Abstract
Gene therapy is a promising approach with enormous potential for treatment of neurodegenerative disorders. Viral vectors derived from canine adenovirus type 2 (CAV-2) present attractive features for gene delivery strategies in the human brain, by preferentially transducing neurons, are capable of efficient axonal transport to afferent brain structures, have a 30-kb cloning capacity and have low innate and induced immunogenicity in pre-clinical tests. For clinical translation, in-depth pre-clinical evaluation of efficacy and safety in a human setting is primordial. Stem cell-derived human neural cells have a great potential as complementary tools by bridging the gap between animal models, which often diverge considerably from human phenotype, and clinical trials.
Herein, we explore helper-dependent CAV-2 (hd-CAV-2) efficacy and safety for gene delivery in a human stem cell-derived 3D neural in vitro model. Assessment of hd-CAV-2 vector efficacy was performed at different multiplicities of infection, by evaluating transgene expression and impact on cell viability, ultrastructural cellular organization and neuronal gene expression.
Under optimized conditions, hd-CAV-2 transduction led to stable long-term transgene expression with minimal toxicity. hd-CAV-2 preferentially transduced neurons, while human adenovirus type 5 (HAdV5) showed increased tropism towards glial cells.
This work demonstrates, in a physiologically relevant 3D model, that hd-CAV-2 vectors are efficient tools for gene delivery to human neurons, with stable long-term transgene expression and minimal cytotoxicity.
Introduction
Neurodegenerative diseases, typically characterized by a progressive nervous system dysfunction, represent a heavy burden both in terms of patient suffering and economic cost. The prevalence of neurologic disorders has dramatically increased over the last decades and continues to increase, mainly due to higher life expectancy of populations, which elicits the urgent need for effective therapeutics1.
With the increasing knowledge on the etiology of these disorders, several genetic mutations have been linked with the sporadic and familial forms of neurodegenerative diseases. The identification of these mutations provides new insight on the molecular mechanisms involved in disease onset and progression, but also promising therapeutic targets for alternative approaches to traditional pharmacological treatments, such as gene therapy2. Briefly, most gene delivery approaches focus either on enzyme replacement, by restoring the enzymatic capacity of the affected brain regions, or on delivery of neurotrophic factors to prevent the progression of the neurodegeneration process3.
Over the last three decades multiple non-viral gene delivery vehicles have been explored, yet viral vectors are still the most efficient tools for in vivo gene transfer. Each viral vector system has unique strengths and specific drawbacks4,5. Therefore, thorough evaluation is required for selection of the optimal vector system for central nervous system (CNS) gene delivery. Most neurological disorders only affect a specific cell type, as is the case of the dopaminergic neurons of the nigrostriatal pathway in Parkinson’s disease (PD)6. Thus, in addition to high transduction efficiency, vector tropism is paramount to achieve targeted gene delivery to the affected cells of a confined brain region, minimizing off-target transduction. To maximize the therapeutic effect and minimize repeated dosage, an ideal vector should be able to sustain long-term expression of the transgene with a single treatment. Most importantly, the vector must be safe and avoid host immune responses or cytotoxicity, which may also hinder the therapeutic effect. Finally, it is important to consider vector manufacturing methods, which should be scalable, allow high titers and high purity5,7,8.
Recombinant adeno-associated viruses (AAV) have been the most widely used and studied vectors for gene delivery in the CNS and peripheral nervous system. The variety of AAV serotypes enables the mixing of viral genomes and capsids creating mosaic “pseudotype” which display a range of tropisms and efficacies, along with low pathogenicity and immunogenicity5,9. However, AAV vectors have a limited cloning capacity (≈4-6 kb), which limits their use in some applications. Recombinant adenovirus (AdV) vectors are an attractive alternative, due to a large cloning capacity (8-30 kb) and long-term transgene expression5,9, being the most explored platform in clinical trials worldwide (http://www.wiley.com/legacy/wileychi/genmed/clinical). Nonetheless, the toxicity and immunogenicity of some AdV types have been widely described5,7,9, limiting the therapeutic efficacy. Non-human AdV vectors, such as canine adenovirus type 2 (CAV-2) vectors emerged as an alternative to human AdV (HAdV), mainly due to lack of immunological memory10. CAV-2 vectors preferentially transduce neurons in the rodent brain and in human organotypic cultures, along with efficient biodistribution via axonal retrograde transport in neurons11,12. The development of helper-dependent CAV-2 vectors (hd-CAV-2)13,14 improved the efficiency and duration of transgene expression, minimizing the adaptive cell-mediated immune response13. hd-CAV-2 vectors have a cloning capacity of 30 kb13 and allow stable transgene levels over 1 year in immunocompetent rat brain without immunosuppression14.
During viral vector development, preclinical testing is crucial to evaluate both efficacy and safety, while understanding vector-cell interactions. Although the traditional primary cultures of rodent brain cells and animal models provide valuable data, these models too often diverge considerably from the human phenotype15, thus not accurately predicting the outcome of clinical trials. In this context, stem cell-derived human neural cells, along with three dimensional (3D) culture systems, have great potential as complementary tools in pre-clinical research, bridging the gap between human clinical studies and animal models15,16. We previously reported the development of a 3D neural cell model based on the differentiation of human midbrain-derived neural progenitor cells (hmNPC) as neurospheres, in a dynamic culture system17,18. Differentiated neurospheres contain glial cells, oligodendrocytes and functional neurons, with enrichment in the dopaminergic phenotype.
In this study, we took advantage of the 3D neural in vitro model17,18 to evaluate hd-CAV-2 vectors gene delivery efficiency and cytotoxicity. We made use of a reporter eGFP-expressing hd-CAV-2, which can be produced in a robust and scalable bioprocess19,20, compatible with pre-clinical and clinical applications. Assessment of hd-CAV-2 transduction was performed at different multiplicities of infection (MOI), by evaluating transgene expression and cell viability, impact on neuronal gene expression and cell morphology. This study show that hd-CAV-2 transduction led to stable long-term transgene expression with low toxicity, demonstrating in a physiologically relevant human in vitro model, that hd-CAV-2 vectors are efficient vehicle for gene transfer to human neurons for treatment of neurodegenerative diseases.
