Abstract
The CXC chemokine receptor 2 (CXCR2) is a G protein coupled receptor mediating interleukin-8 chemotactic signaling and plays an important role in neutrophil mobility and tumor migration. However, efficient CXCR2 signaling requires PDZ domain-mediated scaffolding of signaling complexes at the plasma membrane and functional coupling of the signaling to specific downstream signaling pathways, in which only one PDZ protein has been characterized to interact with CXCR2. Here, we identified five novel CXCR2-binding PDZ-containing proteins, among which PDZ-RhoGEF is of particular interest because this PDZ and RGS-containing guanine nucleotide exchange factor (GEF) is also involved in cell signaling and mobility. To reveal the molecular basis of the interaction, we solved the crystal structure of PDZ-RhoGEF PDZ domain in complex with the CXCR2 C-terminal PDZ binding motif. The structure reveals that the PDZ–CXCR2 binding specificity is achieved by numerous hydrogen bonds and hydrophobic contacts with the last four CXCR2 residues contributing to specific interactions. Structural comparison of CXCR2-binding PDZ domains and PDZ-RhoGEF PDZ bound with different ligands reveals PDZ- and ligand-specific interactions that may underlie the ability of promiscuous CXCR2 binding by different PDZ domains and PDZ binding promiscuity. The structure also reveals an unexpected asymmetric disulfide bond-linked PDZ dimer that allows simultaneous parallel binding of CXCR2 to two PDZ domains. This study provides not only the structural basis for PDZ-mediated CXCR2–PDZ-RhoGEF interaction, but also a new mode of PDZ dimerization, which both could prove valuable in understanding signaling complex scaffolding in CXCR2 signaling and coupling to specific signaling pathways.
Keywords: CXCR2, PDZ-RhoGEF, X-ray crystallography, Scaffold protein, Dimerization, Chemokine signaling
1. Introduction
CXCR2 is a G protein coupled receptor important for cellular mobility and chemotaxis through activation of calcium mobilization and actin polymerization [1]. CXCR2 is central to neutrophil migration to sites of inflammation and involved in wound healing and angiogenesis [1]. It has been shown that disrupting CXCR2 function plays a central role in multiple inflammatory diseases including rheumatoid arthritis, acute respiratory distress syndrome, septic shock, and chronic obstructive pulmonary disease, likely due to the result of excessive release of neutrophils from the bone marrow [2]. CXCR2 also plays a critical role in many cancers such as lung cancer and pancreatic cancer by promoting tumor invasion and metastasis via autocrine and paracrine effects [3]. Elevated expression of CXCR2 enhances cancer cell proliferation and survival and often correlates with aggressive stages of cancer and poor overall prognosis [3].
CXCR2 directing cell trafficking depends on its ability to bind to ELR-positive CXC chemokines [4]. When binding to a chemokine, CXCR2 is capable of initiating G protein dissociation and inducing downstream signaling cascades that drive cell movement along chemokine concentration gradients. However, swift signaling requires direct and indirect interaction of CXCR2 with other membrane receptors, channels, intracellular scaffold proteins, effectors, and cytoskeletal elements, among which PDZ domain-containing proteins play a central role in efficient signaling by scaffolding the formation of macromolecular complexes at the plasma membrane and functionally coupling chemokine signaling to downstream signaling events [1]. In general, PDZ domains mediate protein interaction by recognizing the C-terminal sequence of target proteins and binding to the targets through a canonically and structurally conserved PDZ peptide-binding pocket [5]. The specificity of the interaction is determined mainly by the residues at positions 0 and −2 of the peptides (position 0 referring to the C-terminal residue), whereas other residues do not significantly contribute to the interaction. As a result, PDZ domains are highly promiscuous capable of binding to multiple ligands; single peptides are capable of binding to distinct PDZ domains. Because of this promiscuity, PDZ-mediated interaction can generate complex and interconnected signaling networks that ensure precise and efficient signal transduction via protein-protein interaction.
