Abstract
Background
Vascular endothelial growth factor (VEGF) is a key mediator that plays an important role in angiogenesis, tumor growth, and tumor metastasis. The associations between five polymorphisms of VEGF (rs3025039, rs699947, rs10434, rs1570360, and rs2010963) and renal cell carcinoma (RCC) risk have been extensively investigated, but the currently available results are inconsistent and inconclusive. To obtain a more accurate assessment of the associations, we conducted a meta-analysis in this study.
Materials and methods
Relevant studies were collected systemically from the following three electronic databases: MEDLINE, Web of Science, and CNKI (Chinese National Knowledge Infrastructure). Statistical analyses were performed using Review Manager 5.2 in a fixed- or random-effects model. Pooled odds ratios (ORs) with 95% confidence intervals (95% CIs) were calculated to establish the strength of associations.
Results
A total of eight case–control studies with 1,936 RCC cases and 2,770 controls fulfilling the inclusion criteria were selected for this meta-analysis. The pooled OR indicated that rs699947 polymorphism was significantly associated with RCC risk in all genetic models. A significant association was also found between the rs3025039 polymorphism and RCC risk in a homozygous model (TT vs CC: OR =1.38, 95% CI =1.11–1.72, P=0.004), a dominant model (CT+TT vs CC: OR =1.21, 95% CI =1.05–1.39, P=0.01), and a recessive model (TT vs CC+CT: OR =1.28, 95% CI =1.04–1.57, P=0.02). After a subgroup analysis of ethnicity in the allele contrast model of rs3025039 polymorphism, we found a significant relationship in the allele contrast model (T vs C: OR =1.21, 95% CI =1.05–1.40, P=0.007) in the Asian population. With regard to rs10434 polymorphism, significant association was observed only in a homozygous model (GG vs AA: OR =0.75, 95% CI =0.57–0.98, P=0.03). As to rs1570360 or rs2010963, we did not observe any relationship between the two polymorphisms and RCC risk in our study.
Conclusion
Our meta-analysis confirmed the fact that rs699947, rs3025039, and rs10434 polymorphisms were significantly relevant to elevated RCC risk. In the meanwhile, this study also demonstrated that the allele contrast model of rs3025039 polymorphism was likely to be associated with risk of RCC in the Asian population.
Keywords: renal cell carcinoma, vascular endothelial growth factor, polymorphism, meta-analysis
Introduction
Renal cell carcinoma (RCC) is the most common type of kidney cancer and accounts for >90% of all diagnosed cases.1 It has been reported that RCC has the highest mortality rate among the genitourinary cancers, and the incidence of RCC has increased sequentially, especially in young patients and in patients with high-grade disease.2,3 The current statistical report shows that there are ~209,000 newly diagnosed RCC cases and 102,000 deaths caused by RCC per year worldwide.4 A study indicates that ~40% of RCC patients die of metastatic disease, because metastases are often present at diagnosis and recurrence stage, which are also common after complete resection of the primary tumor.5 It has been revealed that smoking,6,7 overweight and obesity,8,9 hypertension,10 and potential environmental causes11 are risk factors for the development and progression of RCC. However, the pathogenic mechanisms of RCC underlying the established risk factors still remain vague.
Recent molecular studies have suggested that gene polymorphism contributes to tumorigenesis.12–15 Vascular endothelial growth factor (VEGF), an endothelial cell-specific mitogen, has been deemed as a major contributor to the development of RCC.16,17 VEGF is a vital stimulator of pathological and physiological angiogenesis that acts on vascular endothelial cells and promotes human blood vessel growth during tumor growth, enabling invasion and metastasis,18,19 and the role of angiogenesis has been a recurrent denominator of RCC connected to VEGF.20 Therefore, it will be useful to understand better the molecular pathogenesis of RCC, which can help to predict RCC and develop targeted agents to treat this disease.
VEGF is located on chromosome 6p21.3 with 8 exons and 7 introns, spanning ~14 kb.21 It is highly polymorphic, especially in the promoter, the 5′-untranslated region (UTR), and the 3′-UTR.22 There are several common single nucleotide polymorphisms (SNPs) in the VEGF gene, including rs3025039, rs699947, rs10434, rs1570360, and rs2010963 positions, which could increase the risk of developing diseases characterized by deranged angiogen-esis and alter gene expression and protein production.15,23,24 Up to now, many studies have investigated the association between SNPs of the VEGF gene and RCC in different populations.25–32 However, the results have been inconsistent and inconclusive. In 2013, Zhang et al33 conducted a meta-analysis to present the genetic knowledge on the VEGF gene polymorphisms and RCC risk in humans based on the published evidence. However, due to the limitations of the included study, the results of this meta-analysis indicated that the VEGF rs3025039(+936C/T), rs699947(−2578C/A), rs10434(1612G/A), rs1570360(+405C/G), and rs2010963 (−1154G/A) gene polymorphisms are not associated with the risk of RCC. Therefore, in this study, we performed an updated meta-analysis with eight studies for rs3025039, rs699947, rs10434, rs1570360, and rs2010963 polymorphisms to further clarify the associations.
Materials and methods
Literature and search strategy
A comprehensive systematic search was conducted for published articles using MEDLINE, Web of Science, and CNKI (Chinese National Knowledge Infrastructure); the years were limited from 1993 to 2016. The following keywords and MeSH terms were used: (“renal cell carcinoma” OR “renal cell cancer” OR “RCC”) AND (“vascular endothelial growth factor” OR “VEGF”) AND (“polymorphisms” OR “mutations” OR “variants” OR “single nucleotide polymorphisms” OR “SNP”). All included articles were published in English language. At the same time, the reference lists of retrieved papers and recent reviews were manually searched.
