Abstract
Gap junction intercellular communication (GJIC) between cardiomyocytes is essential for synchronous heart contraction and relies on connexin-containing channels. Connexin 43 (Cx43) is a major component involved in GJIC in heart tissue, and its abnormal expression is closely associated with various cardiac diseases. Silica nanoparticles (SNPs) are known to induce cardiovascular toxicity. However, the mechanisms through which GJIC plays a role in cardiomyocytes apoptosis induced by SNPs remain unknown. The aim of the present study is to determine whether SNPs-decreased GJIC promotes apoptosis in rat cardiomyocytes cell line (H9c2 cells) via the mitochondrial pathway using CCK-8 Kit, scrape-loading dye transfer technique, Annexin V/PI double-staining assays, and Western blot analysis. The results showed that SNPs elicited cytotoxicity in H9c2 cells in a time- and concentration-dependent manner. SNPs also reduced GJIC in H9c2 cells in a concentration-dependent manner through downregulation of Cx43 and upregulation of P-Cx43. Inhibition of gap junctions by gap junction blocker carbenoxolone disodium resulted in decreased survival and increased apoptosis, whereas enhancement of the gap junctions by retinoic acid led to enhanced survival but decreased apoptosis. Furthermore, SNPs-induced apoptosis through the disrupted functional gap junction was correlated with abnormal expressions of the proteins involved in the mitochondrial pathway-related apoptosis such as Bcl-2/Bax, cytochrome C, Caspase-9, and Caspase-3. Taken together, our results provide the first evidence that SNPs-decreased GJIC promotes apoptosis in cardiomyocytes via the mitochondrial pathway. In addition, downregulation of GJIC by SNPs in cardiomyocytes is mediated through downregulation of Cx43 and upregulation of P-Cx43. These results suggest that in rat cardiomyocytes cell line, GJIC plays a protective role in SNPs-induced apoptosis and that GJIC may be one of the targets for SNPs-induced biological effects.
Keywords: silica nanoparticles, cardiotoxicity, connexin 43, cell death, gap junction
Introduction
Silica nanoparticles (SNPs) have found extensive applications in biomedical and biotechnological fields,1 such as drug delivery, medical diagnostics, biomolecules detection, gene therapy, photodynamic therapy, and bioimaging. So, the opportunities for industrial exposure to SNPs and their consumer use are obviously high. Epidemiological studies have suggested that silica is a key inorganic component that can increase the morbidity and mortality in cardiovascular diseases.2,3 Our previous study has indicated that intratracheal-instilled SNPs could pass through the alveolar–capillary barrier into systemic circulation.4 Thus, the heart could be directly exposed to SNPs. Accumulating evidence from our laboratory demonstrates that SNPs could induce damage in various organs and cause apoptosis in a variety of cells.5–8 Although SNPs have a correlation with cardiovascular diseases, most of the relevant studies primarily focus on their effects on endothelial cells. Whether SNPs affect cardiomyocytes’ functioning has seldom been studied, and the underlying molecular mechanism of SNPs-induced cardiomyocytes cytotoxicity is still poorly understood. Therefore, it is important to understand the interaction between SNPs and cardiomyocytes.
Intercellular communication through gap junctions plays a critical role in a variety of cellular and biological processes, including regulation of cell growth and differentiation, maintenance of tissue homeostasis, and embryogenesis.9 Gap junctions are intercellular membrane channels between adjacent cells that are formed by two hemichannels each consisting of six connexin proteins. These channels mediate direct cell-to-cell communication, facilitating the passage of molecules and ions <1 kDa,10 including small metabolites, and second messengers such as sodium, potassium, calcium, cAMP/cGMP, and ADP/ATP and their translocation from cell to cell.11,12 Connexins are a family of proteins consisting of 20 connexin isoforms in the mouse genome and 21 in the human genome13 that allow the synthesis of a large number of different connexons and gap junction channels. Connexins, either alone or as components of gap junction complex, participate in processes such as growth control,14 migration,15 cell adhesion,16 and intercellular signaling.17
In cardiac muscle, gap junction greatly contributes to electrical cell-to-cell coupling and impulse propagation between cardiomyocytes. In order to maintain cardiovascular homeostasis, cells communicate with neighboring cells and exchange various small molecules via the gap junctions.18,19 Gap junction channels are critical in maintaining cardiac homeostasis by allowing the free flow of ions and metabolites between cardiomyocytes, which contributes to the synchronized contraction and signal exchange throughout the tissue. Such exchange of molecules is thought to be important in coordinating appropriate response to external stimuli. Modulation in various connexin gene expressions has been shown to regulate gap junctional permeability and trafficking of signaling molecules between adjacent cells. In addition, gap junction and connexins have been reported to conduct both cell survival and cell death signals.20 Cell death through apoptosis, compensatory cell proliferation, and disruption of gap junction intercellular communication (GJIC) are important not only for the fixation of unrepaired DNA damage but also in the selection process of initiated precancerous cells. The major gap junction protein of mammalian cardiomyocytes is connexin 43 (Cx43), which is the major constituent of ventricular gap junction channels involved in ventricular electrical coupling.21 Changes in Cx43 expression and gap junction activity have been associated with a wide variety of pathologic conditions and diseases,22–24 including the ventricular myocardium and cardiac arrhythmia.20,25 These effects were further confirmed by recent studies which showed the disorganization of normal microconduction pathways and arrhythmias that resulted from significant remodeling of the distribution of Cx43.26 In addition, phosphorylation of Cx43 affects connexin transport, assembly, and GJIC.
Most studies on apoptosis have focused predominantly on intracellular signaling pathways, and less on intercellular communication and gap junctions. There is a possible mechanistic link between GJIC and apoptosis. For example, GJIC may facilitate signal transduction needed for the apoptotic process in cells contacting each other in the tissues. Alternatively, inhibition of GJIC may interfere with the apoptotic process.27 So far, there are few reports about the effect of nanoparticles on GJIC. Moreover, the consequence of compromised GJIC induced by SNPs in H9c2 had not been reported. To maintain cardiovascular homeostasis, a delicate balance between the number of cells dying and the number of cells proliferating is essential. Several studies have shown that changes in GJIC can influence cell survival or apoptosis.28–30 In rat ventricular myocytes, reduced Cx43 expression resulted in increased apoptosis.31 Similar effects were found in mammary epithelial cells with lower connexin 26 expression.32 This effect was further confirmed in connexin 45-deficient mice in which massive apoptosis was observed in vascular cells.33 Cx43-null mice exhibited increased programmed cell death,34 and blockage of testicular connexins induced apoptosis in rat seminiferous epithelial cells.35 Overall, these studies indicate that alteration in connexin expression profoundly affects cell survival.
