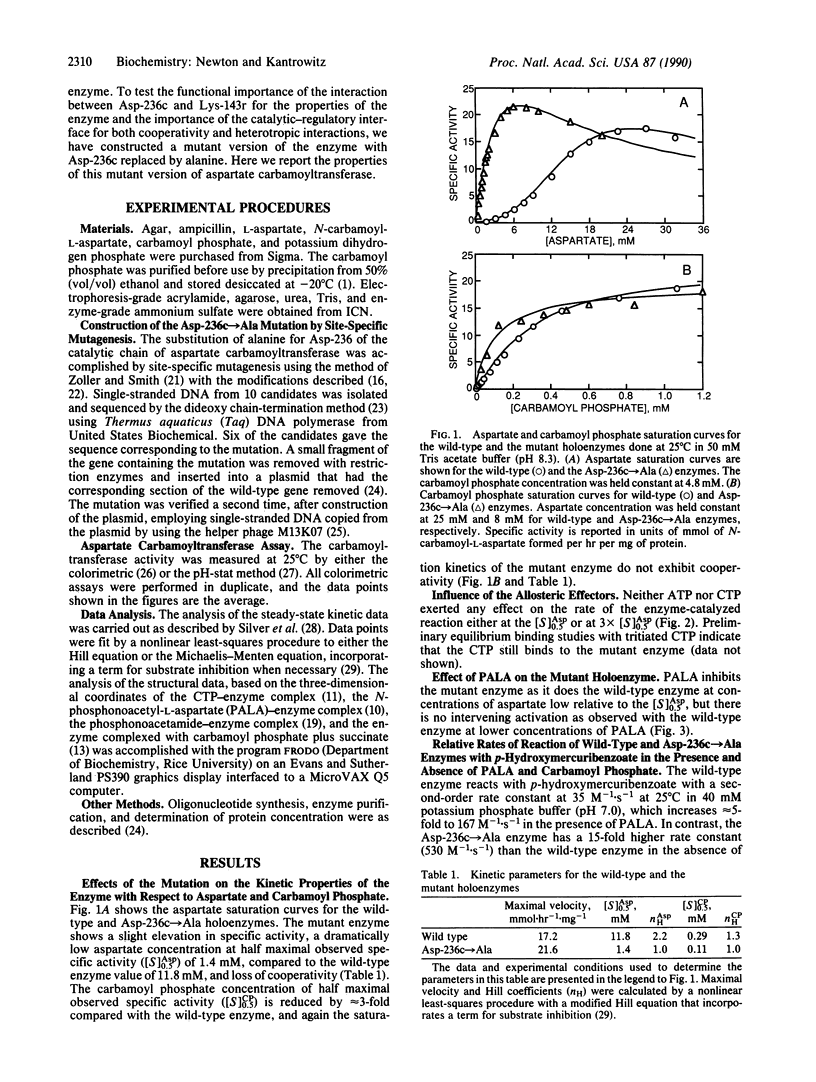
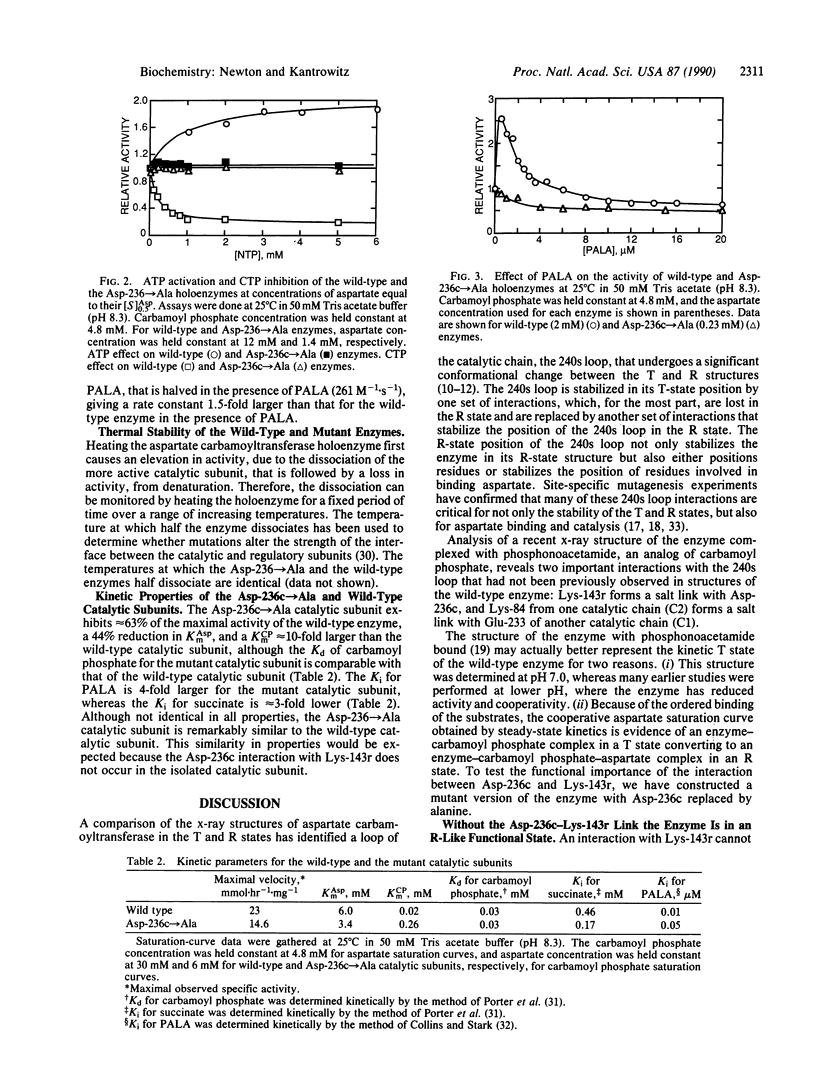
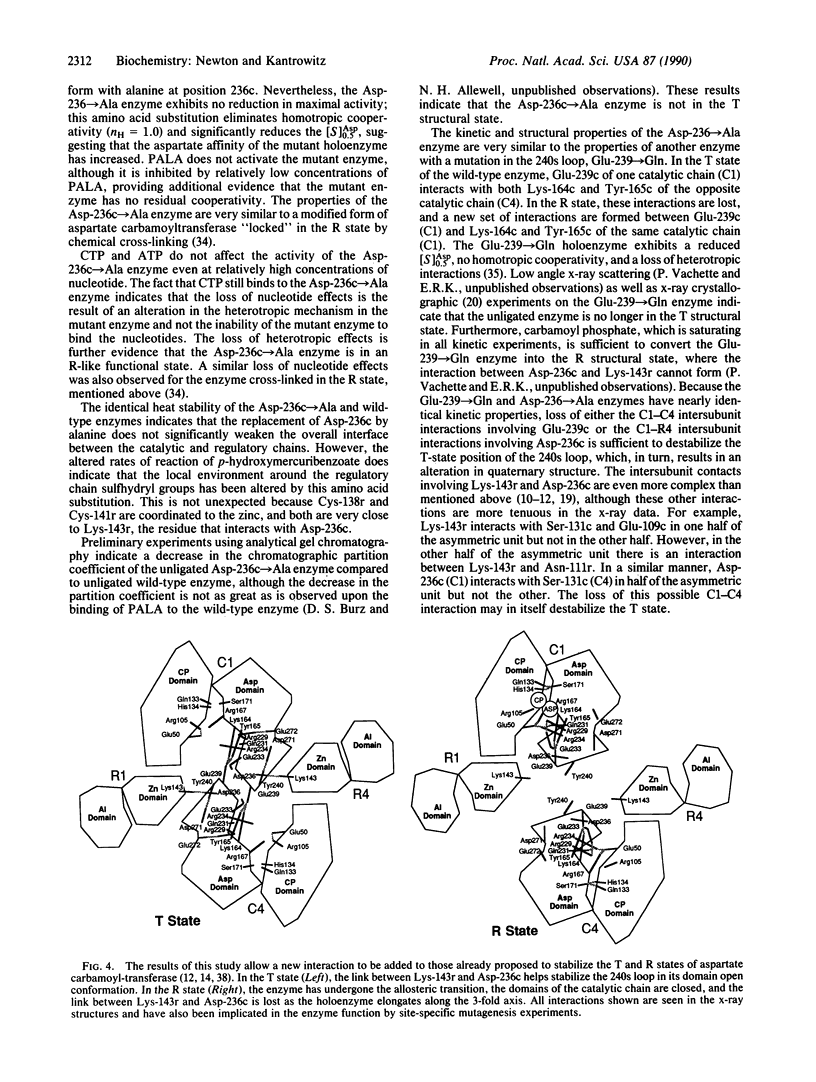
Abstract
A recent x-ray structure of aspartate carbamoyltransferase (carbamoyl-phosphate: L-aspartate carbamoyl-transferase, EC 2.1.3.2) with phosphonoacetamide bound [Gouaux, J. E. & Lipscomb, W. N. (1990) Biochemistry 29, 389-402] shows an interaction between Asp-236 of the catalytic chain and Lys-143 of the regulatory chain. Asp-236 is part of the loop containing residues 230-245 (240s) of the catalytic chain that undergoes a significant conformational change between the tight and the relaxed states of the enzyme. Furthermore, side-chain interactions between the 240s loop and other portions of the enzyme have been shown to be important for the low activity and low affinity of the tight state and the high activity and high affinity of the relaxed state. To determine whether the intersubunit link between Lys-143 of the regulatory chain and Asp-236 of the catalytic chain is important for either homotropic cooperativity and/or the heterotropic interactions in aspartate carbamoyltransferase, site-specific mutagenesis was used to replace Asp-236 with alanine. The mutant enzyme exhibits full activity and a loss of both homotropic cooperativity and heterotropic interactions. Furthermore, the aspartate concentration at half the maximal observed specific activity is reduced by approximately 8-fold. The mutant enzyme exhibits normal thermal stability but drastically altered reactivity toward p-hydroxymercuribenzoate. The catalytic subunit of the mutant and wild-type enzymes have very similar properties. These results, in conjunction with previous experiments, suggest that the intersubunit link involving Asp-236 is involved in the stabilization of the 240s loop in its tight-state position and that the regulatory subunits exert their effect on the catalytic subunits by influencing the position of the 240s loop.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bethell M. R., Smith K. E., White J. S., Jones M. E. Carbamyl phosphate: an allosteric substrate for aspartate transcarbamylase of Escherichia coli. Proc Natl Acad Sci U S A. 1968 Aug;60(4):1442–1449. doi: 10.1073/pnas.60.4.