Abstract
T lymphocytes undergo extensive changes in their metabolic properties during their transition through various differentiation states, from naïve to effector to memory or regulatory roles. The cause and effect relationship between metabolism and differentiation is a field of intense investigation. Many recent studies demonstrate the dependency of T cell functional outcomes on metabolic pathways and the possibility of metabolic intervention to modify these functions. In this review, we describe the basic metabolic features of T cells and new findings on how these correlate with various differentiation fates and functions. We also highlight the latest information regarding the main factors that affect T cell metabolic reprogramming.
Keywords: metabolism, T lymphocytes, differentiation fate and function
INTRODUCTION
Although aerobic glycolysis has been initially identified as a feature of growing cancer cells [1], it is also observed as a predominant metabolic pathway in other physiological states of other cell types, including T cells. Specifically, undifferentiated, naïve T cells utilize oxidative phosphorylation (OXPHOS) for energy generation, but upon T cell receptor (TCR) activation they switch their metabolic program to glycolysis, which, although energetically less efficient, is required to support cell growth, effector differentiation and function [2–4]. Glycolysis has a selective advantage over oxidative phosphorylation during T cell activation, and although it produces less ATP per cycle, it has a higher ATP generation rate, can function under difficult hypoxic and/or acidic microenvironmental conditions, and has higher biosynthetic benefit and better maintenance of redox balance [5]. These properties make glycolysis highly beneficial for T cells undergoing activation and clonal expansion. Besides encounter with antigens, other factors either cell intrinsic or extrinsic also affect the selection of T cell metabolic programs and play critical role as key regulators of immunity. In this review we will describe the basic metabolic features of T cells, how these correlate with various differentiation pathways and functions, and will also describe the main factors that are involved and affect the above processes.
Basic metabolic features of T cells
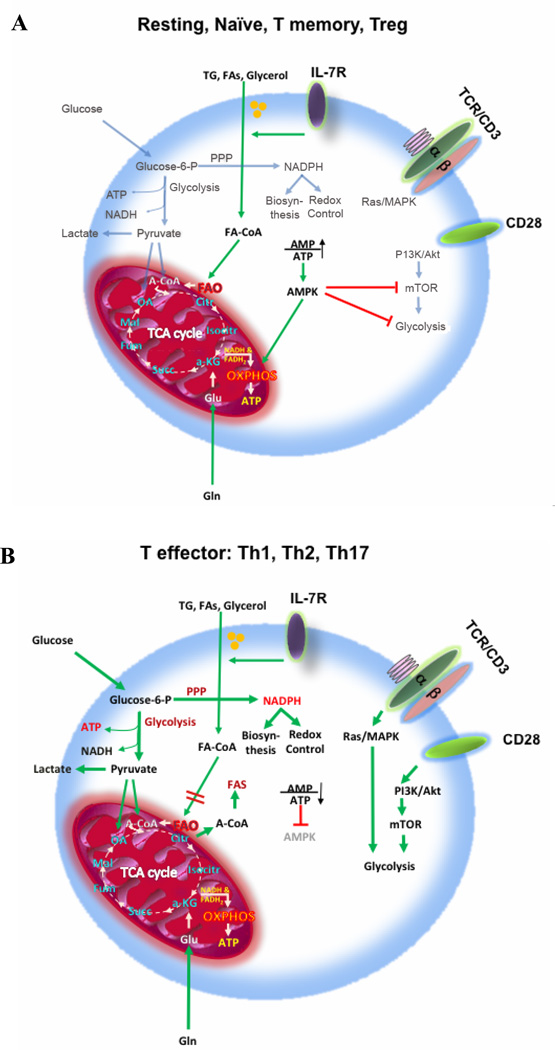
Following maturation process and exit from the thymus, resting T cells have low metabolic requirements and rely predominantly on fatty acid β-oxidation and on pyruvate and glutamine oxidation via the tricarboxylic acid (TCA) cycle. But even resting T cells require cell extrinsic signals, such as IL-7, to maintain this basal energy–generating metabolism and to support their continuous migration through secondary lymphoid tissues on immune surveillance prior to activation (Figure 1A) [2]. Upon encounter with antigens, T cells undergo a dynamic change on their metabolism characterized by extensive proliferation and differentiation into effector T cells (Teff). TCR-mediated signaling promotes the upregulation of glucose and amino acid transporters at the T cell surface and directs the metabolic reprogramming of naïve T cells from oxidative phosphorylation (OXPHOS) to glycolysis (Figure 1B) [6–8]. Intermediate metabolites of glycolysis can be used in the pentose phosphate pathway to support nucleotide, amino acid biosynthesis and generation of reducing capacity in the form of nicotinamide adenine dinucleotide phosphate (NADPH), which is important for anabolic pathways and maintenance of redox balance [5]. Despite the availability of extracellular lipids and the ability of activated T cells to uptake lipids [9], glycolysis also supports de novo fatty acid biosynthesis through generation of pyruvate and TCA cycle-derived citrate which is then transferred to the cytoplasm [10]. On the contrary, catabolic pathways important for ATP production such as fatty acid β-oxidation are actively suppressed due to the upregulation of the transcription factor c-Myc [11]. During the subsequent stages of a T cell response, when the pathogen (or other target antigen) is cleared, most Teff cells die, in a so-called contraction phase, leaving behind a small population of long-lived antigen-specific T cells known as T memory cells (Tm). Tm cells display a characteristic increase in mitochondrial mass and thus a greater mitochondrial spare respiratory capacity (SRC) [2, 12], which is the maximal mitochondrial respiratory capacity available to a cell to produce energy under conditions of increased work or stress. These properties allow Tm cells to respond rapidly to antigen-medicated rechallenge during recall responses.
Figure 1. Basic metabolic features of T cells.
