Abstract
In this review, we provide an update on the current and future applications of saliva for diagnostic purposes. There are many advantages of using saliva as a biofluid. Its collection is fast, easy, inexpensive, and non-invasive. In addition, saliva, as a “mirror of the body,” can reflect the physiological and pathological state of the body. Therefore, it serves as a diagnostic and monitoring tool in many fields of science such as medicine, dentistry, and pharmacotherapy. Introduced in 2008, the term “Salivaomics” aimed to highlight the rapid development of knowledge about various “omics” constituents of saliva, including: proteome, transcriptome, micro-RNA, metabolome, and microbiome. In the last few years, researchers have developed new technologies and validated a wide range of salivary biomarkers that will soon make the use of saliva a clinical reality. However, a great need still exists for convenient and accurate point-of-care devices that can serve as a non-invasive diagnostic tool. In addition, there is an urgent need to decipher the scientific rationale and mechanisms that convey systemic diseases to saliva. Another promising technology called liquid biopsy enables detection of circulating tumor cells (CTCs) and fragments of tumor DNA in saliva, thus enabling non-invasive early detection of various cancers. The newly developed technology—electric field-induced release and measurement (EFIRM) provides near perfect detection of actionable mutations in lung cancer patients. These recent advances widened the salivary diagnostic approach from the oral cavity to the whole physiological system, and thus point towards a promising future of salivary diagnostics for personalized individual medicine applications including clinical decisions and post-treatment outcome predictions.
Impact statement
The purpose of this mini-review is to make an update about the present and future applications of saliva as a diagnostic biofluid in many fields of science such as dentistry, medicine and pharmacotherapy. Using saliva as a fluid for diagnostic purposes would be a huge breakthrough for both patients and healthcare providers since saliva collection is easy, non-invasive and inexpensive. We will go through the current main diagnostic applications of saliva, and provide a highlight on the emerging, newly developing technologies and tools for cancer screening, detection and monitoring.
Keywords: Saliva, diagnostics, trancriptomics, point-of-care, liquid biopsy, biomarkers
Introduction
Saliva (whole saliva [WS], oral fluids [OFs]) is an acidic (pH = 6–7) biological fluid composed of secretions from the three major salivary glands (parotid, submandibular, sublingual) and from minor glands (i.e. labial, buccal, lingual, and palatal tissues), gingival crevicular fluid, cell debris, plaque, bacteria, nasal and bronchial secretions, lining cells, blood and exogenous substances.1,2 It contains 99% water, 0.3% proteins and both 0.2% inorganic and organic substances.3 The most prevalent inorganic components include: sodium, potassium, calcium, magnesium, chloride, and carbonates, while the organic components comprise amylases, peroxidase, lipase, mucins, lysozyme, lactoferrins, kallikreins, cystatins, hormones, and growth factors.4 In a healthy individual, the daily salivary secretion is estimated to be between 0.5 and 1.5 L.5
Saliva plays an important role in many biological functions such as perception of oral sensations (i.e. taste, temperature and touch), lubrication, chewing, swallowing, and digestion. In addition, it enhances remineralization of tooth enamel and prevents demineralization due to its buffering capacity.6,7
Saliva also protects oral mucosa against biological, mechanical, and chemical factors, as well as against bacterial, viral, and fungal infections, thus maintaining the oral cavity ecosystem remain in balance.8,9
There are many advantages of using saliva as a biofluid. Its collection is fast, easy, inexpensive, and non-invasive.10 It is suitable for home use (without the need for medical personal) as well as for epidemiological researches. It is easy to store and ship, does not clot, and can reflect the current physiological state of an individual.1,11 Since there is no need for using needles for sample collection, it is not only more comfortable for patients, since anxiety levels are reduced. Saliva is also beneficial for healthcare providers, as the risk for percutaneous injury and self-contagion is avoided. Saliva, as a “mirror of the body,” can thus reflect the physiological and pathological state of the body.2
In the last few decades, one of the disadvantages of using saliva as a diagnostic tool was the lack of suitable cost-effective technology.12 However, recent publications show that this obstacle will soon be removed. Segal and Wong13 and Wong14 reported that a biological fluid such as saliva could be used as a diagnostic tool for monitoring a disease if it meets the criteria of being easily and non-invasive collected, possesses validated and definitive biomarkers for a specific disease, and is capable of having its biomarkers detected on existing technologies.13,14
In this review, we will present an update on the current approaches of salivary diagnostics, including the discovery of salivary biomarkers for the diagnosis of various human pathological conditions.
Areas of diagnostic application of saliva
Saliva reflects both local and general health of the human body, and thus it has the potential to be used for the detection of essential biomarkers for both oral and systemic diseases.1,10
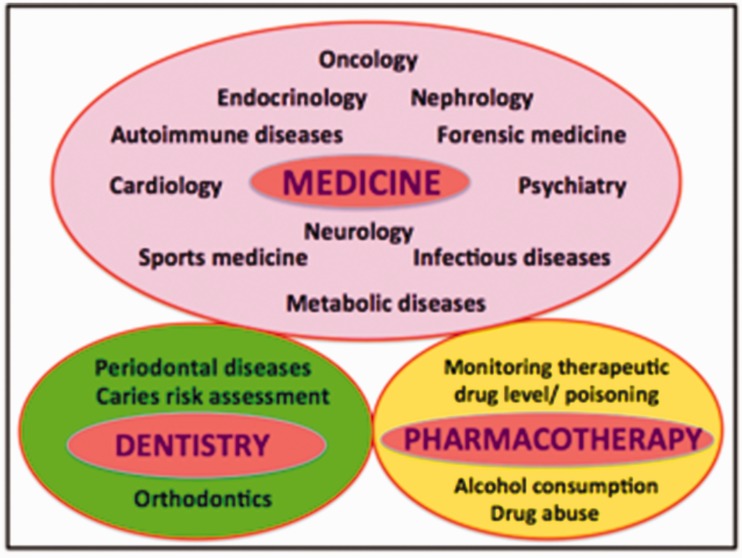
There are many major areas where salivary diagnostics can be applied, including the fields of medicine, dentistry, pharmacotherapy, epidemiology, and bioterrorism (Figure 1).
Figure 1.
Major areas of salivary diagnostics. (A color version of this figure is available in the online journal.)
Medicine
For a better comprehension of some of the main applications of salivary biomarkers in current and prospective medicine, we have subdivided them into several categories according to specific medical specialties:
Oncology
Oncology is a branch of medicine that deals with the prevention, diagnosis, and treatment of cancer. Saliva serves as a useful diagnostic mean in the early detection of various cancers such as oral cancer, pancreatic cancer, breast cancer, lung cancer, or gastric cancer. Anti-p53 antibodies are known tumor markers for esophageal cancer, stomach cancer, and cancer of the large intestine as well as for oral squamous cell carcinoma (OSCC).15 CA-125 is used as a marker for ovarian, endometrial, lung, breast, and gastrointestinal cancers.16 CA 15-3 and c-erbB-2 are used in diagnostics of breast cancer,17 while overexpression of EGFR receptor is observed in pancreatic cancer.18
Oral cancer
The use of salivary biomarkers for early detection of oral cancer, for which the five-year survival rate is still very low (62%), has recently attracted much interest in research studies.19 More than 90% of oral cancers are OSCC and most OSCCs are diagnosed at an advanced stage, thus pointing out the need for clinical diagnostic aids for early detection of OSCC (Figure 2).
Figure 2.
Oral squamous cell carcinoma in the right buccal mucosa of a 50-year-old female patient. Since the lesion was asymptomatic, the patient did not seek for medical consultation earlier and the cancer was diagnosed during a regular check-up appointment. Early diagnosis is preferable due to tendency of the tumor to spread. (A color version of this figure is available in the online journal.)
Currently, more than 100 OSCC biomarkers of various types have been reported in the literature and various technologies were used for their discovery20 (Table 1).
Table 1.
Types of salivary biomarkers used in the diagnosis of OSCC and the technology used for their discovery
| Type of biomarkers (OSCC) | Technology behind |
|---|---|
| Non-organic compound biomarkers | Identified by flame photometry, atomic absorption, and spectrophotometry21 |
| Peptide or protein biomarkers | High-performance liquid chromatography (HPLC)22 Enzyme-linked immunosorbent assay (ELISA)23 Radio-immunoassay24 Two-dimensional gel electrophoresis (2DE) followed by mass spectrometry (MS)25 2DE and reverse-phase liquid chromatography (LC) followed by LC-tandem MS26 Matrix-assisted laser desorption/ionization time-of- flight mass spectrometry (MALDI-TOF MS)27 2DE followed by MALDI-TOF MS28 |
| DNA, mRNA or microRNA biomarkers | Polymerase chain reaction (PCR)29 Quantitative PCR (qPCR)30 Microarrays followed by qPCR31 |
| Metabolomic biomarkers | Capillary electrophoresis TOF MS32 HPLC with quadrupole/TOF MS33 Hydrophilic interaction chromatography-ultraperformance LC-MS analysis34 Reversed phase liquid and hydrophilic interaction chromatography/TOF-MS34 |
| Miscellaneous biomarkers (chemical and enzyme activity) | HPLC35 Colorimetric (mostly commercially available) assays36 |
Three tumor markers (Cyfra 21-1, tissue polypetide antigen [TPA] and cancer antigen CA-125) are significantly increased in saliva of patients diagnosed with OSCC.11,24 Specifically, Cyfra-21-1 was found to be elevated in patients with potentially malignant and malignant disorders of the oral cavity.37 Shpitzer et al.38 reported that cyclin D1 and Ki-67 were increased in correlation with tumor proliferation and the presence of metastasis, thus indicating a poor prognosis for oral cancer patients.
Also, higher levels of MMP-2, MMP-9, and TNFα were observed in oral cancer patients compared to control subjects.38,39
In addition, our group identified five proteins such as M2BP, MRP14, profilin, CD59, and catalase that were able to discriminate oral cancer with greater than 90% clinical accuracy (sensitivity of 90% and specificity of 83%).26
We also developed salivary RNA biomarkers for OSCC by means of microarray analysis including transcripts of IL8, IL-1β, DUSP1, HA3, OAZ1, S100P, and SAT with 91% sensitivity and 91% specificity.31 Similarly, Brinkmann et al.40 and Elashoff et al.41 have recently confirmed that DUSP1, IL-1β, and IL-8 are increased in OSCC.
Furthermore, changes in valine, lactic acid, and phenylalanine yielded high sensitivity (90%) and specificity (83%) with positive predictive value (85%) between healthy, oral cancer and precancerous conditions in the study performed using ultraperformance LC coupled with quadrupole/TOF-MS and multivariate statistical analysis.33 Similarly, recently a study was carried out using hydrophilic interaction chromatography-ultraperformance LC-MS analysis reports significant differences in the concentration of choline, betaine, pipecolinic acid (high in OSCC), and L-carnitine (low in OSCC).34 Finally, the use of a novel system in which reversed phase liquid and hydrophilic interaction chromatography were combined with TOF-MS resulted in the discovery of five new metabolic markers for OSCC, such as propionylcholine, N-acetyl-L-phenylalanine, sphinganine, phytosphingosine, and S-carboxymethyl-L-cysteine.34
Pancreatic cancer
Pancreatic cancer affects ∼44,030 individuals and ∼37,660 succumb to the disease annually in the United States.19 The five-year survival for pancreatic cancer is worst of all human cancers. Our laboratory demonstrated the development of validated salivary extracellular RNA (exRNA) biomarkers for the detection of early resectable pancreatic cancer. The logistic regression model with the combination of four messenger RNA biomarkers (KRAS, MBD3L2, ACRV1 and DPM1) could differentiate pancreatic cancer patients from non-cancer subjects with 90.0% sensitivity and 95.0% specificity (ROC = 0.971).42
Recently, Humeau et al.43 explored that salivary micro-RNA (miRNA) are discriminatory in patients with pancreatic tumors that are not eligible for surgery compared to patients diagnosed with precancerous lesions, inflammatory disease, or cancer-free control subjects.