GJIC has been widely considered to be involved in cardiomyocytes proliferation and differentiation, but the biochemical and molecular mechanisms underlying the accelerated cell loss induced by SNPs have not been well understood. In particular, the upstream events that trigger apoptosis after exposure to SNPs are unclear. The consequence of reduced Cx43 expression and GJIC induced by SNPs in cardiomyocytes has not been determined. We hypothesize that SNPs-induced downregulation of Cx43 expression reduces GJIC and contributes to cardiomyocytes loss by apoptosis. In this study, we have examined SNPs effects on GJIC and expression of Cx43 in H9c2, and then looked at their possible implications in apoptosis.
Our present work provides the first evidence that SNPs-decreased GJIC promotes apoptosis in cardiomyocytes via the mitochondrial pathway. It suggests GJIC downregulation can also be considered as the potential mechanism responsible for the cytotoxic effects in cardiomyocytes. These results suggest that GJIC plays a protective role in SNPs-induced apoptosis and that GJIC may be one of the targets for SNPs-induced biological effects in cardiomyocytes.
Materials and methods
Materials
SNPs with a diameter of 60 nm and a mass concentration of 12.4 g/L were provided by School of Chemistry, Jilin University. The morphology, average size, and distribution of SNPs in serum-free Dulbecco’s Modified Eagle’s Medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) were determined using a transmission electron microscope (JEM-2010; JEOL, Tokyo, Japan). The average hydrodynamic size of SNPs in serum-free DMEM and serum-DMEM was measured by dynamic light scattering (DLS) method. The SNPs at a concentration of 100 μg/mL were dispersed in serum-free DMEM and DMEM containing 10% fetal bovine serum (FBS) for 24 h. Then, the particle size was analyzed using a laser particle size analyzer without or after ultrasonic treatment for 5 min.
Cell culture and exposure to SNPs
The rat cardiomyocytes cell line, H9c2(2-1) (catalog number: GNR 5), was purchased from Cell Resource Center, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. The cells were maintained in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Gibco), 100 U/mL penicillin, and 100 μg/mL streptomycin, and then incubated at 37°C in a 5% CO2 humidified atmosphere.
Cell viability (CCK-8 assay)
The effect of SNPs on cell viability was determined using WST-8 cell counting kit (CCK-8 Kit; Dojindo, Kumamoto, Japan) according to the manufacturer’s instruction. H9c2 cells were exposed to SNPs. Then, 10 μL of CCK-8 solution was added into each well, and the cells were incubated for an additional 1 h at 37°C. Optical density (nm) was detected at 450 nm using a microplate reader (Thermo Fisher Scientific).
Gap junction activity analysis
Scrape-loading dye transfer technique was used to assess GJIC. Briefly, following treatment, the confluent cells on cover slips were washed three times with phosphate-buffered saline (PBS) containing 0.01% Ca2+ and Mg2+. Then, the cells were cut with a surgical scalpel in the presence of lucifer yellow (LY, 0.5 mg/mL) followed by incubation for 3 min at room temperature, and then rinsed three times with PBS containing 0.01% Ca2+ and Mg2+. The cells were fixed with 4% paraformaldehyde in PBS. The level of gap junction activity was measured using a fluorescent microscope (Diaphot; Nikon, Tokyo, Japan) and quantified as the average distance traveled (mm) by the LY dye from the designated cut to the neighboring cells from six different sites in each sample.
Modulation of GJIC
The pharmacological drugs for GJIC modulation were dissolved in dimethylsulfoxide (DMSO) as stock solutions.36 For the potentiation process, the cells were incubated with gap junction activator retinoic acid (RA, 10 μM in DMSO) for 24 h prior to SNPs exposure and throughout treatment. The cells were also incubated with gap junction blocker carbenoxolone disodium (CBX, 50 mM in DMSO) under the same condition of activator treatment. Control cells were incubated with DMSO alone.
Annexin V-FITC/PI apoptosis assay
The quantification of apoptosis induced by SNPs in H9c2 cells was done using a flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) with Annexin V-FITC/PI double-staining assay. Briefly, the cells were harvested after 24 h of exposure to 100 μg/mL of SNPs, and then stained by using Annexin V-FITC/PI apoptosis assay kit (KeyGen, Nanjing, People’s Republic of China) according to the manufacturer’s instructions. At least 10,000 cells were collected and detected by a flow cytometer within 15 min. The apoptotic percentages were analyzed by FACSDiva 4.1 software.
Western blot assay
Western blot was used to determine the expression of GJIC-/apoptosis-related proteins. Immediately after treatment, the cells were harvested, lysed in ice cold radioimmunoprecipitation assay lysis buffer, and then sonicated. The protein content was determined using a BCA protein assay kit (Beyotime, Jiangsu, People’s Republic of China). Protein samples (50 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% resolving gels, 5% stacking gels) and transferred to polyvinylidene difluoride membranes (150 mA, 1.5 h) (EMD Millipore, Billerica, MA, USA). The membrane was incubated with blocking solution containing 5% nonfat milk in Tris-buffered saline with 0.1% Tween-20 (TBST) before incubation with primary antibodies against Cx43 (1:4,000), P-Cx43 (1:1,000), Bax (1:200), Bcl-2 (1:200), cytochrome C (Cyt C) (1:500), Caspase-9 (1:500), Caspase-3 (1:1,000), and GAPDH (1:5,000) overnight at 4°C, followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5,000) at room temperature for 1.5 h. After washing with TBST, the membranes were processed using an enhanced chemiluminescence detection system (ECL detection kit; Pierce) and exposed to XO mat AR film (Eastman Kodak Company, Rochester, NY, USA). The proteins were quantified through densitometric analysis. Experiments were performed in triplicate to ensure reproducibility.
