Abstract
Declining fossil fuel reserves, coupled with environmental concerns over their continued extraction and exploitation have led to strenuous efforts to identify renewable routes to energy and fuels. One attractive option is to convert glycerol, a by-product of the biodiesel industry, into n-butanol, an industrially important chemical and potential liquid transportation fuel, using Clostridium pasteurianum. Under certain growth conditions this Clostridium species has been shown to predominantly produce n-butanol, together with ethanol and 1,3-propanediol, when grown on glycerol. Further increases in the yields of n-butanol produced by C. pasteurianum could be accomplished through rational metabolic engineering of the strain. Accordingly, in the current report we have developed and exemplified a robust tool kit for the metabolic engineering of C. pasteurianum and used the system to make the first reported in-frame deletion mutants of pivotal genes involved in solvent production, namely hydA (hydrogenase), rex (Redox response regulator) and dhaBCE (glycerol dehydratase). We were, for the first time in C. pasteurianum, able to eliminate 1,3-propanediol synthesis and demonstrate its production was essential for growth on glycerol as a carbon source. Inactivation of both rex and hydA resulted in increased n-butanol titres, representing the first steps towards improving the utilisation of C. pasteurianum as a chassis for the industrial production of this important chemical.
Keywords: Clostridium pasteurianum; Rex; HydA; DhaBCE; Butanol; 1,3-propanediol
Highlights
-
•
A comprehensive molecular tool set for Clostridium pasteurianum.
-
•
Abolishment of by-product (1,3-propanediol) formation by dhaBCE KO.
-
•
First reported deletion of main hydrogenase HydA in Clostridium spp.
-
•
Inactivation of rex and hydA leads to increased n-butanol titres.
1. Introduction
Declining fossil fuel reserves, coupled with environmental concerns over their continued extraction and exploitation have led to strenuous efforts to identify renewable routes to energy and fuels. The two most commonly used biofuels are bioethanol and biodiesel (REN21, 2015). Between 2004 and 2014 biodiesel production has increased more than twelvefold, from 2.4 billion litres/year to 29.7 billion litres/year (REN21, 2015). For every ton of biodiesel produced through the transesterification of vegetable oils and animals fats with short-chain alcohols (most commonly methanol) 100 kg of crude glycerol (10% [w/w]), or glycerine, are formed as a by-product. As a consequence, glycerol availability has increased significantly (Johnson and Taconi, 2007, Yang et al., 2012). The glycerol formed is almost always used in its refined or purified form to make a multitude of products, including cosmetics, pharmaceuticals and food and beverages (Quispe et al., 2013). The glut of glycerine that has resulted from the biodiesel industry has impacted crude and refined glycerol prices. Economically, crude glycerol has shifted from by-product to waste-product (Kerr et al., 2011, Quispe et al., 2013). With the associated disposal costs, ways to convert crude glycerol into valuable products are becoming increasingly important (Yazdani and Gonzalez, 2007). Currently crude glycerol is utilised as a supplement for animal feeds, reacting agent in chemical catalytic procedures and as a feedstock for the biological production of chemicals and fuels (Yang et al., 2012). The high degree of reduction of its carbon atoms (Clomburg and Gonzalez, 2013), its wide availability and low cost have made glycerol an attractive feedstock for biorefinery (Werpy et al., 2004; Cherubini, 2009; Dobson et al., 2012), especially for anaerobic fermentation processes (Yazdani and Gonzalez, 2007).
Clostridium pasteurianum is, unlike other well-studied clostridia, capable of converting glycerol directly into the value-added chemicals n-butanol and 1,3-propanediol (PDO) (Biebl, 2001, Jensen et al., 2012, Malaviya et al., 2012). Butanol is an attractive biofuel as it offers a high energy density, low water solubility, low vapour pressure and good blending abilities and can be used in regular combustion engines without the need of modification (Dürre, 2007). The organic compound PDO is used as a precursor for the production of useful polymers such as polyesters, polyurethans and polyethers. The polyester polytrimethylene terephthalate (PTT) accounts for 90.0% of the total PDO market which is expected to be worth $621.2 million by 2021 (Market and Market, 2015). It shares many of the features of its polyester counterpart polybutylene terephthalate (PBT) and polyethylene terephthalate (PET) but offers higher tensile and flexural strengths and stiffness (Zhang, 2004). In addition, PDO is used in composites, adhesives, laminates, coatings, moldings, aliphatic polyesters and antifreeze (Liu et al., 2010).
Under certain growth conditions C. pasteurianum has been shown (Dabrock et al., 1992, Jensen et al., 2012, Moon et al., 2011; Taconi et al., 2009) to predominantly produce n-butanol, ethanol and PDO with trace amounts of organic acids (see Fig. 1). In optimized batch cultures up to 17 g/l n-butanol have been produced from glycerol, in pH auxostat cultures around 7 g/l PDO (Biebl, 2001). Recently, a C. pasteurianum mutant strain was generated producing 17.8 g/l n-butanol and 8.7 g/l PDO from refined glycerol (Malaviya et al., 2012). In comparison, engineered C. acetobutylicum and C. beijerinckii mutant strains have been described to produce 18.9 g/l and 20.9 g/l of butanol (Chen and Blaschek, 1999; Jang et al., 2012). Further increases in yields of n-butanol produced by C. pasteurianum could be accomplished through rational metabolic engineering of the strain. Accordingly, in the present study we have developed the requisite gene modification systems through the implementation of our previously described roadmap for gene tool development (Minton et al., 2016), and then used the developed allelic exchange vectors to make in-frame deletions of spo0A (for exemplification purposes) and a selection of mutants likely to affect solvent yields, namely, dhaBCE (encoding glycerol dehydratase), rex (coding for the redox responsive repressor Rex) and hydA (encoding an iron-coupled hydrogenase).
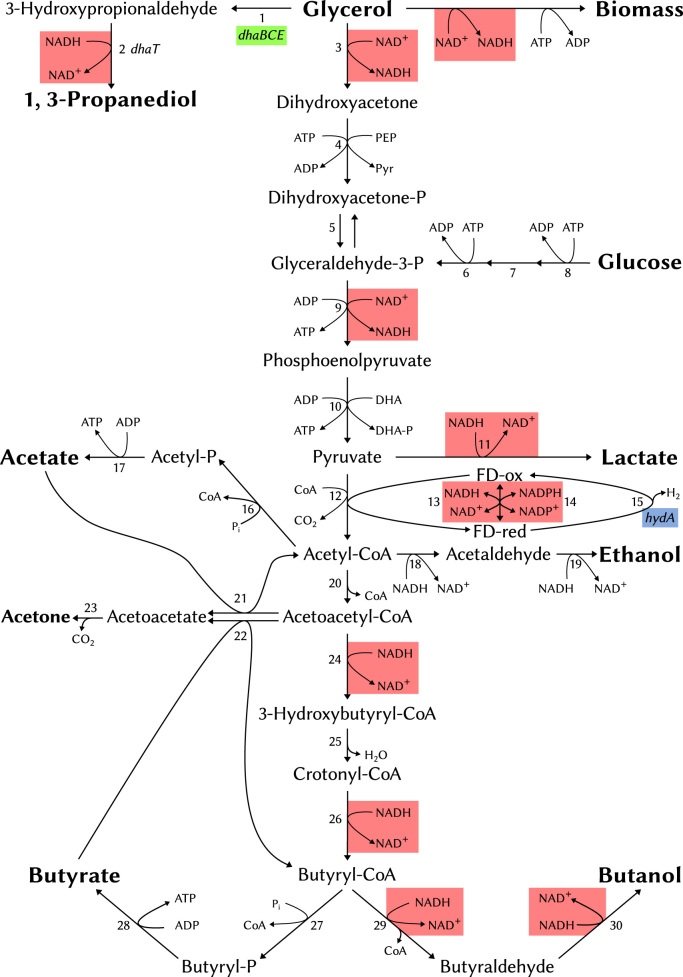
Fig. 1.
Central metabolic energy pathway in C. pasteurianum from glucose and glycerol. Figure based on Malaviya et al. (2012) and Biebl (2001). The knocked out genes (hydA, dhaBCE) and the NADH/NAD+ utilising pathways putatively influenced by Rex are highlighted. Other enzymes involved in the central energy pathway are numbered as follows: 1, glycerol dehydratase; 2, 1,3-propanediol oxydoreductase. 3, glycerol-3-phosphate dehydrogenase; 4, dihydroxyacetone kinase; 5, triose-phosphate isomerase; 6, phosphofructokinase; 7, phosphoglucose isomerase; 8, hexokinase; 9, glyceraldehyde-3-phosphate dehydrogenase; 10, pyruvate kinase; 11, lactate dehydrogenase; 12, pyruvate-ferredoxin oxidoreductase; 13, ferredoxin-NADP reductase; 14, NADPH-ferredoxin oxidoreductase; 15, ferredoxin hydrogenase; 16, phosphate acetyltransferase; 17, acetate kinase; 18, acetaldehyde dehydrogenase; 19, ethanol dehydrogenase; 20, thiolase; 21, acetoacetyl-CoA: acetate:CoA transferase; 22, acetoacetyl-CoA: butyrate:CoA transferase; 23, acetoacetate decarboxylase; 24, β -hydroxybutyryl-CoA dehydrogenase; 25, crotonase; 26, butyryl-CoA dehydrogenase; 27, phosphotransbutyrylase; 28, butyrate kinase; 29, butaraldehyde dehydrogenase; 30, butanol dehydrogenase.