Gastric cancer
Gastric cancer is the fourth most common cancer and the second leading cause of cancer-related death worldwide. 44 Approximately 880,000 people succumb to this malignancy each year. Such high mortality is mainly due to the delayed diagnosis, since early gastric cancers are typically asymptomatic or cause only nonspecific symptoms.45 Three salivary proteins have been linked to gastric cancer: cystatin B (CSTB), triosephosphate isomerase (TPI1) and deleted in malignant brain tumors 1 protein (DMBT1). These markers could be used to differentiate gastric cancer patients from control subjects (p < 0.05) with 85% sensitivity and 80% specificity.46
Future perspectives
Despite the scientific acceptance of salivary biomarkers for the detection of human diseases, the absence of a mechanistic rationale in regards to the communication between the distal tumor and the oral cavity undermines saliva's scientific credibility for clinical utility. Currently, tumor-derived microvesicles (exosomes) are of high interest to researchers, as they might be the key to understanding the communication between cancer and the oral cavity, leading to the development of tumor-specific salivary biomarkers.47 Using a rodent pancreatic cancer model, we have demonstrated that tumor-specific mRNA markers are shed from the pancreatic tumor cells, packaged in exosome and shuttled to salivary gland.48
Infectious bacterial diseases
Helicobacter pylori (H. pylori) is a Gram-negative, microaerophilic bacterial pathogen that usually grows in the stomach mucus. H. pylori infection is the strongest risk factor for developing gastric and duodenal ulcers, the most common disease of the digestive system49 and has been classified as type I carcinogen by the World Health Organization (WHO) due to its known involvement in the development of gastric MALT lymphoma. In the stomach, it can be diagnosed by means of endoscopy and the urea breath test.50 However, this test should be avoided for diagnosing oral H. pylori infection, since in the oral cavity there are many Campillobacter-like and Streptoccocus bacteria with the urease-positive capability, which may contribute to false positive results.51 Because of these false positives, the detection of H. pylori in the saliva is more effectively detected by means of polymerase chain reaction (PCR).52,53
Infectious viral diseases
Numerous viruses including hepatitis A, B, and C viruses,54–57 cytomegalovirus,58,59 Epstein Barr virus,60 virus herpes (1,2,6,7,8)61 and recently Zika virus62 can be isolated from the saliva. Moreover, measuring the level of salivary antibodies enables detection of Morbillivirus infection causing measles (with 97% sensitivity and 100% specificity),63 Paramyxoviridae causing mumps (94% sensitivity and 94% specificity), or Togaviridiae causing rubella (98% sensitivity and 98% specificity).64,65
Infectious fungal diseases
Some fungal local infections, like candidiasis, have been already diagnosed in saliva.66
Autoimmune diseases
Salivary diagnostics is a common tool in detection of autoimmune diseases such as Sjögren's syndrome, celiac disease, and cystic fibrosis.
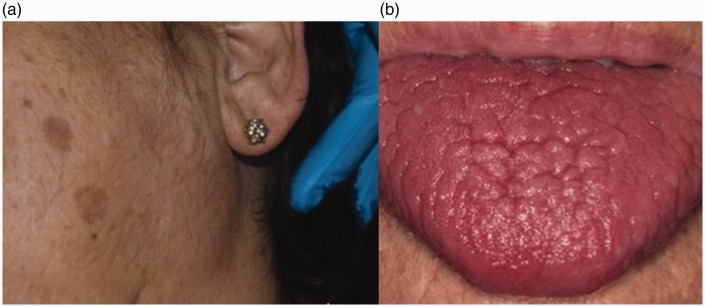
Sjögren's syndrome (SS) is an autoimmune disease of salivary and tear glands (Figure 3). It is characterized by increased levels of salivary interleukins such as IL-2 and IL-6 reduction in stimulated and unstimulated salivary flow and an increase in the concentration of IgA, IgG, IgM, Na, lactoferrin, albumin, β2 microglobulin, cystatin S and C, lipid, prostaglandin E2, and thromboxane B2.67,68 In this line, Streckfus et al.69 analyzed WS in patients with primary and secondary Sjögren's syndrome and those treated with interferon. Their results were consistent with the current literature and confirmed that healthy individuals have lower levels of IL-2 and IL-6 than the affected subjects. In addition, the authors stated that the application of topical interferon may increase the rate of salivary secretion and decrease the presence of these cytokines in saliva.
Figure 3.
55-year-old female patient with xerostomia, xeroftalmia, and non-tumoral, non-inflammatory bilateral enlargement of the parotid gland diagnosed as Sjögren's syndrome. (A color version of this figure is available in the online journal.)
In 2007, our group identified a panel of protein and mRNA biomarkers in saliva for the detection of primary Sjögren Syndrome (pSS).70 In the later study by Hu et al., three protein biomarkers (cathepsin D [CPD], α-enolase, and ß2-microglobulin [ß2m]) and three mRNA biomarkers (myeloid cell nuclear differentiation antigen [MNDA], guanylate binding protein 2 [GBP-2], and low-affinity IIIb receptor for the Fc fragment of IgG) were significantly elevated in patients with primary SS compared with both systemic lupus erythematosus (SLE) patients and healthy controls. The combination of three protein biomarkers (CPD, α-enolase and ß2m) yielded a receiver operating characteristic (ROC) value of 0.99 in distinguishing primary SS from healthy controls. The combination of protein biomarker ß2m and 2 mRNA biomarkers, MNDA, and GBP-2, reached a ROC of 0.95 in discriminating primary SS from SLE.71 We have also discovered salivary autoantibody biomarkers for primary SS using a protein microarray approach that reflects damaged glandular cells and an activated immune response. These results have the potential to lead to the development of a low-cost clinical tool for simple, non-invasive detection of pSS.72
In celiac disease, the ingestion of gluten leads to the damage of salivary anti-gliadin antibodies in the small intestine of genetically predisposed patients.73,74 Bonamico et al.75 demonstrated the presence of IgA and anti-tissue transglutaminase (tTG) in saliva of children diagnosed with celiac disease, while Dane et al.76 reported about decreased salivary flow rate and buffering capacity compared to non-celiac controls.
Cystic fibrosis patients showed increased levels of prostaglandin PGE2 and decreased activity of protease and EGF in saliva.77–79
Endocrinology
In endocrinology, saliva has proven to be a useful diagnostic tool in measuring the concentration of unconjugated steroid hormones and melatonin, even substituting other biological fluids used until now (such as plasma, serum, and urine).80 Currently, salivary diagnostics allow for monitoring of the cycle of hormonal secretions, endocrine functions using dynamic tests (Dexamethasone), controlling the concentration and metabolism of hormones used as drugs (hormone replacement therapy) and determining the free fraction of many hormones. The consolidation of these techniques could significantly reduce the costs of expensive hormonal endocrine studies.81,82
Thus, saliva is used to monitor the levels of aldosterone,83 parathyroid hormone,84 glucose,85 and insulin.86,87
In addition, the levels of dehydroepiandrosterone are measured in the diagnosis of hirsutism, adrenal tumors, and adrenal genital syndrome,88,89 progesterone in diagnosis of menstruation disorders or infertility,90 estriol in diagnosis of fetal maturation,91 or cortisol in diagnosis of Cushing's syndrome.92–94 Saliva is also used for determining of levels of estradiol and testosterone in the diagnosis of hirsutism, menstrual disorders, adrenal and testicular tumors, steroidogenesis disorders, and for control of antiandrogenic therapy outcomes.95 Taking into consideration that testosterone in saliva is free and unbound with proteins (sex-hormone-binding globulins [SHBGs]), the superiority of its determination is unquestionable compared to other biofluids.96,97
Psychiatry
Several authors have demonstrated the changes in hormones such as cortisol and alpha-amylase in anxiety disorders.98–102 For instance, Richter et al.103 studied salivary cortisol levels in a population of pregnant women with anxiety disorder.103 In this line, Rai et al.104 conducted a study on periodontal patients with depression. Also, a wide range of stressors have been already explored in occupational and environmental medicine.105 Recently, measurements of testosterone are widely used in assessing the degree of aggression, depression, violence, and antisocial behavior in psychiatry.106,107
Nephrology
Currently, the levels of salivary creatinine in saliva can be determined in order to monitor renal functions, and to ascertain the efficacy of dialysis in patients with end-stage terminal renal disease.108,109
Venkatapathy et al.110 studied the correlation between serum and salivary creatinine in chronic renal failure patients. Their results showed that creatinine was higher both in serum and saliva of diseased patients. With a cut-off value of 0.2mg/dL, a sensitivity of 97.1% and specificity of 86% was reported.
Cardiology
Salivary diagnostics also plays an important role in cardiology, i.e. in risk assessment for cardiovascular diseases in people with insulin resistance111,112 or for acute miocardial infarction.113 Thus, salivary alpha-amylase was reported as an independent diagnostic factor for acute myocardial infarction in patients suffering from precordial pain less than 4 h.114 In addition, the use of nano-biochips based on salivary proteins (including C-reactive protein, myoglobin and myeloperoxidase) in patients with acute myocardial infarction was reported to be an effective technique for screening purposes.115
In this way, salivary analysis may contribute to a better therapeutic management of acute cardiac events.
Metabolic diseases
Salivary biomarkers have been recently explored as a useful screening tool in patients diagnosed with metabolic disorders such as obesity116 or diabetes mellitus.86,117–119
Diabetes mellitus is a metabolic disorder characterized by elevated blood glucose levels and it is one of the most important health problems faced by the humankind today. It leads to morphological changes in the salivary glands and in the composition of saliva. According to Malicka et al.,120 myeloperoxidase and IgA were correlated with a poor periodontal status in diabetic patients.120
In addition, in diabetes mellitus type 2, Aitken-Saavedra et al.121 reported a positive correlation between HbA1 and total amount of proteins, and an indirect correlation between HbA1 and pH in saliva. Since diabetes is a common chronic disease with many associated comorbilities, measuring the qualitative and quantitative salivary alterations could be a promising and cheaper way of monitoring affected patients.
Neurology
Salivary diagnostics is also used in some neurodegenerative diseases like Alzheimer's disease, where Shi et al.122 found elevated levels of phosphorilated and total TAU proteins compared to healthy individuals.
In addition, Jang et al.123 observed increased levels of nerve growth factor (NGF) and sensory neuropeptides (including substance P and calcitonin gene-related peptide [CGRP]) strongly correlated with the severity of pain in patients diagnosed with chronic migraine.123
Sports medicine
Saliva analysis enables to monitor the metabolic response of sportsmen during physical training. Thus, they can avoid overtraining and lessen the risk of injuries. Moreover, they can accordingly modify their plan of training, in terms of duration, frequency, and intensity of exercises. Gatti and de Palo124 have published a thorough review of the salivary components and their changes in relation to physical workouts. Also, Zauber et al.125 studied changes in protein and metabolite levels in saliva during excessive exercising.