Results
hd-CAV-2 impact on cell viability and neuronal population
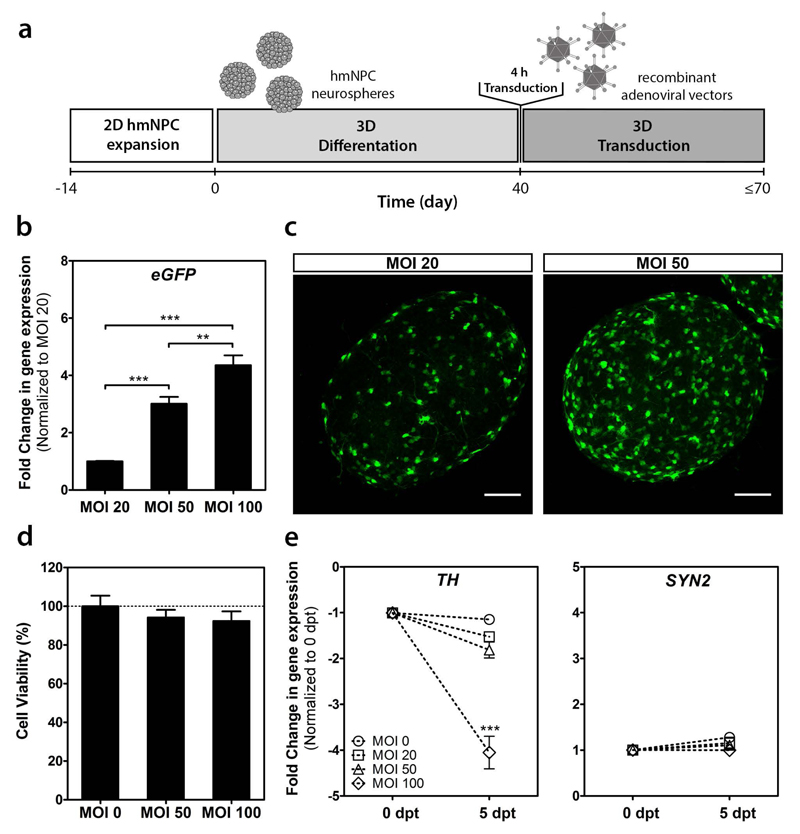
To determine the most suitable transduction parameters, three MOIs were tested on the human 3D neural model (Figure 1a). We assessed gene delivery efficiency by evaluating transgene expression levels, and cytotoxic effects exerted by the vectors, measuring cell viability and expression levels of neuronal and synaptic markers. The MOIs (20, 50 and 100 infectious particles/cell) were selected based on data previously generated with an E1-deleted CAV-2 vector expressing GFP17. Neurosphere transduction showed an increase in transgene mRNA levels according to the MOI increase, with a 3- and 4.5-fold increase relatively to MOI 20 for MOI 50 and 100, respectively, at 5 days post-transduction (dpt) (Figure 1b). In agreement with these observations, the fraction of GFP-positive cells also increased with the MOI, as observed by confocal microscopy (Figure 1c).
Figure 1.
hd-CAV-2 transduction of differentiated human midbrain-derived neural precursor cells (hmNPC) neurospheres. (a) Timeline of cell differentiation and transduction. (b) Fold changes of eGFP expression for multiplicities of infection (MOI) 20, 50 and 100 infectious particles per cell (normalized to MOI 20). (c) Confocal microscopy of whole neurospheres. Maximum intensity z-projections of 38 (MOI 20) and 33 (MOI 50) optical sections of 1 μm. Scale bars, 100 μm. (d) Cell viability assessment, normalized for control (MOI 0). (e) Fold changes in tyrosine hydroxylase (TH) and synapsin II (SYN2) gene expression at 0 and 5 days post-transduction (dpt) (normalized for 0 dpt). Each group was performed in triplicates. Data are mean ± s.e.m. of 3 independent experiments. Asterisks indicate significant difference (*P < 0.05, **P < 0.01, ***P < 0.001).
Cytotoxicity assessment showed no significant decrease in cell viability for MOI 50 and 100, relatively to control (Figure 1d). Moreover, no significant modulation in the pre-synaptic marker synapsin II (SYN2) gene expression was observed for the different MOIs (Figure 1e). Still, a 4-fold decrease in the expression of the dopaminergic marker tyrosine hydroxylase (TH) was observed for MOI 100 at 5 dpt, relatively to the neurospheres before hd-CAV-2 transduction. These results suggest that higher hd-CAV-2 MOIs, although allowing higher gene delivery efficiency, can affect specific neuronal populations, such as dopaminergic neurons. These observations emphasize the need for a careful dose assessment in pre-clinical studies to identify toxic effects, while sustaining an efficient gene delivery to target cells. In this study, considering the level of transgene expression and the absence of modulation for the neuronal markers evaluated, MOI 20 and 50 were selected for further characterization of hd-CAV-2 transduction.
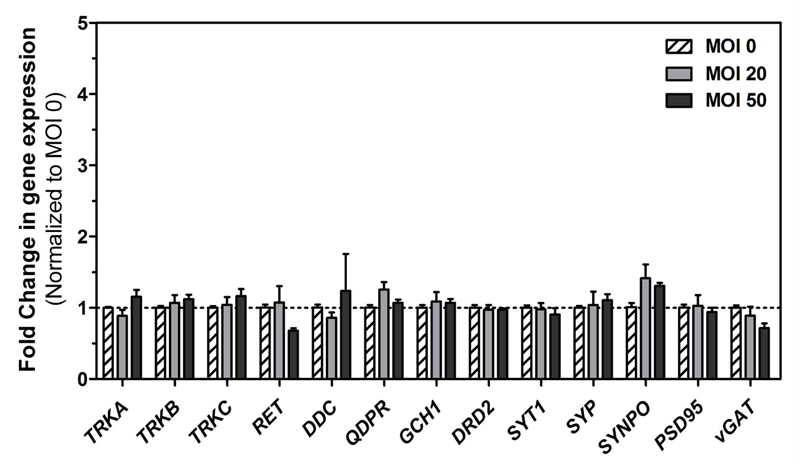
The impact of hd-CAV-2 transduction on the neurospheres was further addressed by analyzing the expression of an enlarged set of neuronal markers (Figure 2), comprising neurotrophic receptors (TrkA, TrkB, TrkC and RET), dopamine biosynthesis pathway enzymes (DDC, QDPR, GCH1), dopamine receptor (DRD2), pre-synaptic (SYT1, SYP, SYNPO and vGAT) and post-synaptic (PSD95) proteins. Compared with the control (MOI 0), an MOI of 20 or 50 did not induce significant modulation on these markers. Thus, given the 3-fold increase in transgene expression with an MOI 50 (Figure 1b) and the comparable low cytotoxic effects, the MOI 50 was selected for more comprehensive studies addressing the impact of hd-CAV-2 transduction on ultrastructural cell morphology, vector tropism and duration of transgene expression.
Figure 2.