2. Methods
2.1. Bacterial strains, growth and maintenance conditions
Bacterial strains used are listed in Table 1. E. coli was grown aerobically at 30 °C or 37 °C in Luria-Bertani (LB) broth or agar supplemented with 25 µg/ml chloramphenicol (LBCm25) or 100 µg/ml kanamycin (LBKm100), respectively. C. pasteurianum was routinely grown at 37 °C in an anaerobic workstation (Don Whitley, Yorkshire, UK) in 2x YTG broth (16 g/l tryptone, 10 g/l yeast extract, 5 g/l NaCl, 5 g/l glucose, pH 6.2) or on RCM agar (Oxoid Ltd) supplemented with 15 µg/ml thiamphenicol (2x YTGTm15, RCMTm15), 20 µg/ml erythromycin (2x YTGEm20, RCMEm20) or 40 µg/ml uracil (2x YTGUra40, RCMUra40), if required. Selections using 5-fluoroorotic acid (FOA, 600 µg/ml; Sigma-Aldrich, Dorset, UK) and uracil (5 µg/ml) were carried out in modified clostridial basal medium containing 0.5% (w/v) CaCO3 and 5% (w/v) glucose (CBM-S) (Steiner et al., 2012) or on standard clostridial basal medium (CBM) agar (O'Brien and Morris, 1971). All solidified media contained 1.5% [w/v] agar. Solvent/ acid profiling was undertaken in CGM (Sandoval et al., 2015), Biebl medium (Biebl, 2001) or 2x YT media supplemented with 60 g/l glycerol or glucose. Biebl medium was additionally supplemented with 1 g/l yeast extract for glycerol fermentations Table 2.
Table 1.
Strains and plasmids used in this study. C. pa. = C. pasteurianum.
| Name | Designation | Properties | Source |
|---|---|---|---|
| C. pa. ATCC 6013 | CRG4080 | wild type, type strain | ATCC |
| C. pa. DSM 525 | CRG4091 | wild type, type strain | DSMZ |
| C. pa. DSM 525-H1 | CRG4111 | hypertransformable strain based on DSM 525 | This study |
| C. pa. DSM 525-H1ΔpyrE | CRG4273 | ACE pyrE truncation mutant | This study |
| C. pa. DSM 525-H1ΔpyrEΔspo0A | CRG5514 | ACE spo0A deletion mutant with ΔpyrE background | This study |
| C. pa. DSM 525-H1Δspo0A | CRG5516 | ACE spo0A deletion mutant with pyrE repaired | This study |
| C. pa. DSM 525-H1::spo0A* | CRG5518 | ACE spo0A complementation in pyrE locus | This study |
| C. pa. DSM 525-H1ΔpyrEΔrex | CRG5520 | ACE rex deletion mutant with ΔpyrE background | This study |
| C. pa. DSM 525-H1Δrex | CRG5522 | ACE rex deletion mutant with pyrE repaired | This study |
| C. pa. DSM 525-H1::rex* | CRG5524 | ACE rex complementation in pyrE locus | This study |
| C. pa. DSM 525-H1ΔpyrEΔhydA | CRG5526 | ACE hyd deletion mutant with ΔpyrE background | This study |
| C. pa. DSM 525-H1ΔhydA | CRG5528 | ACE hyd deletion mutant with pyrE repaired | This study |
| C. pa. DSM 525-H1::hydA* | CRG5530 | ACE hyd complementation in pyrE locus | This study |
| C. pa. DSM 525-H1ΔpyrEΔdhaBCE | CRG5532 | ACE dhaBCE deletion mutant with ΔpyrE background | This study |
| C. pa. DSM 525-H1ΔdhaBCE | CRG5534 | ACE dhaBCE deletion mutant with pyrE repaired | This study |
| C. pa. DSM 525-H1::dhaBCE* | CRG5536 | ACE dhaBCE complementation in pyrE locus | This study |
| E. coli Top10 x CR1 | CRG3131 | Strain harbouring plasmid CR1 With M.BepI methylase | This study |
| pMTL85151 | E. coli-Clostridium shuttle vector (pIM13, catP, ColE1 traJ, lacZα ORF/MCS, TCpa fdx) | Heap et al. (2009) | |
| pMTL-AMH101 | catP-pyrE module used for pMTL-KS15 | Liew et al. (2017) | |
| pMTL-KS01 | pMTL85151, 300-bp internal pyrE fragment, 1200-bp fragment immediately downstream of pyrE | This study | |
| pMTL-KS03 | pMTL85151, 300-bp internal pyrE fragment, 1200-bp fragment immediately downstream of pyrE, with TCtet gluRS | This study | |
| pMTL-KS04 | pMTL85151, 937-bp fragment immediately downstream of pyrE, 300-bp internal pyrE fragment, TCtet gluRS, TCpa fdx | This study | |
| pMTL-KS05 | pMTL85151, 548-bp internal pyrE fragment, 1200-bp fragment immediately downstream of pyrE | This study | |
| pMTL-KS08 | pMTL85151, 937-bp fragment immediately downstream of pyrE, 548-bp internal pyrE fragment, TCtet gluRS, TCpa fdx | This study | |
| pMTL-KS10 | pMTL-KS01 features, TCtet gluRS, TCpa fdx,AsiSI | This study | |
| pMTL-KS12 | pMTL85151, 548-bp internal pyrE fragment, 937-bp fragment immediately downstream of pyrE, TCtet gluRS, TCpa fdx,AsiSI | This study | |
| pMTL-KS15 | pMTL85151, pyrE (C. acetobutylicum), TCtet gluRS, TCpa fdx, 300-bp SHA pyrE, lacZα ORF/MCS | This study | |
| pMTL-KS16 | pMTL85151, pyrE (C. acetobutylicum), TCtet gluRS, TCpa fdx, AsiSI, 300-bp SHA pyrE, lacZα ORF/MCS | This study | |
| pMTL-AGH12 | 1748-bp fragment comprising pyrE (35–582 nt) and 1200-bp immediately downstream of pyrE cloned into SbfI/AscI recognition sites of pMTL-KS12 | This study | |
| pMTL-KS12::spo0A | 1095-bp fragment comprising the 267-bp sequence upstream of spo0A and the spo0A gene cloned into the NotI/NheI recognition sites of pMTL-KS12 | This study | |
| pMTL-KS12::rex | 816-bp fragment comprising the 183-bp sequence upstream of rex and the rex gene cloned into the NotI/NheI recognition sites of pMTL-KS12 | This study | |
| pMTL-KS12::hyd | 2118-bp fragment comprising the 393-bp sequence upstream of hyd and the hyd gene cloned into the NotI/NheI recognition sites of pMTL-KS12 | This study | |
| pMTL-KS12::dhaBCE | 2984-bp fragment comprising the 294-bp sequence upstream of dhaB and the dhaBCE genes cloned into the NotI/NheI recognition sites of pMTL-KS12 | This study | |
| pMTL-KS15::KO_spo0A | 1400-bp KO out cassette for spo0A | This study | |
| pMTL-KS15::KO_rex | 1364-bp KO out cassette for rex | This study | |
| pMTL-KS16::KO_hyd | 2002-bp KO out cassette for hyd | This study | |
| pMTL-KS15::KO_dhaBCE | 1602-bp KO out cassette for dhaBCE | This study |
Table 2.
Solvent and acid yields of C. pasteurianum DSM 525 (WT) and its various mutant derivatives when grown in bioreactors on either glucose or glycerol as the carbon source. Mutants were: C. pasteurianum DSM 525-H1Δrex (Δrex, CRG5522); C. pasteurianum DSM 525-H1Δhyd (Δhyd, CRG5528), and; C. pasteurianum DSM 525-H1ΔdhaBCE (ΔdhaBCE, CRG5534). Abbreviations used: butanol (BuOH), ethanol (EtOH), 1,3-propanediol (PDO), solvents (EtOH, BuOH, PDO), acids (acetate, butyrate, lactate). Carbon recovery was calculated by assuming 3.5 g/l dry-weight per 10 OD values (Sarchami et al., 2016), carbon dioxide desorption as described by Percheron et al. (1995) and the assumption that 46.2% of dry-weight is carbon (Papoutsakis and Meyer, 1985). The carbon fraction of yeast extract was neglected. *Fermentation of ΔdhaBCE was undertaken in serum bottles along with a wild type control.