Study inclusion and exclusion criteria
Studies eligible for inclusion in our meta-analysis should meet the following criteria: 1) must be independent case–control or cohort design studies; 2) the article pertained to the abovementioned VEGF polymorphisms (rs3025039, rs699947, rs10434, rs1570360, or rs2010963) and RCC risk; 3) patients have clinically confirmed RCC; 4) the studies provided the number of cases and controls for various genotypes and sufficient data for calculating odds ratios (ORs) with 95% confidence intervals (CIs); and 5) genotype distributions of polymorphism of the control population were consistent with Hardy–Weinberg equilibrium (HWE). Accordingly, the exclusion criteria of the meta-analysis were 1) meta-analyses, reviews, case reports, or no healthy control population; 2) animal studies; 3) non-conformity with the criteria for RCC; 4) there are no sufficient data to estimate the ORs and 95% CIs; and 5) duplication of previous publications.
Data extraction
According to the inclusion and exclusion criteria, the following information was independently extracted from eligible studies by two investigators (Yu Tian and JingJing Song): name of the first author, year of publication, ethnicity (such as Asian or Caucasian), genotype method, sample sizes of cases and controls, genotype frequency of cases and controls, and P-value for HWE of controls. Any disagreements were figured out by discussion until a consensus was achieved.
Statistical analysis
HWE was assessed with the chi-square test using the genotypes of the controls in each study. The pooled ORs with the corresponding 95% CIs were used to investigate whether there existed associations between VEGF polymorphisms and the risk of RCC based on allele contrast, homozygote, heterozygote, dominant, and recessive models as follows: 1) rs3025039(+936C/T) polymorphism (T vs C, TT vs CC, CT vs CC, TT+CT vs CC, and TT vs CT+CC, respectively); 2) rs699947(−2578C/A) polymorphism (A vs C, AA vs CC, CA vs CC, AA+CA vs CC, and AA vs CA+CC, respectively); 3) rs10434(1612G/A) polymorphism (G vs A, GG vs AA, GA vs AA, GG+GA vs AA, and GG vs GA+AA, respectively); 4) rs1570360(−1154G/A and −1156G/A) polymorphism (A vs G, AA vs GG, GA vs GG, AA+GA vs GG, and AA vs GA+GG, respectively); and 5) rs2010963(+405C/G and −634G/C) polymorphism (C vs G, CC vs GG, CG vs GG, CC+GC vs GG, and CC vs GC+GG, respectively).
Heterogeneity was quantifiably measured using Cochran’s Q test and I 2 statistic combined with the corresponding P-value.34 If I2 value exceeded 50% or P<0.10, the heterogeneity was significant, and a random-effects model was employed. Otherwise, the ORs were calculated by fixed-effects model (I2<50% or P>0.10).35 To consider potential ethnicity variation, subgroup analysis was conducted on the basis of ethnicity. To test the stability of the result, we performed the sensitivity analysis by excluding one study in turn. Visual inspection of asymmetry in Begg’s funnel plots and Egger’s test was carried out to assess the potential publication bias. Those statistical analyses or data syntheses were calculated using Review Manager 5.2 and STATA version 11. All P-values were two sided, and P<0.05 was defined to be statistically significant.
Results
Study characteristics
The initial search yielded 1,304 references. Of these articles, we excluded 1,250 studies based on titles and/or abstracts. Fifty-four texts were then reviewed for a further evaluation. In accordance with the study inclusion criteria, 46 articles were excluded for different reasons: 30 studies were about VEGF as a predictive and/or prognostic biomarker; two studies were review; 12 studies were obviously irrelevant; one study was a meta-analysis, and one study was about animal experiment. Finally, eight studies were included in this meta-analysis.25–32 A flow diagram about the literature search and study selection process is presented in Figure 1.
Figure 1.
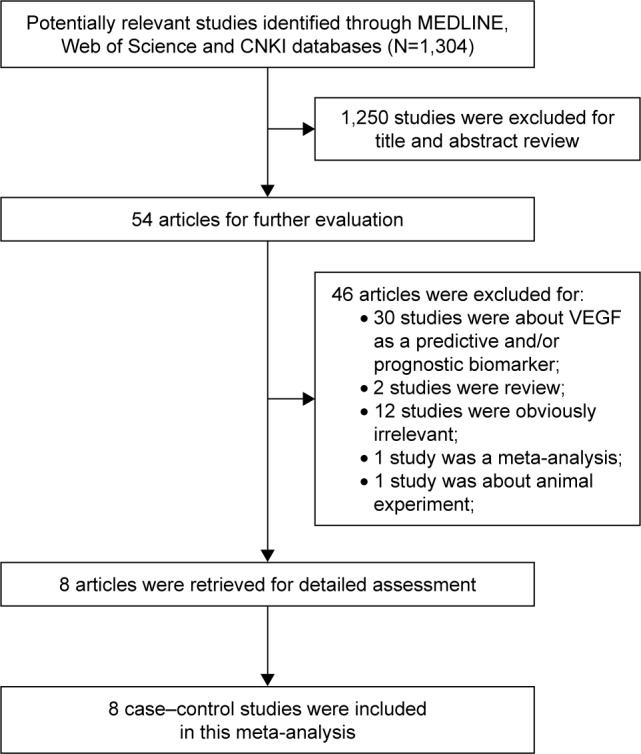
Flow diagram of the study selection process.
Abbreviations: CNKI, Chinese National Knowledge Infrastructure; VEGF, vascular endothelial growth factor.
A total of 4,706 subjects were involved in this meta-analysis, including 1,936 RCC patients and 2,770 healthy controls. For the rs3025039(+936C/T) polymorphism, six literatures with a total number of 1,450 cases and 2,343 controls were included.25–27,29,30,32 For the rs699947(−2578C/A) polymorphism, five literatures with a total number of 1,397 cases and 2,102 controls were included.25–27,29,31 For the rs10434(1612G/A) polymorphism, four literatures with a total number of 985 cases and 1,861 controls were included.25,26,29,32 For the rs2010963(+405C/G and −634G/C) polymorphism, five literatures with a total number of 1,305 cases and 2,198 controls were included.25,27,29,30 While for the rs1570360(−1154G/A and −1156G/A) polymorphism, only three literatures with a total number of 660 cases and 1,055 controls were included.25,28,30 Overall, three of these included studies were conducted in Caucasian population27,28,30 and the other five studies were conducted in Asian population.25,26,29,31,32 DNA samples that are used to detect the VEGF genetic polymorphisms were extracted from blood in all included studies. Methods used for genotyping of the eight case–control studies include Taqman, polymerase chain reaction (PCR), and PCR-restriction fragment length polymorphism (PCR-RFLP). Genotype distributions among the controls of all studies were consistent with HWE. The detailed study characteristics of the included studies are displayed in Table 1.