Gene expression analysis of differentiated neurospheres transduced with hd-CAV-2. Gene expression fold changes (normalized to the MOI 0 control) of neurotrophic receptors (TrkA, TrkB, TrkC and RET), dopamine biosynthesis pathway enzymes (DDC, QDPR, GCH1), dopamine receptor (DRD2), pre-synaptic proteins (SYT1, SYP, SYNPO and vGAT) and post-synaptic protein (PSD95). Each group was performed in triplicates. Data are mean ± s.e.m. of 3 independent experiments.
hd-CAV-2 impact on cell morphology
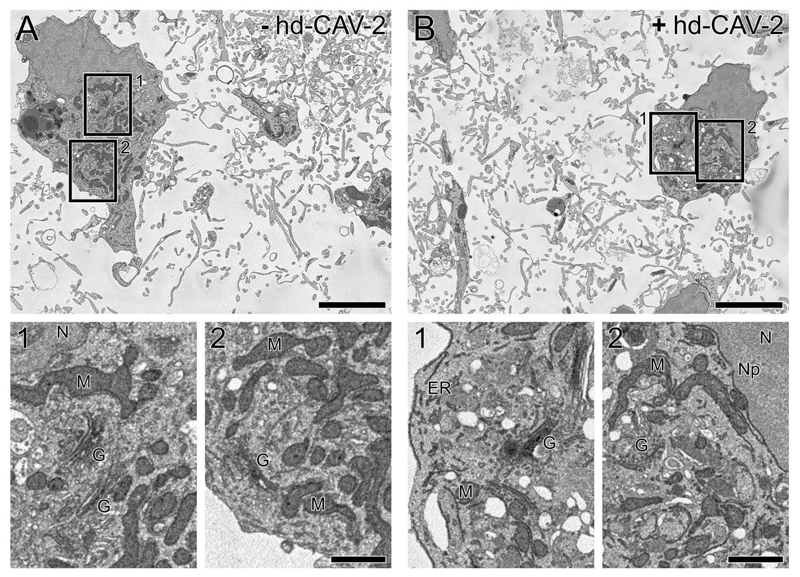
Cellular ultrastructure was analysed both for transduced (MOI 50) and non-transduced neurospheres by serial blockface scanning electron microscopy (SBF-SEM; Figure 3 and supplementary video 1 and 2). The image stacks showed similar numbers and spatial distribution of cell bodies, as well as the complexity of the extending neuronal network. Moreover, comparable cellular ultrastructural details within non-transduced and transduced cells were observed, including clearly defined mitochondria, Golgi stacks, endoplasmic reticulum and other subcellular membrane structures. Altogether, these results suggest that hd-CAV-2 transduction did not induced noticeable structural alterations on human neural cells within the neurospheres.
Figure 3.
Effects of hd-CAV-2 transduction on the ultrastructural cell organization of differentiated neurospheres. Electron micrographs extracted from the SBF SEM image stacks showing the internal structure of non-transduced (a) and hd-CAV-2 transduced (b; MOI 50) neurospheres at 5 days post-transduction. Boxed areas (1, 2) highlight details of ultrastructure within each cell, including mitochondria (M), Golgi stacks (G), endoplasmic reticulum (ER), and other subcellular structures (N: nucleus, Np: nuclear pore). Scale bars, 5 μm (a, b), 1 μm (1, 2).
hd-CAV-2 tropism and long-term transduction dynamics
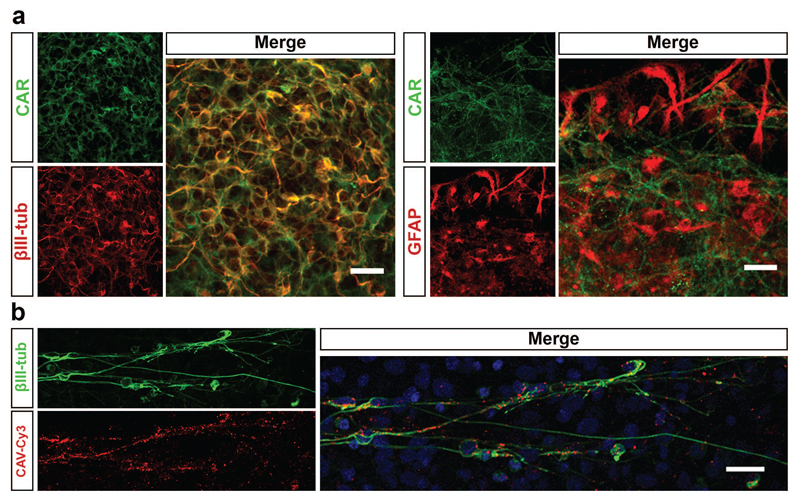
hd-CAV-2 preferential transduction of neurons in rodent has been linked with the expression of coxsackievirus and adenovirus receptor (CAR)11,12,21. Given this, we analyzed the expression and distribution of CAR on human neurospheres by confocal microscopy. As shown in Figure 4a, CAR is highly expressed in βIII-tubulin positive neurons, but not in glial cells labeled with GFAP. To further explore the interaction between CAV-2 vectors and human neurons, we then incubated neurospheres with fluorescent CAV-2 (CAV-Cy3), followed by immunofluorescence labeling of neurons with βIII-tubulin. CAV-Cy3 particles were detected along neuronal cells (Figure 4b), indicating that CAV-2 vectors recognize these cells, both at the soma and branching processes, for attachment and internalization.
Figure 4.
CAR and CAV-2 distribution in differentiated neurospheres. (a) Immunofluorescence analysis of neurospheres stained for CAR, βIII-tubulin and GFAP. Maximum intensity z-projections of 33 (βIII-tubulin) and 14 (GFAP) optical sections of 0.38 μm. Scale bars, 20 μm. (b) Immunofluorescence of neurospheres incubated with CAV-Cy3 particles (red) stained for βIII-tubulin (green). Nuclei were labelled with TO-PRO3. Single optical section. Scale bar, 20 μm.