However, the canonical ligand-binding of PDZ by itself has a limited capacity to scaffold multiprotein arrays within membrane microdomains, as PDZ domains can only bind to their ligands one at a time. Recent data suggest that PDZ dimerization plays an important role in increasing the scaffolding capacity [6]. PDZ dimerization with the same or different PDZ-containing proteins has been shown to amplify the complexity of interacting proteins in signal transduction networks and provide a mechanism to expand the scaffolding capacity in the assembly of multiprotein complexes. Of note, many PDZ domains can dimerize and 30% out of 150 PDZ domains in the mouse genome has been shown to participate in protein-protein interaction [7]. This suggests PDZ domains have evolved as a dual binding module in facilitating complex formation. Therefore, there has been a considerable interest in elucidating the structural basis of PDZ specificity, promiscuity and dimerization and how they can set up a specific interaction network for proper signaling, the nature of which still remains obscure.
Recently, we showed that the PDZ domains of NHERF1 play a pivotal role in CXCR2 signaling during the formation of macromolecular signaling complexes [1, 3]. NHERF1 scaffolds the interaction between CXCR2 and PLCβ2/3 by simultaneously binding to the C-terminal tail of CXCR2 and PLCβ2/3 and physically connecting them through linked PDZ domains or PDZ dimerization. This resulted in a macromolecular complex essential for coupling CXCR2 activation to PLCβ2/3 signaling cascades in neutrophils and pancreatic cancer cells. Disruption of the interaction effectively abolished chemotaxis and transepithelial migration suggesting a functional importance of PDZ-mediated scaffolding. However, NHERF1 is the only PDZ domain-containing protein identified to date to interact with CXCR2. As a result, there is a limited understanding regarding the molecular mechanism of CXCR2 PDZ-binding promiscuity and how binding to different PDZ domains may interconnect different signaling pathways in CXCR2 signaling.
In this study, we sought to identify additional PDZ domains that could interact with the PDZ motif of CXCR2 and to better understand PDZ binding promiscuity and specificity. We identified several novel CXCR2 binding proteins using a PDZ binding array, among which PDZ-RhoGEF is of particular interest because it is also involved in signaling and cellular mobility. PDZ-RhoGEF is a PDZ and RGS-containing protein and belongs to the guanine nucleotide exchange factors family. It is a protein ubiquitously expressed in humans and involved in initiating the Rho signaling pathway for actin organization and cellular mobility [8]. To understand the molecular mechanism of CXCR2 PDZ motif binding to the PDZ domain of PDZ-RhoGEF, we solved the crystal structure of PDZ-RhoGEF PDZ domain in complex with the CXCR2 C-terminal PDZ binding motif. The structure reveals that the CXCR2 peptide binds to PDZ in an extended conformation with the last four residues making specific side chain contacts. Sequence alignment and structural comparison analyses suggest the sequence- and position-specific interactions determine CXCR2 PDZ-binding promiscuity and specificity. Unexpectedly, we identified a disulfide bond-linked PDZ dimer which enables parallel binding of CXCR2 peptides to the well-separated ligand-binding pockets. This new mode of PDZ dimerization demonstrates structural diversity in PDZ–PDZ interaction and could prove valuable for understanding the complex-scaffolding function of PDZ-RhoGEF in CXCR2 signaling.
2. Materials and Methods
2.1. PDZ domain array screen
CXCR2-binding PDZ domains were screened using TranSignal PDZ Domain Array IV (Panomics) according to the manufacture's instruction. His-tagged C-terminal fragment of human CXCR2 (residues 316–360) was used in the assay screen, which was generated by PCR cloning into pET30 and purified using cobalt resins [1]. The purified CXCR2 was incubated with the PDZ Domain Arrays in blocking buffer for 1 h at room temperature, and washed thrice with wash buffer for 5 min. They were then incubated with Anti-histidine horseradish peroxidase (HRP) conjugate for 1 h at room temperature. Antibody complexes were detected by enhanced chemiluminescence and imaged using BioSpectrum 500 (UVP). The array was repeated twice and similar results were observed.