| Carbon source |
60 g/l glycerol |
60 g/l glucose |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Medium |
Biebl plus 1 g/l yeast extract |
2x YT* |
Biebl |
|||||||
| Strain | Δrex | Δhyd | WT | ΔdhaBCE | WT | Δrex | Δhyd | ΔdhaBCE | WT | |
| Growth characteristics | ||||||||||
| Specific growth rate [h−1] | 0.32±0.04 | 0.35±0.01 | 0.42±0.01 | 0.39±0.00 | 0.40±0.00 | 0.15±0.02 | 0.11±0.01 | 0.13±0.03 | 0.16±0.03 | |
| Doubling time [min] | 135±15 | 119±3 | 100±1 | 106±1 | 105±0 | 90±9 | 103±10 | 85±4 | 81±12 | |
| Max. OD | 15.0±0.5 | 17.5±0.6 | 15.3±0.3 | 4.38±0.12 | 4.48±0.01 | 34.9±1.0 | 36.2±0.1 | 34.8±2.5 | 34.3±2.7 | |
| Carbon recovery [%] | 88.9±2.7 | 91.4±1.3 | 90.5±4.4 | 114.4±6.0 | 127.8±5.4 | 70.7±14.3 | 83.5±15.0 | 72.3±13.9 | 73.3±14.9 | |
| Selectivity [M/M] | ||||||||||
| BuOH/Solvents | 0.516±0.002 | 0.312±0.003 | 0.289±0.024 | 0.862±0.002 | 0.795±0.001 | 0.703±0.015 | 0.729±0.021 | 0.635±0.027 | 0.563±0.051 | |
| Yield [M/M] | ||||||||||
| BuOH/C-Source | 0.250±0.005 | 0.166±0.004 | 0.142±0.003 | 0.399±0.019 | 0.431±0.021 | 0.124±0.034 | 0.156±0.029 | 0.041±0.008 | 0.031±0.003 | |
| EtOH/C-Source | 0.053±0.006 | 0.102±0.001 | 0.037±0.000 | 0.064±0.002 | 0.066±0.003 | 0.055±0.018 | 0.061±0.017 | 0.025±0.007 | 0.026±0.008 | |
| PDO/C-Source | 0.182±0.000 | 0.265±0.005 | 0.317±0.034 | 0.000±0.000 | 0.046±0.002 | n.a. | n.a. | n.a. | n.a. | |
| Solvents/C-Source | 0.484±0.011 | 0.533±0.007 | 0.496±0.030 | 0.463±0.021 | 0.542±0.026 | 0.179±0.052 | 0.217±0.046 | 0.066±0.016 | 0.057±0.001 | |
| Acids/C-Source | 0.050±0.006 | 0.106±0.000 | 0.114±0.008 | 0.122±0.009 | 0.136±0.004 | 0.567±0.129 | 0.567±0.153 | 0.686±0.156 | 0.708±0.177 | |
2.2. Standard molecular biology techniques
Standard plasmids used in this study are detailed in Table 1. Electroporation of E. coli was carried out using a Gene-Pulsar (Bio-Rad, Hemel Hempstead, UK) in agreement with the manufacturer's instructions. Plasmid DNA and genomic DNA was isolated using a QIAprep spin miniprep or a DNeasy blood and tissue kit (Qiagen, Manchester, UK), respectively. Restriction digest by endonucleases, ligation reactions and agarose gel electrophoresis were performed according to the manufacturer's instructions. Restriction endonucleases were purchased from ThermoFisher Scientific (Loughborough, UK) and a LigaFast™ Rapid DNA Ligation kit from Promega (Southampton, UK). PCR was carried out using the BIO-X-ACT™ Short Mix (Bioline Reagents, London, UK) in accordance with the manufacturer's instructions. Site-directed mutagenesis (SDM) was carried out using the Quik Change II Site-Directed Mutagenesis Kit (Agilent Technologies, Stockport, UK). Oligonucleotides were ordered from Eurofins Genomics (Ebersberg, Germany) and are given in Supplementary Table S1. Sanger sequencing was carried out by Source BioSciences (Nottingham, UK). Synthetic DNA fragments were ordered either from Biomatik (Cambridge, Ontario/Canada) or GeneArt® Gene Synthesis (Life Technologies, Paisley, UK).
2.3. Plasmid methylation and plasmid transfer into C. pasteurianum
Plasmid DNA for the transformation into C. pasteurianum strain was in vivo methylated by propagation in the dam+, dcm+ E. coli host CR1, an E. coli Top10 strain harbouring the plasmid pCR1, comprising a RSF1030-derived RSF origin of replication (Som and Tomizawa, 1982, Novagen, Merck KGaA, Darmstadt, Germany), a kanamycin resistance marker and a gene encoding the M.BepI methyltransferase of Brevibacterium epidermidis under the transcriptional control of the C. sporogenes fdx promoter (PCsp fdx). Alternatively, in vitro methylation was performed by incubation of plasmid DNA with CpG methyltransferase (M.SssI; NEB, Hitchin, UK), according to the manufacturer's instructions. Methylated plasmids 0.5–5 μg DNA) were electroporated into C. pasteurianum as detailed in Supplementary Information.
2.4. Construction of Allele-Coupled Exchange vectors
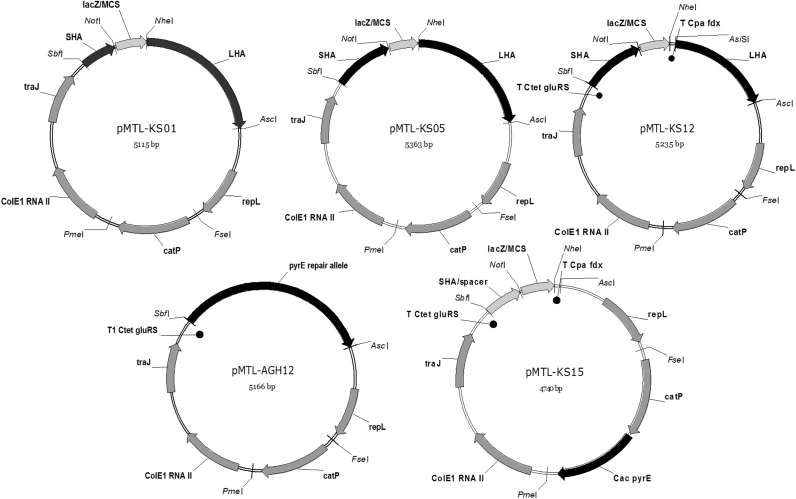
All Allele-Coupled Exchange (ACE) vectors generated in this study were based on the modular plasmid pMTL85151 (Heap et al., 2009) and conform to previous design principles (Heap et al., 2012). The pyrE ACE KO vector (pMTL-KS01) used to generate a ∆pyrE mutant (strain CRG4273) of C. pasteurianum DSM 525 was based on pMTL-JH12 (Heap et al., 2012). The requisite pyrE KO cassette comprised a long (1200 bp) right homology arm (RHA) homologous to a region downstream of pyrE and a short (300 bp) left homology arm (LHA) composed of an internal portion (nt 35–334) of the pyrE gene (full details in Supplementary Information). The pyrE ACE correction (pMTL-AGH12, Fig. 2) and complementation (pMTL-KS12, Fig. 2) vectors were constructed equivalent to pMTL-YN1/YN2 and pMTL-YN1C/YN2C (Ng et al., 2013). Plasmid pMTL-AGH12 was generated through the PCR amplification of a 1748-bp fragment comprising the pyrE nucleotides 35–582 (RHA) and a contiguous region 1200-bp downstream of its stop codon (LHA) using primers AGH0001_pyrE_Fw and AGH0001_pyrE_Rev (Supplementary Table S1) and CRG4111 genomic DNA. Following amplification, the DNA product generated was cleaved with SbfI and AscI, and inserted between the SbfI and AscI sites of pMTL-KS10 (More information in Supplementary Information). Plasmid pMTL-KS12 (Fig. 2) was essentially the same as pMTL-AGH12, except in this case the RHA and LHA were separated by a segment of DNA encompassing a lacZ’-encoding region containing a MCS region. In contrast to pMTL-YN1C/YN2C, however, the RHA was followed by a 77-bp fragment encoding the C. tetani E88 glutamyl-tRNA synthetase terminator (TCtet gluRS), while the LHA was preceded by a 42-bp DNA region encompassing the C. pasteurianum transcriptional terminator of the ferredoxin gene (TCpa fdx). Full details of the construction of pMTL-KS12 is given in Supplementary Information.
Fig. 2.
Plasmid maps of major plasmids used in this study.
For the generation of the complementation vectors target genes including their native promoter were PCR amplified using CRG4111 genomic DNA as template and a gene specific oligonucleotide pair (Fw and Rev) encompassing 5’-NotI and a 3’-NheI recognition sites (see Supplementary Table S1). Following amplification, the DNA fragment was digested with NotI and NheI, purified and ligated into appropriately cleaved pMTL-KS12. Full details of the construction of the complementation vectors is given in Supplementary Information.
2.5. Construction of allelic exchange KO vectors
All generated allelic exchange vectors are based on the modular plasmid pMTL85151 (Heap et al., 2009) designed in accordance with Ng et al. (2013). For the marker-less, in-frame deletion of genes in strain CRG4273 (Table 1), the pyrE gene of C. acetobutylicum was employed as a heterologous counter (negative) selection marker. Therefore, the catP-pyrE module of the plasmid pMTL-AMH101 (Liew et al., 2017) was cloned as a FseI-PmeI-fragment into the equivalent sites of pMTL-KS08. The resulting plasmid, pMTL-KS15 (Table 1, Fig. 2), carried strong transcriptional terminators (5’ TCtet gluRS, 3’ TCpa fdx) that flanked the site of insertion (between SbfI and NheI) of the KO cassette. Cassettes were generated by SOE (splicing by overlap extension) PCR as described previously (Ng et al., 2013) and cloned into pMTL-KS15 via SbfI/NheI sites. Full details of the construction of AE KO plasmids can be found in Supplementary Information.
2.6. Allele‐Coupled Exchange procedure
Allele-Coupled Exchange (ACE) vectors were transformed into C. pasteurianum DSM 525-H1ΔpyrE (CRG4273) and plated onto RCMTm15 agar. Putative single crossover integrants were identified as faster growing, larger colonies (Cartman et al., 2012). These were restreaked onto RCMTm15 agar and PCR screened for the presence of single crossovers, using primers KS004_Cpa_pyrE_gen_Fw and KS007_ 85151_LHA_Rev2 (Supplementary Table S1). In the case of pyrE KO using pMTL-KS01, clones confirmed as single crossovers were re-streaked onto CBMFOA600, Ura5 agar plates and incubated for 24- 28 h. Colonies that developed represented putative double crossover mutants. These were patch plated onto CBM, CBMTm15U5 and CBMU5 agar plates to identify double crossover; uracil auxotrophs that had lost the excised plasmid grow only on the latter media. Deletion mutants were verified by Sanger sequencing of the PCR amplified DNA fragment generated using the above primer pair.