In order for the TCA cycle to function properly, it must be supplied with intermediate metabolites to ensure continuation of its reactions, a process known as anaplerosis. Both, excess of oxaloacetate and acetyl-coA are required for the TCA cycle to continue. Glycolysis can fulfill this requirement by supplying TCA with oxaloacetate and acetyl-coA derived from pyruvate. Moreover, acetyl-coA derived from FAO and ketogenic amino acids can supply the TCA cycle in parallel with pyruvate coming from glycolysis or α-ketoglutarate derived from glutamine. Glycolysis and TCA cycle generate ATP and biosynthetic intermediates. Glucose is necessary not only for glycolysis, but also for the pentose phosphate pathway, which generates intermediates for nucleic acid synthesis and glycolysis and also NADPH. NADPH is a critical co-factor for anabolic reactions such as FA synthesis and also for antioxidant enzymes such as glutathione reductase, and thus important for the maintenance of cellular redox homeostasis. Naïve, resting, Tm and Treg T cells (panel A) depend mainly on OXPHOS, FAO and on either glucose or glutamine to support anaplerosis of the TCA cycle. Tm cells have high spare respiratory capacity and thus are immediately ready for maximal activation upon antigen encounter. In contrast, Teff cells Th1, Th2 and Th17 cells (panel B) depend on high rate of glycolysis and to a lower degree on OXPHOS for maximum energy production and biosynthesis. TG: triglycerides, FAs: fatty acids, FAO: fatty acid oxidation, FAS: fatty acid synthesis, A-CoA: acetyl-coenzyme A, TCA cycle: tricarboxylic acid cycle, PPP: pentose phosphate pathway, OXPHOS: oxidative phosphorylation.
Differentiation of T cell subsets
Besides the metabolic differences of Teff and Tm cells, the metabolic signature of T cells can also vary depending on the differentiation process that leads to the generation of various T cell subsets. CD4+ Teff cells Th1, Th2, and Th17 cells and CD4+ Tregs are the best-defined CD4+ T cell subsets at the metabolic level. It is well known that extracellular queues mediated via specific cytokines determine the differentiation fate of T cell subsets. For example, Th1 cells are induced by type 1 interferons and propagated by IL-12, Th2 cells require IL-4, and Th17 cells are induced by IL-6 and transforming growth factor beta (TGF-β) and propagated by IL-23 and IL-21. The activity of all of these effector T cell types is attenuated by anti-inflammatory Tregs that inhibit T cell proliferation and autoimmune responses [13, 14]. Tregs can be induced from naïve T cells upon exposure to TGF-β and are propagated by IL-2 [15–17]. Metabolism seems to play a significant role in the differentiation outcomes. For example, proinflammatory CD4+ Th1, Th2, and Th17 lineages display a strong bias toward glycolysis over mitochondrial metabolism, whereas induced CD4+ Treg lineage cells display a mixed metabolism involving glycolysis, lipid oxidation, and OXPHOS [18]. Notably, blockade of glycolysis during Th17 differentiation culture favors the formation of Tregs rather than Th17 cells [19]. Interestingly, addition of exogenous fatty acids (FAs) in the culture of T cells activated under skewing conditions strongly inhibits the production of Th1, Th2 and Th17 cytokines, but not the Treg suppressive function. Importantly, inhibition of Teff function in the presence of FAs cannot be rescued by re-addition of Th1-, Th2- and Th17-promoting cytokines [18]. Also, enforcing FA oxidation (FAO) by elevating AMP-activated protein kinase (AMPK) activity or by inhibiting mammalian target of rapamycin (mTOR) results in increased numbers of memory T cells [20, 21]. Although there are pronounced metabolic differences between Teff and Treg cells, distinct metabolic differences within the various Teff subsets have not yet been identified. Furthermore, the cause and effect relationship between metabolism and differentiation of T cells is an active field of investigation.
The above observations point to the convergence of cytokine-mediated signaling and metabolic pathways. The critical pathways that are predominantly activated by the cytokine signals are the Ras/MAPK, the PI3K/Akt/mTOR and the AMPK pathways. The last one promotes lipid oxidation-OXPHOS and counteracts the first two, which promote glycolysis. However, exceptions to the above links exist that suggest a more complex regulation. For example, although it is widely acknowledged that the Akt–mTOR axis is a crucial negative regulator of Treg-cell de novo differentiation [22–26] and expansion [27], and that activation of mTOR delivers a signal for proper activation and differentiation of effector CD4+ T cells [28, 29], another study [30] showed unexpectedly that mTORC1 signaling is a pivotal positive determinant of Treg cell function. Disruption of mTORC1 through Treg-specific deletion of the essential component raptor leads to aberrant expression of lipid metabolism genes and profound loss of Treg cell suppressive activity in vivo resulting in the development of a fatal early onset inflammatory disorder. Regardless of these contradicting findings, it is unequivocal that extracellular signals from the TCR and cytokine receptors are translated into distinct metabolic programs that lead to specific patterns of T cell differentiation and function.
Nutrients and pH
Glucose, amino acids and lipids can support lymphocyte growth. As mentioned above, depending on the state of differentiation, T cells utilize differential nutrient sources and rely largely on glucose for effector functions. Conversely, recent findings support the notion that depending on the type of nutrients present in the microenvironment T cells can acquire differential fate and function [8, 31, 32]. Limiting glucose or glutamine in the culture medium decreases the activation of naive T cells and subsequent T cell proliferation and cytokine production [7, 11, 33–38]. Glutamine can support the TCA cycle through conversion to glutamate via glutaminases, which subsequently leads to production of α-ketoglutarate (α-KG). Importantly, α-KG has been involved in the maintenance of pluripotency of embryonic stem cells by epigenetic mechanisms [39] but it is currently unknown whether α-KG regulates such mechanisms in T cells. Glutamate serves also as a component for glutathione (GSH) synthesis and antioxidant defense [5].