2.2. PDZ protein expression and purification
The cDNA fragment of human PDZ-RhoGEF PDZ (residues 41–123) was cloned into a pSUMO vector containing an N-terminal His6-SUMO tag. The C-terminal extension TSTTL that corresponds to residues 356–360 of human CXCR2 was included in the reverse primer to create a chimeric clone. The clone was transformed into Escherichia coli BL21 Condon Plus (DE3) cells for protein expression. The transformants were grown to an OD600 of 0.4 at 37°C in LB medium, and then induced with 0.1 mM isopropylthio-β-D-galactoside at 15°C overnight. The cells were harvested and lysed by French Press. The soluble fraction was then subjected to Ni2+ affinity chromatography purification, followed by cleavage the His6-SUMO tag with yeast SUMO Protease 1. PDZ proteins were separated from the cleaved tag by a second Ni2+ affinity chromatography and further purified by size-exclusion chromatography. Finally, the proteins were concentrated to 10 mg/ml in a buffer containing 0.1 M sodium acetate pH 4.8., 150 mM NaCl, 5 mM β-mercaptoethanol (BME), and 5% glycerol.
2.3. Crystallization, data collection and structure determination
Crystals were grown by the hanging-drop vapor-diffusion method by mixing the protein (∼10 mg/ml) with an equal volume of a reservoir solution containing 0.1 M sodium acetate pH 4.6, 0.1 M sodium citrate, 25% PEG8000 at 20°C. Crystals were cryoprotected in a solution containing 20% glycerol. Crystal data were collected at 100 K at the Advanced Photon Source (Argonne, IL) at beamline 21-ID-D and processed and scaled using XDS [9]. Crystals belong to the space group C2221 with four molecules in the asymmetric unit (Supplementary Material). The structure was solved by molecular replacement using PDZ-RhoGEF PDZ–PlexinB2 structure (PDB code: 5E6P) as a search model. Structure modeling was carried out in COOT [10] and refinement was performed with PHENIX [11]. The final model was analyzed and validated with Molprobity [11]. All figures of PDZ–CXCR2 structure were made with PyMOL.
2.4. Protein data bank accession number
Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 5TYT.
3. Results
3.1. CXCR2 C-terminus binds directly to PDZ-RhoGEF PDZ domain
Using a PDZ Array screen (Panomics), the CXCR2 C-terminus was identified to directly bind to the PDZ domains of PDZ-RhoGEF, leukemia associated RhoGEF (LARG), disks large homolog 3 (DLG3-D2), alpha-1-syntrophin (SNA1) and SH3/multiple ankyrin repeat domains protein 1 (SHANK1) (SHK1) (Fig. 1A). DLG3-D2 is the second PDZ domain in synapse-associated protein 102 which serves as a post-synaptic scaffold for glutamate receptor signaling in developing cortical neurons. SNA1 is the only PDZ domain in alpha-1-syntrophin which serves as a scaffold for dystrophin protein complexes in Rac1 signaling in skeletal muscles. SHK1 is the first PDZ domain in SHANK1, a scaffolding protein that clusters neurotransmitter receptors necessary for synapse changes and development. PDZ-RhoGEF and LARG were of interest due to their imperative roles in cellular signaling and mobility [8], an analogous function of CXCR2. PDZ-RhoGEF and LARG possess one PDZ domain and were known to bind to PlexinB1/2, LPA1/2, and insulin-like growth factor-1 receptor in addition to CXCR2 [8]. These together raise an interesting question regarding the mechanism of how CXCR2 is recognized by different PDZ domains and how PDZ-RhoGEF PDZ binds to different substrates.
Figure 1.