The procedure was the same for restoration of the pyrE locus to wild type using the pyrE ACE correction vector (pMTL-AGH12) and the pyrE ACE complementation vectors (pMTL-KS12) carrying functional copies of the gene to be complemented, but in this case the faster growing, single crossover integrants were re-streaked twice onto RCMTm15 agar plates to purify and incubated for 16- 24 h. Single colonies were then restreaked onto CBM agar plates, and replica plated onto RCMTm15 agar plates to check for plasmid loss. Following, successive re-streaking (3- 5 times) onto fresh CBM and RCMTm15, clones were identified that exhibited strong growth on CBM but not on RCMTm15 agar plates. Restoration of the pyrE locus to WT was confirmed by Sanger sequencing of the DNA fragment PCR amplified using primers KS004_Cpa_pyrE_gen_Fw and KS004_Cpa_pyrE_gen_Rev (Supplementary Table S1).
2.7. Allelic exchange KO procedure
For the construction of specific gene deletions (spo0A, dhaBCE, hydA and rex) by allelic exchange, appropriate KO vectors based on pMTL-KS15 were transformed into the pyrE deletion strain CRG4273 (Table 1) and plated on RCMTm15 agar. Following 48 h incubation, faster growing colonies were re-streaked twice onto RCMTm15 agar plates and their identity as single crossover integrants confirmed by Sanger sequencing of the PCR amplified DNA fragment using appropriate primer pairs. Confirmed single crossover mutants were grown overnight in 5 ml CBMSFOA600, Ura5 broth to allow the double crossover to occur, centrifuged (10 min, 8500g, RT) and re-suspended in 250 µl PBS. A 100 µl aliquot of the suspended cells was serially diluted (up to 10−7) in PBS and 100 µl of each dilution plated onto RCM agar plates. After a 24 h incubation, 50 single, faster growing colonies were selected and re-streaked in the indicated order onto RCMTm15, CBM and RCM agar plates. Colonies, which lost the plasmid and either reverted back to the WT or carried the desired deletion exhibited no growth on RCMTm15 and CBM agar plates but grew on RCM agar plates. These re-streaks were subjected to colony PCR, using gene specific primers that flanked the intended deletion, and the amplified DNA fragment subjected to Sanger sequencing to confirm the expected genotype. The pyrE deletion of verified mutants was restored to WT using the ACE plasmid pMTL-AGH12 (Fig. 2).
2.8. Next-generation sequencing and resequencing analysis
Illumina sequencing of genomic DNA was done by the Deep Seq: Next Generation Sequencing Facility (University of Nottingham, UK) using a MiSeq Illumina system (Illumina, USA). Paired-end reads were mapped against the published C. pasteurianum DSM 525 (strain CRG4091) genome (Poehlein et al., 2015) CLC Genomics Workbench 8.0.2 (Qiagen, DK). Single nucleotide polymorphisms (SNPs) were analysed using the basic variant caller in CLC Genomics Workbench 8.0.2 (Qiagen, DK).
2.9. Fermentation in serum flasks
To analyse butanol production, strains were grown either in 50 ml CGM (Sandoval et al., 2015), Biebl medium (Biebl, 2001) or 2x YTG broth in serum bottles at 37 °C with a starting pH of 6.2. Media was supplemented with 60 g/l glucose or glycerol as carbon source. Biebl medium was additionally supplemented with 1 g/l yeast extract for glycerol fermentations. Cultures were initiated in an anaerobic workstation (Don Whitley, Yorkshire, UK) by inoculating and re-suspending several colonies from CBM agar plates into 10 ml CGM, Biebl or 2x YTG broth containing 60 g/l glucose. After overnight incubation, 1 ml of culture was used to inoculate 20 ml CGM or 2x YTG or 35 ml Biebl broth supplemented with 60 g/l glucose or glycerol and incubated for 6 h prior to diluting up to 10−3 and incubating in fresh media and growing overnight. Those cultures that were in mid-exponential phase were used to inoculate 50 ml CGM, Biebl or 2x YTG broth supplemented with 60 g/l glucose or glycerol to a starting OD600 of 0.05 (CGM, 2x YTG, Bieblglucose) or 0.075 (Bieblglycerol). At this point, serum bottles were removed from the anaerobic workstation and incubated 37 °C for up to 48 h. The OD600 and pH were monitored and samples for product analysis taken every 3 h initially then at 24 h and close to the end of fermentation. All culturing was carried out in triplicate.
2.10. pH-controlled batch fermentation
To enable optimum butanol production, pH-controlled batch fermentations were carried out in a Multifors 2 parallel reactor system (Infors UK, Reigate, UK) containing 350 ml Biebl broth supplemented with 60g/l glucose or glycerol and 1g/l yeast extract were incubated at 37 °C, 250 rpm and continuous sparging with sterile N2 (1 l/min). The initial pH of the medium was 6.0. Following inoculation the pH was held above 6.0 by the automatic addition of 4 M KOH. Pre-cultures were grown as described above for the serum flask fermentation, with the exception, that the volume of the final 16 h dilution series was 35 ml. The main fermentation broth was inoculated to a starting OD600 of 0.75. Batch fermentations were run for 48 h. Growth was monitored through online optical density (OD) as well as through the external measurement of triplicate samples employing a BioMate 3 spectrophotometer (Thermo Fisher Scientific, Loughborough, UK). Samples for solvent analysis were taken at the same time. Additionally the pH, redox potential and temperature of the culture broth were monitored online.
To enable optimum sporulation, pH-controlled batch fermentations were carried out at 37 °C, 200 rpm, with continuous sparging of sterile N2 (0.02 ml/ml) in the same Multifors 2 parallel reactor system (Infors UK, Reigate, UK) containing 400 ml of CBM broth comprising no CaCO3 but 6% (w/v) glucose. To set up the pH-controlled batch fermentation, the strain of interest was streaked from -80 °C stocks onto RCM agar plates, incubated overnight at 37 °C and subsequently used to inoculate 10 ml CBM-S broth containing 6% (w/v) glucose. The 10 ml culture was grown overnight and used to inoculate 50 ml CBM-S broth at a 2% (v/v) inoculum. A 50 ml aliquot of this pre-culture was used to start the pH-controlled batch culture at an inoculum of 6.5% (v/v). The pH was allowed to drop from 6.5 to 5.5, before being maintained at 5.5 by the addition of 4 M KOH. Batch fermentations were run for 120 h.
2.11. Spore assay
Spore assays were carried out either in flasks for a quick screen or in pH controlled batch cultures (see Section 2.10) to enable efficient sporulation. In flasks, 200 ml CBM-S broth supplemented with 0.25 mM phosphate buffer (pH 7.3, Steiner et al., 2012) were inoculated from CBM-S overnight culture to a final OD600 of 0.1 and grown anaerobically for 120 h at 37 °C. After incubation cultures were shaken thoroughly and 100 µl samples taken in duplicate to be treated at 80 °C for 10 min and plated on RCM agar plates to quantify the number of heat resistant colony forming units (CFUs) per ml. pH-controlled (5.5) batch cultures were grown for 120 h. Cells were normalized to the lowest OD600 in a final volume of 5 ml PBS buffer, centrifuged (10 min, 10,000g, 4 °C), re-suspended in 200 µl PBS buffer and heated to 80 °C for 30 min. Serial dilutions in a total volume of 1 ml PBS were carried out and 10 µl aliquots were spotted onto RCM agar plates. The plates were incubated for 24 h at 37 °C before colonies were enumerated. The sporulation efficiency was determined as number of heat-resistant CFU/ml of culture, that germinate and grow. All results were confirmed microscopically.
2.12. HPLC analysis of glycerol, glucose and metabolites
Samples (2 ml) of C. pasteurianum cultures from bottle fermentations were collected, centrifuged (10 min, 16,000g, 4 °C) and cell-free supernatants stored at -80 °C until analysed. Cell-free supernatants were thawed on ice, mixed with an equal volume (200 µl) of internal standard solution (80 mM valeric acid [Sigma-Aldrich, Dorset, UK] in 0.005 M H2SO4), filtered through a 0.22 µm HPLC certified syringe filter (Whatman® Spartan® 13/0.2 RC; GE Healthcare Life Sciences, Little Chalfont, UK) and transferred into a HPLC vial with a 100 µl insert. Substrate (glucose, glycerol) and fermentation products (acetate, acetone, butanol, butyrate, lactate, ethanol, PDO) were analysed by the use of a Dionex UltiMate 3000 HPLC system (Thermo Fisher Scientific, Loughborough, UK) equipped with a Bio-Rad Aminex HPX-87H (Hertfordshire, UK) column, a refractive index (RI) and diode array detector (DAD) at UV 210 nm at an isocratic flow rate of 0.5 ml/min of 0.005 M H2SO4 as mobile phase and a column temperature of 35 °C for 55 min. The injection volume was 20 µl. If required samples were diluted using reverse osmosis (RO) water. Standard concentrations ranged from 0.98 to 250 mM. For glycerol two additional concentrations of 500 mM and 750 mM were employed. Signal analysis was performed using the Chromeleon 7.2 Chromatography Data System (Thermo Fisher Scientific, Loughborough, UK). Statistical analysis of significance was performed using the PRISM (GraphPad Software, La Jolla, USA) employing Fisher's t-test. The significance level (α-value) was set at 0.05.