Statistical analysis
Data were expressed as mean ± standard deviation, and significance was determined by using one-way analysis of variance followed by least significant difference test to compare the differences between groups. Differences were considered significant at P<0.05.
Results
Characterization of SNPs
The hydrodynamic sizes of SNPs were measured in serum-free DMEM as stock media to reflect their dispersion throughout the experiments. The morphology and average size of SNPs were determined by transmission electron microscopy (TEM). The average size of individual nanoparticle was about 60±3.54 nm. The TEM images (Figure 1) showed that dried SNPs were mostly spherical and well dispersed. Influence of serum on hydrodynamic size of SNPs in the testing solution is summarized in Table 1. The purity of SNPs was higher than 99.80%, which was measured using inductively coupled plasma mass spectrometry system.
Figure 1.
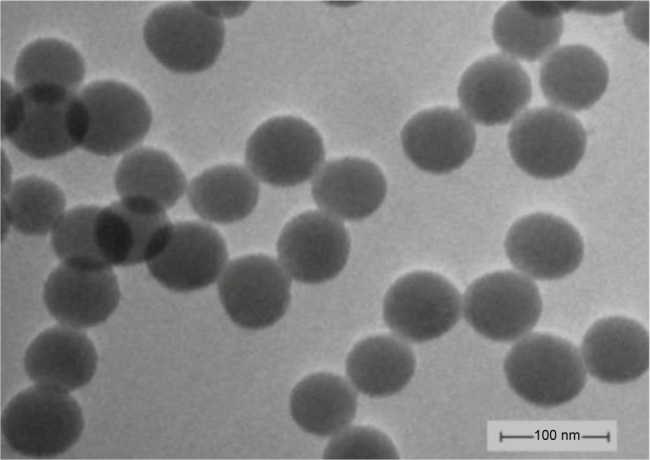
Transmission electron microscopy images of silica nanoparticles in serum-free Dulbecco’s Modified Eagle’s Medium (×100,000).
Table 1.
Influence of serum on hydrodynamic size of SNPs
| Stock media | Groups | Hydrodynamic size (nm) |
|---|---|---|
| SNPs in DMEM | Serum-free DMEM DMEM +10% FBS |
138.51±3.64 321.94±4.58 |
| SNPs in DMEM after ultrasonication | Serum-free DMEM DMEM +10% FBS |
137.99±3.67 200.76±13.78 |
Note: Values represent mean ± standard deviation (n=5).
Abbreviations: SNPs, silica nanoparticles; DMEM, Dulbecco’s Modified Eagle’s Medium; FBS, fetal bovine serum.
Concentration- and time-dependent cytotoxicity of SNPs
To investigate the potential cytotoxicity of the SNPs, we conducted cell viability test in the H9c2 cells treated with SNPs at concentrations of 12.5, 25, 50, 100, and 200 μg/mL for 1, 6, 12, 24, and 48 h. Results (Figure 2) indicated that after exposure to SNPs, the cell viability decreased in a concentration- and time-dependent manner in comparison to control cells. An obvious cytotoxicity of SNPs was observed at the concentration of 100 μg/mL and 24 h under which the viable cells were about 75% of control. We therefore chose SNPs at 100 μg/mL and less for 24 h treatment conditions to study the role of GJIC in apoptosis induced by SNPs.
Figure 2.

Viability of H9c2 cells after exposure to silica nanoparticles at different concentrations and times. The results indicated that viability of H9c2 cells reduced in both concentration- and time-dependent manner (mean, n=6).
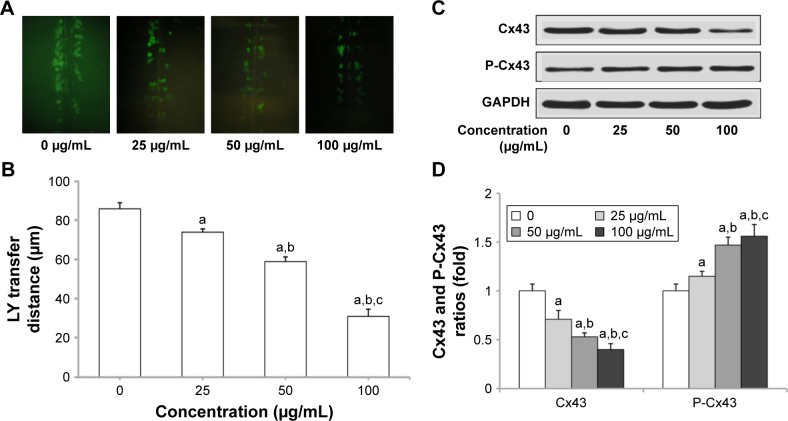
SNPs induce GJIC inhibition in H9c2 cells
Gap junction activity was determined in H9c2 cells following treatment with SNPs. As shown in Figure 3A and B, treatment with SNPs for 24 h resulted in GJIC inhibition in a dose-dependent manner. Furthermore, Cx43 protein expression was downregulated significantly (P<0.05), while the level of P-Cx43 was significantly increased (P<0.05) in a dose-dependent manner after SNPs treatment for 24 h (Figure 3C and D).
Figure 3.
Silica nanoparticles (SNPs)-induced gap junction intercellular communication (GJIC) inhibition in H9c2 cells. (A) SNPs-induced downregulation of GJIC in H9c2 cells. Lucifer yellow (LY, green) transferred to adjacent cells via open gap junctions. (B) The average distance of LY spread from the side of the scraped edge from six different sites in each sample was obtained and quantified. (C and D) Reduced expression of connexin 43 (Cx43) and P-Cx43 after SNPs treatment was detected using Western blot. Values represent the mean ± standard deviation (n=6). aP<0.05, vs control group; bP<0.05, vs 25 μg/mL group; cP<0.05, vs 50 μg/mL group.