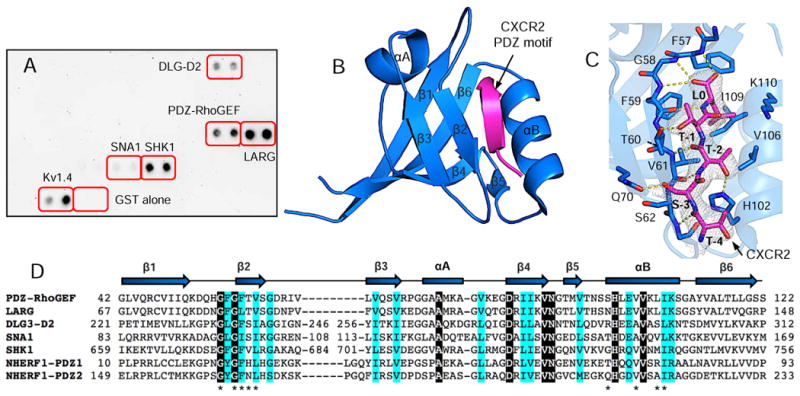
Structure of PDZ-RhoGEF PDZ (rPDZ) in complex with the CXCR2 C-terminal sequence TSTTL. (A) PDZ Array screen of CXCR2-binding PDZ domains. Kv1.4 serves as a positive control for PDZ–peptide binding, and GST alone a negative control. (B) Ribbon diagram of rPDZ–CXCR2 structure. The PDZ is shown in blue and CXCR2 in magenta. Secondary structures are numbered and labeled based on their sequence position. (C) Detailed view of the PDZ ligand-binding site. 2Fo – Fc omit map of CXCR2 peptide was calculated at 2.4 Å and contoured at 1.0 σ. Hydrogen bonds are illustrated as yellow broken lines. (D) Sequence alignment of CXCR2-binding PDZ domains. Identical residues are represented as white on black and similar residues are colored in cyan. Residues important for binding to CXCR2 are indicated by asterisks. The secondary structure elements are labeled according to 1B.
3.2. Binding specificity of the PDZ–CXCR2 interaction
To understand the interaction between PDZ-RhoGEF and CXCR2, we solved the crystal structure of PDZ-RhoGEF PDZ in complex with the C-terminal sequence (TTSTL) of CXCR2. The overall structure of PDZ-RhoGEF PDZ is similar to other PDZ domains [5], consisting of six β strands (β1–β6) and two α-helices (αA and αB) (Fig. 1B). The CXCR2 peptide binds in the cleft between β2 and αB, burying a total solvent-accessible surface area of 507.4 Å2. The binding specificity of the PDZ–CXCR2 interaction is achieved through networks of hydrogen bonds and hydrophobic interactions (Fig. 1C). At the ligand position 0, the side chain of Leu0 is nestled in a deep hydrophobic pocket formed by conserved residues Phe57, Phe59 and Val61 from β2 and Val106 and Ile109 from αB (Fig. 1D). In the pocket, the position of Leu0 is further secured by both a hydrogen bond from its amide nitrogen to the Phe59 carbonyl oxygen and bifurcated hydrogen bonding between the Leu0 carboxylate and the amides of Phe57 and Gly58. Similar interactions have been observed in several other PDZ-mediated complexes which represent the most-conserved binding mode for terminal leucine recognition [5]. Residues at other peptide positions also contribute to the PDZ–CXCR2 complex formation. At position −1, the side chain hydroxyl of Thr-1 forms a hydrogen bond with the Og1 atom of the Thr60 side chain. At position −2, Thr-2 makes one hydrogen bond to the His102 imidazole group and two hydrogen bonds to the highly conserved residue Val61. At the ligand position −3, the interactions with Ser-3 include one hydrogen bond from its side chain hydroxyl to the Oε1 atom of Gln70, and a VDW interaction with the side chain of Ser62. Finally, the peptide residue Thr-4 engages in a main-chain contact with Gly63, but does not participate in any specific side-chain interactions. These observations indicate that the last four residues of CXCR2 contribute to the binding specificity in the PDZ–CXCR2 complex formation.
3.3. CXCR2 and PDZ binding promiscuity
To gain insight into promiscuous CXCR2 binding by different PDZ domains, we compared the structures of all available PDZ domains in complex with CXCR2, including NHERF1 PDZ1, NHERF1 PDZ2 and current PDZ-RhoGEF PDZ (rPDZ) [5, 12]. We also compared the rPDZ– CXCR2 structure to the structure of rPDZ in complex with a PlexinB2 peptide in order to understand PDZ binding promiscuity. These liganded PDZ structures are very similar with pairwise root-mean-square differences (RMSDs) ranging from 0.47 to 0.87 Å for entire Cα atoms (Fig. 2A). The main chains of the bound peptides superimpose well, as do their relative spatial positions to the conserved PDZ motifs. Additionally, the ligand recognition modes at the peptide positions 0 and −2 are virtually indistinguishable, characterized by structurally similar binding sites composed with highly conserved residues (Fig. 2B). This observation is consistent with previous evidence that the 0 and −2 residues of the ligand are critical for determining the binding specificity and affinity of PDZ–peptide interaction [5].