A recent study [40] examined the mechanisms by which nutrient availability impacts Teff cell metabolism and function and revealed a critical role for the key energy sensor AMP-activated protein kinase (AMPK) in the plasticity and adaptation of T cell metabolic reprogramming. This work demonstrated that AMPK is essential for Th1 and Th17 cell development and for primary T cell responses to viral and bacterial organisms in vivo. Besides differentiation, T cell “metabolic fitness” also depends on nutrient availability and is central to the development of effective antitumor immunity. Metabolic fitness is modulated by both the tumor nutrient microenvironment and immune checkpoints [41]. For example, tumor cells overexpress indoleamine-pyrrole 2,3-dioxygenase (IDO) [42, 43], which results in decreased levels of essential amino acids such as L-tryptophan and accumulation of byproducts of amino acid metabolism. These byproducts act in concert with microenvironmental changes induced by hypoxia and acidification induced by the high rate of aerobic glycolysis employed by tumor cells to form a barrier to antitumor immunity. In addition, interaction of T cell inhibitory receptors with their ligands results in down modulation of T cell responses, an effect which although has physiological roles in central and peripheral tolerance, it also has detrimental consequences in disease outcomes. The emerging role of checkpoint inhibition on T cell metabolism is discussed further below.
Role of fatty acids in T cell fate and function
Besides glucose and amino acids, lipids are a good source for energy generation as well as for biosynthetic intermediates [44]. Triglycerides are broken down by various types of phospholipases to glycerol and fatty acids (FAs). Glycerol can enter glycolysis and lead to pyruvate generation while FAs are broken down in mitochondria or peroxisomes. FAs are classified according a) to their backbone lengths (short-, medium-, long- and very long-chain), b) to saturation, i.e. the number of double bonds (unsaturated, mono-, poly-unsaturated), and c) to position of the double bonds. Oxidation of saturated fatty acids gives more ATP molecules than unsaturated fatty acids of the same length. This is because oxidation of saturated FAs requires an extra flavin adenine dinucleotide (FADH)-generating step of double bond formation.
Although not initially aimed to study effects on the immune cells, studies of obesity have revealed important information regarding the effects of lipids on T cell function, which seem to be complexly dependent on diverse types of lipids mentioned above. Here, we will briefly mention representative examples from studies investigating the effects of lipids on T cell fate and function. Generally, FAs are toxic to T cells at high concentrations but when administered at non-toxic concentrations they can affect T cell proliferation [45–47] and can modulate cytokine production [48–50]. FA uptake can occur by passive diffusion through the plasma membrane as well as active transport. Besides using FAs for energy generation like other cell types, breakdown of FAs in T cells also has special importance on T cell differentiation. Specific T cell fates and functional outcomes have been linked to the preferential usage of FA oxidation (FAO) such as the development of CD8+ Tm cells [20, 51] and the induction of Tregs [18], thus leading to the idea that lipid metabolism is a central switch regulating T cell fate decisions. Moreover, Myc, a central transcription factor of growth and proliferation in many cell types, is crucial not only for the activation of glucose metabolizing genes but also for FA synthesis in an mTOR-dependent manner, linking glycolysis to de novo FA synthesis (FAS) [52].
AMPK has a central role in regulating FAO in T cells by multiple mechanisms, including the direct regulation of the enzymatic activity of key lipid metabolizing enzymes [53, 54], the negative regulation of the mTOR pathway [55, 56], and the cellular FA transport [57]. Expression of the FA transporter CD36 is higher in CD8 Teff cells than in CD8+ Tm cells and this correlates with higher ability of Teff cells for FA uptake. Although Tm cells depend on FAO for energy generation, FAs that are used to fuel FAO are not derived from extracellular sources, but are instead produced in a “futile” cell intrinsic manner via fatty acid synthesis (FAS) driven by glycolysis, stored as neutral lipids in lysosomes and mobilized by lysosomal hydrolase and used for FAO [58]. In addition, FAO is crucial for high SRC [51, 59] and mitochondrial biogenesis in CD8+ Tm cells. On CD8+ Tm cells, IL-7 induces a high expression of the glycerol channel Aquaporin 9 (AQP9) and thus contributes to triglyceride synthesis, lipid storage, and effective antiviral responses [60]. Collectively these coordinated processes leading to lipid synthesis and utilization provide two key properties of Tm cells, namely longevity and quiescence.
Role of lactate on T cell fate and function
The hallmark of glycolysis is the lactate degydrogenase (LDH)-mediated conversion of pyruvate to lactate at the expense of one NADH molecule. An important consequence of lactate production is the acidification of the surrounding microenvironment. It has been shown that lactic acid secreted by tumor cells is a proinflammatory mediator that activates the IL-23/IL-17 pathway, thereby inducing inflammation, angiogenesis and tissue remodeling [61]. Studies that have addressed the role of low pH in immunity have mostly focused on the impact of lactate, which promotes the polarization of macrophages toward the M2 suppressive subtype [62]. However, lactate-induced acidification can affect T cell function. For example, lactate-mediated acidification can promote depletion of extracellular arginine levels through Arginase 1, resulting in the inhibition of T cell activation and proliferation [62, 63]. Manipulation of the pH of the tumor microenvironment by the use of proton pump inhibitors results in less dysfunctional tumor infiltrating lymphocytes and increased therapeutic efficacy of both active and adoptive immunotherapy [64]. Lactic acid can suppress the proliferation, cytokine production and cytotoxic activity of human cytotoxic T lymphocytes (CTLs) [65, 66].
A recent study [67] investigated the role of lactate in the regulation of metabolic and inflammatory circuits that control T cell migration and functions in vitro and in vivo and provided evidence that lactate inhibited the motility of chemokine (C-X-C motif) ligand 10 (CXCL10)-activated CD4+ T cells due to a decrease in glycolysis. It makes sense to think that this is a physiological reason why resting mature T cells, which continuously need to migrate, rely more on lipid oxidation and OXPHOS rather than glycolysis. Moreover, in CD4+ T helper cells, lactate induced a switch towards the Th17 differentiation whereas in CD8+ T cells, lactate could suppress cytolytic function [67]. While cancer cells can utilize lactate [68], it is unclear whether T cells also have such capacity. It is also unknown whether utilization of lactate as metabolic substrate might impact Teff or Treg cell development.
