demonstrated in Fig. 2A, the level of miR-21 expression was significantly higher in 10 cases of HS than in NS tissues (P=0.0019). As fibroblasts and myofibroblasts are important in HS development (1), the present study further investigated the expression of miR-21 in 10 samples of NSFBs (normal skin fibroblasts) and HSFBs. As demonstrated in Fig. 2B, miR-21was significantly overexpressed in HSFBs compared with NSFBs (P<0.0001). These data support the suggestion that miR-21 may act as a pro-fibrotic factor in HS.
Figure 2.
miR-21 expression is upregulated in HS. (A) The expression of miR-21 was analyzed by RT-qPCR and normalized to U6 snRNA in 10 pairs of NS and HS tissue samples. (B) The expression of miR-21 in 10 paired NSFB and HSFB samples were analyzed by RT-qPCR. (C) ISH was performed to determine the localization and expression of miR-21 in skin tissue of HS or NS samples. ISH of skin tissues highlighted extensive cytoplasmic staining for miR-21 (red), which localized in epithelial cells and subdermal fibroblasts. The expression of miR-21 in HS tissues was higher compared with NS controls. Bars: 20 µm. miR, microRNA; HS, hypertrophic scar; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NS, normal skin; NSFBs, normal skin fibroblasts; HSFBs, hypertrophic scar fibroblasts; ISH, in situ hybridization; snRNA, small nuclear RNA. **P<0.01.
ISH with an miR-21specific probe was then performed to examine the expression of miR-21 in HS and NS tissues (Fig. 2C). ISH of skin tissues highlighted extensive cytoplasmic staining for miR-21, which was localized to epithelial cells and subdermal fibroblasts. The expression of miR-21 in HS tissues was higher compared with NS tissues. Together, these data demonstrated that miR-21 was overexpressed in HS, suggesting that miR-21 may participate in the pathogenesis of hypertrophic scar development.
miR-21 antagomir inhibits the proliferation of HSFBs
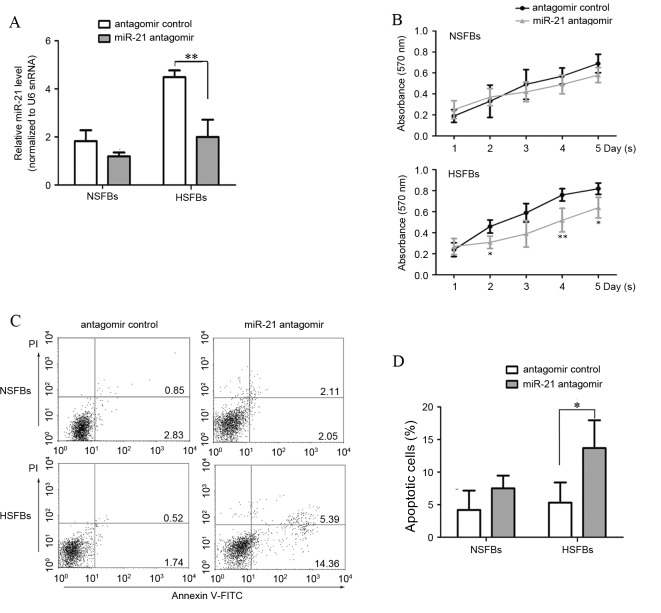
HSFBs are important for the production and deposition of collagen and are the primary effectors in HS formation. To further investigate the potential for miR-21 to be a therapeutic target for HS treatment, HSFBs were transfected with miR-21 antagomir for 48 h andmiR-21 expression levels were detected. An miRNA antagomir control was used as a control. Levels of miR-21 in anti-miR-21-transfected HSFBs were significantly decreased compared with the antagomir control group (Fig. 3A, P<0.0001). The present study then assessed growth and apoptosis in HSFBs and NSFBs following transfection of the miR-21 antagomir. Initially, the MTT assay was used to evaluate the effect of miR-21 on cell growth. The results demonstrated that the miR-21antagomir significantly decreased the proliferation of HSFBs to a nearly normal level (P<0.01; Fig. 3B), suggesting that suppression of miR-21 with an antagomir inhibited HSFB growth. The differences were most significant at day 4, compared with the HSFBs transfected with the antagomir control. Flow cytometric analysis of annexin V and propidium iodide stained cells was performed to assess whether decreased viability was a result of induction of cell apoptosis. Annexin V-positive early apoptotic cells were increased in HSFBs transfected with miR-21antagomir compared with the antagomir control group (Fig. 3C and D). These results indicate that miR-21 affects cell proliferation and apoptosis, and suggests that miR-21 may function as an anti-apoptotic factor in the development of HS.