Forensic medicine
Saliva is a useful diagnostic tool in forensic sciences, where there is a possibility to differentiate individuals, who are still alive, from dead bodies. Interestingly, due to the fast oral tissue turnover, DNA extracted from the OFs is much more valid compared to other possible DNA sources.126
Dentistry
Periodontics
Analysis of saliva may serve as a useful tool in assessment of current periodontal status, monitoring response to treatment and prediction of disease progression. Salivary biomarkers for periodontal diseases include proteins of host origin (i.e. enzymes and immunoglobulins), phenotypic markers, host cells, hormones, bacteria and bacterial products, ions, and volatile compounds.127 The most common periodontal pathogens implicated in periodontal diseases include Tanerella forsythensis, Porphyromonas gingivalis and Treponema denticola, so called “red complex” of bacteria.128,129
Host response and inflammatory mediators in saliva include: IL-1β, IL-6, IL-8, TNF-α, elastase, aspartate, and aminotransferase,130,131 while bone-specific markers of tissue destruction and connective tissue breakdown comprise: collagen telopeptides, MMP-9, osteocalcin, proteoglycans, or fibronectin.12,132,133
In addition, metabolic profiling of saliva can provide a global outlook of the changes associated with periodontal diseases, particularly host enzymes (alkaline phosphatase, esterase, glucuronidase and aminopeptidase), prostaglandin E2, matrix metalloproteinase-8, 8-hydroxy-deoxyguanosine, dipeptides (leucylisoleucine, phenylphenol and serylisoleucine), as well as the fatty acids (arachidonate, arachidate and dihomolinolate).11,134–139
Caries risk assessment
The use of salivary diagnostics for caries risk assessment includes microbiome, proteomic, genomic, and transcriptomic approaches. The most common human dental caries-associated pathogens are Streptococcus mutans (S. mutans), Streptococcus sobrinus (S. sobrinus), and lactobacilli.140,141 Low salivary levels of alpha-defensins HNP1-3 contribute to caries susceptibility in children,142 while salivary mucins (i.e. MUC7) promote agglutination of streptococci.143
Caries risk assessment can be also managed by means of analyzing host-related factors in saliva including salivary flow rate, salivary pH, and buffer capacity.141
Diagnostic tools include culture-based methods such as mitis salivarius bacitracin broth (MSBB),144 dip-slide methods as well as newly emerging molecular technologies such as checkerboard DNA–DNA hybridization, genomic fingerprinting, 16S rRNA gene cloning and sequencing, T-RFLP and DNA sequencing including analysis of bacterial genome data.145,146 Also, PCR-based bacterial identification enables measurements of the cariogenic species in saliva.147
Orthodontics
There are many compelling reasons to use saliva as a diagnostic aid to monitor the risk and the development of root resorption during orthodontic treatment. Nowadays, available methods of clinical evaluation are mostly radiographic. Although an easy and accessible method, they have disadvantages such as limited points of view and radiation exposure.
The composition of saliva may reflect the pathophysiology of many diseases connected with orthodontic treatment. Several studies reported on changes in saliva in patients undergoing orthodontic treatment, including: interleukin-1β and interleukin-1β receptor antagonist,148 proteoglycans,149 regulatory subunit of type II (RII) of cyclic AMP-dependent protein kinase (PKA)150 or anti-HDE sIgA antibodies.151 The results of salivary analysis could provide the evidence for clinical decisions to minimize the risk and severity of developing root resorption.
Pharmacotherapy
Monitoring the therapeutic drug level and poisoning
Salivary diagnostics plays a crucial role in pharmacotherapy to monitor the therapeutic drug levels, assess treatment outcomes of diseases through the use of medicines, to detect overdose as well as to study the biochemical and physiological effects of drugs such as carbamazepine, cisplatin, diazepam, digoxin, ethosuximide, irinotecan, lithium, metoprolol, paracetamol, phenytoin, primidone, procainamide, quinine, theophylline, or valproic acid.152,153 Also, cotinine can be monitored in saliva of smoking subjects.154
Drug abuse
Saliva plays an important role in detection of various drugs in the blood such as amphetamine, cocaine, methadone, phencyclidine, marihuana, or opiates.155–157
Alcohol consumption
Salivary diagnostics serves as a diagnostic tool in alcohol consumption.158
Five salivary diagnostic toolboxes “omics”
The term “Salivaomics” was introduced in 2008 due to the rapid development of knowledge about various “omics” constituents of saliva. Currently, there are known five major salivary diagnostic components such as proteome, transcriptome, micro-RNA (miRNA), metabolome, and microbiome.159
Proteomics
The proteomics is the large-scale screening for proteins, their expression, modifications, and interactions by using high-throughput approaches.160
In the recent times, a great breakthrough appeared in the field of proteomics. From about 40 proteins identified in the early 80s, nowadays more than 3000 various proteins are detected.161
Currently, there are known two principal methods of proteomic analysis: two-dimensional gel electrophoresis (2DE) and mass-spectrometry analysis.162 2DE enables separation of proteins in two dimensions according to the isoelectric point (IE) and the molecular weight (MW). The second method—MS enables identification of proteins and their qualitative (qualitative MS) as well as quantitative (quantitative MS) evaluation.163 Proteins identified by MS can be further analyzed by electrospray ionization (ESI), matrix-assisted laser desorption ionization (MALDI), quadrupole/linear ion trap, time-of-flight (TOF), quadrupole TOF (QTOP), Fourier transform ion cyclotron resonance (FT-ICR), or the OrbiTrap. In addition, post-translationally modified proteins (phosphorylated, glycosylated, acetylated or methylated) can be evaluated by means of dendrimer-associated MS/MS, MALDI-MS, or targeted HPLC-ESI-MS/MS.2
Proteomic analysis has been hampered by the presence of high-abundance proteins that either mask or reduce separation sensitivity. In saliva, those proteins include mainly alpha-amylase, albumin, and proline rich proteins (75% of the total saliva proteome). Those proteins hamper the detection of low-abundance proteins appearing in different disease conditions and as a result should be removed. There are three major methods of high-abundance protein removal164: enzyme-substrate absorption method used for alpha-amylase affinity removal,165 immunodepletion method, and combinatorial peptide ligand library (CPLL).166 Proteomic analysis of saliva is commonly used in the diagnostics of oral diseases as well as general health disorders such as oral candidiasis,167 OSCC,168 glossodynia,169 head and neck squamous cell cancer,170 Sjögren's syndrome 171 HIV,167 autism,172 fibromyalgia,173 breast cancer,174 lung cancer, melanoma,82 or pancreatic cancer.26
Transcriptomics
The transcriptome is composed of all gene transcripts present in a cell, and their quantity, for a specific developmental stage or physiological condition. It helps to reveal the functional elements of the genome as well as molecular components of cells and tissues, development, and disease.175
The main method for identification of salivary transcriptomic biomarkers is microarray technology that can be validated by means of the quantitative real-time PCR (qPCR). Several salivary exRNAs have been already identified to allow the detection of many various diseases176 such as oral cancer,31 Sjögren syndrome,70 resectable pancreatic cancer,42 lung cancer,177 ovarian cancer,178 and breast cancer.179 We have recently obtained proof of concept data that salivary biomarkers possess discriminatory power for the detection of pancreatic cancer with high sensitivity (90.0%) and high specificity (95.0%) (area under curve, AUC = 0.971)42 that paves the way for prediction model validation study followed by a pivotal clinical validation.
Currently, a new high-resolution array from Affymetrix, GeneChip Human Transcriptome Array 2.0 (HTA 2.0) is commonly used that includes all transcript isoforms in the human transcriptome with >6 million probes targeting coding transcripts, exon–exon splice junctions, and non-coding transcripts.180
Due to the limitations in microarray technology, detecting and quantifying coding transcript isoforms, in addition to non-coding trancripts, has been challenging. As a result, currently, RNA sequencing (RNA-Seq) has been the preferred newly developed method for characterizing the full human transcriptome by means of deep-sequencing technologies. Compared with microarrays, RNA-Seq is analytically more sensitive in terms of detecting moderately and differentially expressed genes and gives sequence information at each nucleotide position of specific gene.175,181 Therefore, our group is currently working on the development of highly discriminatory and definitively validated salivary exRNA biomarkers for gastric cancer detection by means of RNA Seq analysis.
Micro-RNA-Omics
MicroRNAs (miRNAs) are nucleic acids that are encoded by genes but not translated into proteins. They are non-coding RNAs, in which each primary transcript (pri-miRNA) is processed into a short stem-loop structure called a pre-miRNA and finally into functional miRNA. Mature miRNA molecules cause down-regulation of gene expression.47 They play an important role in cell growth, differentiation, apoptosis, stress and immune response or glucose secretion.182–184
Metabolomics
Metabolome is the complete set of small molecular metabolites of living tissues including metabolic intermediates such as carbohydrates, lipids, amino acids, nucleic acids, hormones, and other signaling molecules.159
Salivary metabolites are important in elucidating the pathways underlying different diseases, thus making it ideal for the early detection of a wide range of diseases, including oral cancer and periodontal diseases.11 A systematic study of metabolites is called metabolomics. The major role of metabolomics is to identify novel metabolic biomarkers from cells, tissues, or body fluids by means of high-performance liquid chromatography-mass spectrometry (HPLC-MS) or two-dimensional gas chromatography MS and nuclear magnetic resonance spectroscopy in conjunction with pattern recognition methods. In this way, it will be possible to monitor and discover metabolic changes related to disease onset or therapeutic interventions. Those techniques have been already applied to chronic renal diseases, hepatocellular carcinoma and colorectal cancers185 as well as to oral cancer and periodontal diseases.11
Microbiomics
New technologies have allowed the scientists to start to unravel the complex interactions between the microorganisms and the human body.159
It was reported that salivary microbiome could be used in the detection of early resectable pancreatic cancer by means of microbial profiling (the Human Oral Microbe Identification Microarray), where two microbial markers (Neisseria elongata and Streptococcus mitis) were successfully developed with 96.4% sensitivity and 82.1% specificity (ROC=0.9).186 Currently, newer microbiome-based technologies have also become available, such as study of microbial sequences by means of RNA or DNA sequencing.
Technologies for salivary diagnostics
Point-of-care diagnostics
Currently, a great need exists for convenient and accurate point-of-care diagnostics that can serve as a non-invasive diagnostic tool.162,187
Novel point-of-care salivary technologies are being developed, which can facilitate biomarker identification without any pre-processing, screening, and non-invasive diagnostic testing such as Oral Fluid NanoSensor Test (OFNASET) for oral cancer detection,188 my PerioPath (OralDNA Labs) for diagnosing of periodontal disease,159 or the OraRisk HPV test (OralDNA Labs) to detect oral human papillomavirus (HPV) infection that could potentially lead to oral cancer.