Role of reactive oxygen species on T cell fate and function
Although reactive oxygen species (ROS) levels are considered as harmful by-products of metabolism, or weapons of phagocytes against pathogens, ROS also function as signaling messengers in a multitude of pathways, in all cells, tissues and organs. T cell activation is paralleled by transient generation of low, physiologically relevant levels of ROS, i.e., an H2O2-mediated oxidative signal, which facilitates activation of ROS-dependent transcription factors NF-kB and AP-1 [69, 70]. This oxidative signal is indispensable for T cell activation. Together with a Ca2+ influx, it constitutes the minimal requirement for activation-induced gene expression (e.g., interleukin 2 [IL-2], IL-4, CD95 ligand) [71]. Different enzymatic sources such as the respiratory chain [72, 73], lipoxygenases [74] and NADPH oxidases (NOX2, DUOX2) [75, 76] have been described as participating in T cell activation-triggered ROS production. The source, the kinetics, and the localization of ROS production, influence cell responses [71, 75, 77, 78]. In phagocytic cells, ROS are produced by the phagocytic NADPH oxidase (PHOX), an enzyme consisting of several subunits. In T cells, ROS can be produced by NADPH oxidase 2 (NOX-2), which is a catalytic subunit of PHOX expressed in the plasma membrane of T cells, or by a cytoplasmic non-phagocytic isoform of NADPH oxidase called dual oxidase I (DUOX-1). ROS can also derive from the electron transport chain of mitochondria. The important involvement of ROS in T cell metabolic fate and function was recently shown by the identification of lymphocyte expansion molecule (LEM). LEM controls the levels of OXPHOS complexes and respiration, resulting in the production of pro-proliferative mitochondrial ROS, which is critical for promoting antigen-dependent CD8+ T cell proliferation, effector function, and long-term protective memory cells in response to infection with lymphocytic choriomeningitis virus [79].
In contrast to the indispensable role of low ROS levels in T cell activation, prolonged exposure to high ROS concentrations can inhibit T-cell proliferation and lead to apoptosis [80]. In addition, incubation of T cells with reactive nitrogen species (RNS) such as peroxynitrite can inhibit proliferation [81]. Oxidative stress-induced modification to selective molecules involved in T-cell receptor (TCR) signaling can render T cells hyporesponsive to activating stimuli [82]. The redox environment also affects T-cell differentiation. Peripheral blood mononuclear cells (PBMC), stimulated with a ROS generator promoted Th2 and inhibited Th1 differentiation [83]. Moreover, products of lipid peroxidation such as 4-hydroxy-2-nonenal (4HNE) and malonyldialdehyde (MDA), promote differentiation towards a Th2 phenotype [84]. Interestingly, NOX-2 deficiency leads to differentiation towards the Th17 lineage [85]. Since ROS can affect critical metabolism-related T cell signaling pathways such as the MAPKs and Akt pathways [75], it is not unexpected that ROS would directly affect T cell differentiation and function. A study of the novel mechanism of Treg-mediated suppression by extracellular redox remodeling showed that murine Tregs suppress GSH synthesis and cysteine release by dendritic cells (DCs) in a cytotoxic T-lymphocyte-associated protein 4 (CTLA-4)-dependent manner, leading to the oxidation of surface thiols, decrease in the major cellular antioxidant GSH, and reduced proliferation of conventional T cells [86, 87].
In a recent work from our group, metabolite analysis of T cells in the presence of the checkpoint inhibitor programmed cell death 1 (PD-1) showed that PD-1 ligation resulted in significantly more pronounced decrease in the levels of reduced GSH. However, T cells receiving PD-1 signals displayed higher levels of cysteine-GSH disulfide and ophthtalmate, a GSH-like product synthesized by the same enzymes. These changes indicate a higher attempt to increase GSH synthesis, which, together with the more pronounced decrease in the levels of reduced GSH, are suggestive of a more oxidative environment in T cells receiving PD-1 signals [88]. A key mediator of oxidative detoxification is the PPARγ coactivator-1α (PGC-1α) [89, 90]. Induction of PGC-1α during caloric restriction mediates adaptations to provide a permissive setting for increased OXPHOS. Such changes include the expression of uncoupling proteins, which have been associated with decreased oxidation-induced damage. Importantly, mTOR, a target of PD-1 downstream of PI3K/Akt inhibition, has a direct effect on PGC-1α expression [91] and thus, in this way PD-1 might impair oxidative detoxification. Consistent with a role of PD-1 in generating a more oxidative environment, another study showed that, following allogeneic bone marrow transplantation, alloreactive T cells simultaneously upregulated PD-1 expression and ROS production derived from FAO, resulting in higher susceptibility of these cells to metabolic inhibition by F1F0-ATP synthase complex inhibitors, and that process could be reversed by antioxidants. PD-1-blockade not only decreased both mitochondrial H2O2 and total cellular ROS levels but also decreased the efficacy of later F1F0-ATP synthase modulation [92]. Further studies will investigate the role of PD-1 and other checkpoint or metabolic inhibitors in regulating T cell oxidative state, which might serve as one of several fine tuning systems of cellular differentiation.
