Abstract
Sampling of mallards in Alaska during September 2014–April 2015 identified low pathogenic avian influenza A virus (subtypes H5N2 and H1N1) that shared ancestry with highly pathogenic reassortant H5N2 and H5N1 viruses. Molecular dating indicated reassortment soon after interhemispheric movement of H5N8 clade 2.3.4.4, suggesting genetic exchange in Alaska or surrounds before outbreaks.
Keywords: influenza A virus, genetic reassortment, bird migration, spillover transmission, agricultural biosecurity, viruses, infectious diseases, mallards, ducks, poultry, wild birds, Alaska, H5 clade 2.3.4.4, avian, zoonoses
The emergence of highly pathogenic avian influenza (HPAI) A virus subtype H5 of clade 2.3.4.4 in East Asia followed by spread into North America in 2014 highlights the importance of ecologic interactions along the Pacific Rim to the incursion of novel viruses. Introduction of influenza A subtype H5N8 into North America is hypothesized to have occurred through wild bird movement across the Bering Strait (1,2); the virus then spread through Canada to the continental United States, concurrently infecting wild birds (ducks, geese, passerines, and raptors) and poultry (turkeys and chickens) (3). Reassortment of H5N8 with low pathogenic avian influenza (LPAI) A virus in North America generated 3 subtypes (H5N8, H5N2, and H5N1, collectively referred to as H5Nx) that followed different trajectories in local bird populations. HPAI H5N2 became the most widespread in US poultry, prompting the culling of ≈49 million chickens and turkeys in 15 states (4). During the outbreaks (November 2014–December 2015), surveillance efforts increased; consequently, later stages of the epidemic were better characterized (5,6) relative to the beginning. Analysis of wild bird viruses from Alaska preceding outbreaks remains one of the few avenues for elucidating how H5N8 entered and reassorted with North American lineage viruses.
Our sampling of mallards (Anas platyrhynchos) from urban ponds in Anchorage, Alaska, during September 2014–April 2015 identified LPAI H5N2 and H1N1 (Technical Appendix Figure 1, Table 1 ). These viruses were the closest relatives for 4 of the 8 North American segments that contributed to the H5Nx reassortants based on the time of most recent common ancestry (tMRCA) analysis (Technical Appendix Figures 2–12). All North American segments of the H5N2 reassortant (basic polymerase protein 1, nucleoprotein, and neuraminidase) shared most recent common ancestry with LPAI H5N2 that circulated in Anchorage mallards and a concurrently sampled wild bird population at Izembek National Wildlife Refuge in western Alaska (1) (Technical Appendix Figures 3, 5, 8, 9). Molecular dating indicated reassortment of H5N8 and LPAI H5N2 (or precursors) shortly after the interhemispheric movement of H5N8. We estimated that ancestors of the H5N2 reassortant emerged among wild birds in Alaska during August 2013–May 2014, based on tMRCAs of multiple segments (Figure 1, panel A), followed by emergence of the H5N2 reassortant during May–September 2014. Our analysis refines the hypothesis of Beringia introduction (1,2) by indicating that H5N8 reassorted with viruses shed by waterfowl in Alaska (or nearby high latitudes) shortly after introduction into North America (May 2014–October 2014, 95% highest posterior density January 2014–January 2015) (Figure 1, panel A). Accuracy of molecular dating hinges on the availability of virus sequences from relevant hosts, which were lacking from North American poultry and wild birds during the year preceding outbreaks (Figure 1, panel B). Our sampling was fortuitous, being one of the few in Alaska conducted before the outbreaks, underscoring the importance of routine sampling where interhemispheric mixing of viruses occurs with high frequency.
Figure 1.