Figure 3.
miR-21 antagomir inhibits proliferation and induces apoptosis in HSFBs. HSFBs and NSFBs were treated with miR-21 antagomir or miR-21 antagomir control for 48 h. (A) At 48 h after transfection, miR-21 expression levels were evaluated by reverse transcription-quantitative polymerase chain reaction analysis. (B) MTT assays were performed at 24 h intervals for five days. *P<0.05, **P<0.01 vs. the antagomir control. (C) Apoptosis was measured by flow cytometric analysis using Annexin V and PI staining. Right lower quadrant, early apoptotic cells; right upper quadrant, late apoptotic cells. (D) The rate of apoptosis in different groups was calculated and compared. miR, microRNA; HSFBs, hypertrophic scar fibroblasts; NSFBs, normal skin fibroblasts; PI, propidium iodide; snRNA, small nuclear RNA. *P<0.05, **P<0.01.
miR-21 antagomir decreases the expression of fibrosis-associated genes and collagen synthesis in HSFBs
HS is characterized by excessive collagen synthesis. Thus, the present study investigated the effects of altered miR-21 expression on fibrogenesis. The mRNA expression of fibrosis-associated genes, including COL1A1, COL1A2 and Fn, following treatment of HSFBs and NSFBs with miR-21 antagomir or antagomir control was investigated. Compared with NSFBs, mRNA levels of COL1A1, COL1A2 and Fn were elevated in HSFBs, and were significantly decreased by miR-21 antagomir transfection in HSFBs (P<0.01; Fig. 4A). Additionally, the H3-proline incorporation assay demonstrated that HSFBs exhibited increased collagen synthesis compared with NSFBs in the antagomir control groups. However, after 48 h of treatment with miR-21 antagomir, collagen synthesis by HSFBs was significantly decreased compared with HSFBs treated with the antagomir control (P<0.0001), whereas collagen synthesis by NSFBs changed only marginally (Fig. 4B). These results suggest that miR-21 has a pathological role in skin fibrosis and inflammation by promoting the synthesis of collagen, and targeting miR-21 may be a potential therapeutic strategy to block the progression of abnormal scar formation.
Figure 4.
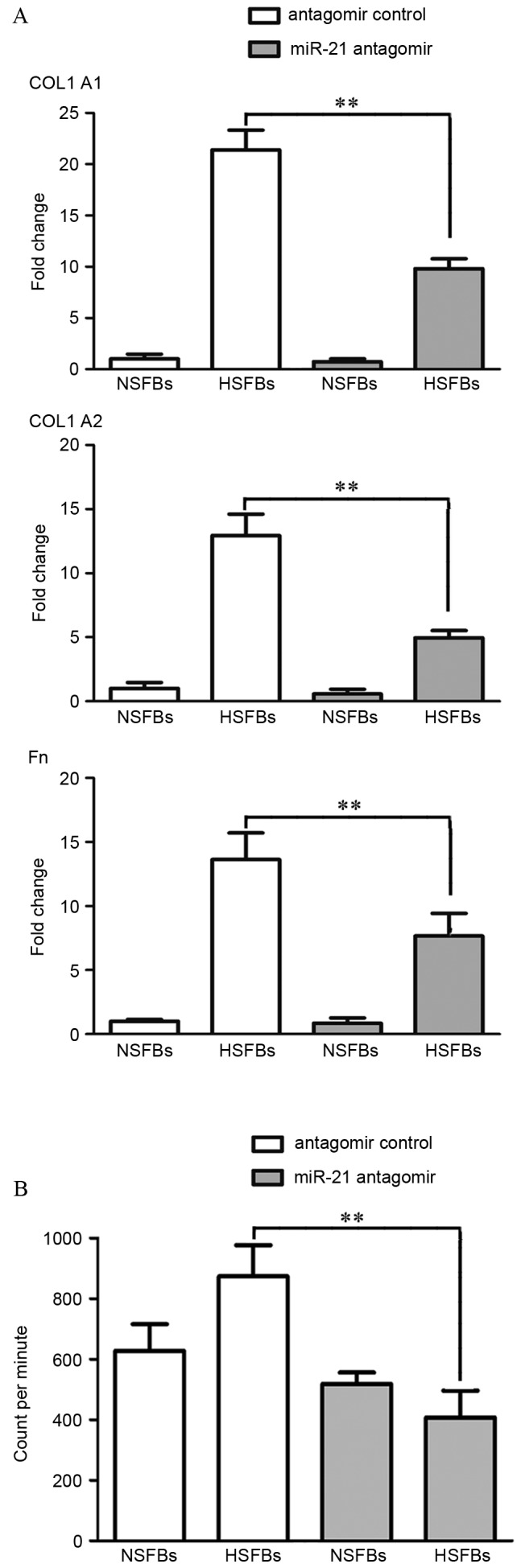
Expression of fibrosis-associated genes is decreased following inhibition of miR-21 by miR-21 antagomir in HSFBs. (A) NSFBs and HSFBs were treated with miR-21 antagomir or miR-21 antagomir control for 48 h. Messenger RNA levels of COL1A1, COL1A2 and Fn were determined by reverse transcription-quantitative polymerase chain reaction. (B) Inhibition of collagen synthesis was measured by H3-proline incorporation assay. **P<0.01, comparison indicated by brackets. Experiments were repeated at least three times. Values are presented as the mean ± standard error of the mean. miR, microRNA; HSFBs, hypertrophic scar fibroblasts; NSFBs, normal skin fibroblasts; COL1A1, collagen type I α 1 chain; COL1A2, collagen type I α 2 chain; Fn, fibronectin.