Role of hypoxia on T cell fate and function
Hypoxia is a condition that occurs physiologically in various microenvironments, such as in primary lymphoid organs, bone marrow and thymus [93, 94] and plays a critical role for thymocyte survival and development [95]. However, most prevalently, hypoxia occurs in tissue microenvironments under pathological conditions such as in the cases of cancer, inflammation, infection, necrosis and autoimmunity. In the hypoxic microenvironment, all cells, including T lymphocytes need to adapt their metabolic profiles to these survival- and growth-unfavorable conditions [6, 11, 96, 97]. Lymphocytes sense hypoxia via the transcription factor hypoxia-inducible factor 1α (HIF-1α), which mediates the metabolic switch from OXPHOS to aerobic glycolysis [98]. A recent study showed that HIF-1α also directly regulates the Th17/Treg balance [99]. Specifically, HIF-1-deficient T cells display a marked reduction in the expression of IL-17 and Th17-signature genes. Mice with HIF-1α-deficient T cells are resistant to induction of Th17-dependent experimental autoimmune encephalitis associated with diminished Th17 and increased Treg cells. On the one hand, HIF-1α activates Th17 development through RORγt and p300, but attenuates Treg development by binding to Foxp3 and targeting it for proteasomal degradation. In support to this finding, it was reported that blocking glycolysis during Th17 cell differentiation reduced the development of Th17 cells and favored the formation of Tregs [19]. Another study [100] showed that deletion of von Hippel–Lindau tumor suppressor (VHL), the main negative regulator of HIFs, altered the differentiation of effector and memory CD8+ T cells. Moreover, in that system, hypoxia modulated the expression of pivotal transcription factors, effector molecules, costimulatory receptors and activation-induced inhibitory receptors in a HIF-1α- and HIF-2α-dependent manner. In addition, the VHL-deficient cytotoxic CD8+ lymphocytes (CTLs) had a high glycolytic and low OXPHOS metabolic profile. The above findings highlight the critical implications of local oxygen concentration to the fate and function of T cells.
Role of PD-1 and checkpoint inhibition on T cell metabolic reprogramming
During activation, in addition to multiple activating receptors and positive costimulatory molecules, T cells upregulate a diversity of inhibitory receptors mostly known as checkpoint inhibitors, which play a physiological role in keeping a balance between stimulatory and inhibitory signals, thereby maintaining central and peripheral tolerance. However, checkpoint inhibitors have detrimental effects on immune T cell functions such as preventing anti-tumor immunity and viral clearance. Particularly in chronic viral infections, T cells enter a condition termed “exhaustion” characterized by progressive and hierarchical loss of effector functions, sustained upregulation and coexpression of multiple inhibitory receptors, altered expression and usage of key transcription factors, and failure to transition to quiescence and to acquire memory T cell homeostatic responsiveness [101].
PD-1 is one of the most extensively studied checkpoint inhibitors and its therapeutic exploitation has proved extremely effective in a number of cancers. In a recent study from our group, we examined the outcome of PD-1 ligation on T-cell metabolic reprogramming [88]. We determined that T cells receiving PD-1 signals were unable to engage in glycolysis, glutaminolysis or metabolism of branched-chain amino acids but displayed an increased rate of FAO. PD-1 largely blocked glucose uptake and glycolysis as evidenced by unchanged levels of glucose and diminished products of glycolysis. This effect of PD-1 was due to inhibition of Glut 1 expression and glucose transport and also due to inhibition of hexokinase 2 (HK2), which catalyzes the first step of glycolysis by converting glucose to glucose-6-phosphate. Besides an impact on glycolysis, PD-1 largely blocked glutamine uptake by impairing the upregulation of glutamine transporters [88]. PD-1 also inhibited glutaminolysis, as determined by higher intracellular levels of glutamine and glutamate. In spite of the abrogated glycolysis and amino acid utilization, these T cells remained viable and metabolically active. A critical pathway targeted by PD-1 is the PI3K/Akt pathway [102]. Inhibition of this pathway by growth factor deprivation in haematopoietic cells activates lipid metabolism through induction of carnitine palmitoyltransferase 1A (CPT1A), the rate-limiting enzyme of mitochondrial FAO, which plays an important role in the utilization of fatty acids as an energy source [103]. Indeed, in T cells PD-1 promoted FAO of endogenous lipids by increasing CPT1A expression and by inducing lipolysis as determined by the increase of the major triacylglycerol (TG) hydrolase desnutrin/adiposite triglyceride lipase (ATGL) and release of fatty acids and glycerol. Concomitantly, PD-1 caused decrease in lipid biosynthesis by abrogation of FA synthase (FASN). Consistent with the increased rate of FAO, PD-1 induced a significant elevation of the ketone body 3-hydroxybutyrate, which is produced during FAO. In addition to increased FAO, activated T cells receiving PD-1 signals had lower extracellular acidification rate (ECAR) and oxygen consumption rate (OCR), which are indicators of glycolysis and oxidative phosphorylation, respectively, but had higher OCR/ECAR ratio compared with T cells stimulated without PD-1 ligation. These findings indicated that in contrast to proliferating T cells, which preferentially use glycolysis for energy production, T cells receiving PD-1 signals are rather metabolically quiescent and preferentially use oxidative phosphorylation than glycolysis as indicated by the higher OCR/ECAR ratio. Moreover, T cells stimulated in the presence of PD-1 ligation possessed substantial SRC. These findings indicated that PD-1 ligation alters the metabolic reprogramming induced upon T cell activation by inhibiting glycolysis and promoting FAO.
In contrast to PD-1, although CTLA-4 inhibited the expression of glutamine and glucose transporters, it did not augment CPT1A and FAO, suggesting that CTLA-4 maintains immune quiescence by preserving the metabolic profile of non-stimulated cells. Since enforcing FAO by pharmacologic means promotes the generation of Treg cells [18], PD-1 might promote Treg development [104] by reprogramming the metabolism of activated T cells from glycolysis to FAO. As burning fat has a strong association with longevity in many cell types [105–107] these unexpected findings indicate that PD-1 ligation enables T cells to survive as long-lived cells by utilizing a fat-based metabolism.
Concluding remarks
Almost six decades since the discovery of the main metabolic features of cancer cells by Otto Warburg, only now we start to appreciate the importance of metabolic reprogramming in normal T cell differentiation and function. The recent advancements in understanding the role of metabolism in T cell fate and function has been made feasible by novel technologies, which allow for the generation of detailed information in real time about how T cells sense extracellular queues that impact their decisions about utilization of specific metabolic programs. Further developments in basic research of T cell metabolism are expected to unravel the complexity of the metabolically driven T cell fate and function and to reveal new targets for therapeutic intervention.
Acknowledgments
This work was supported by NIH grants CA183605, CA183605S1 and AI098129-01 (to VAB), and by the DoD grant PC140571 (to VAB and PS).