Table 1.
Baseline characteristics of the included studies in the meta-analysis
| Gene polymorphism | Author (reference) | Year | Ethnicity | Genotype method | Case | Control | Genotype*
|
P-value for HWE |
|---|---|---|---|---|---|---|---|---|
| Case control | ||||||||
| rs3025039 | ||||||||
| +936C/T | Abe et al32 | 2002 | Asian | PCR-RFLP | 145 | 145 | 7/4/97 3/52/90 |
0.146 |
| +936C/T | Bruyere et al30 | 2010 | Caucasian | PCR | 51 | 202 | 1/17/29 2/53/141 |
0.124 |
| +936C/T | Saenz-Lopez et al27 | 2013 | Caucasian | TaqMan | 216 | 280 | 2/57/156 7/73/200 |
0.912 |
| +936C/T | Lu et al29 | 2015 | Asian | PCR-RFLP | 412 | 824 | 59/91/262 105/166/554 |
0.124 |
| +936C/T | Shen et al26 | 2015 | Asian | PCR-RFLP | 360 | 360 | 55/81/224 46/73/240 |
<0.001 |
| +936C/T | Xian et al25 | 2015 | Asian | PCR-RFLP | 266 | 532 | 69/127/70 100/236/196 |
0.06 |
| rs699947 | ||||||||
| −2578C/A | Ajaz et al31 | 2011 | Asian | PCR-RFLP | 143 | 106 | 32/81/30 21/41/44 |
0.053 |
| −2578C/A | Saenz-Lopez et al27 | 2013 | Caucasian | TaqMan | 216 | 280 | 48/114/54 53/142/77 |
0.388 |
| −2578C/A | Lu et al29 | 2015 | Asian | PCR-RFLP | 412 | 824 | 67/174/171 95/332/397 |
0.06 |
| −2578C/A | Shen et al26 | 2015 | Asian | PCR-RFLP | 360 | 360 | 61/149/150 41/141/178 |
0.11 |
| −2578C/A | Xian et al25 | 2015 | Asian | PCR-RFLP | 266 | 532 | 48/119/99 64/225/243 |
0.29 |
| rs10434 | ||||||||
| +1612G/A | Abe et al32 | 2002 | Asian | PCR-RFLP | 145 | 145 | 113/31/1 109/33/3 |
0.788 |
| +1612G/A | Lu et al29 | 2015 | Asian | PCR-RFLP | 412 | 824 | 172/191/49 365/375/85 |
0.43 |
| +1612G/A | Shen et al26 | 2015 | Asian | PCR-RFLP | 360 | 360 | 152/170/39 166/164/30 |
0.23 |
| +1612G/A | Xian et al25 | 2015 | Asian | PCR-RFLP | 266 | 532 | 113/123/30 248/243/41 |
0.08 |
| rs1570360 | ||||||||
| −1154G/A | Ricketts et al28 | 2009 | Caucasian | PCR | 343 | 321 | 134/143/47 146/130/38 |
0.281 |
| −1154G/A | Bruyere et al30 | 2010 | Caucasian | PCR | 51 | 202 | 27/17/5 94/83/25 |
0.322 |
| −1156G/A | Xian et al25 | 2015 | Asian | PCR-RFLP | 266 | 532 | 39/112/115 80/220/232 |
0.18 |
| rs2010963 | ||||||||
| +405C/G | Bruyere et al30 | 2010 | Caucasian | PCR | 51 | 202 | 15/25/8 86/92/20 |
0.522 |
| +405C/G | Saenz-Lopez et al27 | 2013 | Caucasian | TaqMan | 216 | 280 | 101/93/20 129/118/32 |
0.528 |
| −634G/C | Lu et al29 | 2015 | Asian | PCR-RFLP | 412 | 824 | 139/194/79 299/377/148 |
0.13 |
| −634G/C | Shen et al26 | 2015 | Asian | PCR-RFLP | 360 | 360 | 121/170/69 134/163/63 |
0.27 |
| − 634G/C | Xian et al25 | 2015 | Asian | PCR-RFLP | 266 | 532 | 30/132/104 49/256/227 |
0.06 |
Notes:
Genotype: for rs3025039(+936C/T), TT/CT/CC; for rs699947(−2578C/A), AA/AC/CC; for rs10434(+1612G/A), GG/GA/AA; for rs1570360(−1154G/A and −1156G/A), GG/GA/AA; for rs2010963(+405C/G and −634G/C), GG/GC/CC.
Abbreviations: HWE, Hardy–Weinberg equilibrium; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism.
Meta-analysis results
On the whole, the pooled ORs and 95% CIs of RCC were considered under allele contrast, homozygous, heterozygous, dominant, and recessive genetic models. A summary of our meta-analysis results for the five studied polymorphisms and RCC risk is provided in Table 2.
Table 2.