Inhibition of miR-21 in vivo diminishes the HS formation in rabbit ears
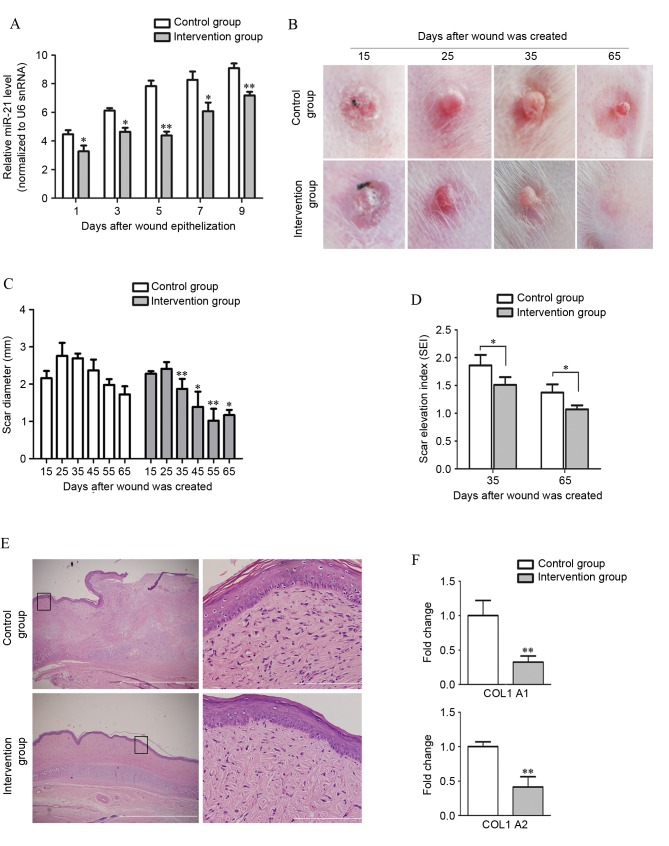
To investigate whether inhibition of miR-21 has therapeutic potential in the treatment of HS, miR-21 antagomir or antagomir control were administered by subcutaneous injection following complete healing of wounds (day 15) in a HS model in rabbit ears. miR-21 continued to increase over 9 days following wound epithelization. However, the increase in miR-21 was significantly suppressed by miR-21 antagomir (Fig. 5A). The extent of skin fibrosis was subsequently analyzed. Macroscopic images of scar samples were taken at each time point by digital camera (Fig. 5B). At 10 days after complete healing (day 25), the scars in the control group were severely congested and appeared purple and swollen. By contrast, the perfusion of scars in the intervention group was dramatically reduced and these scars appeared pink and less swollen following injection of miR-21 antagomir. At 20 days after healing (day 35), the intra-scar microvascular perfusion in both groups had decreased and the scars had become less red. However, scars in the control group remained thick, and had become hardened, while scars in the intervention group were contracted and had softened. The diameter of scars in both groups was measured with a slide gauge. Between days 35 and 65, the diameter of scars in the miR-21 antagomir treated group was reduced compared with the control group (P<0.05; Fig. 5C). The thickness of scars, measured as SEI, in both groups decreased over time. However, the decrease was faster in the intervention group compared with the control group and significant statistical differences in the SEI were observed between the control and intervention group on days 35 and 65 (P<0.05; Fig. 5D). Compared with the control group, administration of miR-21 antagomir led to an ~80% reduction of the SEI in rabbit ear HS model on day 65 (P<0.0001).
Figure 5.