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P., Bedouelle H., Winter G. Improved oligonucleotide site-directed mutagenesis using M13 vectors. Nucleic Acids Res. 1985 Jun 25;13(12):4431–4443. doi: 10.1093/nar/13.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. D., Stark G. R. Aspartate transcarbamylase. Interaction with the transition state analogue N-(phosphonacetyl)-L-aspartate. J Biol Chem. 1971 Nov;246(21):6599–6605. [PubMed] [Google Scholar]
- Eisenstein E., Markby D. W., Schachman H. K. Changes in stability and allosteric properties of aspartate transcarbamoylase resulting from amino acid substitutions in the zinc-binding domain of the regulatory chains. Proc Natl Acad Sci U S A. 1989 May;86(9):3094–3098. doi: 10.1073/pnas.86.9.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enns C. A., Chan W. W. Stabilization of the relaxed state of aspartate transcarbamoylase by modification with a bifunctional reagent. J Biol Chem. 1978 Apr 25;253(8):2511–2513. [PubMed] [Google Scholar]
- GERHART J. C., PARDEE A. B. The enzymology of control by feedback inhibition. J Biol Chem. 1962 Mar;237:891–896. [PubMed] [Google Scholar]
- Gouaux J. E., Lipscomb W. N. Crystal structures of phosphonoacetamide ligated T and phosphonoacetamide and malonate ligated R states of aspartate carbamoyltransferase at 2.8-A resolution and neutral pH. Biochemistry. 1990 Jan 16;29(2):389–402. doi: 10.1021/bi00454a013. [DOI] [PubMed] [Google Scholar]
- Gouaux J. E., Lipscomb W. N. Three-dimensional structure of carbamoyl phosphate and succinate bound to aspartate carbamoyltransferase. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4205–4208. doi: 10.1073/pnas.85.12.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouaux J. E., Stevens R. C., Ke H. M., Lipscomb W. N. Crystal structure of the Glu-239----Gln mutant of aspartate carbamoyltransferase at 3.1-A resolution: an intermediate quaternary structure. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8212–8216. doi: 10.1073/pnas.86.21.