We apologize to the authors of the many studies that were not discussed or cited in this review because of limited space.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have no competing conflict of interest.
REFERENCES
- 1.Warburg O. Science. 1956;123:309. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.MacIver NJ, Michalek RD, Rathmell JC. Annu. Rev. Immunol. 2013;31:259. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frauwirth KA, Thompson CB. J. Immunol. 2004;172:4661. doi: 10.4049/jimmunol.172.8.4661. [DOI] [PubMed] [Google Scholar]
- 4.Rathmell JC, Vander Heiden MG, Harris MH, Frauwirth KA, Thompson CB. Mol. Cell. 2000;6:683. doi: 10.1016/s1097-2765(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 5.Cairns RA, Harris IS, Mak TW. Nat. Rev. Cancer. 2011;11:85. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 6.Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, Thompson CB. Immunity. 2002;16:769. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 7.Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, Turay AM, Frauwirth KA. J. Immunol. 2010;185:1037. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA. Nat. Immunol. 2013;14:500. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki K. Clin. Exp. Immunol. 1994;99:479. doi: 10.1111/j.1365-2249.1995.tb05576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. Cell Metab. 2008;7:11. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, Green DR. Immunity. 2011;35:871. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gubser PM, Bantug GR, Razik L, Fischer M, Dimeloe S, Hoenger G, Durovic B, Jauch A, Hess C. Nat. Immunol. 2013;14:1064. doi: 10.1038/ni.2687. [DOI] [PubMed] [Google Scholar]
- 13.Barnes MJ, Powrie F. Immunity. 2009;31:401. doi: 10.1016/j.immuni.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Cell. 2008;133:775. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. J. Exp. Med. 2003;198:1875. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson TS, DiPaolo RJ, Andersson J, Shevach EM. J. Immunol. 2007;178:4022. doi: 10.4049/jimmunol.178.7.4022. [DOI] [PubMed] [Google Scholar]
- 17.Rajewsky K, von Boehmer H. Curr. Opin. Immunol. 2008;20:127. doi: 10.1016/j.coi.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, Maciver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. J. Immunol. 2011;186:3299. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. J. Exp. Med. 2011;208:1367. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Nature. 2009;460:103. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. Nature. 2009;460:108. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. Immunity. 2009;30:832. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haxhinasto S, Mathis D, Benoist C. The Journal of Experimental Medicine. 2008;205:565. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, Knight ZA, Cobb BS, Cantrell D, O'Connor E, Shokat KM, Fisher AG, Merkenschlager M. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7797. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu G, Burns S, Huang G, Boyd K, Proia RL, Flavell RA, Chi H. Nat. Immunol. 2009;10:769. doi: 10.1038/ni.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu G, Yang K, Burns S, Shrestha S, Chi H. Nat. Immunol. 2010;11:1047. doi: 10.1038/ni.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Battaglia M, Stabilini A, Roncarolo MG. Blood. 2005;105:4743. doi: 10.1182/blood-2004-10-3932. [DOI] [PubMed] [Google Scholar]
- 28.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. Nat. Immunol. 2011;12:295. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. Immunity. 2010;32:743. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. Nature. 2013;499:485. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, Anderson SM, Abel ED, Chen BJ, Hale LP, Rathmell JC. Cell Metab. 2014;20:61. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, Blonska M, Lin X, Sun SC. Immunity. 2014;40:692. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ardawi MS, Newsholme EA. Biochem. J. 1983;212:835. doi: 10.1042/bj2120835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brand K. Biochem. J. 1985;228:353. doi: 10.1042/bj2280353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacobs SR, Herman CE, Maciver NJ, Wofford JA, Wieman HL, Hammen JJ, Rathmell JC. J. Immunol. 2008;180:4476. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cham CM, Driessens G, O'Keefe JP, Gajewski TF. Eur. J. Immunol. 2008;38:2438. doi: 10.1002/eji.200838289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cham CM, Gajewski TF. J. Immunol. 2005;174:4670. doi: 10.4049/jimmunol.174.8.4670. [DOI] [PubMed] [Google Scholar]
- 38.Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O'Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, Weber JD, Pearce EJ, Jones RG, Pearce EL. Cell. 2013;153:1239. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Nature. 2015;518:413. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blagih J, Coulombe F, Vincent EE, Dupuy F, Galicia-Vazquez G, Yurchenko E, Raissi TC, van der Windt GJ, Viollet B, Pearce EL, Pelletier J, Piccirillo CA, Krawczyk CM, Divangahi M, Jones RG. Immunity. 2015;42:41. doi: 10.1016/j.immuni.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 41.Siska PJ, Rathmell JC. Trends Immunol. 2015;36:257. doi: 10.1016/j.it.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, Boon T, van den Eynde BJ. Nat. Med. 2003;9:1269. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 43.Jiang T, Sun Y, Yin Z, Feng S, Sun L, Li Z. Future Med. Chem. 2015;7:185. doi: 10.4155/fmc.14.151. [DOI] [PubMed] [Google Scholar]