Figure 2.
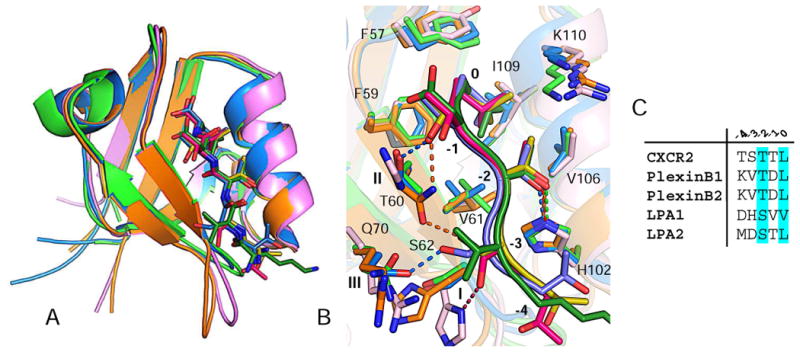
Structural comparison between PDZ–ligand complexes. (A) Superposition of rPDZ– CXCR2 (blue), rPDZ–PlexinB2 (green), NHERF1 PDZ1–CXCR2 (pink) and NHERF1 PDZ2– CXCR2 (orange). PDZ domains are represented by ribbon, and ligand residues are displayed as sticks. (B) Superposition of PDZ ligand-binding sites. Colors are identical from 2A. (C) Sequence alignment of last five residues of known rPDZ binding targets.
Large differences were found in ligand recognition at the peptide positions −1 and −3. In CXCR2-binding PDZ domains, residues that recognize these two positions are not conserved; in fact, the residues that recognize the −3 position are not even structurally equivalent (Fig. 1D and 2B). At position −1, the binding conformation of Thr-1 is nearly identical in different PDZ structures. The side chain of Thr-1 is oriented towards the same direction facing a residue equivalent to rPDZ Thr60. As a result, Thr-1 is recognized by different residues from the equivalent position. In NHERF1 PDZ1, the side chain hydroxyl of Thr-1 is stacked by the imidazole ring of a histidine residue. In NHERF1 PDZ2, the side chain hydroxyl of Thr-1 forms a hydrogen bond with the side chain nitrogen of an asparagine. In current rPDZ structure, the equivalent hydroxyl group forms a hydrogen bond to the side chain of Thr60. In SHK1 and DLG3-D2, Thr-1 may interact with a valine and serine respectively (Fig. 1D). This demonstrates Thr-1 can be recognized by different residues via different interactions without the need for significant structural changes. This is also consistent with previous data that −1 residue in the peptide ligands is less stringently specified by individual PDZ domains than the residues at the 0 and −2 positions, thereby allowing binding promiscuity [5].
At position −3, the peptide binding is more PDZ specific, facilitated by the rotameric flexibility of Ser-3 (Fig. 2B). In the binding, Ser-3 adopts different rotamers and each rotamer is able to bind to a unique position in PDZ domains. In NHERF1 PDZ1, the side chain hydroxyl of Ser-3 points to the N-terminus of the peptide forming a hydrogen bond to a histidine residue at the receptor position 1. In NHERF1 PDZ2, the side chain hydroxyl of Ser-3 points towards the C-terminus forming a hydrogen bond to an asparagine at the receptor position 2. In current rPDZ, the pointing direction of the Ser-3 side chain is perpendicular to the peptide direction which enables hydrogen bonding with a glutamine residue at the receptor position 3. Of note, all these receptor positions have been suggested to contribute to a high degree of selectivity in PDZ ligand recognition and the ability for the PDZ domain family to bind to different sequences [13]. The present study is extending the role of these receptor positions in determining binding diversity, and the fact that one of such positions can be specifically selected for interacting with −3 residue makes it possible for different PDZ domains to recognize the ligand residues of same sequence, providing an explanation for promiscuous CXCR2 binding.