Meta-analysis results for the five studied polymorphisms and RCC risk
| Gene polymorphism | Inherited model | Heterogeneity test
|
Analysis model | Pooled OR (95% CI) | P-value | |
|---|---|---|---|---|---|---|
| P for Q test | I2 (%) | |||||
| rs3025039 | Allele contrast (T vs C) | 0.02 | 61 | REM | 1.13 (0.93, 1.37) | 0.22 |
| Homozygous (TT vs CC) | 0.24 | 25 | FEM | 1.38 (1.11, 1.72) | 0.004 | |
| Heterozygous (CT vs CC) | 0.24 | 25 | FEM | 1.17 (1.00, 1.37) | 0.06 | |
| Dominant (TT+CT vs CC) | 0.15 | 39 | FEM | 1.21 (1.05, 1.39) | 0.01 | |
| Recessive (TT vs CT+CC) | 0.45 | 0 | FEM | 1.28 (1.04, 1.57) | 0.02 | |
| rs699947 | Allele contrast (A vs C) | 0.60 | 0 | FEM | 1.31 (1.19, 1.45) | <0.00001 |
| Homozygous (AA vs CC) | 0.77 | 0 | FEM | 1.69 (1.37, 2.07) | <0.00001 | |
| Heterozygous (CA vs CC) | 0.11 | 47 | FEM | 1.31 (1.12, 1.52) | 0.0006 | |
| Dominant (AA+CA vs CC) | 0.19 | 35 | FEM | 1.39 (1.21, 1.61) | <0.00001 | |
| Recessive (AA vs CA+CC) | 0.78 | 0 | FEM | 1.43 (1.19, 1.73) | 0.0002 | |
| rs10434 | Allele contrast (G vs A) | 0.60 | 0 | FEM | 0.89 (0.80, 1.00) | 0.06 |
| Homozygous (GG vs AA) | 0.53 | 0 | FEM | 0.75 (0.57, 0.98) | 0.03 | |
| Heterozygous (GA vs AA) | 0.65 | 0 | FEM | 0.82 (0.63, 1.07) | 0.15 | |
| Dominant (GG+GA vs AA) | 0.57 | 0 | FEM | 0.79 (0.61, 1.01) | 0.06 | |
| Recessive (GG vs GA+AA) | 0.57 | 0 | FEM | 0.89 (0.77, 1.04) | 0.14 | |
| rs1570360 | Allele contrast (A vs G) | 0.26 | 25 | FEM | 1.05 (0.90, 1.22) | 0.55 |
| Homozygous (AA vs GG) | 0.47 | 0 | FEM | 1.10 (0.81, 1.50) | 0.55 | |
| Heterozygous (GA vs GG) | 0.40 | 0 | FEM | 1.07 (0.83, 1.37) | 0.60 | |
| Dominant (AA+GA vs GG) | 0.29 | 19 | FEM | 1.08 (0.86, 1.36) | 0.52 | |
| Recessive (AA vs GA+GG) | 0.64 | 0 | FEM | 1.04 (0.81, 1.32) | 0.78 | |
| rs2010963 | Allele contrast (C vs G) | 0.21 | 31 | FEM | 1.04 (0.94, 1.15) | 0.49 |
| Homozygous (CC vs GG) | 0.23 | 28 | FEM | 1.07 (0.86, 1.32) | 0.56 | |
| Heterozygous (CG vs GG) | 0.68 | 0 | FEM | 1.09 (0.92, 1.28) | 0.32 | |
| Dominant (CC+CG vs GG) | 0.41 | 0 | FEM | 1.09 (0.93, 1.27) | 0.29 | |
| Recessive (CC vs CG+GG) | 0.44 | 0 | FEM | 1.00 (0.84, 1.19) | 1.00 | |
Note: P-value of overall effect.
Abbreviations: OR, odds ratio; CI, confidence interval; REM, random effects model; FEM, fixed-effects model; RCC, renal cell carcinoma.
Pooled effects for the rs3025039(+936C/T) polymorphism and RCC risk
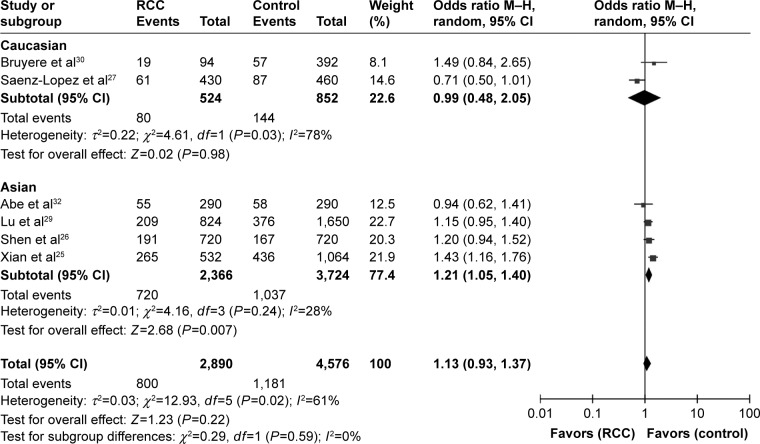
The pooled results of all analyses showed that a significant association between the rs3025039(+936C/T) polymorphism and RCC risk was demonstrated under homozygous, dominant, and recessive models (TT vs CC: OR =1.38, 95% CI =1.11–1.72, I2=25%, P =0.004; CT+TT vs CC: OR =1.21, 95% CI =1.05–1.39, I2=39%, P=0.001; TT vs CC+CT: OR =1.28, 95% CI =1.04–1.57, I2=0%, P=0.02). And there was no relationship in the other two models (T vs C: OR =1.13, 95% CI =0.93–1.37, I2=61%, P=0.61; CT vs CC: OR =1.17, 95% CI =1.00–1.37, I2=25%, P=0.06). According to the heterogeneity (I2.50%) in the allele contrast model, we performed a subgroup analysis of ethnicity. The result of this subgroup analysis was shown as follows: Caucasian (T vs C: OR =0.99, 95% CI =0.48–2.05, I2=78%, P=0.98) and Asian (T vs C: OR =1.21, 95% CI =1.05–1.40, I2=28%, P=0.007). We surprisingly found that rs3025039(+936C/T) polymorphism was significantly associated with RCC for Asians in the allele contrast model (Table 2; Figure 2). Furthermore, no publication bias was found, showing that the results are statistically robust (T vs C, P=0.468; TT vs CC, P=0.877; CT vs CC, P=0.765; TT+CT vs CC, P=0.707; TT vs CT+CC, P=0.881).
Figure 2.
Forest plot of the association between the rs3025039(+936C/T) polymorphism and RCC risk in the allele contrast model (T vs C).
Abbreviations: RCC, renal cell carcinoma; CI, confidence interval.