2.13. Identification of putative Rex boxes in C. pasteurianum
The C. pasteurianum genome was analysed for the presence of Rex binding boxes using the Rex box consensus sequence (5’-TTGTTAANNNNTTAACAA) reported by Ravcheev et al. (2012) with the ’Virtual Footprint’ algorithm (Münch et al., 2005) allowing 2 mismatches in a similar fashion to Wietzke and Bahl (2012). As the C. pasteurianum genome was not available in the PRODORIC database (Münch et al., 2003) to which ’Virtual Footprint’ is linked, the results were analysed manually. Each ‘hit’ was searched against the genome of C. pasteurianum DSM 525 (Poehlein et al., 2015) using the Artemis genome browser (Rutherford et al., 2000). The information extracted was the locus tag, gene name if applicable, protein function and location of the target sequence in respect of the genome context. Only target regions in intergenic regions with a putatively regulated gene up- or downstream were considered. For every entry in the list the distance to the closest start codon was measured and a frequency plot with a bin size of 10 bp was considered. The result is a positive skewed distribution with the maximal target site occurrence between 80 bp and 90 bp from the start codon which dropped to near zero occurrences after 250 bp from the start codon. This observation led to further filtering the list with a threshold of 250 bp distance to the start codon. The resulting list comprised 40 sequences which were used in an iterative approach to run ’Virtual Footprint’ with a new Position Weight Matrix (PWM). This led to a list of 113 targets which were filtered to exclude hits in coding regions of genes and in intergenic regions with antidromic genes. The final list comprised 47 targets which putatively regulate downstream genes (Supplementary Table S2).
3. Results
3.1. Improved transformation of C. pasteurianum DSM 525
C. pasteurianum ATCC 6013 has been shown to be transformable with several pMTL80000 modular vectors (Heap et al., 2009) at efficiencies of up to 7.5×104 transformants μg-1 DNA (Pyne et al., 2013). This high frequency was reliant on circumventing the activity of the endogenous CpaAII restriction-modification system by in vivo methylation of its recognition site (5’-CGCG-3’) using the M.FnuDII methyltransferase (Pyne et al., 2013). As the E.coli host employed (Top10) was dam+, the activity of the restriction enzyme CpaII, a MboI/DpnII-type restriction endonuclease previously identified as CpaI (Richards et al., 1988), was of no consequence. Here we used either in vitro or in vivo methylation by M.SssI or M.BepI, respectively. The former methylates all cytosine residues within the double-stranded recognition site 5’-mCG-3’ (Renbaum et al., 1990), whereas similar to M.FnuDII, M.BepI methylates the external cytosine (5’-mCGCG-3’) of the CpaAII recognition site (Lunnen et al., 1988). Whilst either methylation procedure allowed transformation of both ATCC 6013 and DSM 525 (equivalent to ATCC 6013) to be obtained with plasmids pMTL83151 (pCB102 replicon) and pMTL85151 (pIM13 replicon) (Heap et al., 2009) using our published method (Cooksley et al., 2012), the success rate and frequencies obtained was very low, 1.5×101 transformants μg-1 DNA. After adapting the method published by Pyne et al. (2013) with added sucrose in growth medium and extra step of glycine wash, the transformation frequency was improved to 1.6×102 transformants μg-1 DNA, still two orders of magnitude lower than reported by Pyne et al. (2013) of which up to 7.5×104 transformants μg-1 DNA was achieved. Plasmids pMTL82151 (pBP1 replicon) and pMTL84151 (pCD6 replicon) could not be transformed.
We hypothesised that one reason for the low number of transformants was that those cells that were competent for the acquisition of plasmid DNA represented rare mutant cells present within the population. This hypothesis was tested by curing the plasmid of a randomly selected transformant of DSM 525 through repeated subculture in the absence of the selective antibiotic, and then retesting the cured strain to see if the plasmid-free strain was more amenable to electroporation. The strain was found to transform at frequencies of 2.6×105 transformants μg-1 DNA. Moreover, similar frequencies were observed with plasmids pMTL85151, pMTL82151 and pMTL84151 (Heap et al., 2009). Whole genome sequencing of the strain with higher frequency C. pasteurianum DSM 525-H1 (designated CRG4111) and compared to the parental strain C. pasteurianum DSM 525 (Poehlein et al., 2015) indicated that two SNPs had arisen within two distinct open reading frames, that of CLPA_c13710 and CLPA_c33080 (Poehlein et al., 2015). Unexpectedly, neither encoded protein was obviously associated with restriction/ modification. Both SNPs resulted in frame-shifts in the encoded sequence. CLPA_c13710 encodes a predicted β-lysine N6-acetyltransferase (ablB) which was reduced from 283 amino acids to 176 residues in the hypertransformable mutant. CLPA_c33080 encoded a histidine kinase (ResE9) and leads to a frameshift that reduces the protein from 615 to 160 amino acids in length. To ascertain whether the general phenotypic properties of the CRG4111 mutant strain had been affected by the SNPs, comparative growth experiments were performed on glycerol as the carbon source. The growth and glycerol consumption rates of the mutant were similar to the parental strain. Aside from a small increase in the levels of PDO and a small reduction in n-butanol, all other measured metabolites (ethanol, lactate, acetate and butyrate) were essentially the same between the two strains. On this basis, the strain was taken forward for metabolic engineering studies.
3.2. Implementation of a gene system roadmap
We have previously described the implementation of allelic exchange, gene KO systems in specifically generated pyrE mutants of Clostridium acetobutylicum (Ehsaan et al., 2016) and Clostridium difficile (Ng et al., 2013), and most recently Geobacillus thermoglucosidasius (Sheng et al., 2017). The generation of mutants by allelic exchange is facilitated by the use of replication defective, or pseudo-suicide vectors (Cartman and Minton, 2010), and the incorporation into the vector of a functional copy of a heterologous pyrE gene (encoding orotate phosphoribosyl transferase) that serves as a counter/ negative selection marker. This approach serves as a general roadmap for the implementation of gene KO systems in clostridia (Minton et al., 2016). Pivotal are the generation of a pyrE truncation mutant using Allele-Coupled Exchange (ACE) technology (Heap et al., 2012) and the identification of an appropriately replication defective (pseudo-suicide) vector.
Segregation stability studies (see Supplementary Information) on transformed cells carrying the various pMTL80000 modular vectors established that plasmids based on the pIM13 replicon were suitably defective, with 99% of the cells losing the plasmid after eight 12 h subcultures (approximately 51 generations) in media lacking antibiotic selection. Accordingly, a pIM13-based vector (pMTL-KS01) was constructed (Fig. 2 and Materials and Methods) equivalent to the ACE plasmids pMTL-YN18 (Ng et al., 2013) and pMTL-JH12 (Heap et al., 2012) and a pyrE (CLPA_c26850) truncation mutant generated as previously described (Heap et al., 2012, Ng et al., 2013). In essence, thiamphenicol (Tm) resistant (R) transformants in which pMTL-KS01 had integrated into the CRG4111 genome via a longer Right Homology Arm (RHA) were identified as larger, faster growing colonies on RCM media supplemented with 15 µg/ml Tm (RCMTm15) and purified by restreaking onto the same selective media. Of the 24 selected colonies, all were shown to be pure single crossover mutants through PCR screening using appropriate primers (see Supplementary Table S1). To select for a subsequent double crossover excision event involving the shorter Left Homology Arm (LHA) and the generation of the required 5-fluoroorotic acid (FOA), resistant pyrE deletion mutant, single crossover integrants were restreaked onto CBM media supplemented with 5 µg/ml uracil (Ura) and FOA at the experimentally determined MIC (600 µg/ml) of the parental strain (CBMUra5, FOA600). From a total of eight FOAR clones obtained, subsequent restreaking and testing of phenotype on appropriately supplemented media demonstrated that in addition to being FOAR, three were uracil auxotrophs (required supplementation of CBM media with exogenous uracil for growth) and sensitive (S) to Tm (could not grow if Tm was present). From PCR screening using primers flanking the pyrE gene and the subsequent sequencing of the amplified DNA fragment, two of the three clones were shown to carry the desired modification. This equated to a deletion extending from nt 335–582 of CLPA c26850, and the concomitant insertion of the lacZα/MCS originating from pMTL-KS01. One strain was selected for further use and designated CRG4273. To demonstrate that CRG4273 could be restored to wildtype (WT) through correction of the pyrE allele an ACE, pyrE repair vector was made (pMTL-AGH12) equivalent to pMTL-YN1/2 (Ng et al., 2013) and pMTL-ME6 (Ehsaan et al., 2016) (see Fig. 2). The region of homologous DNA in this plasmid essentially comprises a contiguous region of DNA from the pyrE locus that includes a complete copy of the pyrE gene and downstream region equivalent to the 1200 bp RHA of plasmid pMTL-KS01. This plasmid was transformed into CRG4273 and single crossover integrants identified as larger, faster growing colonies on CBMTm15, Ura5 agar plates. Growth from several representative colonies were thereafter restreaked onto CBM agar lacking any supplementation, and single colonies patch plated onto RCM and RCMTm15 agar plates to identify those clones in which the excised plasmid had been lost. The pyrE locus of three representative clones was amplified by PCR using appropriate PCR primers (KS004_Cpa_pyrE_gen_Fw and KS004_Cpa_pyrE_gen_Rev, Supplementary Table S1) and subjected to Sanger sequencing. This data confirmed that the locus had been restored to WT in all three cases. One was chosen for storage and designated CRG5570 (Table 1).