Local treatment with miR-21 antagomir significantly decreases scar formation. (A) miR-21 antagomir administration in the intervention group significantly suppressed the upregulation of miR-21 in scar tissue (n=3) following wound epithelialization, as compared with the miRNA antagomir control administration group. (B) Representative images of scar formation in the rabbit ear model. Scars in the group withanti-miR-21 intervention exhibited a delay in formation process compared with the control group. (C) Quantification of formed HS. The diameter of scars in both groups was measured with a slide gauge. Between day 25 and day 65, the diameter of scars in the miR-21 antagomir intervention group was significantly smaller compared with the control group. (D) Comparison of SEI on days 35 and 65. The degree of dermal hypertrophy of each scar was expressed as the SEI. This index is the ratio of the area of newly formed dermis of the scar to the area of surrounding normal dermis. (E) Pathological analyses by hematoxylin and eosin staining confirmed that scar formation was inhibited in the miR-21 antagomir-treated group when comparing the collagen distribution on day 65. The distribution of collagen fibers in the control group were dense, enlarged and disordered, with more cells in the epidermis and dermis of HS, while collagen fibers were more mature and well arranged with few cells in the intervention group. Boxes in the left column indicate the areas that are enlarged in the corresponding panels in the right column. Scale bars: 1 µm (right) and 50 µm (left). (F) Expression of COL1A1 and COL1A2 in different groups of scars was measured and compared by reverse transcription-quantitative polymerase chain reaction analysis. miR, microRNA; miRNA, microRNA; HS, hypertrophic scar; SEI, scar elevation index; snRNA, small nuclear RNA; COL1A1, collagen type I α 1 chain; COL1A2, collagen type I α 2 chain. *P<0.05, **P<0.01 vs. the control group.
On day 35 post-wounding, scar samples were harvested and collagen distribution was observed by hematoxylin and eosin staining (Fig. 5E). Collagen fibers in the control group were dense, enlarged and disordered, with more cells in the epidermis and dermis of HS, while collagen fibers were more mature and well arranged with few cells in the intervention group, which suggested downregulation of proliferative activity in HS by miR-21 antagomir administration. Finally, mRNA expression of COLA1 and COLA2 in scars was measured by RT-qPCR and the expression levels of COLA1 (P<0.0001) and COLA2 (P<0.0001) were significantly repressed by miR-21 antagomir treatment (Fig. 5F). These differences between the two groups suggest a therapeutic effect of miR-21 antagomir in HS.