There are several currently available or newly emerging technologies based on salivary diagnostics and development of microfluidics or micro/nanoelectromechanical systems (MEMS/NEMS). They are composed of mechanical, electrical, and functional elements such as sensors, actuators, and microelectronics that are made using the techniques of microfabrication. Those technologies enable to measure proteins, DNA, transcripts (mRNA), electrolytes, and small molecules in saliva.162 Currently developed tools include electrochemical sensing,189 on-chip qRT-PCR,190 fiber optic microsphere-based arrays,191 high-throughput DNA microarrays, 192 surface plasmon resonance-based fiber optic sensors,193 and microchip-based electrophoretic immunoassay.137
The new avenue of point-of-care diagnostics for “lab-on-a-chip” provides a new facet of point-of-care diagnostics, because it concurrently enables the detection of multiple biomarkers, and thus simultaneous diagnosis of many diseases. It seeks to integrate and automate all the complexities of a laboratory procedure into a device of the size of a computer chip.194
Liquid biopsy
Liquid biopsy tests are non-invasive biofluid tests (i.e. serum, urine, saliva) that detect CTCs and fragments of tumor DNA shed into the bloodstream by cells undergoing apoptosis or necrosis. The amount of circulating cell-free DNA (cfDNA) corresponds to tumor staging and prognosis. Nowadays, liquid biopsy enables a variety of clinical and investigational applications such as early detection, assessment of molecular heterogeneity of general disease, monitoring of tumor dynamics (in melanoma, breast, ovarian or colon cancers), identification of genetic determinants for targeted therapy, evaluation of early treatment response, monitoring of minimal residual disease, or assessment of resistence evolution in real time.195
There are various ways of detection and quantification of ctDNA (circulating tumor DNA) in blood such as Sanger sequencing, pyrosequencing, next generation sequencing, PCR-based technology, HPLC, mutant-enriched liquid chips, amplification refractory mutation system (ARMS), beads, emulsion, amplification and magnetics (BEAMing), or pyrophosphorolysis-activated polymerization (PAP).195
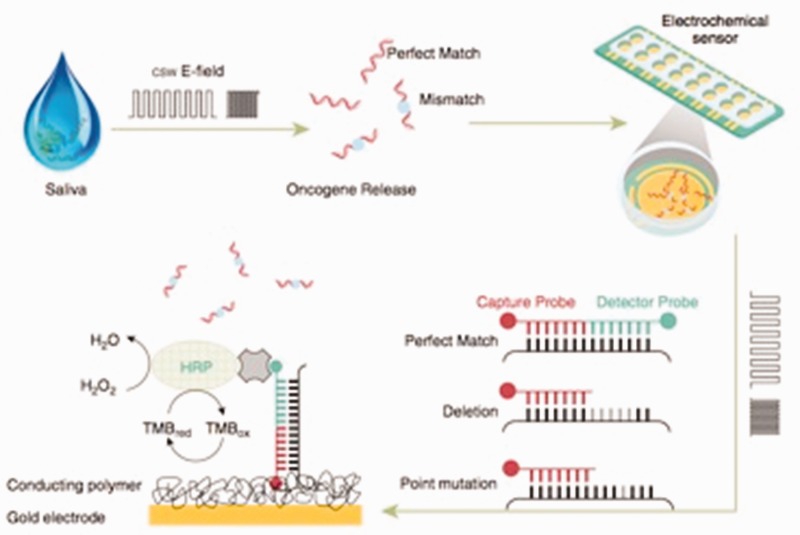
However, a new technique was developed at the University of California at Los Angeles (UCLA), School of Dentistry, called EFIRM.196 This technique enables disrupting and releasing the contents from exosomes and on-site monitoring of the released exosomal RNA/proteins biomarkers for a specific cancer. The first step involves pipetting of a sample of biofluid to the surface of a biosensor and applying a pulsed electric field to release molecular content from the exosome. Biorecognition of these molecules is carried out concurrently through a series of probes on the surface of the biosensor. Following the capture of the molecular targets, a series of reporting molecules are added to the biosensor, thus allowing the amount of a protein, DNA, or RNA target to be measured through electrochemical measurement of the current.197 This technique allows direct detection in plasma and in saliva of specific mutations present in cancers, i.e. detection of epidermal growth factor receptor (EGFR) mutation in non-small cell lung cancer (NSCLC), thus eliminating the need for quantitative PCR196 (Figure 4).
Figure 4.
Electric field-induced release and measurement (EFIRM) technology for the detection of epidermal growth factor receptor (EGFR) mutations in bodily fluids of patients with lung cancer. (reproduction from Wei et al.196). (A color version of this figure is available in the online journal.)
The results of performed experiments demonstrate that tumor-shed exosomes can be detected not only in blood, but also in saliva, thus launching a new venue for the non-invasive detection of tumor-specific proteins, micro RNAs, mRNAs, as well as gene mutations in saliva.197
Conclusion
Non-invasiveness is the Holy Grail for early detection of diseases. While saliva diagnostics is widely recognized for human diseases, the absence of discriminatory and definitively validated biomarkers that reached the regulatory FDA approval undermines saliva's clinical utility. However, newly emerging and rapidly developing technologies such as recent point-of-care systems, RNA sequencing or liquid biopsy have the potential to deliver novel diagnostic solutions in the salivary field. These recent advances have broadly widened the salivary diagnostic approach from the oral cavity to the whole physiological system, thus making salivary diagnostics a clinical reality that can be highly accurate and feasibly used to make an assessment of health and disease status. Their outcome will provide clinical and scientific credibility for saliva that might translate it into improvement of treatment outcomes and advancing prevention in human oral and general diseases.
Authors' contributions
KEKU, CMCP, KA, and MT wrote the manuscript, while FGG and DTWW revised, corrected, and supervised the content of the manuscript. All authors read and approved the manuscript prior to submission.
Declaration of Conflicting Interests
David Wong is the co-founder of RNAmeTRIX Inc., a molecular diagnostic company. He holds equity in RNAmeTRIX and serves as a company Director and Scientific Advisor. The University of California also holds equity in RNAmeTRIX. Intellectual property that David Wong invented and which was patented by the University of California has been licensed to RNAmeTRIX. Additionally, he is a consultant to PeriRx.
None of the other authors have a conflict of interest in relation to this study.
Funding
This work was supported by the Public Health Service (PHS) grants from the National Institute of Health/National Institute of Dental and Craniofacial Research (NIH/NIDCR) in United States: UH3 TR000923, R90 DE022734 and U01 DE17593. The research was also supported from Colgate-Palmolive Company grants (No. 20130399 and No. 20140855) and Delta Dental grants (No. 20143014 and 20164106). DTWW acknowledges the donation from the Stand Up To Cancer.
References
- 1.Kaufman E LI. The diagnostic applications of saliva. A review. Crit Rev Oral Biol Med 2002, pp. 197–212. [DOI] [PubMed] [Google Scholar]
- 2.Lee YH, Wong DT. Saliva: an emerging biofluid for early detection of diseases. Am J Dent 2009; 22: 241–8. [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, Duan Y. Saliva: a potential media for disease diagnostics and monitoring. Oral Oncol 2012; 48: 569–77. [DOI] [PubMed] [Google Scholar]
- 4.Chiappin S, Antonelli G, Gatti R, De Palo EF. Saliva specimen: a new laboratory tool for diagnostic and basic investigation. Clin Chim Acta 2007; 383: 30–40. [DOI] [PubMed] [Google Scholar]
- 5.Mese H, Matsuo R. Salivary secretion, taste and hyposalivation. J Oral Rehabil 2007; 34: 711–23. [DOI] [PubMed] [Google Scholar]
- 6.Hicks J, Garcia-Godoy F, Flaitz C. Biological factors in dental caries: role of saliva and dental plaque in the dynamic process of demineralization and remineralization (part 1). J Clin Pediatr Dent 2003; 28: 47–52. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Godoy F, Hicks MJ. Maintaining the integrity of the enamel surface: the role of dental biofilm, saliva and preventive agents in enamel demineralization and remineralization. J Am Dent Assoc 2008; 139(Suppl 5): 25S–34S. [DOI] [PubMed] [Google Scholar]
- 8.Amerongen AV, Veerman EC. Saliva – the defender of the oral cavity. Oral Dis 2002; 8: 12–22. [DOI] [PubMed] [Google Scholar]
- 9.Jankowska AK, Waszkiel D, Kowalczyk A. Saliva as a main component of oral cavity ecosystem. Part I. Secretion and function. Wiad Lek 2007; 60: 148–54. [PubMed] [Google Scholar]
- 10.Buczko P, Zalewska A, Szarmach I. Saliva and oxidative stress in oral cavity and in some systemic disorders. J Physiol Pharmacol 2015; 66: 3–9. [PubMed] [Google Scholar]
- 11.Mikkonen JJ, Singh SP, Herrala M, Lappalainen R, Myllymaa S, Kullaa AM. Salivary metabolomics in the diagnosis of oral cancer and periodontal diseases. J Periodontal Res 2016; 51: 431–7. [DOI] [PubMed] [Google Scholar]
- 12.Giannobile WV, McDevitt JT, Niedbala RS, Malamud D. Translational and clinical applications of salivary diagnostics. Adv Dent Res 2011; 23: 375–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segal A, Wong DT. Salivary diagnostics: enhancing disease detection and making medicine better. Eur J Dent Educ 2008; 12(Suppl 1): 22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong DT. Salivary diagnostics: scientific and clinical frontiers. Adv Dent Res 2011; 23: 350–2. [DOI] [PubMed] [Google Scholar]
- 15.Tavassoli M, Brunel N, Maher R, Johnson NW, Soussi T. P53 Antibodies in the saliva of patients with squamous cell carcinoma of the oral cavity. Int J Cancer 1998; 78: 390–1. [DOI] [PubMed] [Google Scholar]
- 16.Malathi N, Mythili S, Vasanthi HR. Salivary diagnostics: a brief review. ISRN Dent 2014; 2014: 158786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Streckfus C, Bigler L, Tucci M, Thigpen JT. A preliminary study of CA15-3, c-erbB-2, epidermal growth factor receptor, cathepsin-D, and p53 in saliva among women with breast carcinoma. Cancer Invest 2000; 18: 101–9. [DOI] [PubMed] [Google Scholar]
- 18.Dumbrigue HB, Sandow PL, Nguyen KH, Humphreys-Beher MG. Salivary epidermal growth factor levels decrease in patients receiving radiation therapy to the head and neck. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2000; 89: 710–6. [DOI] [PubMed] [Google Scholar]
- 19.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013; 63: 11–30. [DOI] [PubMed] [Google Scholar]
- 20.Cheng YS, Rees T, Wright J. A review of research on salivary biomarkers for oral cancer detection. Clin Transl Med 2014: 33-1326-3-3. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shpitzer T, Bahar G, Feinmesser R, Nagler RM. A comprehensive salivary analysis for oral cancer diagnosis. J Cancer Res Clin Oncol 2007; 133: 613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Contucci AM, Inzitari R, Agostino S, Vitali A, Fiorita A, Cabras T, Scarano E, Messana I. Statherin levels in saliva of patients with precancerous and cancerous lesions of the oral cavity: a preliminary report. Oral Dis 2005; 11: 95–9. . [DOI] [PubMed] [Google Scholar]