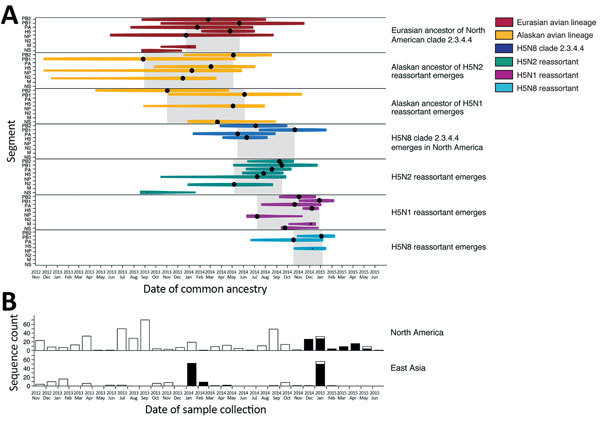
Molecular dating of the emergence of H5 clade 2.3.4.4 influenza A virus in Eurasia and North America and concurrent trends in surveillance effort. A) Events contributing to the evolution of H5 clade 2.3.4.4 estimated using multiple influenza segments. Time of most recent common ancestry (indicated by a black circle) is size-scaled by the posterior probability (0.0–1.0), and the 95% highest posterior density is color-coded by lineage. Gray shading indicates time of most recent common ancestry of multiple segments with a posterior probability >0.85. B) Surveillance effort estimated by the number of hemagglutinin sequences (high and low pathogenicity) available in the Influenza Research Database (https://www.fludb.org). Black bars indicate surveillance effort for H5 clade 2.3.4.4; white bars indicate surveillance effort for other clades. M, matrix gene; N2, neuraminidase 2 gene; NP, nucleoprotein gene; PA, polymerase acidic, PB1, basic polymerase protein 1 gene; PB2, basic polymerase protein 2 gene; tMRCA, time of most recent common ancestry.
The rapid reassortment of H5N8 clade 2.3.4.4 within North America is premised on the movement of at least a single infected bird across the Bering Strait followed by infection of a host population at high latitude. Interhemispheric movement during spring 2014 (or earlier) is most plausible given the circulation of the Eurasian ancestor of North American clade 2.3.4.4 during December 2013–May 2014 (95% highest posterior density October 2013–January 2015) (Figure 1, panel A), an event that preceded introduction. The presence of overwintering birds in Alaska, a known area for influenza exchange between East Asia and North America (7,8), might enhance opportunities for viruses originating in Eurasia to reassort with LPAI in local bird populations. Mallards from this study are a prime example of an overwintering population, occupying urban ponds that remained thawed because of human activity, which allows some birds to remain in southcentral Alaska from September through April, when many migratory waterfowl have since flown south. We found evidence that LPAI H5N2 shed by overwintering mallards from Anchorage (south-central Alaska) and wild birds from Izembek (western Alaska) were highly related and formed monophyletic clades (Technical Appendix Figures 5, 6, 8–10). This provided evidence of regional dispersal of LPAI in Alaska concurrent with the proposed timing of H5N8 introduction and reassortment.
Anchorage mallards shed viruses that shared ancestry with 2 of 4 North American segments (basic polymerase protein 1 and nonstructural) of the H5N1 reassortant (Technical Appendix Figures 2, 5, 12). Emergence of the H5N1 reassortant probably occurred after July 2014 (Figure 1, panel A), after the H5N2 reassortant emerged. Molecular dating of tMRCA of H5N8 reassortants was confounded by long branch lengths of parental lineages indicative of unsampled ancestors; however, estimates based on 2 segments suggest emergence after October 2014. The H5N1 and H5N8 reassortants possessed a highly similar polymerase acidic segment (Technical Appendix Figure 6), suggesting a similar evolutionary trajectory of the two subtypes that later diverged. Our tMRCA estimates for H5N1 are consistent with reassortment during or after the breeding season for mallards in Alaska followed by southward dispersal along the Pacific Flyway during autumn (Figure 2). These results suggest that H5N1 is a multiple reassortant that acquired PB1 and NS segments in Alaska (or surrounds) followed by polymerase acidic and neuraminidase 2 from different host populations before detection in Washington (9) and Oregon (4) in early 2015.
Figure 2.