Effect of GJIC on the SNPs-induced apoptosis
We investigated the effect of GJIC on the cytotoxicity of SNPs. Gap junction functions were manipulated in two ways: enhancement of junctional channels by RA and inhibition of junctional channels by CBX. For investigating this, the cells were incubated with 10 μM RA or 50 mM CBX for 24 h prior to SNPs exposure and during SNPs treatment. The viability of cells incubated with RA or CBX 24 h prior to SNPs exposure was not affected, whereas CBX combined with 100 μg/mL SNPs caused significant cytotoxicity compared with the control group (Figure 4). Notably, RA combined with 100 μg/mL SNPs increased the cell viability significantly compared with the SNPs treatment alone, yielding substantially decreased cytotoxicity (Figure 4).
Figure 4.
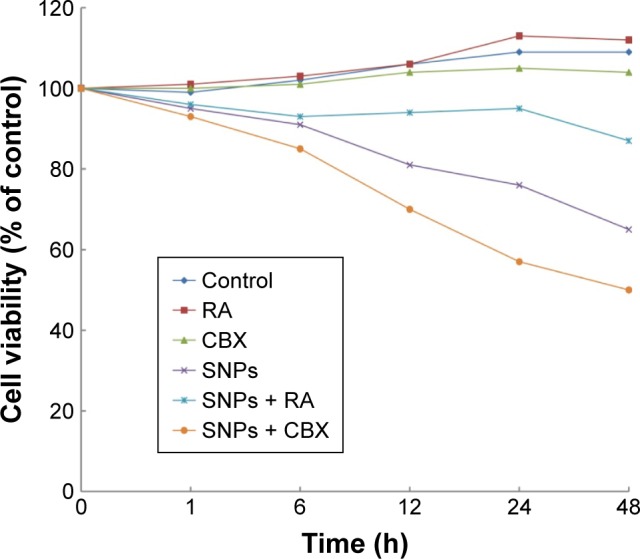
Effect of gap junction activator retinoic acid (RA) and inhibitor carbenoxolone disodium (CBX) on the survival of cells treated with silica nanoparticles (SNPs) (mean, n=6).
The effect of gap junctions activator RA and inhibitor CBX on GJIC of H9c2 cells was also investigated. Figure 5A shows that treatment with 10 μM RA increased GJIC in the H9c2 cells, while treatment with 50 mM CBX decreased GJIC. Compared with 100 μg/mL SNPs alone, RA combined with 100 μg/mL SNPs significantly upregulated GJIC, while CBX combined with 100 μg/mL SNPs significantly downregulated GJIC (P<0.05) (Figure 5B).
Figure 5.
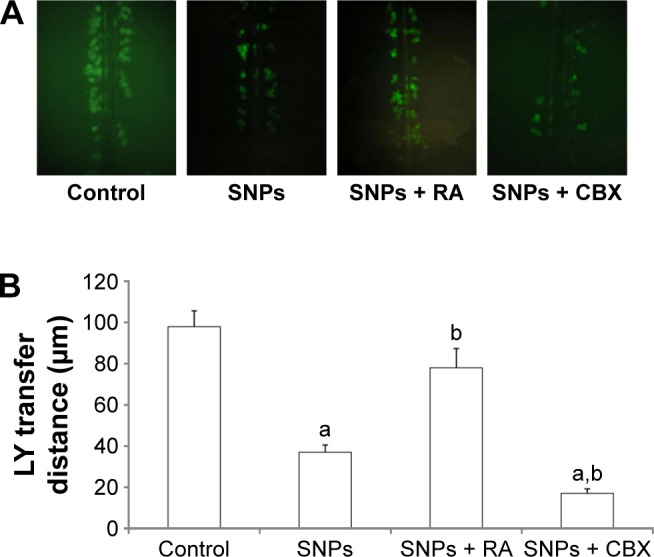
Effect of gap junctions modulator (activator retinoic acid [RA] and inhibitor carbenoxolone disodium [CBX]) on gap junction intercellular communication (GJIC) of H9c2 cells. (A) Modulation of GJIC in H9c2 cells. Lucifer yellow (LY, green) transferred to adjacent cells via open gap junctions. (B) The average distance of LY spread from the side of the scraped edge from six different sites in each sample was obtained for quantification. Values represent the mean ± standard deviation (n=6). aP<0.05, vs control group; bP<0.05, vs silica nanoparticles (SNPs) group.
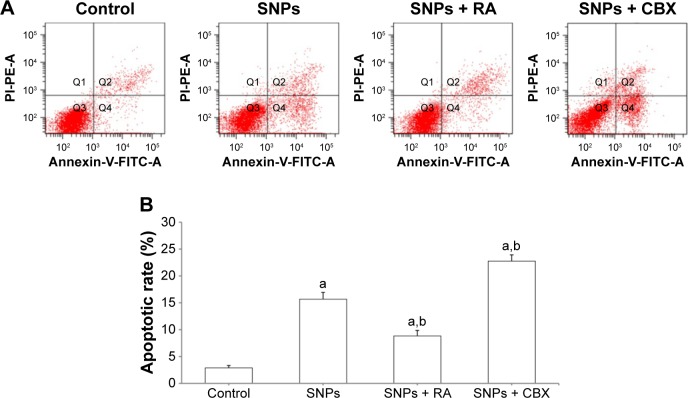
Annexin V/PI double staining was used to assess the effect of GJIC on SNPs-induced apoptosis, which was detected after 24-h incubation with SNPs at the concentration of 100 μg/mL. As shown in Figure 6A and B, all the three treatments induced apoptosis in H9c2 cells evidenced by the significantly increased apoptotic rate (P<0.05). RA decreased the rate of SNPs-induced apoptosis significantly (P<0.05), while CBX significantly increased the rate of SNPs-induced apoptosis (P<0.05).
Figure 6.
Effect of gap junction intercellular communication on silica nanoparticles (SNPs)-induced apoptosis in H9c2 cells determined by flow cytometry. (A) The apoptotic rate after treatment with 100 μg/mL SNPs for 24 h was determined by flow cytometry using Annexin V/PI double staining. (B) Effect of gap junction modulators (activator retinoic acid [RA] and inhibitor carbenoxolone disodium [CBX]) on the apoptotic rates. Values represent the mean ± standard deviation (n=6). aP<0.05, vs control group; bP<0.05, vs SNPs group.
Abbreviations: FITC, fluorescein isothiocyanate; PI, propidium iodide.