3.3. Exemplification of the knock-out vector pMTL-KS15
The availability of the pyrE deletion mutant CRG4273 is a prerequisite for the use of a functional heterologous pyrE gene as a counter selection marker in gene knock-out (KO) by allelic exchange. Accordingly, a KO vector broadly equivalent to pMTL-YN3/4 (Ng et al., 2013), pMTL-ME3 (Ehsaan et al., 2016) and was made, pMTL-KS15 (Fig. 2), and exemplified through the in-frame deletion of the master regulator of sporulation, spo0A. Precise details of the vector are given in Materials and Methods, but is essentially based on the pIM13 replicon and incorporates the C. perfringens catP selectable marker and a heterologous pyrE gene derived from C. acetobutylicum (Liew et al., 2017), as opposed to the C. sporogenes pyrE gene used in the previous studies (Ehsaan et al., 2016, Ng et al., 2013).
The KO cassette for spo0A (CLPA c19180) was generated by SOE PCR as described in Materials and Methods and cloned between the SbfI and NheI sites of pMTL-KS15. Following transformation of the resultant plasmid (pMTL-KS15::spo0A, Table 1) into CRG4273 and plating on RCMTm15 agar plates, putative single crossover integrants were identified as faster growing, larger colonies. Their identity as pure single crossover integrants was confirmed by appropriate PCR (primers KS005_spo0A_genome_Fw, KS005_rex_genome_Rev, Supplementary` Table S1) and then one of the twelve identified grown overnight in CBMSFOA600, Ura5 broth to allow the double crossover event to take place. After the overnight incubation the culture was centrifuged, re-suspended and serial dilutions plated to single colonies on RCM agar plates. Selected colonies were patch plated onto RCMTm15 and un-supplemented RCM agar plates and onto CBM media lacking uracil. Growth on RCM alone confirmed loss of the plasmid, and the encoded catP and pyrE genes, following its excision. PCR screening with flanking primers (KS005_spo0A_genome_Fw and KS005_spo0A_genome_Rev, Supplementary Table S1), was then used to distinguish between those excision events that had generated the desired deletion as opposed to return of the cell to WT. A total of 17 of the 50 putative KO clones screened generated a smaller DNA fragment consistent with the intended deletion in spo0A (Supplementary Fig. S1). Through Sanger sequencing of the amplified DNA fragment, the presence of the expected spo0A deletion was confirmed in all 17 strains. One of the clones was selected (CRG5514) and restored to uracil prototrophy using the ACE vector pMTL-AGH12 as previously described. The final strain, carrying only the spo0A mutation, was designated CRG5516. In parallel, a derivative of pMTL-KS12 was constructed (pMTL-KS12::spo0A*, Table 1) in which a functional copy of the C. pasteurianum spo0A gene, together with its native promoter, was inserted into the MCS between the LHA and RHA. Use of this strain to restore the pyrE mutation of CRG5514 to WT led to insertion of spo0A into the chromosome for complementation studies. The genotype of the strain generated, CRG5518, was confirmed by PCR amplification of the pyrE locus and inserted spo0A gene and nucleotide sequencing of the DNA fragment amplified.
3.4. Phenotypic analysis of spo0A deletion mutant
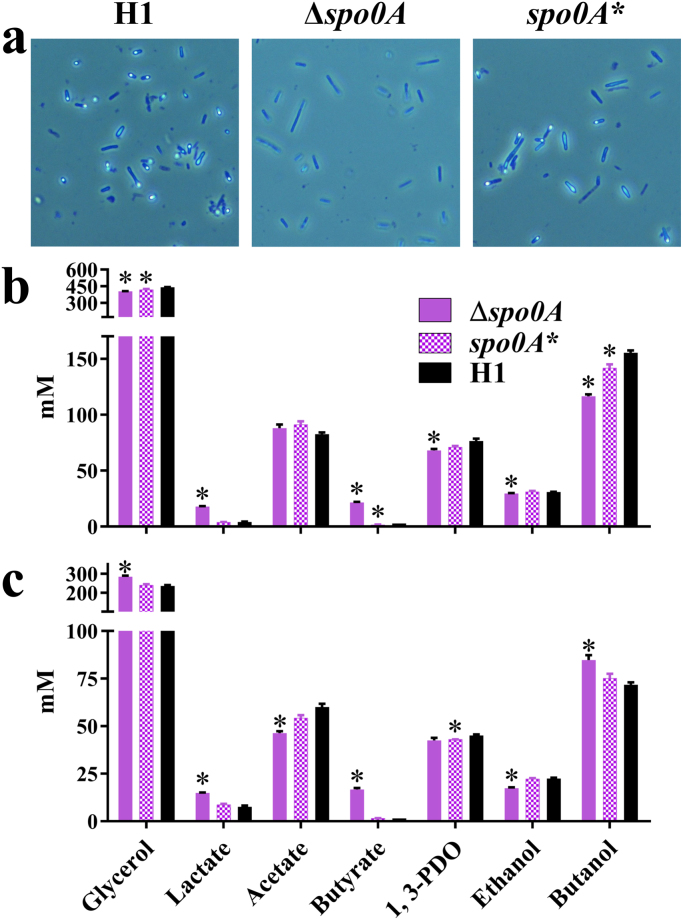
As expected, after 5 days of growth in a pH-controlled batch cultures (pH 5.5), no heat resistant CFUs were obtained after plating on RCM of the spo0A mutant strain CRG5516. In contrast, cultures of the WT (CRG4111) and the complemented mutant strain CRG5518 yielded 1×108 heat resistant CFU/ml. In keeping with these measurements, spores were easily detected using phase contrast microscopy in the cell suspensions of CRG4111 (parent) and CRG5518 (spo0A, complemented), whereas none could be detected in cultures of CRG5514 (spo0A) (Fig. 3a). To analyse the effects of the spo0A deletion on solvent profiles, strains CRG4111 (parent), CRG5516 (spo0A) and CRG5518 (spo0A, complemented) were grown in 50 ml Biebl medium supplemented with 60 g/l glycerol (Fig. 3b). All strains exhibited a similar growth rate but the pH of the spo0A mutant (CRG5516) dropped to a lower level than the parent and complemented strain, presumably due to increased production of lactate and butyrate. Glycerol uptake, acetate production and reassimilation and ethanol production are similar in all strains.
Fig. 3.
Comparison of sporulation and fermentation phenotypes of C. pasteurianum-H1 (H1), C. pasteurianum-H1Δspo0A (Δspo0A) and C. pasteurianum-H1-spo0A complementation (spo0A*). a) Spores can be observed in H1 and spo0A* whereas Δspo0A as expected is incapable of producing spores. b), c) Pure glycerol fermentation in serum bottles with 60 g/l glycerol in Biebl medium (b) and CGM (c) was carried out for 48 h with fermentation being visibly completed after 24 h which is the time point shown for glucose consumption and product formation. * indicate statistical significance in t-test α>0.05 of deletion or complementation strain against H1. Error-bars indicate standard error of three replicates.
Butanol and PDO levels were slightly lower in the mutant in Biebl medium. In contrast, the complemented spo0A mutant strain (CRG5518) produced slightly higher levels of these solvents. The data obtained was at some variance from that of Sandoval et al. (2015) who undertook a similar analysis of a spo0A mutant of C. pasteurianum. However, these authors used rich media (CGM) as opposed to the minimal media employed here. We, therefore, repeated our analysis using CGM (Fig. 3c). In this medium, the production of lactate and butyrate by the spo0A mutant was significantly increased compared to the parent strain, CRG4111. Interestingly, the mutant (CRG5516) produced much more acetate in the initial stages of the fermentation, but then reassimilated the acid from 12 h onwards. No equivalent reassimilation was seen in either the parental (CRG4111) or complemented (CRG5518) cultures. In common with the observations of Sandoval et al. (2015), compared to the parental strain (CRG4111), glycerol consumption was significantly increased together with the production of higher levels of n-butanol. In contrast to Sandoval et al. (2015), however, the PDO levels of the spo0A mutant (CRG5516) were not reduced compared to the wildtype but remained unaltered.
3.5. Construction of deletion mutants in genes involved in solvent production
To investigate whether solvent profiles could be altered in favour of n-butanol production we targeted genes encoding the redox-responsive regulator Rex, the main hydrogenase (hydA) and the glycerol dehydratase (dhaBCE).
Rex has been previously shown to control the expression of n-butanol biosynthetic genes in response to the cellular NADH/NAD+ ratio in C. acetobutylicum where its disruption led to reduced acid production and increased solvents yields (Wietzke and Bahl, 2012). In C. acetobutylicum the rex gene resides immediately 5’ to the crt-bcd-etfAB-hbd operon responsible for butyryl-CoA biosynthesis (Wietzke and Bahl, 2012). An equivalent gene (CLPA_c28640) is found in the same context in C. pasteurianum and encodes a 213 amino acid protein exhibiting 76% amino acid sequence identity to the C. acetobutylicum Rex. In the case of hydrogenase, a number of different [FeFe]-hydrogenases and [NiFe]-hydrogenase exist in C. pasteurianum but little information is available concerning their specific individual roles in hydrogen formation and redox balance. BLAST analysis showed that the protein encoded by CLPA_c00280 exhibits 71% identity to the well described C. acetobutylicum hydrogenase, hydA (Santangelo et al., 1995). Accordingly, KO cassettes to bring about the deletion of CLPA_c28640 (Rex) and CLPA_c00280 (Hyd) were generated by SOE PCR, cloned into pMTL-KS15 and the resultant mutants generated by allelic exchange using our standard procedure (Table 1, Supplementary Fig. S1 and Materials and Methods).