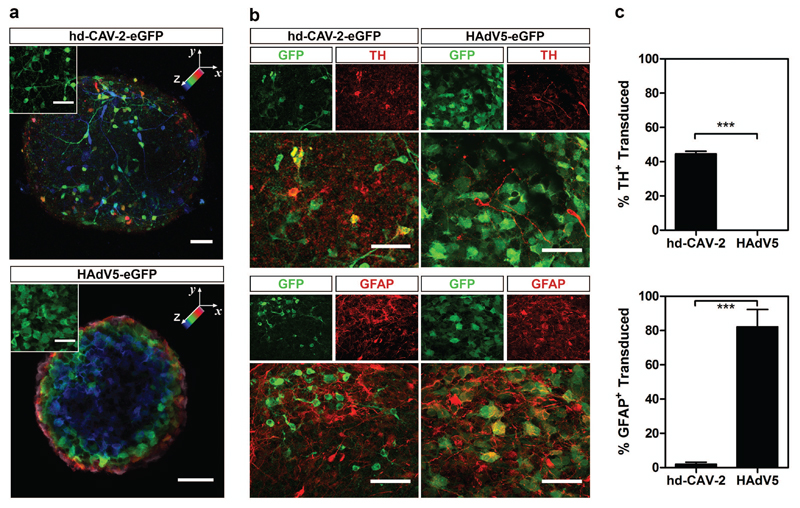
For a more detailed study on the tropism of hd-CAV-2 in human neural cells, we then compared the phenotype of hd-CAV-2 transduced cells to a vector derived from human adenovirus type 5 (HAdV5), which has been widely explored in gene therapy22,23. Differences both in terms of morphology and distribution of transduced cells along the neurospheres’ volume were observed (Figure 5a). The typical morphology of hd-CAV-2 transduced cells was neuronal-like, with long processes extending from the soma. In contrast, HAdV5-transduced cells presented glial-like morphology, with larger cytoplasmic volume and no neurites. Interestingly, upon hd-CAV-2 transduction, it was possible to identify multiple GFP-positive cells across different layers towards the inner core of the neurospheres (Figure 5a). Sections were plotted in blue progressing to green and red/pink across a 41-μm-volume depth. On the other hand, upon HAdV5 transduction, the presence of GFP-positive cells was restricted to the neurospheres’ surface, presenting a radial pattern from blue cells in the center towards red/pink cells only in the periphery.
Figure 5.
Characterization of hd-CAV-2 and HAdV5 tropism. (a) Confocal microscopy of hd-CAV-2 and HAdV5 transduced neurospheres. Spatial distribution of transduced cells is highlighted by a depth lookup table. Maximum intensity z-projections of 41 optical sections of 1 μm, where blue indicates 0 μm and pink 41 μm. Inset depicts typical morphology of transduced cells. Scale bars, 50 μm. (b) Immunofluorescence analysis of neurospheres transduced with hd-CAV-2 and HAdV5, stained for TH and GFAP. Maximum intensity z-projections of 55 (TH – hd-CAV-2), 19 (TH – HAdV5), 42 (GFAP – hd-CAV-2) and 23 (GFAP – HAdV5) optical sections of 0.33 μm. Scale bars, 50 μm. (c) Quantification of TH+ and GFAP+ transduced cells for hd-CAV-2 and HAdV5, expressed as percentage of total TH+ or GFAP+ cells. Data are mean ± s.e.m. of 2 independent experiments.
To address the identity of hd-CAV-2 and HAdV5 transduced cells using another approach, the cell lineages were identified by immunodetection: TH staining for dopaminergic neurons and GFAP staining for glia (Figure 5 b and c). This assay revealed that hd-CAV-2 efficiently transduced dopaminergic neurons (TH-positive cells), with up to 50% of total TH-positive cells being GFP-positive (Figure 5c). Glial cells (GFAP-positive cells) were poorly transduced by hd-CAV-2. These data are consistent with CAR expression on neurons. By contrast, HAdV5 preferentially transduced glial cells, with more than 80% GFAP-positive cells also GFP-positive (Figure 5c), while poorly transducing TH-positive cells.
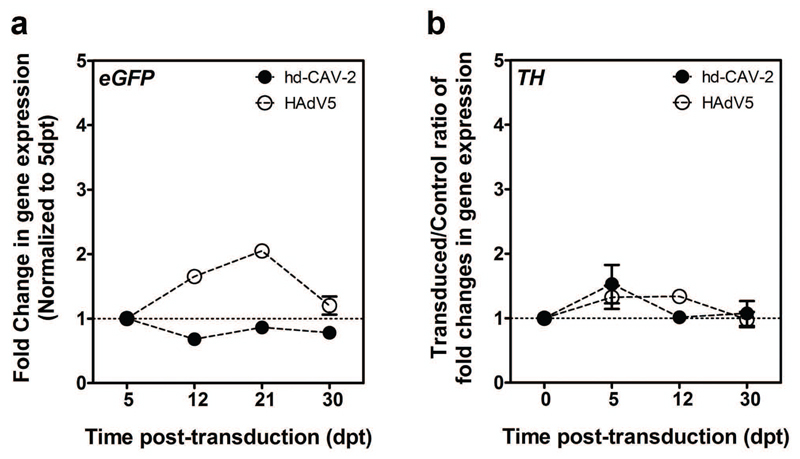
In addition to tropism, we also analyzed hd-CAV-2-driven transgene expression. Following incubation with hd-CAV-2 the neurospheres were maintained for at least 30 dpt. Compared to controls, we detected no significant change in cell viability. hd-CAV-2 transduction led to stable eGFP expression during this culture time (Figure 6). TH expression dynamics revealed no significant modulations over the 30 dpt, suggesting low long-term cytotoxic effects of hd-CAV-2 on dopaminergic neurons at the MOI used in this experiments. These analyses were performed in parallel for HAdV5 transduction, which resulted in slightly less stable transgene expression levels with a 2-fold increase during the first 21 dpt, followed by a decrease at 30 dpt to levels similar to those observed at 5 dpt. Similarly to hd-CAV-2, no significant modulation was observed for HAdV5 in terms of transduction impact on TH expression.
Figure 6.
hd-CAV-2 and HAdV5 transgene expression dynamics. Gene expression fold changes of eGFP (a) and TH (b) expression in hd-CAV-2 and HAdV5 transduced neurospheres, along 30 dpt. Each group was performed in triplicate. Data are mean ± s.e.m. of 2 independent experiments.
Discussion
In the rodent brain CAV-2 vectors efficiently and preferentially transduce neurons, undergoing fast retrograde axonal transport, allow long-term transgene expression and induce negligible levels of immunogenicity12,14. Given the fundamental differences between rodent and human cells, it is critical to demonstrate the efficacy and safety of such vectors in human cells at early stages of development. In this study, we assayed hd-CAV-2 vectors efficacy in a human 3D in vitro CNS model based on differentiation of hmNPC. During differentiation, these cells recapitulate the specific midbrain developmental program, resulting in a human cell model enriched in functional dopaminergic neurons18.
Here, we found that hd-CAV-2 vectors efficiently transduce human neural cells. Our screening detected cytotoxic effects at high MOIs (100 ip/cell). These effects were more evident on the dopaminergic population, as determined by a significant decrease in TH expression levels and no modulation on the synaptic gene SYN2. Notably, differential hd-CAV-2 transduction efficacy has also been observed in the rodent brain, where the higher efficiency was observed for dopaminergic neurons, followed by noradrenergic neurons and serotonergic neurons (Schwarz et al Nature (in press)). Dopaminergic neurons are more susceptible to GFP toxicity, relative to other neuronal populations24. In this study, transduction of rodent brain with high doses of an AAV8-GFP vector induced a significant loss in TH+ neurons compared to an empty vector. By contrast, no loss of pyramidal or granular neurons was observed in the hippocampus. Together, these data indicate different transduction efficiencies or transgene related effects in different neuronal subtypes, which can lead to differential survival.