Effect of GJIC on the expression of apoptosis-related proteins following SNPs treatment
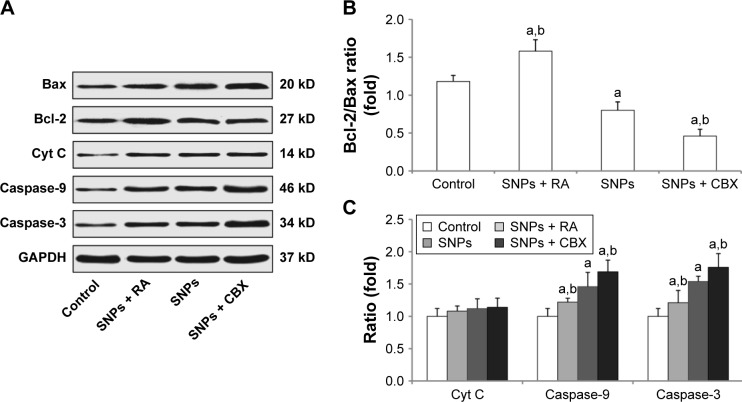
We performed Western blot analysis to measure the expression of apoptosis-related proteins in order to determine the possible factors involved in SNPs-induced apoptosis in the cardiomyocytes. Figure 7A and B shows that the ratio of Bcl-2/Bax was increased significantly in the RA-pretreated group (P<0.05), while it was decreased significantly in the CBX group (P<0.05) compared to that of the 100 μg/mL SNPs-treated group. Figure 7A and C shows that the treatment with 100 μg/mL SNPs and RA or CBX for 24 h activated Cyt C, Caspase-9, and Caspase-3 in H9c2 cells. Compared with SNPs-treated group, the expression of Caspase-3 and Caspase-9 was downregulated significantly in the group treated with SNPs and RA (P<0.05), while it was upregulated significantly in the group treated with SNPs and CBX (P<0.05). These results suggest that GJIC modulates the mitochondrial pathway in apoptosis induced by SNPs.
Figure 7.
Effect of gap junction intercellular communication on the expression of apoptosis-related proteins following treatment with silica nanoparticles (SNPs) as determined by Western blot analysis. (A) Expression of apoptosis-related proteins in H9c2 cells following treatment with SNPs, retinoic adid (RA) combined with SNPs, and carbenoxolone disodium (CBX) combined with SNPs. (B and C) Columns are the means from densitometric scanning from blots, and bars are the standard deviation (SD). Values represent the mean ± SD (n=6). aP<0.05, vs control group; bP<0.05, vs SNPs group.
Discussion
The purpose of this study was to assess the potential role of GJIC in SNPs-induced apoptosis. Hence, SNPs (60 nm) were studied in cultured rat cardiomyocytes cell line (H9c2 cells) to illustrate the relationship between GJIC effect and SNPs-induced apoptosis. It is recommended that prior to the toxicity research, nanomaterials should be appropriately characterized not only initially but also repeatedly if possible during the experiment in order to better interpret and compare the toxic effect. In this study, we characterized the SNPs by TEM and DLS. TEM reflected the morphology and original diameter of the particles (Figure 1). DLS mainly measured the hydrodynamic size of the particles in dispersion media (Table 1). Cell culture medium contains a large amount of serum proteins and a variety of organic and inorganic components. Nanoparticles can adsorb and interact with some components in the culture medium to form agglomerates or aggregates. This can lead to uncertain biological effects and false evaluation of exposure to nanoparticles. In this study, how non-modified SNPs change in serum-free DMEM and whether serum can affect the particle properties were assessed to interpret their toxicity and biological effects in H9c2 cells. SNPs did not aggregate to form larger particles which could not be dispersed by ultrasonic means. Moreover, the size of SNPs in serum-free DMEM could be affected by components in DMEM due to the formation of cluster agglomerates which can be dispersed by ultrasonic means (Table 1). Thus, the SNPs in serum-free DMEM were selected to possess uniform shape and structure along with relatively favorable dispersibility, which were conducive to our study of the role of GJIC in SNPs-induced apoptosis.
The cell viability test results in this study indicated that SNPs induced cytotoxicity in a concentration- and time-dependent manner in H9c2 cells (Figure 2). Yet, the underlying mechanism of SNPs-induced cardiotoxicity remains unclear. In this study, we determined that SNPs could promote apoptosis in H9c2 cells possibly through reducing GJIC and inducing downregulation of Cx43 expression. In line with our observation, other research has reported increased apoptosis in different cell types with reduced GJIC and connexin expression.35 To the best of our knowledge, our study is the first to show that reduced GJIC and downregulation of Cx43 expression may be one of the mechanisms by which SNPs contribute to the induction of apoptosis in H9c2 cells. Additionally, we found that in H9c2 cells, inhibition of GJIC by SNPs increased toxicity, whereas enhancement of GJIC by SNPs decreased toxicity. Furthermore, GJIC-mediated apoptosis in H9c2 cells following treatment with SNPs was associated with expression of Bcl-2/Bax, Cyt C, Caspase-9, and Caspase-3.
When cells are connected via gap junction channels, they can exchange ions and small metabolites and form a cell–cell communication network through GJIC that contributes to the maintenance and regulation of homeostasis. Much toxicological research focuses on single cells, ignoring the influence of adjacent cells on cell survival or death. It has been well described that functional loss of gap junctions can result in apoptosis, necrosis, and carcinogenesis.37–39 In addition to allowing the diffusion of small and hydrophilic substances, GJIC can spatially extend apoptosis through the communication of cell death signals from apoptotic cells to healthy cells.40 When cells are exposed to hazardous stimulants, GJIC downregulation occurs causing injured cells to lose rescue signals provided by healthy cells, which results in a loss of normal growth regulation by the surrounding cells and growth independence.41 Loss of GJIC has been observed in a few other studies after particle exposure. For example, one study found that exposure of rat lung epithelial cells to carbon nanoparticles induced loss of GJIC and delocalization of Cx43.42 Carbon nanotubes-induced reduction in GJIC was found to be involved in inhibiting transfer of cell survival signals between cells.43 In addition, the regulation of the gap junction of human mesenchymal stem cells through the internalization of CdSe/ZnS quantum dots (QDs) was found to play a key role in QD cytotoxicity.44 Furthermore, in rat liver cells, inhibition of GJIC and autophagy were involved in cadmium-induced apoptosis.45 However, one study had opposite results that silver nanoparticles could increase the GJIC in A549 cells through upregulation of Cx43 protein level, suggesting that silver nanoparticles might target GJIC and Cx43 for their biological effects.46 In the present study, the SNPs used had a negative effect on GJIC. We found that SNPs inhibit GJIC in a concentration-dependent manner in H9c2 cells, in addition to causing downregulation of the gap junctional channel protein Cx43 in a concentration-dependent manner after SNPs treatment with upregulation of the level of P-Cx43.