- 23.Vucicevic Boras V, Cikes N, Lukac J, Virag M, Cekic-Arambasin A. Salivary and serum interleukin 6 and basic fibroblast growth factor levels in patients with oral squamous cell carcinoma. Minerva Stomatol 2005; 54: 569–73. [PubMed] [Google Scholar]
- 24.Nagler R, Bahar G, Shpitzer T, Feinmesser R. Concomitant analysis of salivary tumor markers–a new diagnostic tool for oral cancer. Clin Cancer Res 2006; 12: 3979–84. [DOI] [PubMed] [Google Scholar]
- 25.Jessie K, Jayapalan JJ, Ong KC, Abdul Rahim ZH, Zain RM, Wong KT, Hashim OH. Aberrant proteins in the saliva of patients with oral squamous cell carcinoma. Electrophoresis 2013; 34: 2495–502. [DOI] [PubMed] [Google Scholar]
- 26.Hu S, Arellano M, Boontheung P, Wang J, Zhou H, Jiang J, Elashoff D, Wei R, Loo JA, Wong DT. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res 2008; 14: 6246–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jou YJ, Lin CD, Lai CH, Tang CH, Huang SH, Tsai MH, Chen SY, Kao JY, Lin CW. Salivary zinc finger protein 510 peptide as a novel biomarker for detection of oral squamous cell carcinoma in early stages. Clin Chim Acta 2011; 412: 1357–65. [DOI] [PubMed] [Google Scholar]
- 28.Jou YJ, Lin CD, Lai CH, Chen CH, Kao JY, Chen SY, Tsai MH, Huang SH, Lin CW. Proteomic identification of salivary transferrin as a biomarker for early detection of oral cancer. Anal Chim Acta 2010; 681: 41–8. [DOI] [PubMed] [Google Scholar]
- 29.Jiang WW, Masayesva B, Zahurak M, Carvalho AL, Rosenbaum E, Mambo E, Zhou S, Minhas K, Benoit N, Westra WH, Alberg A, Sidransky D, Koch W, Califano J. Increased mitochondrial DNA content in saliva associated with head and neck cancer. Clin Cancer Res 2005; 11: 2486–91. [DOI] [PubMed] [Google Scholar]
- 30.Tang H, Wu Z, Zhang J, Su B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol Med Rep 2013; 7: 761–6. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, Eisele D, Abemayor E, Elashoff D, Park NH, Wong DT. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res 2004; 10: 8442–50. [DOI] [PubMed] [Google Scholar]
- 32.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010; 6: 78–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei J, Xie G, Zhou Z, Shi P, Qiu Y, Zheng X, Chen T, Su M, Zhao A, Jia W. Salivary metabolite signatures of oral cancer and leukoplakia. Int J Cancer 2011; 129: 2207–17. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q, Gao P, Wang X, Duan Y. The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. Sci Rep 2014; 4: 6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almadori G, Bussu F, Galli J, Limongelli A, Persichilli S, Zappacosta B, Minucci A, Paludetti G, Giardina B. Salivary glutathione and uric acid levels in patients with head and neck squamous cell carcinoma. Head Neck 2007; 29: 648–54. [DOI] [PubMed] [Google Scholar]
- 36.Agha-Hosseini F, Mirzaii-Dizgah I, Farmanbar N, Abdollahi M. Oxidative stress status and DNA damage in saliva of human subjects with oral lichen planus and oral squamous cell carcinoma. J Oral Pathol Med 2012; 41: 736–40. [DOI] [PubMed] [Google Scholar]
- 37.Rajkumar K, Ramya R, Nandhini G, Rajashree P, Ramesh Kumar A, Nirmala Anandan S. Salivary and serum level of CYFRA 21-1 in oral precancer and oral squamous cell carcinoma. Oral Dis 2015; 21: 90–6. [DOI] [PubMed] [Google Scholar]
- 38.Shpitzer T, Hamzany Y, Bahar G, Feinmesser R, Savulescu D, Borovoi I, Gavish M, Nagler RM. Salivary analysis of oral cancer biomarkers. Br J Cancer 2009; 101: 1194–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhodus NL, Cheng B, Myers S, Miller L, Ho V, Ondrey F. The feasibility of monitoring NF-kappaB associated cytokines: TNF-alpha, IL-1alpha, IL-6, and IL-8 in whole saliva for the malignant transformation of oral lichen planus. Mol Carcinog 2005; 44: 77–82. [DOI] [PubMed] [Google Scholar]
- 40.Brinkmann O, Kastratovic DA, Dimitrijevic MV, Konstantinovic VS, Jelovac DB, Antic J, Nesic VS, Markovic SZ, Martinovic ZR, Akin D, Spielmann N, Zhou H, Wong DT. Oral squamous cell carcinoma detection by salivary biomarkers in a Serbian population. Oral Oncol 2011; 47: 51–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elashoff D, Zhou H, Reiss J, Wang J, Henson B, Hu S, Arellano M, Sinha U, Le A, Messadi D, Wang M, Nabili V, Lingen M, Morris D, Randolph T, Feng Z, Akin D, Kastratovic DA, Chia D, Abemayor E, Wong DT. Prevalidation of salivary biomarkers for oral cancer detection. Cancer Epidemiol Biomarkers Prev 2012; 21: 664–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Farrell JJ, Zhou H. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 2010; 138: 949–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Humeau M, Vignolle-Vidoni A, Sicard F, Martins F, Bournet B, Buscail L, Torrisani J, Cordelier P. Salivary MicroRNA in pancreatic cancer patients. PLoS One 2015; 10: e0130996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol 2006; 12: 354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maconi G, Manes G, Porro GB. Role of symptoms in diagnosis and outcome of gastric cancer. World J Gastroenterol 2008; 14: 1149–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao H, Zhang Y, Kim Y, Kim S, Kim JJ, Kim KM, Yoshizawa J, Fan LY, Cao CX, Wong DT. Differential proteomic analysis of human saliva using tandem mass tags quantification for gastric cancer detection. Sci Rep 2016; 6: 22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spielmann N, Wong DT. Saliva: diagnostics and therapeutic perspectives. Oral Dis 2011; 17: 345–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lau C, Kim Y, Chia D, Spielmann N, Eibl G, Elashoff D, Wei F, Lin YL, Moro A, Grogan T, Chiang S, Feinstein E, Schafer C, Farrell J, Wong DT. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem 2013; 288: 26888–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gisbert JP. Helicobacter pylori-associated diseases. Gastroenterol Hepatol 2015; 38(Suppl 1): 39–48. [DOI] [PubMed] [Google Scholar]
- 50.Martinez Torres A, Martinez Gaensly M. Helicobacter pylori: a new cardiovascular risk factor? Rev Esp Cardiol 2002; 55: 652–6. [DOI] [PubMed] [Google Scholar]
- 51.Ishihara K, Miura T, Ebihara Y, Hirayama T, Kamiya S, Okuda K. Shared antigenicity between Helicobacter pylori and periodontopathic Campylobacter rectus strains. FEMS Microbiol Lett 2001; 197: 23–7. [DOI] [PubMed] [Google Scholar]
- 52.Anand PS, Kamath KP, Anil S. Role of dental plaque, saliva and periodontal disease in Helicobacter pylori infection. World J Gastroenterol 2014; 20: 5639–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young SH, Luo JC. Will saliva test be a good method to detect Helicobacter pylori in H. pylori-infected patients? J Chin Med Assoc 2016; 79: 351–2. [DOI] [PubMed] [Google Scholar]
- 54.Gholami Parizad E, Gholami Parizad E, Khosravi A, Amraei M, Valizadeh A, Davoudian A. Comparing HBV viral load in serum, cerumen, and saliva and correlation with HBeAg serum status in patients with chronic hepatitis B infection. Hepat Mon 2016; 16: e30385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khadse SV, Bajaj G, Vibhakar P, Nainani P, Ahuja R, Deep G. Evaluation of specificity and sensitivity of oral fluid for diagnosis of hepatitis B. J Clin Diagn Res 2016; 10: BC12–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amado Leon LA, de Almeida AJ, de Paula VS, Tourinho RS, Villela DA, Gaspar AM, Lewis-Ximenez LL, Pinto MA. Longitudinal study of hepatitis A infection by saliva sampling: the kinetics of HAV markers in saliva revealed the application of saliva tests for hepatitis A study. PLoS One 2015; 10: e0145454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghosh M, Nandi S, Dutta S, Saha MK. Detection of hepatitis B virus infection: a systematic review. World J Hepatol 2015; 7: 2482–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cardoso ES, Jesus BL, Gomes LG, Sousa SM, Gadelha SR, Marin LJ. The use of saliva as a practical and feasible alternative to urine in large-scale screening for congenital cytomegalovirus infection increases inclusion and detection rates. Rev Soc Bras Med Trop 2015; 48: 206–7. [DOI] [PubMed] [Google Scholar]
- 59.Pinninti SG, Ross SA, Shimamura M, Novak Z, Palmer AL, Ahmed A, Tolan RW Jr, Bernstein DI, Michaels MG, Sୣhez PJ, Fowler KB, Boppana SB. Comparison of saliva PCR assay versus rapid culture for detection of congenital cytomegalovirus infection. Pediatr Infect Dis J 2015; 34: 536–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwok H, Chan KW, Chan KH, Chiang AK. Distribution, persistence and interchange of Epstein-Barr virus strains among PBMC, plasma and saliva of primary infection subjects. PLoS One 2015; 10: e0120710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corstjens PL, Abrams WR, Malamud D. Saliva and viral infections. Periodontol 2000 2016; 70: 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bonaldo MC, Ribeiro IP, Lima NS, Dos Santos AA, Menezes LS, da Cruz SO, de Mello IS, Furtado ND, de Moura EE, Damasceno L, da Silva KA, de Castro MG, Gerber AL, de Almeida LG, Lourenço-de-Oliveira R, Vasconcelos AT, Brasil P. Isolation of Infective Zika Virus from Urine and Saliva of Patients in Brazil. PLoS Negl Trop Dis 2016; 10: e0004816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sheikhakbari S, Mokhtari-Azad T, Salimi V, Norouzbabaei Z, Abbasi S, Zahraei SM, Shahmahmoodi S. The use of oral fluid samples spotted on filter paper for the detection of measles virus using nested rt-PCR. J Clin Lab Anal 2012; 26: 215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vainio K, Samdal HH, Anestad G, Wedege E, Skutlaberg DH, Bransdal KT, Mundal R, Aaberge IS. Detection of measles- and mumps-specific IgG antibodies in paired serum and oral fluid samples from Norwegian conscripts. Eur J Clin Microbiol Infect Dis 2008; 27: 461–5. [DOI] [PubMed] [Google Scholar]