To confirm the absence of cytotoxicity for MOIs of 20 and 50 ip/cell, expression of a panel of neuronal genes, including neurotrophic receptors, dopamine biosynthesis pathway enzymes, dopamine receptors and synaptic proteins was evaluated at 5 dpt. No significant changes were observed for both MOIs in comparison with the control. These results are in agreement with data on the toxicogenomic profile of hd-CAV-2 transduction of 2D hmNPC cultures, showing no significant vector-induced modulation of the neuron morphogenesis pathways and an activation of pro-survival genes25. Moreover, hd-CAV-2 transduction of neurospheres (MOI 50) did not induce overt ultrastructural alterations, either in terms of the spatial distribution within neurospheres or at intracellular level.
These results show the importance of MOI screening and optimization to attain efficient levels of gene delivery, while maintaining cytotoxic effects to a minimum. Nevertheless, when considering an in vivo setting, one must not exclude possible immunological complications often associated with a diseased brain and gene transfer vectors, which in case of AdV are related with innate and memory immunity to the wild-type pathogen. This can lead to adverse effects upon administration, such as acute inflammatory reactions, posing a safety risk for the patient. Previous reports have shown negligible levels of neutralizing CAV-2 antibodies in healthy humans26, as well as lower immunogenicity than HAdV in immunologically naïve rodents14,27. As with all vectors, this should be a matter of careful scrutiny in potential clinical trials to ensure patient safety and the success of therapy. Moreover, data obtained from rodents may not accurately reflect the human setting. A recent study showed that HAdV5 and murine coagulation factor X (FX) complexes stimulated an innate inflammatory response via Toll-like receptor 4 in murine macrophages28. However, HAdV5 and human FX complexes do not stimulate an innate response in human mononuclear phagocytes29.
Our results showed in a 3D human cell model that hd-CAV-2 vectors preferentially transduce neurons rather than glial cells, in agreement with previous reports on animal models and ex vivo human brain slices12,14. In contrast, transduction with HAdV5 resulted in preferential glial transduction, further confirming the distinct vector tropism of CAV-2 and HAdV5 in spite of sharing many characteristics, such as a very similar capsid structure and genomic organization30. The molecular basis for CAV-2 neuronal tropism relies on the exclusive binding of CAR and CAR-mediated internalization. Notably, the CAV-2 capsid does not contain a readily identifiable integrin-interacting motif (e.g. RGD motif)21. Hence, CAR expression is essential for CAV-2 transduction, which in this human model was restricted to the neuronal population, in agreement with previous reports in rodent models12. Thus, our results in a human context further support the CAR-dependent CAV-2 binding and internalization hypothesis, which leads to increased neuronal tropism of CAV-2 and poor glia transduction. Although HAdV5 is also a “CAR-tropic” virus and can transduce neurons, these vectors preferentially transduce glia12. We can only speculate as to why HAdV5 does not efficiently use neuronal CAR. The different CAR binding efficiencies reported for the two vectors, where HAdV5 presents lower CAR affinity than CAV-2 (Kd values of 7.9 and 1.1 nM, respectively)31 could explain the lower neuronal transduction efficiency of HAdV5. Also, it is conceivable that integrin-associate internalization of HAdV5 is poorly functional in CAR-rich lipid rafts of the neuron membrane, or that intracellular transport of HAdV5 is inefficient because it is released from CAR-positive vesicles prematurely during axonal transport, precluding its delivery to the soma. Clearly, our results showed an accumulation of CAV-Cy3 particles along neuronal processes, which indicates that CAV-2 vectors can bind and be internalized along neurites and not only in the soma. This observation, in addition to the presence of transduced neurons in the inner layers of the neurospheres, is consistent with the trafficking mechanism reported for murine primary neurons11,32. These studies demonstrate that after CAR binding and subsequent internalization, CAV-2 vectors are transported in axons via a CAR-associated mechanism. Retrograde axonal transport of CAV-2 in murine neurons is 10-fold greater compared to HAdV512, allowing long distance targeting that provides means of targeting cells or brain regions that are typically difficult to reach. In addition to CAV-2 tropism, one can further explore strategies for neuronal specific transgene expression, such as neuronal specific promoters33,34 that can reduce potential off-target transduction and/or restrict transgene expression to a neuronal subpopulation (e.g. dopaminergic neurons).
This work demonstrates, in a physiologically relevant 3D model, that hd-CAV-2 vectors are efficient vehicles for gene transfer to human neurons, with stable transgene expression and minimal cytotoxicity. In the long term, this study addresses the potential of hd-CAV-2 vectors for gene therapy of human neurodegenerative diseases, such as PD, further supporting the valuable data previously obtained from pre-clinical animal models.
Materials and Methods
hmNPC expansion and differentiation
Human midbrain-derived neural progenitor cells (hmNPC) derived from aborted fetal brain tissue 12 to 14 weeks post-fertilization17,35 were kindly provided by Dr. Johannes Schwarz (Technical University of Munich, Germany), within the scope of the EU project BrainCAV (FP7-222992). Tissue was obtained with mother’s consent and in accordance with the Ethics Committee of the University of Leipzig and the German state and federal laws. Expansion and differentiation of hmNPC was performed as previously described17,18. Briefly, hmNPC were expanded on poly-L-ornithine-fibronectin (PLOF)-coated surfaces and serum-free medium17. Differentiation was performed as neurospheres in shake flasks (Corning, New York, USA) under constant orbital shaking18. Cells were maintained in a multi-gas cell incubator (Panasonic Biomedical, Leicestershire, UK) at 37°C, in a humidified atmosphere of 5% CO2 and 3% O2 in air. Cells were routinely screened to exclude mycoplasm contamination.
Viral stock production
hd-CAV-2 are vectors derived from CAV-2 strain Toronto A 26/61, GenBank J04368. HAdV5 vectors are E1-deleted adenovirus type 5, based on plasmid pGS66, containing viral genome sequences from nucleotide 1 to 440 and 3523 to 3593536. Both vectors contain an eGFP expression cassette consisting in a human cytomegalovirus immediate early promoter, cDNA coding for eGFP and the SV40 polyA signal. hd-CAV-2 stocks were produced by co-infection with JBΔ5 helper virus of E1-complementing dog kidney cells expressing Cre recombinase (DKCre) and purified by CsCl gradients, as previously described14,26,37. HAdV5 were produced using human embryonic kidney cells (HEK293) followed by CsCl purification, according to previous reports38,39. Stocks titration were performed by infectivity assay for number of infectious particles/mL (ip/mL)19,38. The obtained preparations had a physical to infectious particles (pp/ip) unit ratio of 100:1 for hd-CAV-2 and 15:1 for HAdV5. hd-CAV-2 had a helper virus contamination level of 1.8%. In this study, multiplicity of infection (MOI) was determined by the number of infectious viral particles per cell (ip/cell).