Pooled effects for the rs699947(−2578C/A) polymorphism and RCC risk
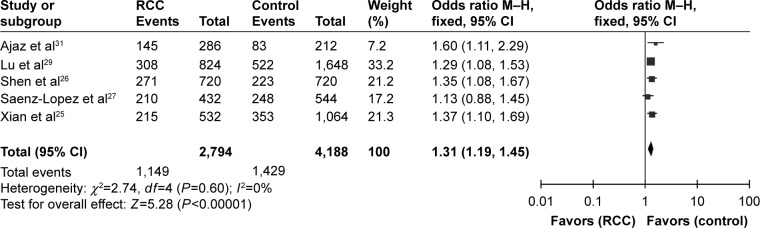
When it came to the rs699947(−2578C/A) polymorphism, we detected significant associations under all genetic models (A vs C: OR =1.31, 95% CI =1.19–1.45, I2=0%, P<0.00001; AA vs CC: OR =1.69, 95% CI =1.37–2.07, I2=0%, P<0.00001; CA vs CC: OR =1.31, 95% CI =1.12–1.52, I2=47%, P=0.0006; CA+AA vs CC: OR = 1.39, 95% CI = 1.21–1.61, I2=35%, P<0.00001; AA vs CC+CA: OR =1.43, 95% CI =1.19–1.73, I2=0%, P=0.0002), as indicated in Table 2 and Figure 3. Funnel plot symmetry was performed to estimate publication bias, and the results were validated by Egger’s test (A vs C, P=0.520; AA vs CC, P=0.642; CA vs CC, P=0.142; AA+CA vs CC, P=0.209; AA vs CA+CC, P=0.305).
Figure 3.
Forest plot of the association between the rs699947(−2578C/A) polymorphism and RCC risk in the allele contrast model (A vs C).
Abbreviations: RCC, renal cell carcinoma; CI, confidence interval.
Pooled effects for the rs10434(1612G/A) polymorphism and RCC risk
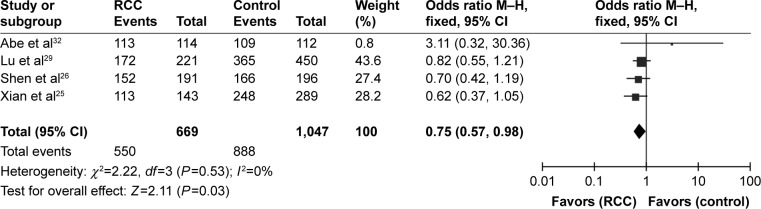
The meta-analysis results showed that the rs10434(1612G/A) polymorphism increased the risk of RCC in the homozygous model (GG vs AA: OR =0.75, 95% CI =0.57–0.98, I2=0%, P=0.03), but no significant associations were found in the heterozygous and dominant models where the results were as follows: G vs A: OR =0.89, 95% CI =0.8–1.00, I2=0%, P=0.06; GA vs AA: OR =0.82, 95% CI =0.63–1.07, I2=0%, P=0.15; GA+GG vs AA: OR =0.79, 95% CI =0.61–1.01, I2=0%, P=0.06; GG vs GA+AA: OR =0.89, 95% CI =0.77–1.04, I2=0%, P=0.14 (Table 2; Figure 4). No publication bias was detected in the funnel plot and Egger’s test (G vs A, P=0.274; GG vs AA, P=0.348; GA vs AA, P=0.355; GG+GA vs AA, P=0.331; GG vs GA+AA, P=0.235).
Figure 4.
Forest plot of the association between the rs10434(+1612G/A) polymorphism and RCC risk in the homozygous model (GG vs AA).
Abbreviations: RCC, renal cell carcinoma; CI, confidence interval.
Pooled effects for the rs1570360(−1154G/A and −1156G/A) polymorphism and RCC risk
No associations were observed between rs1570360(−1154G/A and −1156G/A) polymorphism and RCC risk in any genetic models. The results were as follows: A vs G: OR =1.05, 95% CI =0.9–1.22, I2=25%, P=0.55; AA vs GG: OR =1.10, 95% CI =0.81–1.50, I2=0%, P=0.55; GA vs GG: OR =1.07, 95% CI =0.83–1.37, I2=0%, P=0.60; GA+AA vs GG: OR =1.08, 95% CI =0.86–1.36, I2= 19%, P=0.52; AA vs GG+GA: OR =1.04, 95% CI =0.81–1.32, I2=0%, P =0.78. Moreover, the results of Egger’s test suggested no publication (A vs G, P=0.487; AA vs GG, P=0.557; GA vs GG, P=0.272; AA+GA vs GG, P=0.499; AA vs GA+GG, P=0.920).
Pooled effects for the rs2010963(+405C/G and −634G/C) polymorphism and RCC risk
Our meta-analysis did not show any significant correlations between the rs2010963(+405C/G and −634G/C) polymorphism and RCC risk in all the genetic models in the total population (C vs G: OR =1.04, 95% CI =0.94–1.15, I2=31%, P=0.49; CC vs GG: OR =1.07, 95% CI =0.86–1.32, I2=28%, P=0.56; GC vs GG: OR =1.09, 95% CI =0.92–1.28, I2=0%, P=0.32; GC+CC vs GG: OR =1.09, 95% CI =0.93–1.27, I2=0%, P=0.29; CC vs GC+GG: OR =1.00, 95% CI =0.84–1.19, I2 =0%, P=1.00). Besides, the results of Egger’s test were as follows: C vs G, P=0.587; CC vs GG, P=0.819; CG vs GG, P=0.180; CC+GC vs GG, P=0.909; CC vs GC+GG, P=0.740.
Sensitive analysis
To assess whether the single study influenced the pooled results, we conducted the sensitivity analysis by excluding each single study one by one. The results of pooled ORs indicated that the overall significance of the ORs was not altered by any single study for the rs3025039(+936C/T) polymorphisms (Figure 5). It is demonstrated that the pooled result of our study was of relatively high reliability and stability.
Figure 5.