Discussion
HS is one of the most common skin disorders and is characterized by pathological, excessive dermal fibrosis. Patients with HS experience a severe reduction in quality of life due to physical, psychological and social sequelae (1,14). There is currently no efficient treatment for HS, partially due to the incomplete understanding of the molecular pathogenesis of this disease. Recently, it has emerged that miRNAs have critical roles in various pathophysiological processes and have been reported to participate in tissue development, differentiation, repair and fibrosis (3–6). Strategies that interfere with miRNA function are considered to offer novel opportunities for clinical treatment. However, little is known about the expression and functions of miRNAs in HS formation and how miRNAs regulate fibrosis in HS remains elusive.
In the present study, miRNA expression profiling was performed and a unique miRNA signature associated with HS development was identified. miRNA microarray profiling identified at least 152 miRNAs with altered expression in HS, including 82 upregulated and 70 downregulated miRNAs. The majority of these miRNAs have been reported to function as oncogenes or tumor suppressors (5), which supports the assumptions regarding the similarity of excess proliferation in HS and cancer cells. Certain other miRNAs were reported to be closely associated with the major events of organ fibrosis. For example, miR-31-5p was reported to be upregulated in keloid (15) and systemic sclerosis (16), miR-503 inhibits epithelial-to-mesenchymal transition in gastric cancer cells (17) and miR-34a has a role in the progression of cardiac tissue fibrosis by directly targeting SMAD family member 4 (18), which suggests that miR-34a may be a novel marker for cardiac fibrosis progression. Microarray analysis in the present study indicated that certain miRNAs are involved in the pathological process of HS formation, and subgroup analysis of miRNAs should be performed to further investigate thisassociation.
miRNA expression analysis in other fibrotic processes has also been reported. Zarjou et al (19) identified a unique miRNA signature associated with renal fibrosis, including upregulated miR-21, miR-142-3p, miR-142-5p, miR-214 and miR-223, and significantly downregulated miR-101a, miR-193 and miR-218. It was reported that miRNA alteration drove the acute and late stages of radiation-induced fibrosis in a murine skin model (20). In a keloid study, 293 differentially expressed miRNAs were identified (21). Several differentially expressed miRNAs identified in the present study were also reported in these studies, including miR-21, miR-214 and miR-34, suggesting that these miRNAs may have an important role in the regulation of fibrosis formation. However, several differentially expressed miRNAs identified in the present study were distinct from the study on keloid (21). The reason for the discrepancy may depend on the miRNAs that were screened in the experiments, and also on the differences between keloid and HS. Although both involve persistent fibrosis of the skin, HS and keloid have pathophysiological and clinical differences (1). HS are typically raised, red or pink, and sometimes pruritic, however they do not exceed the margins of the original wound, whereas keloids infiltrate surrounding normal tissue and rarely regress. Additionally, HS usually subsides over time, while keloid continues to evolve without a quiescent or regressive phase. Accordingly, in the present study, the expression of miR-30c-1-3p and miR-23c was downregulated in HS, whereas in keloid, miR-30a-5p and miR-23b-3p were significantly upregulated in a comparative study of miRNA profiling performed by Li et al (15). It is assumed that the differences in the expression of these miRNAs may partially explain why keloid and HS have different clinical presentations and prognosis. Further investigation is required to verify this assumption.
In addition tithe increased expression of miR-21 in HS, as identified in the microarray analysis performed by the present study, evidence suggests that miR-21 may participate in numerous fibrotic processes, including renal fibrosis (22), systemic sclerosis (23) and idiopathic pulmonary fibrosis (24). The results of these studies indicate that miR-21 has an important role in fibrogenesis. Hsa-miR-21 is located on human chromosome 17q23.2 and is transcribed to produce primary-miR-21. The miR-21 strand of the duplex (miR-21-5p) is incorporated into the RNA-induced silencing complex (RISC), while miR-21-3p (previously termed miR-21*) separates from the duplex and is rapidly degraded. miR-21 then facilitates the binding of RISC to the 3-UTR of the target mRNA, thus, inhibiting protein translation or promoting degradation (25). In the present study, RT-qPCR confirmed that miR-21 was upregulated in HS tissue and HSFBs. ISH also highlighted extensive expression of miR-21 in epithelial cells and dermal fibroblasts. Similarly, Wang et al (26) reported a similar miR-21 expression pattern in epithelial cells of the epidermis and mesenchymal cells of the dermis in the wound. These results indicated that miR-21 may have multiple roles during the course of fibrosis.