- 65.Vijaylakshmi P, Muthukkaruppan VR, Rajasundari A, Korukluoglu G, Nigatu W, Warrener LA, Samuel D, Brown DW. Evaluation of a commercial rubella IgM assay for use on oral fluid samples for diagnosis and surveillance of congenital rubella syndrome and postnatal rubella. J Clin Virol 2006; 37: 265–8. [DOI] [PubMed] [Google Scholar]
- 66.Weerasekera MM, Sissons CH, Wong L, Anderson SA, Holmes AR, Cannon RD. Use of denaturing gradient gel electrophoresis (DGGE) for the identification of mixed oral yeasts in human saliva. J Med Microbiol 2013. 62: 319–330. [DOI] [PubMed] [Google Scholar]
- 67.Riega-Torres JC, Villarreal-Gonzalez AJ, Cecenas-Falcon LA, Salas-Alanis JC. Sjogren's syndrome (SS), a review of the subject and saliva as a diagnostic method. Gac Med Mex 2016; 152: 371–80. [PubMed] [Google Scholar]
- 68.Asashima H, Inokuma S, Nakachi S, Matsuo Y, Rokutanda R, Hagiwara K, Kobayashi S. Extremely high salivary beta(2) -microglobulin and Na(+) levels in a Sjogren syndrome patient. Int J Rheum Dis 2012; 15: e31–3. [DOI] [PubMed] [Google Scholar]
- 69.Streckfus C, Bigler L, Navazesh M, Al-Hashimi I. Cytokine concentrations in stimulated whole saliva among patients with primary Sjogren's syndrome, secondary Sjogren's syndrome, and patients with primary Sjogren's syndrome receiving varying doses of interferon for symptomatic treatment of the condition: a preliminary study. Clin Oral Investig 2001; 5: 133–5. [DOI] [PubMed] [Google Scholar]
- 70.Hu S, Wang J, Meijer J, Ieong S, Xie Y, Yu T, Zhou H, Henry S, Vissink A, Pijpe J, Kallenberg C, Elashoff D, Loo JA, Wong DT. Salivary proteomic and genomic biomarkers for primary Sjogren's syndrome. Arthritis Rheum 2007; 56: 3588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu S, Gao K, Pollard R, Arellano-Garcia M, Zhou H, Zhang L, Elashoff D, Kallenberg CG, Vissink A, Wong DT. Preclinical validation of salivary biomarkers for primary Sjogren's syndrome. Arthritis Care Res 2010; 62: 1633–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu S, Vissink A, Arellano M, Roozendaal C, Zhou H, Kallenberg CG, Wong DT. Identification of autoantibody biomarkers for primary Sjogren's syndrome using protein microarrays. Proteomics 2011; 11: 1499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Acar S, Yetkiner AA, Ersin N, Oncag O, Aydogdu S, Arikan C. Oral findings and salivary parameters in children with celiac disease: a preliminary study. Med Princ Pract 2012; 21: 129–33. [DOI] [PubMed] [Google Scholar]
- 74.Adornetto G, Fabiani L, Volpe G, De Stefano A, Martini S, Nenna R, Lucantoni F, Bonamico M, Tiberti C, Moscone D. An electrochemical immunoassay for the screening of celiac disease in saliva samples. Anal Bioanal Chem 2015; 407: 7189–7196. [DOI] [PubMed] [Google Scholar]
- 75.Bonamico M, Nenna R, Montuori M, Luparia RP, Turchetti A, Mennini M, Lucantoni F, Masotti D, Magliocca FM, Culasso F, Tiberti C. First salivary screening of celiac disease by detection of anti-transglutaminase autoantibody radioimmunoassay in 5000 Italian primary schoolchildren. J Pediatr Gastroenterol Nutr 2011; 52: 17–20. [DOI] [PubMed] [Google Scholar]
- 76.Dane A, Gurbuz T. Clinical evaluation of specific oral and salivary findings of coeliac disease in eastern Turkish paediatric patients. Eur J Paediatr Dent 2016; 17: 53–6. [PubMed] [Google Scholar]
- 77.Livnat G, Bentur L, Kuzmisnsky E, Nagler RM. Salivary profile and oxidative stress in children and adolescents with cystic fibrosis. J Oral Pathol Med 2010; 39: 16–21. [DOI] [PubMed] [Google Scholar]
- 78.Zetterquist W, Marteus H, Kalm-Stephens P, Nas E, Nordvall L, Johannesson M, Alving K. Oral bacteria – the missing link to ambiguous findings of exhaled nitrogen oxides in cystic fibrosis. Respir Med 2009; 103: 187–193. [DOI] [PubMed] [Google Scholar]
- 79.Goncalves AC, Augusto de Lima Marson F, Maria de Holanda Mendonca R, Ribeiro JD, Ribeiro AF, Paschoal IA, Levy CE. Saliva as a potential tool for cystic fibrosis diagnosis. Diagn Pathol 2013; 8: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Almeida EA, Di Mascio P, Harumi T, Spence DW, Moscovitch A, Hardeland R, Cardinali DP, Brown GM, Pandi-Perumal SR. Measurement of melatonin in body fluids: standards, protocols and procedures. Childs Nerv Syst 2011; 27: 879–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Inder WJ, Dimeski G, Russell A. Measurement of salivary cortisol in 2012 - laboratory techniques and clinical indications. Clin Endocrinol 2012; 77: 645–51. [DOI] [PubMed] [Google Scholar]
- 82.Gao K, Zhou H, Zhang L, Lee JW, Zhou Q, Hu S, Wolinsky LE, Farrell J, Eibl G, Wong DT. Systemic disease-induced salivary biomarker profiles in mouse models of melanoma and non-small cell lung cancer. PLoS One 2009; 4: e5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Manolopoulou J, Mulatero P, Maser-Gluth C, Rossignol P, Spyroglou A, Vakrilova Y, Petersenn S, Zwermann O, Plouin PF, Reincke M, Bidlingmaier M. Saliva as a medium for aldosterone measurement in repeated sampling studies. Steroids 2009; 74: 853–8. [DOI] [PubMed] [Google Scholar]
- 84.Agha-Hosseini F, Mirzaii-Dizgah I, Mansourian A, Zabihi-Akhtechi G. Serum and stimulated whole saliva parathyroid hormone in menopausal women with oral dry feeling. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2009; 107: 806–10. [DOI] [PubMed] [Google Scholar]
- 85.Jurysta C, Bulur N, Oguzhan B, Satman I, Yilmaz TM, Malaisse WJ, Sener A. Salivary glucose concentration and excretion in normal and diabetic subjects. J Biomed Biotechnol 2009; 2009: 430426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lakshmi PV, Sridevi E, Sai Sankar AJ, Manoj Kumar MG, Sridhar M, Sujatha B. Diagnostic perspective of saliva in insulin dependent diabetes mellitus children: An in vivo study. Contemp Clin Dent 2015; 6: 443–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hartman ML, Goodson JM, Shi P, Vargas J, Yaskell T, Stephens D, Cugini M, Hasturk H, Barake R, Alsmadi O, Al-Mutawa S, Ariga J, Soparkar P, Behbehani J, Behbehani K, Welty F. Unhealthy phenotype as indicated by salivary biomarkers: glucose, insulin, VEGF-A, and IL-12p70 in obese Kuwaiti adolescents. J Obes 2016; 2016: 6860240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Filaire E, Lac G. Dehydroepiandrosterone (DHEA) rather than testosterone shows saliva androgen responses to exercise in elite female handball players. Int J Sports Med 2000; 21: 17–20. [DOI] [PubMed] [Google Scholar]
- 89.Labsy Z, Prieur F, Le Panse B, Do MC, Gagey O, Lasne F, Collomp K. The diurnal patterns of cortisol and dehydroepiandrosterone in relation to intense aerobic exercise in recreationally trained soccer players. Stress 2013; 16: 261–5. [DOI] [PubMed] [Google Scholar]
- 90.Mirzaii-Dizgah I, Agha-Hosseini F. Stimulated and unstimulated saliva progesterone in menopausal women with oral dryness feeling. Clin Oral Investig 2011; 15: 859–62. [DOI] [PubMed] [Google Scholar]
- 91.Marrs CR, Ferraro DP, Cross CL. Salivary hormones and parturition in healthy, primigravid women. Int J Gynaecol Obstet 2007; 99: 59–60. [DOI] [PubMed] [Google Scholar]
- 92.Elias PC, Martinez EZ, Barone BF, Mermejo LM, Castro M, Moreira AC. Late-night salivary cortisol has a better performance than urinary free cortisol in the diagnosis of Cushing's syndrome. J Clin Endocrinol Metab 2014; 99: 2045–51. [DOI] [PubMed] [Google Scholar]
- 93.Antonelli G, Ceccato F, Artusi C, Marinova M, Plebani M. Salivary cortisol and cortisone by LC-MS/MS: validation, reference intervals and diagnostic accuracy in Cushing's syndrome. Clin Chim Acta 2015; 451: 247–51. [DOI] [PubMed] [Google Scholar]
- 94.Ceccato F, Boscaro M. Cushing's syndrome: screening and diagnosis. High Blood Press Cardiovasc Prev 2016; 23: 209–15. [DOI] [PubMed] [Google Scholar]
- 95.Celec P, Ostanikova D, Skoknova M, Hodosy J, Putz Z, Kudela M. Salivary sex hormones during the menstrual cycle. Endocr J 2009; 56: 521–3. [DOI] [PubMed] [Google Scholar]
- 96.Amatoury M, Lee JW, Maguire AM, Ambler GR, Steinbeck KS. Utility of salivary enzyme immunoassays for measuring estradiol and testosterone in adolescents: a pilot study. Int J Adolesc Med Health. Epub ahead of print 9 April 2016. DOI: 10.1515/ijamh-2015-0126. [DOI] [PubMed] [Google Scholar]
- 97.Agha-Hosseini F, Moosavi MS, Mirzaii-Dizgah I. Salivary flow, testosterone, and femur bone mineral density in menopausal women with oral dryness feeling. Oral Surg Oral Med Oral Pathol Oral Radiol 2013. 115:612–6. [DOI] [PubMed] [Google Scholar]
- 98.Bali A, Jaggi AS. Clinical experimental stress studies: methods and assessment. Rev Neurosci 2015; 26: 555–79. [DOI] [PubMed] [Google Scholar]
- 99.Gullander M, Grynderup M, Hansen AM, Hogh A, Persson R, Kolstad HA, Mors O, Kaerlev L, Bonde JP. Are changes in workplace bullying status related to changes in salivary cortisol? A longitudinal study among Danish employees. J Psychosom Res 2015; 79: 435–42. [DOI] [PubMed] [Google Scholar]
- 100.Tanaka Y, Ishitobi Y, Maruyama Y, Kawano A, Ando T, Okamoto S, Kanehisa M, Higuma H, Ninomiya T, Tsuru J, Hanada H, Kodama K, Isogawa K, Akiyoshi J. Salivary alpha-amylase and cortisol responsiveness following electrical stimulation stress in major depressive disorder patients. Prog Neuropsychopharmacol Biol Psychiatry 2012; 36: 220–4. [DOI] [PubMed] [Google Scholar]
- 101.Granger DA, Kivlighan KT, el-Sheikh M, Gordis EB, Stroud LR. Salivary alpha-amylase in biobehavioral research: recent developments and applications. Ann N Y Acad Sci 2007; 1098: 122–44. [DOI] [PubMed] [Google Scholar]
- 102.Kivlighan KT, Granger DA. Salivary alpha-amylase response to competition: relation to gender, previous experience, and attitudes. Psychoneuroendocrinology 2006; 31: 703–14. [DOI] [PubMed] [Google Scholar]
- 103.Richter J, Bittner A, Petrowski K, Junge-Hoffmeister J, Bergmann S, Joraschky P, Weidner K. Effects of an early intervention on perceived stress and diurnal cortisol in pregnant women with elevated stress, anxiety, and depressive symptomatology. J Psychosom Obstet Gynaecol 2012. 33:162–70. [DOI] [PubMed] [Google Scholar]
- 104.Rai B, Kaur J, Anand SC, Jacobs R. Salivary stress markers, stress, and periodontitis: a pilot study. J Periodontol 2011; 82: 287–92. [DOI] [PubMed] [Google Scholar]