The structural comparison between rPDZ–CXCR2 and rPDZ–PlexinB2 provides some insight into PDZ binding promiscuity. The residues at the ligand positions −1 and −3 are highly variable across rPDZ binding targets (Fig. 2C) indicating an ability of rPDZ to bind to ligands with different −1 and −3 side chains. At position −1, Thr-1 of CXCR2 forms a hydrogen bond with Thr60, whereas most of the side chain atoms of Asp-1 in PlexinB2 are disordered suggesting no stable interaction between Asp-1 and rPDZ; and the −1 position of PlexinB2 may not significantly contribute to the binding specificity (Fig. 2B). At position −3, Val-3 of PlexinB2 forms less discriminative VDW interactions with Gln70 and Ser62, differing from Ser-3 of CXCR2 which forms a specific hydrogen bond with Gln70. This difference indicates that rPDZ is able to form different types of interactions with −3 residues, which may underlie its flexibility to accommodate ligands with different −3 side chains.
3.4. Disulfide bond linked PDZ dimer
The most intriguing finding in current rPDZ structure is an asymmetric disulfide bond-linked PDZ dimer found in the asymmetric unit of the crystal (Fig. 3A). PDZ dimerization has been well appreciated as an important mechanism for improving PDZ scaffolding capacity during the formation of multiprotein complexes [7]. The dimerization is usually formed by non-covalent PDZ–PDZ interactions that put two canonical binding sites in a close proximity to facilitate a parallel or antiparallel nucleation of interacting proteins. However, the nature and significance of the less common disulfide bond-linked dimers remain largely elusive, though a few studies have suggested that the formation of disulfide bonds between proteins can be triggered by reactive oxygen species during cellular signaling [14]. In current rPDZ structure, two Cys47 on the outer surface of the β1 strand form an intermolecular disulfide bond responsible for PDZ dimerization (Fig. 3A). The dimer is an asymmetric dimer with the upper side having a pseudo 2-fold symmetry generated by parallel stacking of two copies of strands β1 and β6, and the lower side being asymmetric generated by the interaction between αA and β4 from different monomers. The buried surface area at the dimer interface is 777.6 Å2.
Figure 3.
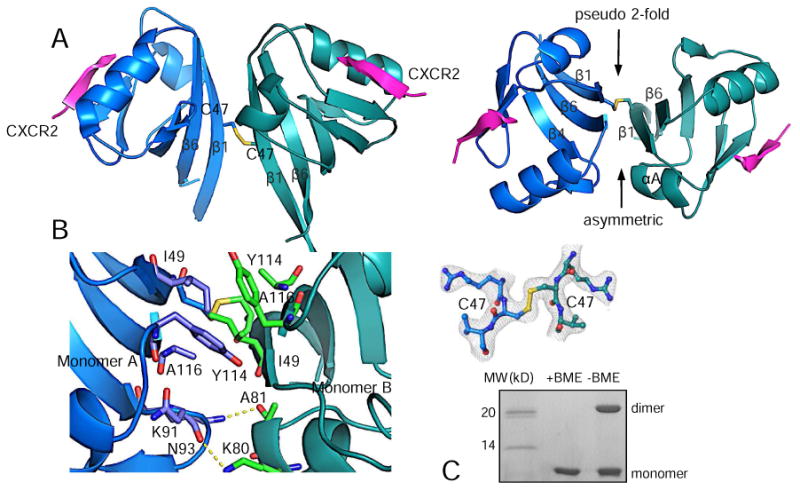
Disulfide bond linked PDZ dimer. (A) Overall view of PDZ dimer (side view, left; top view, right). (B) Close-up view of the dimer interface. 2Fo – Fc omit map of the disulfide bond was calculated at 2.4 Å and contoured at 1.0 σ. (C) SDS-PAGE of 5 μg of rPDZ–CXCR2 fusion protein with/without BME.