It is commonly accepted that HS is caused by high cell proliferation rates and excessive collagen deposition, and abnormal HSFBs are involved in this process. The in vitro experiments performed in this study demonstrated that inhibition of miR-21 significantly inhibited the growth of HSFBs by inducing apoptosis (27). In addition, knockdown of miR-21 reduced the expression of fibrosis-associated genes COL1A1, COL1A2 and Fn. These data suggest that an miR-21 antagomir may be used as a strategy to inhibit HS development clinically. To test this hypothesis, an in vivo rabbit ear scar model of hypertrophic scarring was applied (28). In the present study, miR-21 antagomir was injected directly into HS and, subsequently, miR-21 antagomir significantly decreased the thickness of scars. Additionally, scars in the intervention group were flat and softened compared with the control group. Collagen deposition was also reduced following miR-21 antagomir administration and the distribution of fibers was less disordered. These in vivo and in vitro results indicated that miR-21-induced cell proliferation and collagen deposition are important in the development of HS. The experiments performed in the present study suggest that targeting miR-21 may be a potential therapeutic strategy in HS prevention and treatment, and miR-21 antagomir may be an effective agent to decrease miR-21 expression and reduce excessive fibrosis (29).
It is important to note that miRNAs modulate biological functions via multiple target mRNAs. It is likely that miR-21 participates in fibrotic events in kidney, lung, heart or other organs by regulating a unique array of targets. Liu et al (30) reported that miR-21 promoted collagen deposition by targeting SMAD family member 7, whilst Hatley et al (31) reported that miR-21 drove tumorigenesis through inhibition of negative regulators of the Ras/MEK/extracellular signal-regulated kinase pathway. A previous study reported that loss of PTEN, a well-established miR-21 target, in dermal fibroblasts caused skin fibrosis (32). Our previous study also observed that PTEN expression was significantly decreased in HS, and that PTEN inhibited the proliferation and function of HSFBs through the Akt serine/threonine kinase (AKT) pathway (10). Furthermore, Zhu et al (33) recently reported that miR-21 regulated human telomerase reverse transcriptase expression via the PTEN/phosphatidylinositol-3 kinase/AKT signaling pathway by directly targeting PTEN, thus, controlling growth of HSFBs. Therefore, it is possible that the anti-fibrotic effect of the miR-21 antagomir may be achieved by stabilizing PTEN in HSFBs.
In conclusion, the present study revealed a unique miRNA expression signature associated with the development of HS by microarray analysis. Furthermore, experiments demonstrated that an miR-21 antagomir reversed the phenotype of fibrotic fibroblasts in vitro and attenuated HS formation in an in vivo rabbit ear HS model. The results of the present studydemonstrated that miR-21 has an important role in HS formation and suggest that the miR-21 antagomir is a potential agent for the treatment of HS and other fibrotic skin conditions, including keloid.
Acknowledgements
This research was supported by grants from the National Natural Science Foundation of China (grant nos. 81301638 and 81301639), the State Key Laboratory of Trauma, Burn and Combined Injury of China development program (grant no. SKLZZ201122), and the Grant of Wuhan Young and Middle-aged Medical Personnel Training Project. This manuscript has been edited and proofread by Medjaden Bioscience, Ltd.