Simplistically, one way to increase butanol titres when growing on glycerol might be to eliminate PDO production. The conversion of glycerol to PDO involves a two-step transformation by glycerol dehydratase (DhaBCE), encoded by dhaBCE (CLPA_c22770-CLPA_c22760-CLPA_c22750) and PDO dehydrogenase (DhaT), encoded by dhaT (CLPA_c22740). Given the inability of Pyne et al. (2016) to isolate a clean mutant of dhaT in the presence of the toxic intermediate 3-hydroxypropionaldehyde (3-HPA) (Barbirato et al., 1996, Maervoet et al., 2014), we chose to ablate PDO production by eliminating the synthesis of glycerol dehydratase through deletion of the entire dhaBCE operon. Accordingly, a KO cassette to achieve this was generated by SOE PCR, cloned into pMTL-KS15 and the resultant KO plasmid used to generate a dhaBCE mutant (CRG5532) by allelic exchange using our standard procedure (Table 1, Supplementary Fig. S1 and Materials and Methods).
In all three cases (rex, hyd and dhaBCE), the mutants were restored to uracil prototrophy, through ACE-mediated correction of the pyrE mutation, and complemented through the integration of a functional copy of the deleted gene with the native promoter concomitant with restoration of the mutant pyrE allele to WT (Materials and Methods). Phenotypic analysis of all mutants, and their complemented equivalents, involved the determination of carbon utilisation as well as the concentration of the following fermentation products: acetate, butyrate, lactate, acetone, ethanol, PDO and n-butanol. All studies, with the exception of the glycerol fermentation of the dhaBCE mutant (CRG5534), were carried out in batch culture in bioreactors using Biebl medium supplemented with 1 g/l yeast extract and 6% (w/v) glycerol or with glucose omitting yeast extract. Fermentation profiles are shown in Fig. 4, Fig. 5, Fig. 6.
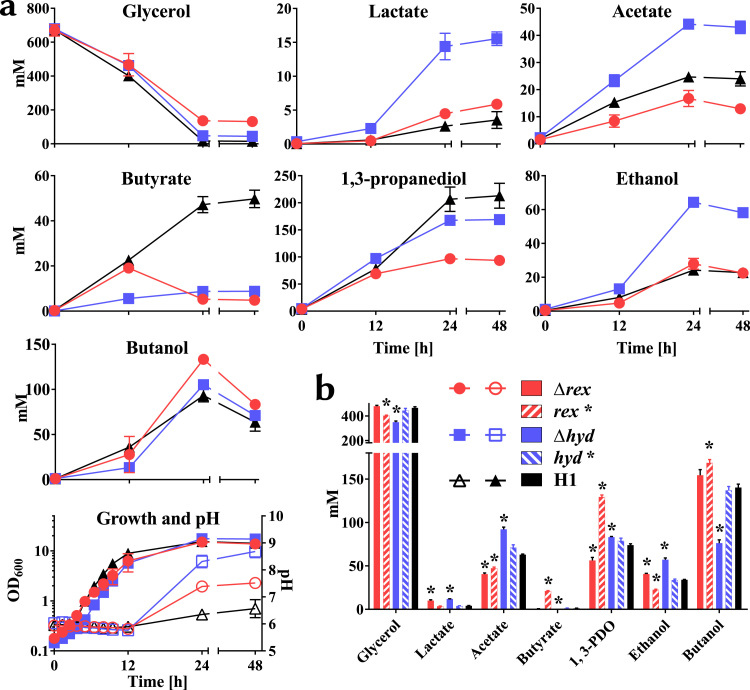
Fig. 4.
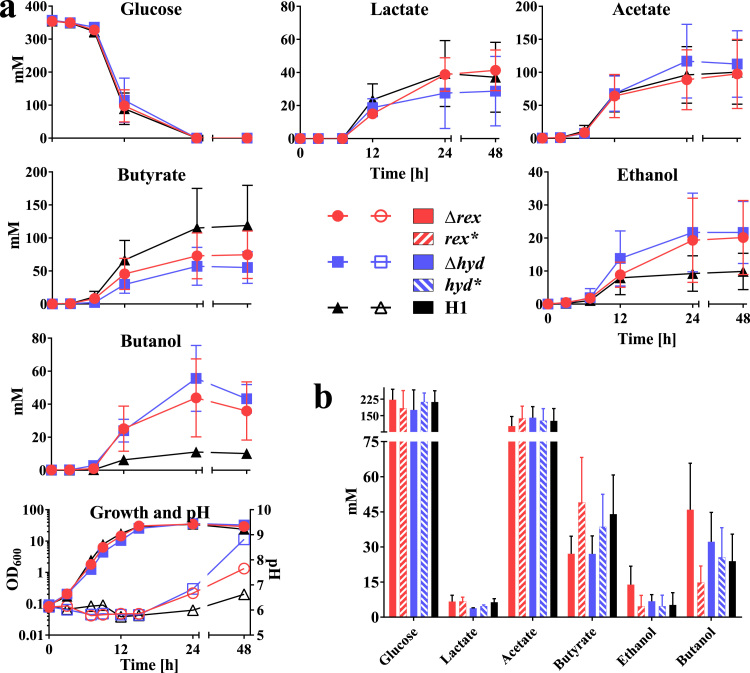
Pure glycerol fermentations of C. pasteurianum-H1 (H1), C. pasteurianum-H1Δrex (Δrex), C. pasteurianum-H1-rex complementation (rex*), C. pasteurianum-H1Δhyd (Δhyd) and C. pasteurianum-H1-hyd complementation (hyd*). a) Bioreactor fermentation with 60 g/l glycerol in Biebl medium with 1 g/l yeast extract at pH 6 was carried out for 48 h with fermentation being visibly completed after 24 h. Error-bars indicate range of two fermentations. b) Histogram showing product formation of serum bottle fermentation of deletion strains and complementations. Glycerol usage and product formation is shown after 24 h. * indicate statistical significance in a t-test α>0.05 of deletion or complementation strain against H1. Error-bars indicate standard error of three replicates.
Fig. 5.
Glucose fermentations of C. pasteurianum-H1 (H1), C. pasteurianum-H1Δrex (Δrex), C. pasteurianum-H1-rex complementation (rex*), C. pasteurianum-H1Δhyd (Δhyd) and C. pasteurianum-H1-hyd complementation (hyd*). a) Bioreactor fermentation with 60 g/l glucose in Biebl medium at pH 6 was carried out for 48 h with fermentation being visibly completed after 24 h. Error-bars indicate range of two fermentations. b) Histogram showing product formation of serum bottle fermentation of deletion strains and complementations. Glucose usage and product formation is shown after 24 h. Error-bars indicate standard error of three replicates.
Fig. 6.
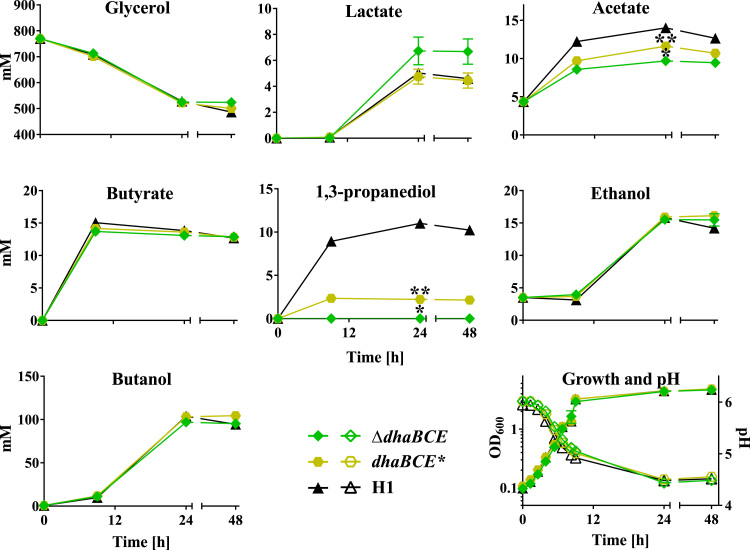
Pure glycerol fermentations of C. pasteurianum-H1 (H1), C. pasteurianum-H1ΔdhaBCE (ΔdhaBCE) and C. pasteurianum-H1-dhaBCE complementation (dhaBCE*). Serum bottle fermentation with 60 g/l glycerol in 2x YT medium was carried out for 48 h with fermentation being visibly completed after 24 h. * indicates statistical significance of ΔdhaBCE and ** of dhaBCE* in a t-test α>0.05 against H1. Error-bars indicate standard error of three replicates.
3.6. Phenotypic analysis of the rex and hyd mutants
In glycerol media (Fig. 4), both mutants grew at broadly equivalent growth rates compared to the WT, but achieved higher final pH values, being 8.7±0.2 and 7.5±0.1 after 48 h for the hydA and rex mutant, respectively compared to 6.6±0.3 in the WT culture. Almost complete glycerol consumption took place in the WT and hydA culture (2% and 7% remaining, respectively), with the rex mutant culture containing 20% after 48 h. The hydA mutant produced the highest titres of both lactate and acetate, but significantly reduced amounts of butyrate. A slightly greater reduction in butyrate production was observed with the rex mutant compared to the hydA mutant. In terms of lactate and acetate production, the former were slightly increased compared to the WT, whereas the latter marginally decreased. In the case of solvents, PDO production was reduced in both mutants, with the largest reduction occurring in the rex mutant, which decreased by 53% compared to the WT, with only a 19% reduction being evident in the hydA mutant. Ethanol formation in the hydA mutant was significantly elevated (64.3±3.2 mM after 24 h) over both the WT and rex mutant which, with respective titres of 24.2±1.4 mM and 28.0±3.2 mM, were broadly equivalent. Butanol formation was highest in the rex mutant, with the titres achieved after 24 h being 133.3±1.8 mM compared to 105.1±0.0 mM in the hydA mutant and 93.2±5 mM in the WT. After 24 h the ethanol and butanol levels decreased, most likely due to solvent extraction by gas stripping from the nitrogen bubbling through the reactor.