Transduction
Following hmNPC neurospheres differentiation, transduction was performed with 50% reduction of the working volume. Total cell number was determined by counting cell nuclei using a Fuchs-Rosenthal hemacytometer after digestion with lysis buffer (0.1 M citric acid, 1% Triton X-100 (w/w) and 0.1% crystal violet (w/v))40. At 4 h post-transduction a complete media exchange was performed. Neurospheres were maintained in culture up to 30 days post-transduction, with a 75% medium exchange performed every 3 days.
Cell viability
Cell viability was assessed using the metabolic indicator PrestoBlue™ (Life Technologies, Paisley, UK), according to the manufacture’s recommendation. Briefly, neurospheres were plated on PLOF-coated multi-well plates and allowed to attach for 2 hours. Cells were incubated with fresh medium containing 10% (v/v) PrestoBlue, for 2 hours. Supernatant’s fluorescence was measured in 96-well plates using a microwell plate fluorescence reader (FluoroMax-4, Horiba JobinYvon, Kyoto, Japan) and neurospheres were harvested for total protein quantification, using the Micro BCA Protein Assay Kit (Life Technologies). Fluorescence intensity was normalized for total protein and evaluated relatively to control (MOI 0).
Fluorescence microscopy
Neurospheres were plated on PLOF-coated glass coverslips and allowed to attach for 2 days, fixed in 4% (w/v) paraformaldehyde (PFA) + 4% (w/v) sucrose in PBS for 30 min and processed for immunostaining as previously described41. Primary and secondary antibodies were used as follows: mouse anti-βIII-tubulin (1:200; Millipore Darmstadt, Germany, MAB1637); mouse anti-GFP (1:200; Sigma-Aldrich, St Louis, MO, USA, G6539); rabbit anti-TH (1:100; Santa Cruz Biotechnology, Dallas, TX, USA, sc-14007); rabbit anti-GFAP (1:200; Millipore, AB5804); rabbit anti-CAR (1:200; gift from Joseph Zabner), mouse anti-CAR (1:200; Millipore, 05-644); AlexaFluor® 488 goat anti-mouse IgG (1:500; Life Technologies, A11001) and AlexaFluor® 594 goat anti-rabbit IgG (1:500; Life Technologies, A11012). Cell nuclei were counterstained with DAPI or TO-PRO-3 (Life Technologies). Samples were visualized using point scan confocal microscopy (SP5, Leica, Wetzlar, Germany). Merge between channels and maximum z-projections, as well as linear brightness and contrast adjustments of the images, were performed using the ImageJ software.
Electron microscopy
Neurospheres were fixed in 2.5% (w/v) glutaraldehyde and 4% (w/v) formaldehyde in 0.1 M phosphate buffer (pH 7.4) and then processed for serial blockface scanning electron microscopy (SBF SEM). Samples were embedded in Durcupan resin following the method of NCMIR42. Small groups of neurospheres were mounted on pins and trimmed for SBF SEM. Images were acquired using a 3View2XP (Gatan Pleasanton, CA, USA) attached to a Sigma VP SEM (Zeiss, Cambridge, UK), at a resolution of 8,192 x 8,192 pixels (horizontal frame width of 64.29 μm; pixel size of 7.8 nm) with 2 μs dwell time and 35 nm slice thickness. SEM was operated in high vacuum, with high current mode active, at an indicated magnification of 4,000. The 20 μm aperture was used, at an accelerating voltage of 1.4 kV (hd-CAV-2 transduced) or 1.2 kV (control). Alignment of the image stack was accomplished using the ‘Register virtual stack slices’ plugin in Fiji43, with translation based extraction and registration models to minimize distortion of the dataset. Aligned image stacks were then calibrated for pixel dimensions. Movies were generated using Amira (FEI Visualization Sciences Group, Merignac, France) and Quicktime Pro, showing 500 slices from the center of each dataset (total dataset for hd-CAV-2 transduced = 1,450 slices, representing a total volume of 209,760 μm2; total dataset for control = 533 slices, representing a total volume of 77,105 μm2).
qRT-PCR
Neurospheres were sedimented by centrifugation at 500 xg for 5 min, washed with PBS and the dry pellet snap-freezed by immersion in liquid nitrogen. Samples were stored at -80 °C until RNA extraction. Total RNA was extracted with High Pure RNA Isolation Kit (Roche Applied Science, Mannheim, Germany), according to the manufacturer’s instructions. RNA was quantified in a NanoDrop 2000c (Life Technologies) and used for cDNA synthesis. Reverse transcription was performed with High Fidelity cDNA Synthesis Kit (Roche Applied Science), using Anchored-oligo(dT)18 Primer (Roche) or with the Super Script III First Strand synthesis system (Life Technologies), using random hexamers (Life Technologies). qPCRs were performed in triplicates using LightCycler 480 SYBR Green I Master Kit (Roche Applied Science) with the following primers: eGFP (GFP fwd 5’-CAACAGCCACAACGTCTATATCATG-3’ and GFP rev 5’-ATGTTGTGGCGGATCTTGAAG-3’), tyrosine hydroxylase (TH fwd 5’-AGCCCTACCAAGACCAGACG-3’ and TH rev 5’-GCGTGTACGGGTCGAACTT-3’), synapsin II (SYN2 fwd 5’-TGGAACAGGCAGAATTTTCA-3’ and SYN2 rev 5’-GGACAACCTTTGTGCCATTC-3’) and ribosomal protein L22 (RPL22 fwd 5’-CACGAAGGAGGAGTGACTGG-3’ and RPL22 rev 5’-TGTGGCACACCACTGACATT-3’). As alternative TaqMan Universal PCR Master Mix (Life Technologies), and the following TaqMan® Gene Expression Assays (Life Technologies): TRKA (ID: Hs01021011_m1); TRKB (ID: Hs00178811_m1); TRKC (ID:Hs00176797_m1); RET (ID:Hs01120030_m1); DDC (ID: Hs01105048_m1); QDPR (ID: Hs00165610_m1), GCH1 (ID: Hs00609198_m1); DRD2 (ID: Hs00241436_m1); SYT1 (ID: Hs00194572_m1); SYP (ID: Hs00300531_m1); SYNPO (ID: Hs00702468_s1); PSD95 (ID: Hs00176354_m1); vGAT (ID: Dm01823909_g1). The reactions were performed with Applied Biosystems 7300 Real Time PCR system or LightCycler 480 Instrument II 96-well block (Roche Applied Science). Quantification cycle values (Cq’s) and melting curves were determined using LightCycler 480 Software version 1.5 (Roche Applied Science). All data were analyzed using the 2-ΔΔCt method for relative gene expression analysis44. Changes in gene expression were normalized using the housekeeping gene RPL22 (ribosomal protein L22) as internal control.