References
- 1.Wolfram D, Tzankov A, Pülzl P, Piza-Katzer H. Hypertrophic scars and keloids-a review of their pathophysiology, risk factors, and therapeutic management. Dermatol Surg. 2009;35:171–181. doi: 10.1111/j.1524-4725.2008.34406.x. [DOI] [PubMed] [Google Scholar]
- 2.Karagoz H, Bayram Y, Sever C, Sahin C, Kulahci Y, Ulkur E. A review of the prevention and treatment of hypertrophic scars: Part II experimental studies. Arch Clin Exp Surg. 2013;2:49–58. doi: 10.5455/aces.20120218125115. [DOI] [Google Scholar]
- 3.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 4.Backes C, Keller A. Reanalysis of 3,707 novel human microRNA candidates. Proc Natl AcadSci USA. 2015;112:E2849–E2850. doi: 10.1073/pnas.1505017112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouyssou JM, Manier S, Huynh D, Issa S, Roccaro AM, Ghobrial IM. Regulation of microRNAs in cancer metastasis. Biochim Biophys Acta. 2014;1845:255–265. doi: 10.1016/j.bbcan.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lei W, Li G, Zheng J, Shui X, Huang S, Chen C. Roles of microRNA in vascular diseases in cardiac and pulmonary systems. Pharmazie. 2014;69:643–647. [PubMed] [Google Scholar]
- 7.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 8.Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- 9.Tandara AA, Mustoe TA. The role of the epidermis in the control of scarring: Evidence for mechanism of action for silicone gel. J Plast ReconstrAesthet Surg. 2008;61:1219–1225. doi: 10.1016/j.bjps.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 10.Guo L, Chen L, Bi S, Chai L, Wang Z, Cao C, Tao L, Li S. PTEN inhibits proliferation and functions of hypertrophic scar fibroblasts. Mol Cell Biochem. 2012;361:161–168. doi: 10.1007/s11010-011-1100-2. [DOI] [PubMed] [Google Scholar]
- 11.Ou H, Li Y, Kang M. Activation of miR-21 by STAT3 induces proliferation and suppresses apoptosis in nasopharyngeal carcinoma by targeting PTEN gene. PLoS One. 2014;9:e109929. doi: 10.1371/journal.pone.0109929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duffield JS, Grafals M, Portilla D. MicroRNAs are potential therapeutic targets in fibrosing kidney disease: Lessons from animal models. Drug Discov Today Dis Models. 2013;10:e127–e135. doi: 10.1016/j.ddmod.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong S, Ma W, Hao B, Hu F, Yan L, Yan X, Wang Y, Chen Z, Wang Z. microRNA-21 promotes cardiac fibrosis and development of heart failure with preserved left ventricular ejection fraction by up-regulating Bcl-2. Int J Clin Exp Pathol. 2014;7:565–574. [PMC free article] [PubMed] [Google Scholar]
- 14.Gabriel V. Hypertrophic scar. Phys Med Rehabil Clin N Am. 2011;22:301–310. doi: 10.1016/j.pmr.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Li C, Bai Y, Liu H, Zuo X, Yao H, Xu Y, Cao M. Comparative study of microRNA profiling in keloid fibroblast and annotation of differential expressed microRNAs. Acta Biochim Biophys Sin (Shanghai) 2013;45:692–699. doi: 10.1093/abbs/gmt057. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Li Y, Qu S, Luo H, Zhou Y, Wang Y, Zhao H, You Y, Xiao X, Zuo X. MicroRNA expression abnormalities in limited cutaneous scleroderma and diffuse cutaneous scleroderma. J Clin Immunol. 2012;32:514–522. doi: 10.1007/s10875-011-9647-y. [DOI] [PubMed] [Google Scholar]
- 17.Peng Y, Liu YM, Li LC, Wang LL, Wu XL. microRNA-503 inhibits gastric cancer cell growth and epithelial-to-mesenchymal transition. Oncol Lett. 2014;7:1233–1238. doi: 10.3892/ol.2014.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Qi Y, Du JQ, Zhang DF. MicroRNA-34a regulates cardiac fibrosis after myocardial infarction by targeting Smad4. Expert Opin Ther Targets. 2014;18:1355–1365. doi: 10.1517/14728222.2014.961424. [DOI] [PubMed] [Google Scholar]