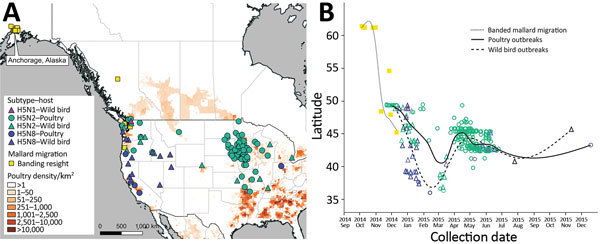
Spatial and temporal distribution of H5 clade 2.3.4.4 influenza A virus outbreaks among wild birds and poultry across North America. A) Spatial distribution of H5 clade 2.3.4.4 influenza A virus outbreaks in wild birds (triangles) and poultry (circles) across North America, color-coded by subtype, relative to poultry density. The location of mallards from Anchorage, Alaska, based on resighting of banded birds, is indicated. B) Temporal distribution of H5 clade 2.3.4.4 influenza A virus detections during the course of the outbreaks relative to the migration of mallards banded in Anchorage.
Arrival of mallards banded in Anchorage at the high-density poultry region of the Fraser Valley, British Columbia, Canada, in November 2014 is compatible with the chronology of evolution and subsequent detection of the H5Nx subtypes (Figure 2). The migration chronology of banded mallards might be broadly representative of other dabbling duck species that breed in Alaska, such as the American green-winged teal (Anas carolinensis), American wigeon (Anas americana), and northern pintail (Anas acuta), in which H5Nx was detected at lower latitudes. Consequently, mallards and other waterfowl species probably were involved in the southward dispersal and reassortment of H5Nx followed by spillover to poultry. Observations of wild birds congregating at water bodies on poultry farms in the Fraser Valley support the scenario of indirect transmission from migratory birds to poultry, seeding outbreaks at lower latitudes (10). Later divergence of H5N2 into multiple lineages during May–November 2014 (Technical Appendix Figures 8) implies that outbreaks were seeded by different H5N2 strains, although the mode of dispersal through wild bird migration, farm-to-farm poultry movement, or poultry workers remains unclear.
Since introduction of HPAI H5 of clade 2.3.4.4 into the Pacific Northwest in late 2014, little evidence exists for additional reassortment, despite continued spread of H5N2 and H5N8 until late 2015 (Figure 2, panel B). Lack of further reassortment implies a change from wild bird–mediated dispersal to intermittent spillover between wild birds and poultry or indirect transmission among poultry farms via fomites, wind, or other undetermined vectors. The spatiotemporal pattern of outbreaks in wild birds and poultry appeared correlated during this later phase (Figure 2, panel B). Correlation might be a function of outbreak investigation procedures that require concurrent sampling of poultry and wild birds inhabiting the control zone. However, our phylogenetic analysis lends support for frequent spillover given that lineages of H5Nx were mixed by host, rather than poultry and wild birds clustering separately (Technical Appendix Figures 4–12). Our analysis and the August 2016 detection of HPAI H5N2 in mallards from Fairbanks, Alaska (11), an area lacking commercial poultry, implicates waterfowl as playing an important role in reassortment, spread, and possibly long-term circulation of H5Nx viruses.
Methods used for study of rapid reassortment of high and low pathogenic influenza A virus among wild birds in Alaska before H5 clade 2.3.4.4 outbreaks, including sampling of wild birds, banding data and outbreak analysis, virus isolation and sequencing, multiple sequence alignment, down-sampling before tree phylogenetic reconstruction, Bayesian phylogenetic analysis, and molecular dating analysis.
Acknowledgments
This study was supported by the National Institutes of Health Center of Excellence in Influenza Research and Surveillance (contract no. HHSN272201400008C) and the J. Craig Venter Institute Genomic Centers for Infectious Diseases Program (NIH/NIAID grant no. U19 AI110819).
Capture and processing of wild mallards was approved by the Institutional Animal Care and Use Committee at the University of Alaska Fairbanks (protocol no. 358515-11/662280-3) and was authorized by US Federal Bird Banding and Marking Permits (nos. 08350 and 23191). None of the authors have any financial interests or conflict of interest with this article. Any use of trade names is for descriptive purposes only and does not imply endorsement by the U.S. Government. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Biography
Dr. Hill is a postdoctoral associate at the Massachusetts Institute of Technology, Cambridge, Massachusetts, USA. Her primary research interests include the ecology and evolution of influenza viruses in natural and man-made systems.