Statistical Analysis
All values are presented as means ± standard error of the mean (s.e.m.) of independent experiments as indicated in the figure captions. Sample size was chosen without accounting for a pre-specified effect size. Data were tested for normality of distribution with the D’Agostino & Pearson omnibus normality test. Statistical significance between groups was assessed using Student’s t-tests. Before choosing the adequate type of t-test, Levene’s test for equal variances was performed. P<0.05 was chosen as the level of significance. All comparisons were made using two-tailed statistical tests.
Supplementary Material
Supplementary information is available at Gene Therapy's website
Acknowledgements
We gratefully acknowledge Dr. Johannes Schwarz for the supply of hmNPC within the scope of the EU project BrainCAV (FP7-222992). This work was supported by BrainCAV (FP7-222992) and Brainvectors (FP7-286071), funded by the EU, PTDC/EBB-BIO/112786/2009 and PTDC/EBB-BIO/119243/2010, funded by Fundação para a Ciência e Tecnologia, Portugal and Cancer Research UK. DS, ACP and PF were recipients of a PhD fellowship from FCT, Portugal (SFRH/BD/78308/2011, PD/BD/52202/2013 and SFRH/BD/70810/2010, respectively).
Footnotes
Conflict of interest
The authors declare no conflict of interest
References
- 1.Forman MS, Trojanowski JQ, Lee VM. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat Med. 2004;10:1055–63. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- 2.Ulusoy A, Kirik D. Development of advanced therapies based on viral vector-mediated overexpression of therapeutic molecules and knockdown of disease-related genes for Parkinson’s disease. Ther Deliv. 2011;2:37–50. doi: 10.4155/tde.10.95. [DOI] [PubMed] [Google Scholar]
- 3.Björklund T, Kirik D. Scientific rationale for the development of gene therapy strategies for Parkinson’s disease. Biochim Biophys Acta. 2009;1792:703–13. doi: 10.1016/j.bbadis.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Gan Y, Jing Z, Stetler RA, Cao G. Gene delivery with viral vectors for cerebrovascular diseases. Front Biosci (Elite Ed) 2013;5:188–203. doi: 10.2741/e607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lentz TB, Gray SJ, Samulski RJ. Viral vectors for gene delivery to the central nervous system. Neurobiol Dis. 2012;48:179–88. doi: 10.1016/j.nbd.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toulouse A, Sullivan AM. Progress in Parkinson’s disease-where do we stand? Prog Neurobiol. 2008;85:376–92. doi: 10.1016/j.pneurobio.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Gray SJ, Woodard KT, Samulski RJ. Viral vectors and delivery strategies for CNS gene therapy. Ther Deliv. 2010;1:517–34. doi: 10.4155/tde.10.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng LR, Maguire-zeiss KA. Gene Therapy in Parkinson’s Disease: Rationale and Current Status. CNS Drugs. 2010;24:177–92. doi: 10.2165/11533740-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manfredsson FP, Mandel RJ. Development of gene therapy for neurological disorders. Discov Med. 2010;9:204–11. [PubMed] [Google Scholar]
- 10.Perreau M, Kremer EJ. The conundrum between immunological memory to adenovirus and their use as vectors in clinical gene therapy. Mol Biotechnol. 2006;34:247–56. doi: 10.1385/MB:34:2:247. [DOI] [PubMed] [Google Scholar]
- 11.Salinas S, Bilsland LG, Henaff D, Weston AE, Keriel A, Schiavo G, et al. CAR-associated vesicular transport of an adenovirus in motor neuron axons. PLoS Pathog. 2009;5:e1000442. doi: 10.1371/journal.ppat.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soudais C, Laplace-Builhe C, Kissa K, Kremer EJ. Preferential transduction of neurons by canine adenovirus vectors and their efficient retrograde transport in vivo. FASEB J. 2001;15:2283–5. doi: 10.1096/fj.01-0321fje. [DOI] [PubMed] [Google Scholar]
- 13.Bru T, Salinas S, Kremer EJ. An Update on Canine Adenovirus Type 2 and Its Vectors. Viruses. 2010;2:2134–53. doi: 10.3390/v2092134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soudais C, Skander N, Kremer EJ. Long-term in vivo transduction of neurons throughout the rat CNS using novel helper-dependent CAV-2 vectors. FASEB J. 2004;18:391–3. doi: 10.1096/fj.03-0438fje. [DOI] [PubMed] [Google Scholar]
- 15.Schüle B, Pera RaR, Langston JW. Can cellular models revolutionize drug discovery in Parkinson’s disease? Biochim Biophys Acta. 2009;1792:1043–51. doi: 10.1016/j.bbadis.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839–45. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 17.Brito C, Simão D, Costa I, Malpique R, Pereira CI, Fernandes P, et al. 3D cultures of human neural progenitor cells: dopaminergic differentiation and genetic modification. Methods. 2012;56:452–60. doi: 10.1016/j.ymeth.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Simão D, Pinto C, Piersanti S, Weston A, Licursi V, Collinson L, et al. Modeling human neural functionality in vitro: 3D culture for dopaminergic differentiation. Tissue Eng Part A. 2015;21:654–68. doi: 10.1089/ten.TEA.2014.0079. [DOI] [PubMed] [Google Scholar]
- 19.Fernandes P, Peixoto C, Santiago VM, Kremer EJ, Coroadinha AS, Alves PM. Bioprocess development for canine adenovirus type 2 vectors. Gene Ther. 2013;20:353–60. doi: 10.1038/gt.2012.52. [DOI] [PubMed] [Google Scholar]
- 20.Fernandes P, Simão D, Guerreiro MR, Kremer EJ, Coroadinha aS, Alves PM. Impact of adenovirus life cycle progression on the generation of canine helper-dependent vectors. Gene Ther. 2015;22:40–9. doi: 10.1038/gt.2014.92. [DOI] [PubMed] [Google Scholar]
- 21.Soudais C, Boutin S, Hong SS, Chillon M, Danos O, Bergelson JM, et al. Canine Adenovirus Type 2 Attachment and Internalization : Coxsackievirus-Adenovirus Receptor, Alternative Receptors, and an RGD-Independent Pathway. J Virol. 2000;74:10639–49. doi: 10.1128/jvi.74.22.10639-10649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh SS, Gopinath P, Ramesh A. Adenoviral vectors: a promising tool for gene therapy. Appl Biochem Biotechnol. 2006;133:9–29. doi: 10.1385/abab:133:1:9. [DOI] [PubMed] [Google Scholar]