Gap junctions have been described to modulate cell death and survival.47 In this study, culturing with CBX only did not affect cell viability, but CBX combined with SNPs caused significant cytotoxicity compared with the SNPs treatment alone. Notably, RA combined with SNPs increased the cell viability significantly compared with the SNPs treatment alone. So, the involvement of GJIC in the decrease of SNPs-induced cytotoxicity in cardiomyocytes suggests that GJIC transmits particular survival signals or inhibits death signals that are activated when cells are exposed to SNPs. However, the exact properties of death signal or survival signal remain to be elucidated. The candidate signals that were generated by treatment with SNPs and transmitted through gap junction composed of Cx43 in H9c2 cells are probably first or second messengers, such as Ca2+, ATP, IP3, and cAMP. Some reports refer to a distinct role of gap junctional communication in promoting apoptosis. For example, experiments performed with heavy ions or Cyt C as inducers of apoptosis demonstrated cell death in untreated neighboring cells, which points towards a role of gap junctions in modulating apoptosis.38 The Annexin V/PI double-staining apoptosis assay using FACS flow cytometer was used to determine the possible factor in GJIC’s role in SNPs-induced apoptosis in the H9c2 cells. Moreover, GJIC modulation due to SNPs treatment was consistent with the results of other studies. We found that GJIC was involved in SNPs-induced apoptosis in H9c2 cells. CBX significantly increased the rate of SNPs-induced apoptosis compared with the SNPs treatment alone, but RA combined with SNPs exhibited an opposite effect. Overall, these results demonstrate that GJIC may play an important role in SNPs-induced apoptosis in H9c2 cells.
GJIC modulation due to SNPs treatment correlated with the expression of Bcl-2, Bax, Cyt C, Caspase-9, and Caspase-3 determined by Western blotting. These proteins are the apoptosis-related factors of mitochondrial apoptotic pathway. The Bcl-2 family is known as crucial regulators of intrinsic apoptosis. In the Bcl-2 family, Bcl-2 and Bax exert an opposite effect on apoptosis induction. Bcl-2 is the apoptotic inhibition protein that protects cells from apoptosis by inhibiting Caspase-3-dependent proteolytic cascade.47 However, the apoptosis regulator Bax promotes apoptosis by binding to and antagonizing the Bcl-2 protein. A previous study has demonstrated that the Bax/Bcl-2 ratio may upregulate Caspase-3 expression and modulate apoptosis associated with disease progression. In addition, a Bax/Bcl-2 ratio of less than one might be a reasonable “marker” for long-term outcome, as it was correlated with complete stable remission in that study.48 Our results showed that, in H9c2 cells, the downregulation of GJIC decreased the ratio of Bcl-2/Bax, while its upregulation increased the ratio. For the downstream of Bcl-2/Bax, caspase activation was the most important executor of induced apoptosis. The caspase cascade system is crucial in the induction, transduction, and amplification of intracellular apoptotic signals. In the mitochondrial pathway, the release of Cyt C from mitochondria activates Caspase-9, and downstream, Caspase-3 is subsequently activated. In this study, the mitochondrial pathway-related Cyt C expression, Caspase-9 expression, and downstream Caspase-3-activated cleavages were enhanced after GJIC inhibition by SNPs, whereas upregulation of GJIC exerted the opposite effect on Cyt C expression, Caspase-9 expression, and Caspase-3 activation. Taken together, cellular apoptosis through mitochondrial pathway mediated by GJIC was elucidated as one of the toxic mechanisms induced by SNPs exposure in H9c2 cells.
In summary, the present work has demonstrated that SNPs induce GJIC inhibition in addition to apoptosis. The important implication is that subdued GJIC amplifies cytotoxicity in H9c2 cells, whereas enhanced GJIC exerts off-target injury or protection. It suggests GJIC downregulation can also be considered as the potential mechanism for the cytotoxic effects in H9c2 cells.
Acknowledgments
The authors would like to thank Dr Yu Zhang and Dr Shangya Chen from Shandong Academy of Occupational Health and Occupational Medicine, Shandong Academy of Medical Sciences for the preparation of SNPs. This work was supported by the National Natural Science Foundation of China (No 81470145 and 81602893), the Natural Science Foundation of Shandong Province (No ZR2015YL049), the Projects of Medical and Health Technology Development Program of Shandong Province (No 2016WS0540), and the Innovation Project of Shandong Academy of Medical Sciences.
Footnotes
Disclosure
The authors declare that there is no conflict of interest in this work.