- 19.Zarjou A, Yang S, Abraham E, Agarwal A, Liu G. Identification of a microRNA signature in renal fibrosis: Role of miR-21. Am J Physiol Renal Physiol. 2011;301:F793–F801. doi: 10.1152/ajprenal.00273.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simone BA, Ly D, Savage JE, Hewitt SM, Dan TD, Ylaya K, Shankavaram U, Lim M, Jin L, Camphausen K. microRNA alterations driving acute and late stages of radiation-induced fibrosis in a murine skin model. Int J Radiat Oncol Biol Phys. 2014;90:44–52. doi: 10.1016/j.ijrobp.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu ZY, Lu L, Liang J, Guo XR, Zhang PH, Luo SJ. Keloid microRNA expression analysis and the influence of miR-199a-5p on the proliferation of keloid fibroblasts. Genet Mol Res. 2014;13:2727–2738. doi: 10.4238/2014.April.14.2. [DOI] [PubMed] [Google Scholar]
- 22.Glowacki F, Savary G, Gnemmi V, Buob D, Van der Hauwaert C, Lo-Guidice JM, Bouyé S, Hazzan M, Pottier N, Perrais M, et al. Increased circulating miR-21 levels are associated with kidney fibrosis. PLoS One. 2013;8:e58014. doi: 10.1371/journal.pone.0058014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu H, Luo H, Li Y, Zhou Y, Jiang Y, Chai J, Xiao X, You Y, Zuo X. MicroRNA-21 in scleroderma fibrosis and its function in TGF-β-regulated fibrosis-related genes expression. J Clin Immunol. 2013;33:1100–1109. doi: 10.1007/s10875-013-9896-z. [DOI] [PubMed] [Google Scholar]
- 24.Li P, Li J, Chen T, Wang H, Chu H, Chang J, Zang W, Wang Y, Ma Y, Du Y, et al. Expression analysis of serum microRNAs in idiopathic pulmonary fibrosis. Int J Mol Med. 2014;33:1554–1562. doi: 10.3892/ijmm.2014.1712. [DOI] [PubMed] [Google Scholar]
- 25.da Costa Martins PA, De Windt LJ. miR-21: A miRaculous Socratic paradox. Cardiovasc Res. 2010;87:397–400. doi: 10.1093/cvr/cvq196. [DOI] [PubMed] [Google Scholar]
- 26.Wang T, Feng Y, Sun H, Zhang L, Hao L, Shi C, Wang J, Li R, Ran X, Su Y, Zou Z. miR-21 regulates skin wound healing by targeting multiple aspects of the healing process. Am J Pathol. 2012;181:1911–1920. doi: 10.1016/j.ajpath.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 27.Buscaglia LE, Li Y. Apoptosis and the target genes of microRNA-21. Chin J Cancer. 2011;30:371–380. doi: 10.5732/cjc.30.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris DE, Wu L, Zhao LL, Bolton L, Roth SI, Ladin DA, Mustoe TA. Acute and chronic animal models for excessive dermal scarring: Quantitative studies. Plast Reconstr Surg. 1997;100:674–681. doi: 10.1097/00006534-199709000-00021. [DOI] [PubMed] [Google Scholar]
- 29.Mattes J, Yang M, Foster PS. Regulation of microRNA by antagomirs: A new class of pharmacological antagonists for the specific regulation of gene function? Am J Respir Cell Mol Biol. 2007;36:8–12. doi: 10.1165/rcmb.2006-0227TR. [DOI] [PubMed] [Google Scholar]
- 30.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parapuram SK, Shi-wen X, Elliott C, Welch ID, Jones H, Baron M, Denton CP, Abraham DJ, Leask A. Loss of PTEN expression by dermal fibroblasts causes skin fibrosis. J Invest Dermatol. 2011;131:1996–2003. doi: 10.1038/jid.2011.156. [DOI] [PubMed] [Google Scholar]
- 33.Zhu HY, Li C, Bai WD, Su LL, Liu JQ, Li Y, Shi JH, Cai WX, Bai XZ, Jia YH, et al. MicroRNA-21 regulates hTERT via PTEN in hypertrophic scar fibroblasts. PLoS One. 2014;9:e97114. doi: 10.1371/journal.pone.0097114. [DOI] [PMC free article] [PubMed] [Google Scholar]