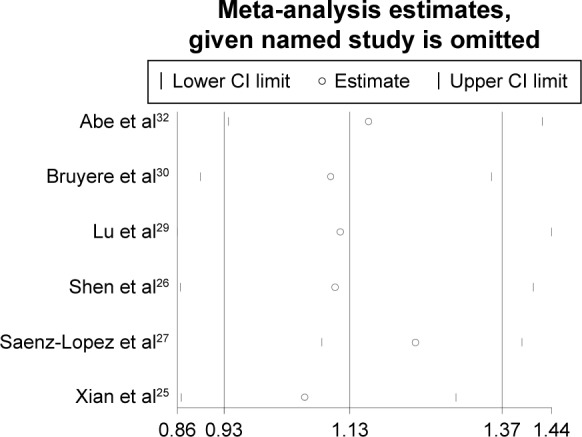
Sensitivity analysis for the associations between rs3025039(+936C/T) polymorphism and RCC risk.
Abbreviations: RCC, renal cell carcinoma; CI, confidence interval.
Publication bias
No significant publication bias was detected for the studies on rs3025039, rs699947, rs10434, rs1570360, and rs2010963 polymorphisms. On one hand, the shapes of the funnel plots were symmetrical by visual inspection. On the other hand, the P-value of all genetic models exceeded 0.05 in the Egger’s test.
Discussion
VEGF is one of the most important cytokines in angiogenesis, which has attracted extensive attention. High expression of VEGF in the primary tumor has been found in certain types of malignant tumors such as breast cancer, gastric cancer, and glioma.36–38 The high expression of VEGF was demonstrated by the vast majority of fresh frozen tumor samples in RCC as well,39 and it was confirmed that von Hippel–Lindau (VHL) tumor suppressor gene inactivation could lead to VEGF overexpression in the majority of clear cell RCC tumors.40
A large number of studies are focusing on the associations among protein expression/activity, gene variants (particularly SNPs), and tumor formation. In 2002, Abe et al32 conducted the first case–control study of the association between SNPs in the 3′-UTR of the VEGF gene and RCC in the Japanese population and indicated that the C702T, C936T, and G1612A polymorphisms in the 3′-UTR of the VEGF gene are not associated with the risk of RCC. Since then, a series of case–control studies have been conducted, but the available results are inconsistent and inconclusive. In 2013, Zhang et al33 conducted a meta-analysis with only five case–control studies and observed that polymorphisms or haplotypes in the VEGF gene did not modify the risk of RCC. To date, three additional studies have revealed the connections between VEGF polymorphisms and RCC risk.25,26,29 Our present meta-analysis aims to update the previous meta-analysis and to provide a more reliable and credible conclusion on the associations between five common functional polymorphisms (rs3025039, rs699947, rs10434, rs1570360, and rs2010963) in VEGF gene and RCC susceptibility.
In our meta-analysis, eight eligible case–control studies were included with a total number of 1,936 RCC patients and 2,770 healthy controls. When all the eligible studies were pooled into the meta-analysis, the results showed that rs699947(−2578C/A) and rs3025039(+936C/T) polymorphisms significantly increased the risk of RCC. One possible reason for these inconsistent results could be explained that rs699947(−2578C/A) and rs3025039(+936C/T) polymorphisms were more influential than other SNPs on VEGF gene expression and protein production and indicate that various polymorphisms yield various effects on gene function, even when they are located at the same unit. Furthermore, in the subgroup analysis by ethnicity for the rs3025039(+936C/T) polymorphism, a statistically significantly increased RCC risk in Asians was found in the allele contrast model of rs3025039(+936C/T) polymorphism. For ethnicity variation, the possible reasons may be great disparities in common VEGF gene polymorphisms and the natural selection and genetic drift that are most likely to influence the risk of RCC. For one thing, most of our included studies were conducted in Asian population, and only three studies were performed in Caucasian population. For the other thing, the small sample size of some included studies could also lead to the discrepancy. In addition, we explain that the reasons why there was no relationship between rs3025039 polymorphism and RCC risk in the heterozygous model might be the insufficient studies and limited sample sizes. Therefore, we need further studies conducted in more different ethnicities, and experiments of enough samples are necessary to evaluate the associations between VEGF polymorphisms and RCC risk. As to rs10434(1612G/A) polymorphism, a significant association was observed in a homozygous model although all ORs in the separate studies were not statistically significant, which may due to the new research data and a large sample size.
Some possible limitations of our meta-analysis should be taken into consideration. First, even though a comprehensive search strategy was applied to determine eligible studies, it was probable that some eligible studies were not brought into, which could make our results have bias. Second, only eight eligible studies were collected, so the limited number of studies may have an influence on the analysis of the correlation between the five VEGF polymorphisms and the risk of RCC. Third, as a retrospective study, the meta-analysis may encounter recall or selection bias; thereby it is a possible reason to affect the reliability of our results. Fourth, for the rs10434(1612G/A) polymorphism, all studies included were conducted in Asian population. With a single Asian race was examined, ethnicity bias may exist and the conclusion may not be the same with other races. Fifth, various risk factors are crucial determining factors of tumor recurrence, such as smoking, hypotension, and other genetic factors. Thus, the unadjusted databases in this meta-analysis could not explain the inherent pathogenic mechanisms clearly.
Accumulating evidence indicates that the pathogenic mechanisms of many diseases including RCC were determined by a unique combination of endogenous and exogenous factors, resulting in various molecular and pathological subtypes of this disease.41 Molecular pathological epidemiology (MPE) is an integrative field that is based on the disease continuum theory and the unique disease principle, which can give clues to analyze the risk factors of diseases.42,43 For example, an MPE study assessed the joint effects of genetic polymorphisms in the mammalian target of rapamycin pathway and epidemiologic risk factors on RCC risk and showed that obesity played a vital role in enhancing RCC risk during the life course.44 Together, the MPE research can provide etiologic and pathogenic insights to precision medicine and public health, which will be applied to reveal the carcinogenic mechanism as a future direction of the field.
In spite of these potential limitations, this meta-analysis has its advantages and innovations. First, extensive search strategy and manual search allowed the eligible studies included as possible as we can. Second, compared to previous meta-analysis, our meta-analysis contained new research data and a large sample size. Third, according to the existence of heterogeneity, we conducted a subgroup analysis in our meta-analysis, which is not performed in previous meta-analysis. Therefore, we can consider that the conclusions are credible and reliable to explore the relationship between the VEGF polymorphisms and RCC risk.