References
- 1.Kumar R, Roy I, Ohulchanskky TY, et al. In vivo biodistribution and clearance studies using multimodal organically modified silica nanoparticles. ACS Nano. 2010;4(2):699–708. doi: 10.1021/nn901146y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansen A, Bi P, Nitschke M, et al. Particulate air pollution and cardiorespiratory hospital admissions in a temperate Australian city: a case-crossover analysis. Sci Total Environ. 2012;416:48–52. doi: 10.1016/j.scitotenv.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 3.Hildebrandt K, Rückerl R, Koenig W, et al. Short-term effects of air pollution: a panel study of blood markers in patients with chronic pulmonary disease. Part Fibre Toxicol. 2009;6:25. doi: 10.1186/1743-8977-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du Z, Zhao D, Jing L, et al. Cardiovascular toxicity of different sizes amorphous silica nanoparticles in rats after intratracheal instillation. Cardiovasc Toxicol. 2013;13(3):194–207. doi: 10.1007/s12012-013-9198-y. [DOI] [PubMed] [Google Scholar]
- 5.Yu Y, Li Y, Wang W, et al. Acute toxicity of amorphous silica nanoparticles in intravenously exposed ICR mice. PLoS One. 2013;8(4):e61346. doi: 10.1371/journal.pone.0061346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Sun L, Jim M, et al. Size-dependent cytotoxicity of amorphous silica nanoparticles in human hepatoma HepG2 cells. Toxicol In Vitro. 2011;25(7):1343–1352. doi: 10.1016/j.tiv.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Sun L, Li Y, Liu X, et al. Cytotoxicity and mitochondrial damage caused by silica nanoparticles. Toxicol In Vitro. 2011;25(8):1619–1629. doi: 10.1016/j.tiv.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Duan J, Yu Y, Li Y, Yu Y, Sun Z. Cardiovascular toxicity evaluation of silica nanoparticles in endothelial cells and zebrafish model. Biomaterials. 2013;34(23):5853–5862. doi: 10.1016/j.biomaterials.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 9.De Maio A, Vega VL, Contreras JE. Gap junctions, homeostasis, and injury. J Cell Physiol. 2002;191(3):269–282. doi: 10.1002/jcp.10108. [DOI] [PubMed] [Google Scholar]
- 10.Lauf U, Giepmans BN, Lopez P, Braconnot S, Chen SC, Falk MM. Dynamic trafficking and delivery of connexons to the plasma membrane and accretion to gap junctions in living cells. Proc Natl Acad Sci U S A. 2002;99(16):10446–10451. doi: 10.1073/pnas.162055899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King TJ, Bertram JS. Connexins as targets for cancer chemoprevention and chemotherapy. Biochim Biophys Acta. 2005;1719(1–2):146–160. doi: 10.1016/j.bbamem.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Jiang JX, Gu S. Gap junction- and hemichannel-independent actions of connexins. Biochim Biophys Acta. 2005;1711(2):208–214. doi: 10.1016/j.bbamem.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Söhl G, Willecke K. An update on connexin genes and their nomenclature in mouse and man. Cell Commun Adhes. 2003;10(4–6):173–180. doi: 10.1080/cac.10.4-6.173.180. [DOI] [PubMed] [Google Scholar]
- 14.Naus CC, Bond SL, Bechberger JF, Rushlow W. Identification of genes differentially expressed in C6 glioma cells transfected with connexin43. Brain Res Brain Res Rev. 2000;32(1):259–266. doi: 10.1016/s0165-0173(99)00087-9. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Li WE, Huang GY, et al. N-cadherin and Cx43alpha1 gap junctions modulates mouse neural crest cell motility via distinct pathways. Cell Commun Adhes. 2001;8(4–6):321–324. doi: 10.3109/15419060109080746. [DOI] [PubMed] [Google Scholar]
- 16.Lin JH, Yang J, Liu S, et al. Connexin mediates gap junction-independent resistance to cellular injury. J Neurosci. 2003;23(2):430–441. doi: 10.1523/JNEUROSCI.23-02-00430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scemes E, Suadicani SO, Spray DC. Intercellular communication in spinal cord astrocytes: fine tuning between gap junctions and P2 nucleotide receptors in calcium wave propagation. J Neurosci. 2000;20(4):1435–1445. doi: 10.1523/JNEUROSCI.20-04-01435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodenough DA, Goliger JA, Paul DL. Connexins, connexons, and intercellular communication. Annu Rev Biochem. 1996;65:475–502. doi: 10.1146/annurev.bi.65.070196.002355. [DOI] [PubMed] [Google Scholar]
- 19.Bruzzone R, White TW, Paul DL. Connections with connexins: the molecular basis of direct intercellular signaling. Eur J Biochem. 1996;238(1):1–27. doi: 10.1111/j.1432-1033.1996.0001q.x. [DOI] [PubMed] [Google Scholar]
- 20.Kanno S, Saffitz JE. The role of myocardial gap junctions in electrical conduction and arrhythmogenesis. Cardiovasc Pathol. 2001;10(4):169–177. doi: 10.1016/s1054-8807(01)00078-3. [DOI] [PubMed] [Google Scholar]
- 21.Severs NJ, Bruce AF, Dupont E, Rothery S. Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc Res. 2008;80(1):9–19. doi: 10.1093/cvr/cvn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang CM, Lincoln J, Cook JE, Becker DL. Abnormal connexin expression underlies delayed wound healing in diabetic skin. Diabetes. 2007;56(11):2809–2817. doi: 10.2337/db07-0613. [DOI] [PubMed] [Google Scholar]
- 23.Lin D, Harris R, Stutzman R, Zampighi GA, Davidson H, Takemoto DJ. Protein kinase C-gamma activation in the early streptozotocin diabetic rat lens. Curr Eye Res. 2007;32(6):523–532. doi: 10.1080/02713680701418124. [DOI] [PubMed] [Google Scholar]
- 24.Sawai K, Mukoyama M, Mori K, et al. Redistribution of connexin43 expression in glomerular podocytes predicts poor renal prognosis in patients with type 2 diabetes and overt nephropathy. Nephrol Dial Transplant. 2006;21(9):2472–2477. doi: 10.1093/ndt/gfl260. [DOI] [PubMed] [Google Scholar]
- 25.van Rijen HV, van Veen TA, Gros D, Wilders R, de Bakker JM. Connexins and cardiac arrhythmias. Adv Cardiol. 2006;42:150–160. doi: 10.1159/000092567. [DOI] [PubMed] [Google Scholar]