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honzatko R. B., Crawford J. L., Monaco H. L., Ladner J. E., Ewards B. F., Evans D. R., Warren S. G., Wiley D. C., Ladner R. C., Lipscomb W. N. Crystal and molecular structures of native and CTP-liganded aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1982 Sep 15;160(2):219–263. doi: 10.1016/0022-2836(82)90175-9. [DOI] [PubMed] [Google Scholar]
- Kantrowitz E. R., Lipscomb W. N. Escherichia coli aspartate transcarbamylase: the relation between structure and function. Science. 1988 Aug 5;241(4866):669–674. doi: 10.1126/science.3041592. [DOI] [PubMed] [Google Scholar]
- Ke H. M., Lipscomb W. N., Cho Y. J., Honzatko R. B. Complex of N-phosphonacetyl-L-aspartate with aspartate carbamoyltransferase. X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms. J Mol Biol. 1988 Dec 5;204(3):725–747. doi: 10.1016/0022-2836(88)90365-8. [DOI] [PubMed] [Google Scholar]
- Kim K. H., Pan Z. X., Honzatko R. B., Ke H. M., Lipscomb W. N. Structural asymmetry in the CTP-liganded form of aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1987 Aug 20;196(4):853–875. doi: 10.1016/0022-2836(87)90410-4. [DOI] [PubMed] [Google Scholar]
- Krause K. L., Volz K. W., Lipscomb W. N. 2.5 A structure of aspartate carbamoyltransferase complexed with the bisubstrate analog N-(phosphonacetyl)-L-aspartate. J Mol Biol. 1987 Feb 5;193(3):527–553. doi: 10.1016/0022-2836(87)90265-8. [DOI] [PubMed] [Google Scholar]
- Krause K. L., Volz K. W., Lipscomb W. N. Structure at 2.9-A resolution of aspartate carbamoyltransferase complexed with the bisubstrate analogue N-(phosphonacetyl)-L-aspartate. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1643–1647. doi: 10.1073/pnas.82.6.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladjimi M. M., Kantrowitz E. R. A possible model for the concerted allosteric transition in Escherichia coli aspartate transcarbamylase as deduced from site-directed mutagenesis studies. Biochemistry. 1988 Jan 12;27(1):276–283. doi: 10.1021/bi00401a042. [DOI] [PubMed] [Google Scholar]