- 105.McGregor BA, Murphy KM, Albano DL, Ceballos RM. Stress, cortisol, and B lymphocytes: a novel approach to understanding academic stress and immune function. Stress 2016; 19: 185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Idris A, Ghazali NB, Said NM, Steele M, Koh D, Tuah NA. Salivary testosterone as a potential indicator for risky behaviour associated with smoking-related peer pressure in adolescents. Int J Adolesc Med Health. Epub ahead of print 9 April 2016. DOI: 10.1515/ijamh-2015-0125. [DOI] [PubMed] [Google Scholar]
- 107.van der Meij L, Schaveling J, van Vugt M. Basal testosterone, leadership and dominance: a field study and meta-analysis. Psychoneuroendocrinology 2016; 72: 72–9. [DOI] [PubMed] [Google Scholar]
- 108.Walt DR, Blicharz TM, Hayman RB, Rissin DM, Bowden M, Siqueira WL, Helmerhorst EJ, Grand-Pierre N, Oppenheim FG, Bhatia JS, Little FF, Brody JS. Microsensor arrays for saliva diagnostics. Ann N Y Acad Sci 2007; 1098: 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tomas I, Marinho JS, Limeres J, Santos MJ, Araujo L, Diz P. Changes in salivary composition in patients with renal failure. Arch Oral Biol 2008; 53: 528–32. [DOI] [PubMed] [Google Scholar]
- 110.Venkatapathy R, Govindarajan V, Oza N, Parameswaran S, Pennagaram Dhanasekaran B, Prashad KV. Salivary creatinine estimation as an alternative to serum creatinine in chronic kidney disease patients. Int J Nephrol 2014; 2014: 742724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ozbay Y, Aydin S, Dagli AF, Akbulut M, Dagli N, Kilic N, Rahman A, Sahin I, Polat V, Ozercan HI, Arslan N, Sensoy D. Obestatin is present in saliva: alterations in obestatin and ghrelin levels of saliva and serum in ischemic heart disease. BMB Rep 2008; 41: 55–61. [DOI] [PubMed] [Google Scholar]
- 112.Chatterton RT, Jr, Vogelsong KM, Lu YC, Ellman AB, Hudgens GA. Salivary alpha-amylase as a measure of endogenous adrenergic activity. Clin Physiol 1996; 16: 433–48. [DOI] [PubMed] [Google Scholar]
- 113.Miller CS, Foley JD, 3rd, Floriano PN, Christodoulides N, Ebersole JL, Campbell CL, Bailey AL, Rose BG, Kinane DF, Novak MJ, McDevitt JT, Ding X, Kryscio RJ. Utility of salivary biomarkers for demonstrating acute myocardial infarction. J Dent Res 2014; 93: 72S–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shen YS, Chen WL, Chang HY, Kuo HY, Chang YC, Chu H. Diagnostic performance of initial salivary alpha-amylase activity for acute myocardial infarction in patients with acute chest pain. J Emerg Med 2012; 43: 553–60. [DOI] [PubMed] [Google Scholar]
- 115.Floriano PN, Christodoulides N, Miller CS, Ebersole JL, Spertus J, Rose BG, Kinane DF, Novak MJ, Steinhubl S, Acosta S, Mohanty S, Dharshan P, Yeh CK, Redding S, Furmaga W, McDevitt JT. Use of saliva-based nano-biochip tests for acute myocardial infarction at the point of care: a feasibility study. Clin Chem 2009; 55: 1530–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hartman ML, Goodson JM, Barake R, Alsmadi O, Al-Mutawa S, Ariga J, Soparkar P, Behbehani J, Behbehani K. Salivary biomarkers in pediatric metabolic disease research. Pediatr Endocrinol Rev 2016; 13: 602–11. [PubMed] [Google Scholar]
- 117.Bencharit S, Baxter SS, Carlson J, Byrd WC, Mayo MV, Border MB, Kohltfarber H, Urrutia E, Howard-Williams EL, Offenbacher S, Wu MC, Buse JB. Salivary proteins associated with hyperglycemia in diabetes: a proteomic analysis. Mol Biosyst 2013; 9: 2785–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kadashetti V, Baad R, Malik N, Shivakumar KM, Vibhute N, Belgaumi U, Gugawad S, Pramod RC. Glucose level estimation in diabetes mellitus by saliva: a bloodless revolution. Rom J Intern Med 2015; 53: 248–52. [DOI] [PubMed] [Google Scholar]
- 119.Indira M, Chandrashekar P, Kattappagari KK, Chandra LP, Chitturi RT, Bv RR. Evaluation of salivary glucose, amylase, and total protein in Type 2 diabetes mellitus patients. Indian J Dent Res 2015; 26: 271–5. [DOI] [PubMed] [Google Scholar]
- 120.Malicka B, Kaczmarek U, Skoskiewicz-Malinowska K. Selected antibacterial factors in the saliva of diabetic patients. Arch Oral Biol 2015; 60: 425–31. [DOI] [PubMed] [Google Scholar]
- 121.Aitken-Saavedra J, Rojas-Alcayaga G, Maturana-Ramirez A, Escobar-Alvarez A, Cortes-Coloma A, Reyes-Rojas M, Viera-Sapiain V, Villablanca-Mart쭥z C, Morales-Bozo I. Salivary gland dysfunction markers in type 2 diabetes mellitus patients. J Clin Exp Dent 2015; 7: e501–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shi M, Sui YT, Peskind ER, Li G, Hwang H, Devic I, Ginghina C, Edgar JS, Pan C, Goodlett DR, Furay AR, Gonzalez-Cuyar LF, Zhang J. Salivary tau species are potential biomarkers of Alzheimer's disease. J Alzheimers Dis 2011; 27: 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jang MU, Park JW, Kho HS, Chung SC, Chung JW. Plasma and saliva levels of nerve growth factor and neuropeptides in chronic migraine patients. Oral Dis 2011; 17: 187–93. [DOI] [PubMed] [Google Scholar]
- 124.Gatti R, De Palo EF. An update: salivary hormones and physical exercise. Scand J Med Sci Sports 2011; 21: 157–69. [DOI] [PubMed] [Google Scholar]
- 125.Zauber H, Mosler S, von Hessberg A, Schulze WX. Dynamics of salivary proteins and metabolites during extreme endurance sports - a case study. Proteomics 2012; 12: 2221–35. [DOI] [PubMed] [Google Scholar]
- 126.Khare P, Raj V, Chandra S, Agarwal S. Quantitative and qualitative assessment of DNA extracted from saliva for its use in forensic identification. J Forensic Dent Sci 2014; 6: 81–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kaufman E, Lamster IB. Analysis of saliva for periodontal diagnosis–a review. J Clin Periodontol 2000; 27: 453–65. [DOI] [PubMed] [Google Scholar]
- 128.Loesche WJ, Lopatin DE, Giordano J, Alcoforado G, Hujoel P. Comparison of the benzoyl-DL-arginine-naphthylamide (BANA) test, DNA probes, and immunological reagents for ability to detect anaerobic periodontal infections due to Porphyromonas gingivalis, Treponema denticola, and Bacteroides forsythus. J Clin Microbiol 1992; 30: 427–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lanza E, Magan-Fernandez A, Bermejo B, de Rojas J, Marfil-Alvarez R, Mesa F. Complementary clinical effects of red complex bacteria on generalized periodontitis in a Caucasian population. Oral Dis 2016; 22: 430–7. [DOI] [PubMed] [Google Scholar]
- 130.Knight ET, Liu J, Seymour GJ, Faggion CM, Jr, Cullinan MP. Risk factors that may modify the innate and adaptive immune responses in periodontal diseases. Periodontol 2000 2016; 71: 22–51. [DOI] [PubMed] [Google Scholar]
- 131.Castro CE, Koss MA, Lopez ME. Biochemical markers of the periodontal ligament. Med Oral 2003; 8: 322–8. [PubMed] [Google Scholar]
- 132.Taba M, Jr, Kinney J, Kim AS, Giannobile WV. Diagnostic biomarkers for oral and periodontal diseases. Dent Clin North Am 2005; 49: 551–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Giannobile WV. C-telopeptide pyridinoline cross-links. Sensitive indicators of periodontal tissue destruction. Ann N Y Acad Sci 1999; 878: 404–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet 2005; 366: 1809–20. [DOI] [PubMed] [Google Scholar]
- 135.Lamster IB, Kaufman E, Grbic JT, Winston LJ, Singer RE. Beta-glucuronidase activity in saliva: relationship to clinical periodontal parameters. J Periodontol 2003; 74: 353–9. [DOI] [PubMed] [Google Scholar]
- 136.Takane M, Sugano N, Iwasaki H, Iwano Y, Shimizu N, Ito K. New biomarker evidence of oxidative DNA damage in whole saliva from clinically healthy and periodontally diseased individuals. J Periodontol 2002; 73: 551–4. [DOI] [PubMed] [Google Scholar]
- 137.Herr AE, Hatch AV, Throckmorton DJ, Tran HM, Brennan JS, Giannobile WV, Singh AK. Microfluidic immunoassays as rapid saliva-based clinical diagnostics. Proc Natl Acad Sci U S A 2007; 104: 5268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dede FO, Ozden FO, Avci B. 8-hydroxy-deoxyguanosine levels in gingival crevicular fluid and saliva in patients with chronic periodontitis after initial periodontal treatment. J Periodontol 2013; 84: 821–8. [DOI] [PubMed] [Google Scholar]
- 139.Barnes VM, Ciancio SG, Shibly O, Xu T, Devizio W, Trivedi HM, Guo L, Jönsson TJ. Metabolomics reveals elevated macromolecular degradation in periodontal disease. J Dent Res 2011; 90: 1293–7. [DOI] [PubMed] [Google Scholar]
- 140.Banas JA. Virulence properties of Streptococcus mutans. Front Biosci 2004; 9: 1267–1277. [DOI] [PubMed] [Google Scholar]
- 141.Guo L, Shi W. Salivary biomarkers for caries risk assessment. J Calif Dent Assoc 2013; 41: 107–9, 112–8. [PMC free article] [PubMed] [Google Scholar]
- 142.Tao R, Jurevic RJ, Coulton KK, Tsutsui MT, Roberts MC, Kimball JR, Wells N, Berndt J, Dale BA. Salivary antimicrobial peptide expression and dental caries experience in children. Antimicrob Agents Chemother 2005; 49: 3883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kirstila V, Hakkinen P, Jentsch H, Vilja P, Tenovuo J. Longitudinal analysis of the association of human salivary antimicrobial agents with caries increment and cariogenic micro-organisms: a two-year cohort study. J Dent Res 1998; 77: 73–80. [DOI] [PubMed] [Google Scholar]
- 144.Matsukubo T, Ohta K, Maki Y, Takeuchi M, Takazoe I. A semi-quantitative determination of Streptococcus mutans using its adherent ability in a selective medium. Caries Res 1981; 15: 40–5. [DOI] [PubMed] [Google Scholar]
- 145.Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA, Asnani K, Griffen AL. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol 2010; 48: 4121–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lazarevic V, Whiteson K, Hernandez D, Francois P, Schrenzel J. Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics 2010; 11: 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Akiyama T, Miyamoto H, Fukuda K, Sano N, Katagiri N, Shobuike T, Kukita A, Yamashita Y, Taniguchi H, Goto M. Development of a novel PCR method to comprehensively analyze salivary bacterial flora and its application to patients with odontogenic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2010; 109: 669–76. [DOI] [PubMed] [Google Scholar]