- 44.Calder PC. Prostaglandins Leukot. Essent. Fatty Acids. 2008;79:101. doi: 10.1016/j.plefa.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 45.Zurier RB, Rossetti RG, Seiler CM, Laposata M. Prostaglandins Leukot. Essent. Fatty Acids. 1999;60:371. doi: 10.1016/s0952-3278(99)80015-5. [DOI] [PubMed] [Google Scholar]
- 46.Gorjao R, Cury-Boaventura MF, de Lima TM, Curi R. Cell Biochem. Funct. 2007;25:305. doi: 10.1002/cbf.1388. [DOI] [PubMed] [Google Scholar]
- 47.Ioan-Facsinay A, Kwekkeboom JC, Westhoff S, Giera M, Rombouts Y, van Harmelen V, Huizinga TW, Deelder A, Kloppenburg M, Toes RE. Eur. J. Immunol. 2013;43:1578. doi: 10.1002/eji.201243096. [DOI] [PubMed] [Google Scholar]
- 48.Stentz FB, Kitabchi AE. Biochem. Biophys. Res. Commun. 2006;346:721. doi: 10.1016/j.bbrc.2006.05.159. [DOI] [PubMed] [Google Scholar]
- 49.Fernanda Cury-Boaventura M, Cristine Kanunfre C, Gorjao R, Martins de Lima T, Curi R. Clin. Nutr. 2006;25:1004. doi: 10.1016/j.clnu.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Szamel M, Rehermann B, Krebs B, Kurrle R, Resch K. Journal of Immunology. 1989;143:2806. [PubMed] [Google Scholar]
- 51.van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL. Immunity. 2012;36:68. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Mol. Cell. 2010;39:171. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jager S, Handschin C, St-Pierre J, Spiegelman BM. Proc. Natl. Acad. Sci. USA. 2007;104:12017. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee WJ, Kim M, Park HS, Kim HS, Jeon MJ, Oh KS, Koh EH, Won JC, Kim MS, Oh GT, Yoon M, Lee KU, Park JY. Biochem. Biophys. Res. Commun. 2006;340:291. doi: 10.1016/j.bbrc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 55.Rolf J, Zarrouk M, Finlay DK, Foretz M, Viollet B, Cantrell DA. Eur. J. Immunol. 2013;43:889. doi: 10.1002/eji.201243008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacIver NJ, Blagih J, Saucillo DC, Tonelli L, Griss T, Rathmell JC, Jones RG. J. Immunol. 2011;187:4187. doi: 10.4049/jimmunol.1100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glatz JF, Luiken JJ, Bonen A. Physiol. Rev. 2010;90:367. doi: 10.1152/physrev.00003.2009. [DOI] [PubMed] [Google Scholar]
- 58.O'Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, Qiu J, Smith AM, Lam WY, DiPlato LM, Hsu FF, Birnbaum MJ, Pearce EJ, Pearce EL. Immunity. 2014;41:75. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Windt GJ, O'Sullivan D, Everts B, Huang SC, Buck MD, Curtis JD, Chang CH, Smith AM, Ai T, Faubert B, Jones RG, Pearce EJ, Pearce EL. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14336. doi: 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cui G, Staron MM, Gray SM, Ho PC, Amezquita RA, Wu J, Kaech SM. Cell. 2015;161:750. doi: 10.1016/j.cell.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shime H, Yabu M, Akazawa T, Kodama K, Matsumoto M, Seya T, Inoue N. J. Immunol. 2008;180:7175. doi: 10.4049/jimmunol.180.11.7175. [DOI] [PubMed] [Google Scholar]
- 62.Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM, Cline GW, Phillips AJ, Medzhitov R. Nature. 2014;513:559. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohashi T, Akazawa T, Aoki M, Kuze B, Mizuta K, Ito Y, Inoue N. Int. J. Cancer. 2013;133:1107. doi: 10.1002/ijc.28114. [DOI] [PubMed] [Google Scholar]
- 64.Calcinotto A, Filipazzi P, Grioni M, Iero M, De Milito A, Ricupito A, Cova A, Canese R, Jachetti E, Rossetti M, Huber V, Parmiani G, Generoso L, Santinami M, Borghi M, Fais S, Bellone M, Rivoltini L. Cancer Res. 2012;72:2746. doi: 10.1158/0008-5472.CAN-11-1272. [DOI] [PubMed] [Google Scholar]
- 65.Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, Gottfried E, Schwarz S, Rothe G, Hoves S, Renner K, Timischl B, Mackensen A, Kunz-Schughart L, Andreesen R, Krause SW, Kreutz M. Blood. 2007;109:3812. doi: 10.1182/blood-2006-07-035972. [DOI] [PubMed] [Google Scholar]
- 66.Mendler AN, Hu B, Prinz PU, Kreutz M, Gottfried E, Noessner E. Int. J. Cancer. 2012;131:633. doi: 10.1002/ijc.26410. [DOI] [PubMed] [Google Scholar]
- 67.Haas R, Smith J, Rocher-Ros V, Nadkarni S, Montero-Melendez T, D'Acquisto F, Bland EJ, Bombardieri M, Pitzalis C, Perretti M, Marelli-Berg FM, Mauro C. PLoS Biol. 2015;13:e1002202. doi: 10.1371/journal.pbio.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kennedy KM, Scarbrough PM, Ribeiro A, Richardson R, Yuan H, Sonveaux P, Landon CD, Chi JT, Pizzo S, Schroeder T, Dewhirst MW. PLoS One. 2013;8:e75154. doi: 10.1371/journal.pone.0075154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Droge W. Physiol. Rev. 2002;82:47. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 70.Kaminski M, Kiessling M, Suss D, Krammer PH, Gulow K. Mol. Cell. Biol. 2007;27:3625. doi: 10.1128/MCB.02295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Devadas S, Zaritskaya L, Rhee SG, Oberley L, Williams MS. J. Exp. Med. 2002;195:59. doi: 10.1084/jem.20010659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yi JS, Holbrook BC, Michalek RD, Laniewski NG, Grayson JM. J. Immunol. 2006;177:852. doi: 10.4049/jimmunol.177.2.852. [DOI] [PubMed] [Google Scholar]