Conclusion
Our meta-analysis reevaluates the relationship between VEGF polymorphisms and RCC risk and suggests that there is a significant association between rs699947(−2578C/A)/rs3025039(+936C/T) polymorphism and RCC. Moreover, our study also demonstrates that the allele contrast model of rs3025039 polymorphism is likely to be associated with risk of RCC in Asian population and the homozygous model of rs10434 polymorphism is statistically significantly relevant to elevated RCC risk, while no significant association is detected between rs1570360(−1154G/A and −1156G/A) or rs2010963(+405C/G and −634G/C) polymorphism and risk of RCC. Considering the limitations mentioned earlier, further well-designed studies including more different ethnicities and larger sample sizes are needed to confirm our results and explore these associations.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Mickley A, Kovaleva O, Kzhyshkowska J, Gratchev A. Molecular and immunologic markers of kidney cancer-potential applications in predictive, preventive and personalized medicine. EPMA J. 2015;6:20. doi: 10.1186/s13167-015-0042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King SC, Pollack LA, Li J, King JB, Master VA. Continued increase in incidence of renal cell carcinoma, especially in young patients and high grade disease: United States 2001 to 2010. J Urol. 2014;191(6):1665–1670. doi: 10.1016/j.juro.2013.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns P. Renal cell carcinoma. Cancer Biomark. 2010;9(1–6):461–473. doi: 10.3233/CBM-2011-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan D, Xu L, Liu H, et al. Interleukin-11 receptor predicts post-operative clinical outcome in patients with early-stage clear-cell renal cell carcinoma. Jpn J Clin Oncol. 2015;45(2):202–209. doi: 10.1093/jjco/hyu194. [DOI] [PubMed] [Google Scholar]
- 5.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373(9669):1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 6.Hunt JD, van der Hel OL, McMillan GP, Boffetta P, Brennan P. Renal cell carcinoma in relation to cigarette smoking: meta-analysis of 24 studies. Int J Cancer. 2005;114(1):101–108. doi: 10.1002/ijc.20618. [DOI] [PubMed] [Google Scholar]
- 7.Yuan JM, Castelao JE, Gago-Dominguez M, Yu MC, Ross RK. Tobacco use in relation to renal cell carcinoma. Cancer Epidemiol Biomarkers Prev. 1998;7(5):429–433. [PubMed] [Google Scholar]
- 8.van Dijk BA, Schouten LJ, Kiemeney LA, Goldbohm RA, van den Brandt PA. Relation of height, body mass, energy intake, and physical activity to risk of renal cell carcinoma: results from the Netherlands Cohort Study. Am J Epidemiol. 2004;160(12):1159–1167. doi: 10.1093/aje/kwh344. [DOI] [PubMed] [Google Scholar]
- 9.Bjorge T, Tretli S, Engeland A. Relation of height and body mass index to renal cell carcinoma in two million Norwegian men and women. Am J Epidemiol. 2004;160(12):1168–1176. doi: 10.1093/aje/. [DOI] [PubMed] [Google Scholar]
- 10.McLaughlin JK, Chow WH, Mandel JS, et al. International renal-cell cancer study. VIII. Role of diuretics, other anti-hypertensive medications and hypertension. Int J Cancer. 1995;63(2):216–221. doi: 10.1002/ijc.2910630212. [DOI] [PubMed] [Google Scholar]
- 11.McCredie M, Pommer W, McLaughlin JK, et al. International renal-cell cancer study. II. Analgesics. Int J Cancer. 1995;60(3):345–349. doi: 10.1002/ijc.2910600312. [DOI] [PubMed] [Google Scholar]
- 12.Meng FD, Ma P, Sui CG, Tian X, Jiang YH. Association between cytochrome P450 1A1 (CYP1A1) gene polymorphisms and the risk of renal cell carcinoma: a meta-analysis. Sci Rep. 2015;5:8108. doi: 10.1038/srep08108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Wang Z, Wei M, Ren Y, et al. miR149 rs71428439 polymorphism and risk of clear cell renal cell carcinoma: a case-control study. Tumour Biol. 2014;35(12):12127–12130. doi: 10.1007/s13277-014-2517-5. [DOI] [PubMed] [Google Scholar]
- 14.Meng F, Ma P, Sui C, et al. The association between VDR polymorphisms and renal cell carcinoma susceptibility: a meta-analysis. Tumour Biol. 2014;35(6):6065–6072. doi: 10.1007/s13277-014-1803-6. [DOI] [PubMed] [Google Scholar]
- 15.Ma N, Li LW, Cheng JL. Predictive value of vascular endothelial growth factor polymorphisms on the clinical outcome of renal cell carcinoma patients. Oncol Lett. 2015;9(2):651–656. doi: 10.3892/ol.2014.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang SM, Huang CY, Shiue HS, et al. Joint effect of urinary total arsenic level and VEGF-A genetic polymorphisms on the recurrence of renal cell carcinoma. PLoS One. 2015;10(12):e0145410. doi: 10.1371/journal.pone.0145410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph RW, Parasramka M, Eckel-Passow JE, et al. Inverse association between programmed death ligand 1 and genes in the VEGF pathway in primary clear cell renal cell carcinoma. Cancer Immunol Res. 2013;1(6):378–385. doi: 10.1158/2326-6066.CIR-13-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar B, Ray KB, Reddy GV, et al. VEGF-C differentially regulates VEGF-A expression in ocular and cancer cells; promotes angiogenesis via RhoA mediated pathway. Angiogenesis. 2011;14(3):371–380. doi: 10.1007/s10456-011-9221-5. [DOI] [PubMed] [Google Scholar]
- 19.Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69(suppl 3):4–10. doi: 10.1159/000088478. [DOI] [PubMed] [Google Scholar]
- 20.Badal SA, Aiken WD, Chin SN. Molecular targets and angiogenesis in renal cell carcinoma, a multi-target approach: mini review. Curr Drug Targets. 2016 May 2; doi: 10.2174/1389450117666160502152518. Epub. [DOI] [PubMed] [Google Scholar]