- 23.Räty JK, Pikkarainen JT, Wirth T, Ylä-Herttuala S. Gene therapy: the first approved gene-based medicines, molecular mechanisms and clinical indications. Curr Mol Pharmacol. 2008;1:13–23. doi: 10.2174/1874467210801010013. [DOI] [PubMed] [Google Scholar]
- 24.Klein RL, Dayton RD, Leidenheimer NJ, Jansen K, Golde TE, Zweig RM. Efficient neuronal gene transfer with AAV8 leads to neurotoxic levels of tau or green fluorescent proteins. Mol Ther. 2006;13:517–27. doi: 10.1016/j.ymthe.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piersanti S, Astrologo L, Licursi V, Costa R, Roncaglia E, Gennetier A, et al. Differentiated neuroprogenitor cells incubated with human or canine adenovirus, or lentiviral vectors have distinct transcriptome profiles. PLoS One. 2013;8:e69808. doi: 10.1371/journal.pone.0069808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kremer EJ, Boutin S, Chillon M, Danos O. Canine Adenovirus Vectors : an Alternative for Adenovirus-Mediated Gene Transfer. J Virol. 2000;74:505–12. doi: 10.1128/jvi.74.1.505-512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keriel A, René C, Galer C, Zabner J, Kremer EJ. Canine Adenovirus Vectors for Lung-Directed Gene Transfer : Efficacy , Immune Response , and Duration of Transgene Expression Using Helper-Dependent Vectors Canine Adenovirus Vectors for Lung-Directed Gene Transfer : Efficacy , Immune Response , and Durat. J Virol. 2006;80:1487–96. doi: 10.1128/JVI.80.3.1487-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doronin K, Flatt JW, Di Paolo NC, Khare R, Kalyuzhniy O, Acchione M, et al. Coagulation factor X activates innate immunity to human species C adenovirus. Science. 2012;338:795–8. doi: 10.1126/science.1226625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eichholz K, Mennechet FJD, Kremer EJ. Human Coagulation Factor X-Adenovirus Type 5 Complexes Poorly Stimulate an Innate Immune Response in Human Mononuclear Phagocytes. J Virol. 2015;89:2884–91. doi: 10.1128/JVI.03576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soudais C, Boutin S, Kremer EJ. Characterization of cis-acting sequences involved in canine adenovirus packaging. Mol Ther. 2001;3:631–40. doi: 10.1006/mthe.2001.0263. [DOI] [PubMed] [Google Scholar]
- 31.Seiradake E, Lortat-Jacob H, Billet O, Kremer EJ, Cusack S. Structural and mutational analysis of human Ad37 and canine adenovirus 2 fiber heads in complex with the D1 domain of coxsackie and adenovirus receptor. J Biol Chem. 2006;281:33704–16. doi: 10.1074/jbc.M605316200. [DOI] [PubMed] [Google Scholar]
- 32.Salinas S, Zussy C, Loustalot F, Henaff D, Menendez G, Morton PE, et al. Disruption of the coxsackievirus and adenovirus receptor-homodimeric interaction triggers lipid microdomain- and dynamin-dependent endocytosis and lysosomal targeting. J Biol Chem. 2014;289:680–95. doi: 10.1074/jbc.M113.518365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang D, Desbois A, Hou ST. A novel adenoviral vector which mediates hypoxia-inducible gene expression selectively in neurons. Gene Ther. 2005;12:1369–76. doi: 10.1038/sj.gt.3302538. [DOI] [PubMed] [Google Scholar]
- 34.Namikawa K, Murakami K, Okamoto T, Okado H, Kiyama H. A newly modified SCG10 promoter and Cre/loxP-mediated gene amplification system achieve highly specific neuronal expression in animal brains. Gene Ther. 2006;13:1244–50. doi: 10.1038/sj.gt.3302779. [DOI] [PubMed] [Google Scholar]
- 35.Storch A, Paul G, Csete M, Boehm BO, Carvey PM, Kupsch A, et al. Long-term proliferation and dopaminergic differentiation of human mesencephalic neural precursor cells. Exp Neurol. 2001;170:317–25. doi: 10.1006/exnr.2001.7706. [DOI] [PubMed] [Google Scholar]
- 36.Schiedner G, Hertel S, Kochanek S. Efficient transformation of primary human amniocytes by E1 functions of Ad5: generation of new cell lines for adenoviral vector production. Hum Gene Ther. 2000;11:2105–16. doi: 10.1089/104303400750001417. [DOI] [PubMed] [Google Scholar]
- 37.Fernandes P, Santiago VM, Rodrigues AF, Tomás H, Kremer EJ, Alves PM, et al. Impact of E1 and Cre on adenovirus vector amplification: developing MDCK CAV-2-E1 and E1-Cre transcomplementing cell lines. PLoS One. 2013;8:e60342. doi: 10.1371/journal.pone.0060342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferreira TB, Perdigão R, Silva AC, Zhang C, Aunins JG, Carrondo MJT, et al. 293 cell cycle synchronisation adenovirus vector production. Biotechnol Prog. 2009;25:235–43. doi: 10.1002/btpr.64. [DOI] [PubMed] [Google Scholar]
- 39.Silva AC, Simão D, Küppers C, Lucas T, Sousa MFQ, Cruz P, et al. Human amniocyte-derived cells are a promising cell host for adenoviral vector production under serum-free conditions. Biotechnol J. 2015;10:760–71. doi: 10.1002/biot.201400765. [DOI] [PubMed] [Google Scholar]
- 40.Alves P, Moreira J, Rodrigues J, Aunins J, Carrondo M. Two-dimensional versus three-dimensional culture systems: Effects on growth and productivity of BHK cells. Biotechnol Bioeng. 1996;52:429–32. doi: 10.1002/(SICI)1097-0290(19961105)52:3<429::AID-BIT9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 41.Serra M, Correia C, Malpique R, Brito C, Jensen J, Bjorquist P, et al. Microencapsulation technology: a powerful tool for integrating expansion and cryopreservation of human embryonic stem cells. PLoS One. 2011;6:e23212. doi: 10.1371/journal.pone.0023212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deerinck TJ, Bushong EA, Thor A, Ellisman MH. NCMIR methods for 3D EM: a new protocol for preparation of biological specimens for serial block face scanning electron microscopy. 2010 http://ncmir.ucsd.edu/sbfsem-protocol.pdf.
- 43.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.