In addition to the disulfide bond, the dimer interface is further stabilized by several hydrogen bonds and hydrophobic interactions (Fig. 3B). At the upper side of the interface, there is a continuous hydrophobic core formed by pairs of residues Ile49, Tyr114 and Ala116, which is located above the disulfide bond bridge. At the lower side of the interface, two asymmetric hydrogen bonds are formed: Lys91 from the β4–β5 loop in monomer A forms a hydrogen bond with Ala81 carbonyl oxygen from the αA helix in monomer B; Lys80 from the αA helix from monomer B forms a hydrogen bond with Asn93 carbonyl oxygen from the β4–β5 loop in monomer A. Of note, all these residues including the disulfide bond forming residue Cys47 are highly conserved in the PDZ and RGS-containing GEF protein family suggesting a conserved function of this disulfide bond-linked dimer (Fig. 1D). Reducing SDS-PAGE indicates that the disulfide bonds contribute to the formation of a dimer in solution (Fig. 3C). Previous studies have also demonstrated that rPDZ dimerization linked by Cys47 disulfide bond regulates the canonical ligand binding and enhances in vitro binding to a bivalent PlexinB2 PDZ motif [15].
4. Discussion
In this study, we identified several CXCR2 interacting PDZ domains including PDZ-RhoGEF PDZ. We solved the crystal structure of PDZ-RhoGEF PDZ in complex with the CXCR2 C-terminal tail that provides the molecular basis of the interaction. The crystal structure also reveals an unexpected asymmetric disulfide bond-linked PDZ dimer that allows simultaneous parallel binding of CXCR2 to two PDZ domains. While the functional link between CXCR2 and PDZ-RhoGEF in signaling and cellular mobility requires future investigation, the identification of new CXCR2-binding PDZ domains is enforcing the view that PDZ domains play important roles in CXCR2 signaling processes capable of scaffolding complex interaction networks and coupling CXCR2 signaling to specific signaling pathways, potentially Rho signaling and Rac1 signaling through interacting with PDZ-RhoGEF and SNA1 respectively [8]. The identified interactions also provide additional models that enable further understanding of PDZ and CXCR2 binding promiscuity and specificity. The structural comparison is able to reveal the residues at the ligand positions −1 and −3 conferring PDZ- and ligand-specific recognition that may underlie the ability of CXCR2 to be bound by different PDZ domains and PDZ-RhoGEF PDZ to bind to different ligands. Additionally, the finding of the unexpected disulfide bond-linked PDZ dimer further demonstrates the structural diversity of PDZ dimerization. Diverse PDZ–PDZ interactions have been optimized as a mechanism in scaffolding the formation of distinct multiprotein complexes [7]. This non-canonical binding mode has been suggested to contribute more to defining the precise composition of protein complexes than does the canonical binding mode due to the structural diversity [7]. Therefore, there has been a continuous interest in revealing the specific nature of PDZ–PDZ interactions and their selectivity in precise scaffolding of temporal and spatial signaling networks. The current study provides an additional example of how PDZ domains may dimerize, and the asymmetric interface and rare disulfide bond linkage effectively define a new mode of PDZ dimerization, which is different from any reported structures [12]. Together with PDZ binding promiscuity, the new mode of dimerization could provide a reactive oxygen species-sensitive molecular scaffold for assembly of distinct CXCR2 signaling networks in actin polymerization and cell mobility.
Supplementary Material
Highlights.