- 148.Iwasaki LR, Haack JE, Nickel JC, Reinhardt RA, Petro TM. Human interleukin-1 beta and interleukin-1 receptor antagonist secretion and velocity of tooth movement. Arch Oral Biol 2001; 46: 185–9. [DOI] [PubMed] [Google Scholar]
- 149.Waddington RJ, Embery G. Proteoglycans and orthodontic tooth movement. J Orthod 2001; 28: 281–90. [DOI] [PubMed] [Google Scholar]
- 150.Burke JC, Evans CA, Crosby TR, Mednieks MI. Expression of secretory proteins in oral fluid after orthodontic tooth movement. Am J Orthod Dentofacial Orthop 2002; 121: 310–5. [DOI] [PubMed] [Google Scholar]
- 151.Ramos Sde P, Ortolan GO, Dos Santos LM, Tobouti PL, Hidalgo MM, Consolaro A, Itano EN. Anti-dentine antibodies with root resorption during orthodontic treatment. Eur J Orthod 2011; 33: 584–91. [DOI] [PubMed] [Google Scholar]
- 152.Dwivedi R, Gupta YK, Singh M, Joshi R, Tiwari P, Kaleekal T, et al. Correlation of saliva and serum free valproic acid concentrations in persons with epilepsy. Seizure 2015; 25: 187–90. [DOI] [PubMed] [Google Scholar]
- 153.Yamada E, Takagi R, Sudo K, Kato S. Determination of abacavir, tenofovir, darunavir, and raltegravir in human plasma and saliva using liquid chromatography coupled with tandem mass spectrometry. J Pharm Biomed Anal 2015; 114: 390–7. [DOI] [PubMed] [Google Scholar]
- 154.Etter JF. A longitudinal study of cotinine in long-term daily users of e-cigarettes. Drug Alcohol Depend 2016; 160: 218–21. [DOI] [PubMed] [Google Scholar]
- 155.Cone EJ, Clarke J, Tsanaclis L. Prevalence and disposition of drugs of abuse and opioid treatment drugs in oral fluid. J Anal Toxicol 2007; 31: 424–33. [DOI] [PubMed] [Google Scholar]
- 156.Huestis MA, Cone EJ. Methamphetamine disposition in oral fluid, plasma, and urine. Ann N Y Acad Sci 2007; 1098: 104–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Christodoulides N, De La Garza R, 2nd, Simmons GW, McRae MP, Wong J, Newton TF, Kosten TR, Haque A, McDevitt JT. Next generation programmable bio-nano-chip system for on-site detection in oral fluids. J Drug Abuse 2015; 1: 1–6. [PMC free article] [PubMed] [Google Scholar]
- 158.Criado-Garcia L, Ruszkiewicz DM, Eiceman GA, Thomas CL. A rapid and non-invasive method to determine toxic levels of alcohols and gamma-hydroxybutyric acid in saliva samples by gas chromatography-differential mobility spectrometry. J Breath Res 2016; 10: 017101. [DOI] [PubMed] [Google Scholar]
- 159.Wong DT. Salivaomics. J Am Dent Assoc 2012; 143: 19S–24. [DOI] [PubMed] [Google Scholar]
- 160.Anderson NL, Anderson NG. Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis 1998; 19: 1853–61. [DOI] [PubMed] [Google Scholar]
- 161.Amado FM, Ferreira RP, Vitorino R. One decade of salivary proteomics: current approaches and outstanding challenges. Clin Biochem 2013; 46: 506–17. [DOI] [PubMed] [Google Scholar]
- 162.Wong DT. Salivary diagnostics powered by nanotechnologies, proteomics and genomics. J Am Dent Assoc 2006; 137: 313–21. [DOI] [PubMed] [Google Scholar]
- 163.Kossowska B, Dudka I, Gancarz R, Antonowicz-Juchniewicz J. Proteomic analysis of protein profiles in some pathological stages of the human organism. Postepy Hig Med Dosw 2009; 63: 549–63. [PubMed] [Google Scholar]
- 164.Krief G, Deutsch O, Zaks B, Wong DT, Aframian DJ, Palmon A. Comparison of diverse affinity based high-abundance protein depletion strategies for improved bio-marker discovery in oral fluids. J Proteomics 2012; 75: 4165–75. [DOI] [PubMed] [Google Scholar]
- 165.Deutsch O, Fleissig Y, Zaks B, Krief G, Aframian DJ, Palmon A. An approach to remove alpha amylase for proteomic analysis of low abundance biomarkers in human saliva. Electrophoresis 2008; 29: 4150–7. [DOI] [PubMed] [Google Scholar]
- 166.Krief G, Deutsch O, Gariba S, Zaks B, Aframian DJ, Palmon A. Improved visualization of low abundance oral fluid proteins after triple depletion of alpha amylase, albumin and IgG. Oral Dis 2011; 17: 45–52. [DOI] [PubMed] [Google Scholar]
- 167.Sweet SP, Denbury AN, Challacombe SJ. Salivary calprotectin levels are raised in patients with oral candidiasis or Sjogren's syndrome but decreased by HIV infection. Oral Microbiol Immunol 2001; 16: 119–23. [DOI] [PubMed] [Google Scholar]
- 168.Stott-Miller M, Houck JR, Lohavanichbutr P, Mendez E, Upton MP, Futran ND, Schwartz SM, Chen C. Tumor and salivary matrix metalloproteinase levels are strong diagnostic markers of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 2011; 20: 2628–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Loeb LM, Naffah-Mazzacoratti MG, Porcionatto MA, Martins JR, Kouyoumdjian M, Weckx LM, Nadera HB. Chondroitin sulfate and kallikrein in saliva: markers for glossodynia. Int Immunopharmacol 2008; 8: 1056–8. [DOI] [PubMed] [Google Scholar]
- 170.Drake RR, Cazare LH, Semmes OJ, Wadsworth JT. Serum, salivary and tissue proteomics for discovery of biomarkers for head and neck cancers. Expert Rev Mol Diagn 2005; 5: 93–100. [DOI] [PubMed] [Google Scholar]
- 171.Fleissig Y, Deutsch O, Reichenberg E, Redlich M, Zaks B, Palmon A, Aframian DJ. Different proteomic protein patterns in saliva of Sjogren's syndrome patients. Oral Dis 2009; 15: 61–8. [DOI] [PubMed] [Google Scholar]
- 172.Castagnola M, Messana I, Inzitari R, Fanali C, Cabras T, Morelli A, Pecoraro AM, Neri G, Torrioli MG, Gurrieri F. Hypo-phosphorylation of salivary peptidome as a clue to the molecular pathogenesis of autism spectrum disorders. J Proteome Res 2008; 7: 5327–32. [DOI] [PubMed] [Google Scholar]
- 173.Bazzichi L, Ciregia F, Giusti L, Baldini C, Giannaccini G, Giacomelli C, Sernissi F, Bombardieri S, Lucacchini A. Detection of potential markers of primary fibromyalgia syndrome in human saliva. Proteomics Clin Appl 2009; 3: 1296–1304. [DOI] [PubMed] [Google Scholar]
- 174.Streckfus CF, Bigler LR, Zwick M. The use of surface-enhanced laser desorption/ionization time-of-flight mass spectrometry to detect putative breast cancer markers in saliva: a feasibility study. J Oral Pathol Med 2006; 35: 292–300. [DOI] [PubMed] [Google Scholar]
- 175.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 2015; 61: 221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang L, Xiao H, Zhou H, Santiago S, Lee JM, Garon EB, Yang J, Brinkmann O, Yan X, Akin D, Chia D, Elashoff D, Park NH, Wong DT. Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell Mol Life Sci 2012; 69: 3341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lee YH, Kim JH, Zhou H, Kim BW, Wong DT. Salivary transcriptomic biomarkers for detection of ovarian cancer: for serous papillary adenocarcinoma. J Mol Med 2012; 90: 427–34. [DOI] [PubMed] [Google Scholar]
- 179.Zhang L, Xiao H, Karlan S, Zhou H, Gross J, Elashoff D, Akin D, Yan X, Chia D, Karlan B, Wong DT. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS One 2010; 5: e15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Palermo M, Driscoll H, Tighe S, Dragon J, Bond J, Shukla A, Vangala M, Vincent J. Expression profiling smackdown: human transcriptome array HTA 2.0 vs. RNA. Seq. Journal of Biomolecular Techniques 2014; 25: S20–1. [Google Scholar]
- 181.Spielmann N, Ilsley D, Gu J, Lea K, Brockman J, Heater S, Setterquist R, Wong DT. The human salivary RNA transcriptome revealed by massively parallel sequencing. Clin Chem 2012; 58: 1314–21. [DOI] [PubMed] [Google Scholar]
- 182.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–8. [DOI] [PubMed] [Google Scholar]
- 183.Stadler BM, Ruohola-Baker H. Small RNAs: keeping stem cells in line. Cell 2008; 132: 563–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: tiny players in a big field. Immunity 2007; 26: 133–7. [DOI] [PubMed] [Google Scholar]
- 185.Beger RD. A review of applications of metabolomics in cancer. Metabolites 2013; 3: 552–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, Paster BJ, Joshipura K, Wong DT. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012; 61: 582–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Koneru S, Tanikonda R. Salivaomics - A promising future in early diagnosis of dental diseases. Dent Res J (Isfahan) 2014; 11: 11–5. [PMC free article] [PubMed] [Google Scholar]
- 188.Gau V, Wong D. Oral fluid nanosensor test (OFNASET) with advanced electrochemical-based molecular analysis platform. Ann N Y Acad Sci 2007; 1098: 401–10. [DOI] [PubMed] [Google Scholar]
- 189.Bartlett CA, Taylor S, Fernandez C, Wanklyn C, Burton D, Enston E, Raniczkowska A, Black M, Murphy L. Disposable screen printed sensor for the electrochemical detection of methamphetamine in undiluted saliva. Chem Cent J 2016; 10: 3-016-0147-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang J, Chen Z, Corstjens PL, Mauk MG, Bau HH. A disposable microfluidic cassette for DNA amplification and detection. Lab Chip 2006; 6: 46–53. [DOI] [PubMed] [Google Scholar]
- 191.Blicharz TM, Siqueira WL, Helmerhorst EJ, Oppenheim FG, Wexler PJ, Little FF, Walt DR. Fiber-optic microsphere-based antibody array for the analysis of inflammatory cytokines in saliva. Anal Chem 2009; 81: 2106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Abraham JE, Maranian MJ, Spiteri I, Russell R, Ingle S, Luccarini C, Earl HM, Pharoah PP, Dunning AM, Caldas C. Saliva samples are a viable alternative to blood samples as a source of DNA for high throughput genotyping. BMC Med Genomics 2012; 5: 19-8794-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Microfluidic diagnostic technologies for global public health. Nature 2006; 442: 412–8. [DOI] [PubMed] [Google Scholar]
- 194.Srinivasan V, Pamula VK, Fair RB. An integrated digital microfluidic lab-on-a-chip for clinical diagnostics on human physiological fluids. Lab Chip 2004; 4: 310–5. [DOI] [PubMed] [Google Scholar]
- 195.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014; 32: 579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wei F, Lin CC, Joon A, Feng Z, Troche G, Lira ME, Chia D, Mao M, Ho CL, Su WC, Wong DT. Noninvasive saliva-based EGFR gene mutation detection in patients with lung cancer. Am J Respir Crit Care Med 2014; 190: 1117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wei F, Yang J, Wong DT. Detection of exosomal biomarker by electric field-induced release and measurement (EFIRM). Biosens Bioelectron 2013; 44: 115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]