- 21.Vincenti V, Cassano C, Rocchi M, Persico G. Assignment of the vascular endothelial growth factor gene to human chromosome 6p21.3. Circulation. 1996;93(8):1493–1495. doi: 10.1161/01.cir.93.8.1493. [DOI] [PubMed] [Google Scholar]
- 22.Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine. 2000;12(8):1232–1235. doi: 10.1006/cyto.2000.0692. [DOI] [PubMed] [Google Scholar]
- 23.Pasqualetti G, Danesi R, Del Tacca M, Bocci G. Vascular endothelial growth factor pharmacogenetics: a new perspective for anti-angiogenic therapy. Pharmacogenomics. 2007;8(1):49–66. doi: 10.2217/14622416.8.1.49. [DOI] [PubMed] [Google Scholar]
- 24.Bianconi M, Faloppi L, Loretelli C, et al. Angiogenesis genotyping in the selection of first-line treatment with either sunitinib or pazopanib for advanced renal cell carcinoma. Oncotarget. 2016;7(25):37599–37607. doi: 10.18632/oncotarget.9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xian W, Zheng H, Wu WJ. Predictive value of vascular endothelial growth factor polymorphisms on the risk of renal cell carcinomas. Genet Mol Res. 2015;14(3):7634–7642. doi: 10.4238/2015.July.13.8. [DOI] [PubMed] [Google Scholar]
- 26.Shen BL, Qu QS, Miao SZ, Zhang YX. Association between SNPs in vascular endothelial growth factor polymorphisms and risk of renal cell carcinoma: a case-control study. Genet Mol Res. 2015;14(3):11119–11125. doi: 10.4238/2015.September.22.5. [DOI] [PubMed] [Google Scholar]
- 27.Saenz-Lopez P, Vazquez F, Cozar JM, Carretero R, Garrido F, Ruiz-Cabello F. VEGF polymorphisms are not associated with an increased risk of developing renal cell carcinoma in Spanish population. Hum Immunol. 2013;74(1):98–103. doi: 10.1016/j.humimm.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 28.Ricketts C, Zeegers MP, Lubinski J, Maher ER. Analysis of germline variants in CDH1, IGFBP3, MMP1, MMP3, STK15 and VEGF in familial and sporadic renal cell carcinoma. PLoS One. 2009;4(6):e6037. doi: 10.1371/journal.pone.0006037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu G, Dong Y, Zhang Q, Jiao L, Yang S, Shen B. Predictive value of vascular endothelial growth factor polymorphisms on the risk of renal cell carcinomas: a case-control study. Tumour Biol. 2015;36(11):8645–8652. doi: 10.1007/s13277-015-3431-1. [DOI] [PubMed] [Google Scholar]
- 30.Bruyere F, Hovens CM, Marson MN, et al. VEGF polymorphisms are associated with an increasing risk of developing renal cell carcinoma. J Urol. 2010;184(4):1273–1278. doi: 10.1016/j.juro.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 31.Ajaz S, Khaliq S, A bid A, et al. Association of a single-nucleotide polymorphism in the promoter region of the VEGF gene with the risk of renal cell carcinoma. Genet Test Mol Biomarkers. 2011;15(9):653–657. doi: 10.1089/gtmb.2011.0029. [DOI] [PubMed] [Google Scholar]
- 32.Abe A, Sato K, Habuchi T, et al. Single nucleotide polymorphisms in the 3′ untranslated region of vascular endothelial growth factor gene in Japanese population with or without renal cell carcinoma. Tohoku J Exp Med. 2002;198(3):181–190. doi: 10.1620/tjem.198.181. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Li S, Xiao HQ, Hu ZX, Xu YC, Huang Q. Vascular endothelial growth factor gene polymorphisms and renal cell carcinoma: a systematic review and meta-analysis. Oncol Lett. 2013;6(4):1068–1078. doi: 10.3892/ol.2013.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, Xu S, Xu W, et al. Expression of cluster of differentiation 34 and vascular endothelial growth factor in breast cancer, and their prognostic significance. Oncol Lett. 2015;10(2):723–729. doi: 10.3892/ol.2015.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang XL, Ai ZS, Fang JP, Tang RY, Chen XM. Expression of vascular endothelial growth factors (VEGF)-A, -C and -D and their prognostic significance and relationship with angio- and lymphangiogenesis in gastric cancer. Zhonghua Zhong Liu Za Zhi. 2008;30(11):837–843. [PubMed] [Google Scholar]
- 38.Chen W, He D, Li Z, Zhang X, Pan D, Chen G. Overexpression of vascular endothelial growth factor indicates poor outcomes of glioma: a systematic review and meta-analysis. Int J Clin Exp Med. 2015;8(6):8709–8719. [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JS, Kim HS, Jung JJ, Park CS, Lee MC. Expression of vascular endothelial growth factor in renal cell carcinoma and the relation to angiogenesis and p53 protein expression. J Surg Oncol. 2001;77(1):55–60. doi: 10.1002/jso.1066. [DOI] [PubMed] [Google Scholar]
- 40.Neufeld G, Cohen T, Gengrinovitch S, Poltorak Z. Vascular endothelial growth factor (VEGF) and its receptors. FASEB J. 1999;13(1):9–22. [PubMed] [Google Scholar]
- 41.Ogino S, Lochhead P, Chan AT, et al. Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease. Mod Pathol. 2013;26(4):465–484. doi: 10.1038/modpathol.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamada T, Keum N, Nishihara R, Ogino S. Molecular pathological epidemiology: new developing frontiers of big data science to study etiologies and pathogenesis. J Gastroenterol. 2017;52(3):265–275. doi: 10.1007/s00535-016-1272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogino S, Nishihara R, VanderWeele TJ, et al. Review article: the role of molecular pathological epidemiology in the study of neoplastic and non-neoplastic diseases in the era of precision medicine. Epidemiology. 2016;27(4):602–611. doi: 10.1097/EDE.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shu X, Lin J, Wood CG, Tannir NM, Wu X. Energy balance, polymorphisms in the mTOR pathway, and renal cell carcinoma risk. J Natl Cancer Inst. 2013;105(6):424–432. doi: 10.1093/jnci/djt005. [DOI] [PMC free article] [PubMed] [Google Scholar]