Chemokine receptor CXCR2 C-terminus binds to the PDZ domain of PDZ-RhoGEF
The crystal structure of PDZ-RhoGEF PDZ domain in complex with CXCR2
The structure reveals the specificity determinants of PDZ–CXCR2 interaction
Asymmetric disulfide bond-linked PDZ dimer with parallelly oriented CXCR2 peptides
Provides implications for signaling complex scaffolding in CXCR2 signaling
Acknowledgments
This work was supported by the National Institutes of Health grant number 1R01HL128647-01 (to CL and ZY),
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wu Y, Wang S, Farooq SM, Castelvetere MP, Hou Y, Gao JL, Navarro JV, Oupicky D, Sun F, Li C. A chemokine receptor CXCR2 macromolecular complex regulates neutrophil functions in inflammatory diseases. J Biol Chem. 2012;287:5744–5755. doi: 10.1074/jbc.M111.315762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonsiorek W, Fan X, Hesk D, Fossetta J, Qiu H, Jakway J, Billah M, Dwyer M, Chao J, Deno G, Taveras A, Lundell DJ, Hipkin RW. Pharmacological characterization of Sch527123, a potent allosteric CXCR1/CXCR2 antagonist. J Pharmacol Exp Ther. 2007;322:477–485. doi: 10.1124/jpet.106.118927. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Wu Y, Hou Y, Guan X, Castelvetere MP, Oblak JJ, Banerjee S, Filtz TM, Sarkar FH, Chen X, Jena BP, Li C. CXCR2 macromolecular complex in pancreatic cancer: a potential therapeutic target in tumor growth. Transl Oncol. 2013;6:216–225. doi: 10.1593/tlo.13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Addison CL, Daniel TO, Burdick MD, Liu H, Ehlert JE, Xue YY, Buechi L, Walz A, Richmond A, Strieter RM. The CXC chemokine receptor 2, CXCR2, is the putative receptor for ELR+ CXC chemokine-induced angiogenic activity. J Immunol. 2000;165:5269–5277. doi: 10.4049/jimmunol.165.9.5269. [DOI] [PubMed] [Google Scholar]
- 5.Lu G, Wu Y, Jiang Y, Wang S, Hou Y, Guan X, Brunzelle J, Sirinupong N, Sheng S, Li C, Yang Z. Structural insights into neutrophilic migration revealed by the crystal structure of the chemokine receptor CXCR2 in complex with the first PDZ domain of NHERF1. PloS one. 2013;8:e76219. doi: 10.1371/journal.pone.0076219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouassier L, Yun CC, Fitz JG, Doctor RB. Evidence for ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) self-association through PDZ-PDZ interactions. J Biol Chem. 2000;275:25039–25045. doi: 10.1074/jbc.C000092200. [DOI] [PubMed] [Google Scholar]
- 7.Chang BH, Gujral TS, Karp ES, BuKhalid R, Grantcharova VP, MacBeath G. A systematic family-wide investigation reveals that ∼30% of mammalian PDZ domains engage in PDZ-PDZ interactions. Chem Biol. 2011;18:1143–1152. doi: 10.1016/j.chembiol.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mikelis CM, Palmby TR, Simaan M, Li W, Szabo R, Lyons R, Martin D, Yagi H, Fukuhara S, Chikumi H, Galisteo R, Mukouyama YS, Bugge TH, Gutkind JS. PDZ-RhoGEF and LARG are essential for embryonic development and provide a link between thrombin and LPA receptors and Rho activation. The Journal of biological chemistry. 2013;288:12232–12243. doi: 10.1074/jbc.M112.428599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 11.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holcomb J, Jiang Y, Guan X, Trescott L, Lu G, Hou Y, Wang S, Brunzelle J, Sirinupong N, Li C, Yang Z. Crystal structure of the NHERF1 PDZ2 domain in complex with the chemokine receptor CXCR2 reveals probable modes of PDZ2 dimerization. Biochemical and biophysical research communications. 2014;448:169–174. doi: 10.1016/j.bbrc.2014.04.085. [DOI] [PubMed] [Google Scholar]
- 13.Tonikian R, Zhang Y, Sazinsky SL, Currell B, Yeh JH, Reva B, Held HA, Appleton BA, Evangelista M, Wu Y, Xin X, Chan AC, Seshagiri S, Lasky LA, Sander C, Boone C, Bader GD, Sidhu SS. A specificity map for the PDZ domain family, PLoS Biol. 2008;6:e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cumming RC, Andon NL, Haynes PA, Park M, Fischer WH, Schubert D. Protein disulfide bond formation in the cytoplasm during oxidative stress. The Journal of biological chemistry. 2004;279:21749–21758. doi: 10.1074/jbc.M312267200. [DOI] [PubMed] [Google Scholar]
- 15.Paduch M, Biernat M, Stefanowicz P, Derewenda ZS, Szewczuk Z, Otlewski J. Bivalent peptides as models for multimeric targets of PDZ domains. Chembiochem : a European journal of chemical biology. 2007;8:443–452. doi: 10.1002/cbic.200600389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