- Ladjimi M. M., Kantrowitz E. R. Catalytic-regulatory subunit interactions and allosteric effects in aspartate transcarbamylase. J Biol Chem. 1987 Jan 5;262(1):312–318. [PubMed] [Google Scholar]
- Ladjimi M. M., Middleton S. A., Kelleher K. S., Kantrowitz E. R. Relationship between domain closure and binding, catalysis, and regulation in Escherichia coli aspartate transcarbamylase. Biochemistry. 1988 Jan 12;27(1):268–276. doi: 10.1021/bi00401a041. [DOI] [PubMed] [Google Scholar]
- Middleton S. A., Kantrowitz E. R. Function of arginine-234 and aspartic acid-271 in domain closure, cooperativity, and catalysis in Escherichia coli aspartate transcarbamylase. Biochemistry. 1988 Nov 15;27(23):8653–8660. doi: 10.1021/bi00423a022. [DOI] [PubMed] [Google Scholar]
- Middleton S. A., Kantrowitz E. R. Importance of the loop at residues 230-245 in the allosteric interactions of Escherichia coli aspartate carbamoyltransferase. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5866–5870. doi: 10.1073/pnas.83.16.5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton S. A., Stebbins J. W., Kantrowitz E. R. A loop involving catalytic chain residues 230-245 is essential for the stabilization of both allosteric forms of Escherichia coli aspartate transcarbamylase. Biochemistry. 1989 Feb 21;28(4):1617–1626. doi: 10.1021/bi00430a029. [DOI] [PubMed] [Google Scholar]
- Navre M., Schachman H. K. Synthesis of aspartate transcarbamoylase in Escherichia coli: transcriptional regulation of the pyrB-pyrI operon. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1207–1211. doi: 10.1073/pnas.80.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastra-Landis S. C., Evans D. R., Lipscomb W. N. The effect of pH on the cooperative behavior of aspartate transcarbamylase from Escherichia coli. J Biol Chem. 1978 Jul 10;253(13):4624–4630. [PubMed] [Google Scholar]
- Pastra-Landis S. C., Foote J., Kantrowitz E. R. An improved colorimetric assay for aspartate and ornithine transcarbamylases. Anal Biochem. 1981 Dec;118(2):358–363. doi: 10.1016/0003-2697(81)90594-7. [DOI] [PubMed] [Google Scholar]