- 26.Davidson JS, Baumgarten IM. Glycyrrhetinic acid derivatives: a novel class of inhibitors of gap-junctional intercellular communication. Structure-activity relationships. J Pharmacol Exp Ther. 1988;246(3):1104–1107. [PubMed] [Google Scholar]
- 27.Peng YS, Ding HC, Lin YT, Syu JP, Chen Y, Wang SM. Uremic toxin p-cresol induces disassembly of gap junctions of cardiomyocytes. Toxicology. 2012;302(1):11–17. doi: 10.1016/j.tox.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Nakase T, Fushiki S, Naus CC. Astrocytic gap junctions composed of connexin 43 reduce apoptotic neuronal damage in cerebral ischemia. Stroke. 2003;34(8):1987–1993. doi: 10.1161/01.STR.0000079814.72027.34. [DOI] [PubMed] [Google Scholar]
- 29.Krysko DV, Mussche S, Leybaert L, D’Herde K. Gap junctional communication and connexin43 expression in relation to apoptotic cell death and survival of granulosa cells. J Histochem Cytochem. 2004;52(9):1199–1207. doi: 10.1369/jhc.3A6227.2004. [DOI] [PubMed] [Google Scholar]
- 30.Seul KH, Kang KY, Lee KS, Kim SH, Beyer EC. Adenoviral delivery of human connexin37 induces endothelial cell death through apoptosis. Biochem Biophys Res Commun. 2004;319(4):1144–1151. doi: 10.1016/j.bbrc.2004.05.097. [DOI] [PubMed] [Google Scholar]
- 31.Yasui K, Kada K, Hojo M, et al. Cell-to-cell interaction prevents cell death in cultured neonatal rat ventricular myocytes. Cardiovasc Res. 2000;48(1):68–76. doi: 10.1016/s0008-6363(00)00145-0. [DOI] [PubMed] [Google Scholar]
- 32.Bry C, Maass K, Miyoshi K, et al. Loss of connexin 26 in mammary epithelium during early but not during late pregnancy results in unscheduled apoptosis and impaired development. Dev Biol. 2004;267(2):418–429. doi: 10.1016/j.ydbio.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 33.Krüger O, Plum A, Kim JS, et al. Defective vascular development in connexin 45-deficient mice. Development. 2000;127(19):4179–4193. doi: 10.1242/dev.127.19.4179. [DOI] [PubMed] [Google Scholar]
- 34.Furlan F, Lecanda F, Screen J, Civitelli R. Proliferation, differentiation and apoptosis in connexin43-null osteoblasts. Cell Commun Adhes. 2001;8(4–6):367–371. doi: 10.3109/15419060109080755. [DOI] [PubMed] [Google Scholar]
- 35.Lee NP, Leung KW, Wo JY, Tam PC, Yeung WS, Luk JM. Blockage of testicular connexins induced apoptosis in rat seminiferous epithelium. Apoptosis. 2006;11(7):1215–1229. doi: 10.1007/s10495-006-6981-2. [DOI] [PubMed] [Google Scholar]
- 36.Tong X, Dong S, Yu M, Wang Q, Tao L. Role of heteromeric gap junctions in the cytotoxicity of cisplatin. Toxicology. 2013;310:53–60. doi: 10.1016/j.tox.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Zhou JZ, Jiang JX. Gap junction and hemichannel-independent actions of connexins on cell and tissue functions–an update. FEBS Lett. 2014;588(8):1186–1192. doi: 10.1016/j.febslet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kameritsch P, Khandoga N, Pohl U, Pogoda K. Gap junctional communication promotes apoptosis in a connexin-type-dependent manner. Cell Death Dis. 2013;4:e584. doi: 10.1038/cddis.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang FF, Morioka N, Nakashima-Hisaoka K, Nakata Y. Spinal astrocytes stimulated by tumor necrosis factor-α and/or interferon-γ attenuate connexin 43-gap junction via c-jun terminal kinase activity. J Neurosci Res. 2013;91(6):745–756. doi: 10.1002/jnr.23213. [DOI] [PubMed] [Google Scholar]
- 40.Decrock E, De Vuyst E, Vinken M, et al. Connexin 43 hemichannels contribute to the propagation of apoptotic cell death in a rat C6 glioma cell model. Cell Death Differ. 2009;16(1):151–163. doi: 10.1038/cdd.2008.138. [DOI] [PubMed] [Google Scholar]
- 41.Vinken M, Vanhaecke T, Papeleu P, Snykers S, Henkens T, Rogiers V. Connexins and their channels in cell growth and cell death. Cell Signal. 2006;18(5):592–600. doi: 10.1016/j.cellsig.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 42.Ale-Agha N, Albrecht C, Klotz LO. Loss of gap junctional intercellular communication in rat lung epithelial cells exposed to carbon or silica-based nanoparticles. Biol Chem. 2010;391(11):1333–1339. doi: 10.1515/BC.2010.133. [DOI] [PubMed] [Google Scholar]
- 43.Arnoldussen YJ, Anmarkrud KH, Skaug V, Apte RN, Haugen A, Zienolddiny S. Effects of carbon nanotubes on intercellular communication and involvement of IL-1 genes. J Cell Commun Signal. 2016;10(2):153–162. doi: 10.1007/s12079-016-0323-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang JC, Hsu SH, Su HL. The regulation of the gap junction of human mesenchymal stem cells through the internalization of quantum dots. Biomaterials. 2009;30(10):1937–1946. doi: 10.1016/j.biomaterials.2008.12.039. [DOI] [PubMed] [Google Scholar]
- 45.Zou H, Zhuo L, Han T, et al. Autophagy and gap junctional intercellular communication inhibition are involved in cadmium-induced apoptosis in rat liver cells. Biochem Biophys Res Commun. 2015;459(4):713–719. doi: 10.1016/j.bbrc.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 46.Deng F, Olesen P, Foldbjerg R, Dang DA, Guo X, Autrup H. Silver nanoparticles up-regulate Connexin43 expression and increase gap junctional intercellular communication in human lung adenocarcinoma cell line A549. Nanotoxicology. 2010;4(2):186–195. doi: 10.3109/17435390903576451. [DOI] [PubMed] [Google Scholar]
- 47.Swanton E, Savory P, Cosulich S, Clarke P, Woodman P. Bcl-2 regulates a caspase-3/caspase-2 apoptotic cascade in cytosolic extracts. Oncogene. 1999;18(10):1781–1787. doi: 10.1038/sj.onc.1202490. [DOI] [PubMed] [Google Scholar]
- 48.Salakou S, Kardamakis D, Tsamandas AC, et al. Increased Bax/Bcl-2 ratio up-regulates caspase-3 and increases apoptosis in the thymus of patients with myasthenia gravis. In Vivo. 2007;21(1):123–132. [PubMed] [Google Scholar]