Footnotes
Suggested citation for this article: Hill NJ, Hussein ITM, Davis KR, Ma EJ, Spivey TJ, Ramey AM, et al. Reassortment of influenza A viruses in wild birds in Alaska before H5 clade 2.3.4.4 outbreaks. Emerg Infect Dis. 2017 Apr [date cited]. http://dx.doi.org/10.3201/eid2304.161668
References
- 1.Ramey AM, Reeves AB, TeSlaa JL, Nashold S, Donnelly T, Bahl J, et al. Evidence for common ancestry among viruses isolated from wild birds in Beringia and highly pathogenic intercontinental reassortant H5N1 and H5N2 influenza A viruses. Infect Genet Evol. 2016;40:176–85. 10.1016/j.meegid.2016.02.035 [DOI] [PubMed] [Google Scholar]
- 2.Lee DH, Torchetti MK, Winker K, Ip HS, Song CS, Swayne DE. Intercontinental spread of Asian-origin H5N8 to North America through Beringia by migratory birds. J Virol. 2015;89:6521–4. 10.1128/JVI.00728-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.US Department of Agriculture. Avian influenza disease. Animal disease information. Avian health [cited 2016 Aug 5]. https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/animal-disease-information/avian-influenza-disease
- 4.Lee DH, Bahl J, Torchetti MK, Killian ML, Ip HS, DeLiberto TJ, et al. Highly pathogenic avian influenza viruses and generation of novel reassortants, United States, 2014–2015. Emerg Infect Dis. 2016;22:1283–5. 10.3201/eid2207.160048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krauss S, Stallknecht DE, Slemons RD, Bowman AS, Poulson RL, Nolting JM, et al. The enigma of the apparent disappearance of Eurasian highly pathogenic H5 clade 2.3.4.4 influenza A viruses in North American waterfowl. Proc Natl Acad Sci U S A. 2016;113:9033–8. 10.1073/pnas.1608853113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bevins SN, Dusek RJ, White CL, Gidlewski T, Bodenstein B, Mansfield KG, et al. Widespread detection of highly pathogenic H5 influenza viruses in wild birds from the Pacific Flyway of the United States. Sci Rep. 2016;6:28980. 10.1038/srep28980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearce JM, Reeves AB, Ramey AM, Hupp JW, Ip HS, Bertram M, et al. Interspecific exchange of avian influenza virus genes in Alaska: the influence of trans-hemispheric migratory tendency and breeding ground sympatry. Mol Ecol. 2011;20:1015–25. 10.1111/j.1365-294X.2010.04908.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramey AM, Pearce JM, Ely CR, Guy LM, Irons DB, Derksen DV, et al. Transmission and reassortment of avian influenza viruses at the Asian-North American interface. Virology. 2010;406:352–9. 10.1016/j.virol.2010.07.031 [DOI] [PubMed] [Google Scholar]
- 9.Torchetti MK, Killian ML, Dusek RJ, Pedersen JC, Hines N, Bodenstein B, et al. Novel H5 Clade 2.3.4.4 reassortant (H5N1) virus from a green-winged teal in Washington, USA. Genome Announc. 2015;3:e00195–15. 10.1128/genomeA.00195-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu W, Berhane Y, Dubé C, Liang B, Pasick J, VanDomselaar G, et al. Epidemiological and evolutionary inference of the transmission network of the 2014 highly pathogenic avian influenza H5N2 outbreak in British Columbia, Canada. Sci Rep. 2016;6:30858. 10.1038/srep30858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee DH, Torchetti MK, Killian ML, DeLiberto TJ, Swayne DE. Reoccurrence of avian influenza A(H5N2) virus clade 2.3.4.4 in wild birds, Alaska, USA, 2016. Emerg Infect Dis. 2017;23:365–7 . 10.3201/eid2302.161616 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods used for study of rapid reassortment of high and low pathogenic influenza A virus among wild birds in Alaska before H5 clade 2.3.4.4 outbreaks, including sampling of wild birds, banding data and outbreak analysis, virus isolation and sequencing, multiple sequence alignment, down-sampling before tree phylogenetic reconstruction, Bayesian phylogenetic analysis, and molecular dating analysis.