- 73.Kaminski MM, Roth D, Sass S, Sauer SW, Krammer PH, Gulow K. Biochim. Biophys. Acta. 1823:1041. doi: 10.1016/j.bbamcr.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 74.Los M, Schenk H, Hexel K, Baeuerle PA, Droge W, Schulze-Osthoff K. EMBO J. 1995;14:3731. doi: 10.1002/j.1460-2075.1995.tb00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jackson SH, Devadas S, Kwon J, Pinto LA, Williams MS. Nat. Immunol. 2004;5:818. doi: 10.1038/ni1096. [DOI] [PubMed] [Google Scholar]
- 76.Kwon J, Shatynski KE, Chen H, Morand S, de Deken X, Miot F, Leto TL, Williams MS. Sci. Signal. 2010;3:ra59. doi: 10.1126/scisignal.2000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Belikov AV, Schraven B, Simeoni L. J. Biomed. Sci. 2015;22:85. doi: 10.1186/s12929-015-0194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chaudhri G, Clark IA, Hunt NH, Cowden WB, Ceredig R. J. Immunol. 1986;137:2646. [PubMed] [Google Scholar]
- 79.Okoye I, Wang L, Pallmer K, Richter K, Ichimura T, Haas R, Crouse J, Choi O, Heathcote D, Lovo E, Mauro C, Abdi R, Oxenius A, Rutschmann S, Ashton-Rickardt PG. Science. 2015;348:995. doi: 10.1126/science.aaa7516. [DOI] [PubMed] [Google Scholar]
- 80.Thoren FB, Betten A, Romero AI, Hellstrand K. J. Immunol. 2007;179:21. doi: 10.4049/jimmunol.179.1.21. [DOI] [PubMed] [Google Scholar]
- 81.Kasic T, Colombo P, Soldani C, Wang CM, Miranda E, Roncalli M, Bronte V, Viola A. Eur. J. Immunol. 2011;41:1843. doi: 10.1002/eji.201040868. [DOI] [PubMed] [Google Scholar]
- 82.Cemerski S, van Meerwijk JP, Romagnoli P. Eur. J. Immunol. 2003;33:2178. doi: 10.1002/eji.200323898. [DOI] [PubMed] [Google Scholar]
- 83.King MR, Ismail AS, Davis LS, Karp DR. J. Immunol. 2006;176:2765. doi: 10.4049/jimmunol.176.5.2765. [DOI] [PubMed] [Google Scholar]
- 84.Moghaddam AE, Gartlan KH, Kong L, Sattentau QJ. J. Immunol. 2011;187:1626. doi: 10.4049/jimmunol.1003906. [DOI] [PubMed] [Google Scholar]
- 85.Shatynski KE, Chen H, Kwon J, Williams MS. Eur. J. Immunol. 2012;42:3202. doi: 10.1002/eji.201242659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yan Z, Garg SK, Kipnis J, Banerjee R. Nat. Chem. Biol. 2009;5:721. doi: 10.1038/nchembio.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan Z, Garg SK, Banerjee R. J. Biol. Chem. 2010;285:41525. doi: 10.1074/jbc.M110.189944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Patsoukis N, Bardhan K, Chatterjee P, Sari D, Liu B, Bell LN, Karoly ED, Freeman GJ, Petkova V, Seth P, Li L, Boussiotis VA. Nat. Commun. 2015;6:6692. doi: 10.1038/ncomms7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhalla K, Hwang BJ, Dewi RE, Ou L, Twaddel W, Fang HB, Vafai SB, Vazquez F, Puigserver P, Boros L, Girnun GD. Cancer Res. 2011;71:6888. doi: 10.1158/0008-5472.CAN-11-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Cell. 2006;127:397. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 91.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. Nature. 2007;450:736. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 92.Tkachev V, Goodell S, Opipari AW, Hao LY, Franchi L, Glick GD, Ferrara JL, Byersdorfer CA. J. Immunol. 2015;194:5789. doi: 10.4049/jimmunol.1402180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Caldwell CC, Kojima H, Lukashev D, Armstrong J, Farber M, Apasov SG, Sitkovsky MV. J. Immunol. 2001;167:6140. doi: 10.4049/jimmunol.167.11.6140. [DOI] [PubMed] [Google Scholar]
- 94.Hale LP, Braun RD, Gwinn WM, Greer PK, Dewhirst MW. Am. J. Physiol. Heart Circ. Physiol. 2002;282:H1467. doi: 10.1152/ajpheart.00682.2001. [DOI] [PubMed] [Google Scholar]
- 95.Biju MP, Neumann AK, Bensinger SJ, Johnson RS, Turka LA, Haase VH. Mol. Cell. Biol. 2004;24:9038. doi: 10.1128/MCB.24.20.9038-9047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kandalaft LE, Motz GT, Busch J, Coukos G. Curr. Top. Microbiol. Immunol. 2011;344:129. doi: 10.1007/82_2010_95. [DOI] [PubMed] [Google Scholar]
- 97.Berraondo P, Umansky V, Melero I. Cancer Res. 2012;72:5159. doi: 10.1158/0008-5472.CAN-12-1952. [DOI] [PubMed] [Google Scholar]
- 98.Semenza GL. Science’s STKE : Signal Transduction Knowledge Environment. 2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 99.Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, Luo W, Zeller K, Shimoda L, Topalian SL, Semenza GL, Dang CV, Pardoll DM, Pan F. Cell. 2011;146:772. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Doedens AL, Phan AT, Stradner MH, Fujimoto JK, Nguyen JV, Yang E, Johnson RS, Goldrath AW. Nat. Immunol. 2013;14:1173. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wherry EJ, Kurachi M. Nat. Rev. Immunol. 2015;15:486. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Parry RV, Chemnitz JM, Frauwirth KA, Lanfranco AR, Braunstein I, Kobayashi SV, Linsley PS, Thompson CB, Riley JL. Mol. Cell. Biol. 2005;25:9543. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Deberardinis RJ, Lum JJ, Thompson CB. J. Biol. Chem. 2006;281:37372. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- 104.Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, Sharpe AH. J. Exp. Med. 2009;206:3015. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, Huang P, Sawyer SK, Fuerth B, Faubert B, Kalliomaki T, Elia A, Luo X, Nadeem V, Bungard D, Yalavarthi S, Growney JD, Wakeham A, Moolani Y, Silvester J, Ten AY, Bakker W, Tsuchihara K, Berger SL, Hill RP, Jones RG, Tsao M, Robinson MO, Thompson CB, Pan G, Mak TW. Genes Dev. 2011;25:1041. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van der Windt GJ, Pearce EL. Immunol. Rev. 2012;249:27. doi: 10.1111/j.1600-065X.2012.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang MC, O'Rourke EJ, Ruvkun G. Science. 2008;322:957. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]