- Porter R. W., Modebe M. O., Stark G. R. Aspartate transcarbamylase. Kinetic studies of the catalytic subunit. J Biol Chem. 1969 Apr 10;244(7):1846–1859. [PubMed] [Google Scholar]
- Robey E. A., Wente S. R., Markby D. W., Flint A., Yang Y. R., Schachman H. K. Effect of amino acid substitutions on the catalytic and regulatory properties of aspartate transcarbamoylase. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5934–5938. doi: 10.1073/pnas.83.16.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof W. D., Foltermann K. F., Wild J. R. The organization and regulation of the pyrBI operon in E. coli includes a rho-independent attenuator sequence. Mol Gen Genet. 1982;187(3):391–400. doi: 10.1007/BF00332617. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver R. S., Daigneault J. P., Teague P. D., Kantrowitz E. R. Analysis of two purified mutants of Escherichia coli aspartate transcarbamylase with single amino acid substitutions. J Mol Biol. 1983 Aug 25;168(4):729–745. doi: 10.1016/s0022-2836(83)80072-2. [DOI] [PubMed] [Google Scholar]
- Stebbins J. W., Xu W., Kantrowitz E. R. Three residues involved in binding and catalysis in the carbamyl phosphate binding site of Escherichia coli aspartate transcarbamylase. Biochemistry. 1989 Mar 21;28(6):2592–2600. doi: 10.1021/bi00432a037. [DOI] [PubMed] [Google Scholar]
- Turnbough C. L., Jr, Hicks K. L., Donahue J. P. Attenuation control of pyrBI operon expression in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1983 Jan;80(2):368–372. doi: 10.1073/pnas.80.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wild J. R., Loughrey-Chen S. J., Corder T. S. In the presence of CTP, UTP becomes an allosteric inhibitor of aspartate transcarbamoylase. Proc Natl Acad Sci U S A. 1989 Jan;86(1):46–50. doi: 10.1073/pnas.86.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. W., Hammes G. G. Relaxation spectra of aspartate transcarbamylase. Interaction of the native enzyme with an adenosine 5'-triphosphate analog. Biochemistry. 1973 Mar 27;12(7):1400–1408. doi: 10.1021/bi00731a021. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]