On glucose (Fig. 5), the growth rate and glucose utilisation rates were little affected in either mutant compared to the WT. Differences in acid production were also less marked in the two mutants compared to when grown on glycerol. Acetate production in the hydA mutant was slightly increased compared to the WT and rex mutant, which were essentially the same. On the other hand, lactate production in the hydA mutant was only slightly reduced compared to the rex mutant and WT. Butyrate produced was decreased in both mutants compared to the WT, with the larger reduction being seen in the hydA mutant, producing 57.0±14.3 mM compared to 115.0±30.0 mM in WT culture after 24 h. Ethanol and n-butanol titres, and in particular the latter solvent, were increased in both mutants, with hydA demonstrating a 2.3-fold and 5-fold higher final titre than the WT, respectively, at 24 h.
Whilst the fermentation profile of the complemented hydA mutant was essentially restored to that of the WT, complementation of the rex mutant was less clear cut. Thus, whereas the PDO levels of the mutant were reduced in comparison to the WT and the n-butanol levels increased, the complemented strain produced higher amounts of PDO and reduced titres of n-butanol, relative to the WT. Such a phenotype would be indicative of higher than normal expression of rex in the complemented strain, however, the rex gene was inserted at pyrE under the control of its native promoter. In certain instances expression of genes can be affected by the proximity of the gene to the origin of chromosome replication (Sauer et al., 2016). This does not appear to be the case here, as the distance of rex and pyrE to the origin are relatively similar with rex being slightly further away from the origin. In C. kluyveri, rex is self-regulated through the agency of a Rex box, repressor site immediately upstream of the structural gene (Hu et al., 2016). Whilst such an operator site was not evident in the C. pasteurianum rex promoter region used to express the complementing copy of rex, a Rex box was found 340 bp upstream of the rex start codon within the upstream CLPA_c28650 gene (genome position 3068768- 3068786). As the promoter region used in the complemented strain lacks this sequence it is likely that rex expression is deregulated, resulting in the observed produced higher amounts of PDO and reduced titres of n-butanol, relative to the WT.
3.7. Phenotypic analysis of the dhaBCE mutant
Growth of C. pasteurianum on glycerol is not redox balanced because cell biomass (_D, 4.3) is more oxidised than glycerol (_D, 4.67). The requirement to oxidise the excess of reducing equivalents generated is met by the PDO pathway which ensures the net oxidation of 1 mol of NADH per 1 mol of PDO formed. The central role of this pathway in maintaining redox balance suggests that the imbalance caused by its inactivation in C. pasteurianum would lead to an inability to ferment glycerol. Indeed, the mutant (CRG5534) was unable to grow in a bioreactor in Biebl medium when the carbon source was 6% (w/v) glycerol. The mutant was, however, able to grow in 2x YT containing glycerol (6%, w/v) at equivalent rates to the WT (Fig. 6). The most notable change in fermentation profile was the production of PDO, which, as expected, was eliminated in the mutant (Fig. 6). In contrast, the production of metabolites derived from acetyl-CoA (butyrate, ethanol and n-butanol) were largely unaffected, apart from a decrease of acetate production to 9.7 mM at time point 24 h compared to 14 mM in the WT, and an increase in lactic acid production (6.7 mM at time point 48 h compared to 4.6 mM in the WT). The solvent profiles of CRG5534 on glucose where as expected essentially the same as the WT (Fig. S2). In the complemented strain, in which dhaBCE has been integrated at pyrE together with its native promoter, production of PDO was re-instated, although at a lower level than the WT. The lactate and acetate profiles were very similar to that of the WT (Fig. S2).
4. Discussion
The use of pyrE alleles for gene knock-out forms part of a general ‘roadmap’ for manipulating bacterial genomes (for a review see Minton et al., 2016), the implementation of which is reliant on effective gene transfer into the target organism. In the case of C. difficile (Ng et al., 2013) and G. thermoglucosidasius (Sheng et al., 2017) no special measures were required to achieve this, whereas in C. acetobutylicum the rational circumvention of host restriction barriers proved to be necessary by appropriate methylation of the plasmid DNA being transferred (Ehsaan et al., 2016). Here we showed that an alternative approach is possible through the isolation of a highly transformable variant of the C. pasteurianum strain DSM 525, strain CRG4111.
The reason for the increased competence of CRG4111 is unclear. Neither of the two SNPs present are in genes with any obvious association with restriction/ modification. Both are destructive in nature resulting in frame-shifts and premature termination of the encoded proteins. Whether one or the other, or both, are responsible for the phenotype is not immediately apparent. One gene encodes β-lysine N6-acetyltransferase (ablB), which, being involved in lysine degradation seems to have no connection with improved DNA transfer. CLPA_c33080 encodes a histidine kinase (ResE9), but does not appear to be part of a two-component system, as no gene encoding a cognate response regulator was located in the immediate vicinity. Interestingly, however, the gene (CLPA_c33090) immediately adjacent to the ResE9 encodes a homologue of an agr quorum sensing AgrB protein, responsible for processing and secretion of the AgrD signal molecule. Indeed, an unannotated ORF is present immediately downstream of CLPA_c33090 encoding a 57 amino acid peptide that shares significant identity with clostridial AgrD signal peptides, including 58% with those of Clostridium tetanomorphum DSM 665 (KAJ50110.1 and KAJ51453.1), Clostridium scatologenes (AKA70070.1) and Clostridium carboxidivorans P7 (EET84565.1) and 52% identity to homologues in Clostridium botulinum (WP_061325181) and Clostridium sporogenes (WP_053818607.1). All of these clostridial species, and many more besides, share the same structural arrangement of genes, corresponding to agrB>agrD>resE. This gene arrangement, was first noted in C. botulinum, where it was shown to be involved in the regulation of sporulation and neurotoxin production (Cooksley et al., 2010). The possibility exists that in C. pasteurianum at least, this quorum sensing system also regulates a process that can affect the efficiency of DNA uptake.
Regardless of the mechanism responsible for increased competence, our findings have provided a further approach for maximising DNA transfer rates which was used in combination with the developed pyrE-based KO system to make a number of mutants (spo0A, hydA, rex and dhaBCE) that influenced solvent production. A general workflow and schematic for mutant generation is shown in Supplementary Fig. S3. Significantly we were able, for the first time, to completely eliminate PDO production in C. pasteurianum. The formation of PDO from glycerol is mediated by the sequential action of glycerol dehydratase (DhaBCE) and PDO dehydrogenase (DhaT). Here we chose to entirely delete the dhaBCE region, and as a consequence generated a strain which no longer produced PDO under all conditions tested. Previous attempts to ablate the production of either glycerol dehydratase or PDO dehydrogenase through the use of ClosTron technology met with little success (Pyne et al., 2013). Intron mutants of dhaB, dhaC and dhaE could not be isolated, whilst an intron insertion in dhaT was obtained but the intron delivery plasmid and its encoded LtrA protein could not be cured. Such a phenomenon can occur if the intron insertion is in the sense strand of an essential gene, and LtrA-mediated splicing of the transcribed mRNA provides a route for some production of the encoded protein. However, the intron insertion in the dhaT gene was in the antisense orientation. This led to the search for, and apparent discovery of an additional ecotopic, sense strand, intron insertion in speA (encoding arginine decarboxylase) which was hypothesised as being essential. One consequence of these difficulties was that Pyne et al. (2016) were unable to eliminate PDO production.
Glycerol is metabolized both oxidatively and reductively in Clostridium (Malaviya et al., 2012). The pyruvate-generating oxidative pathway leads to the production of CO2, H2, and electrons in the form of the reducing equivalent NADH, which is also released during biomass formation. Glycerol, being a highly reduced substrate, results in twice the amount of reducing equivalents compared to using glucose as fermentation substrate. These electrons require acceptors for redox homeostasis, which is precisely the purpose of glycerol reducing pathway producing the highly reduced product PDO. By deleting dhaBCE in C. pasteurianum, the pathway for funnelling extra reducing power during glycerol fermentation is absent, resulting in the poor growth we observed in the mutant in defined medium. The mutant was, however, able to grow in a rich medium containing yeast extract. The latter ingredient supports growth of the mutant by both providing an alternative source of carbon and acting as a reducing agent. Re-introducing dhaBCE back into the deletion mutant reversed the phenotype, confirming the production of PDO in C. pasteurianum is important for biomass production and redox balance. Our findings support the hypothesis of Johnson and Rehmann (2016), who found that unlike C. acetobutylicum, C. pasteurianum glycerol fermentation does not display strong biphasic behaviour, suggesting that PDO production is regulated and further that its regulation may serve to function in redox homeostasis and allow C. pasteurianum to behave in a non-biphasic manner.
The Rex response regulator directly senses changes in the redox status of the cell, having a much higher affinity for NADH than for the oxidised nucleotide NAD+ (Wang et al., 2008). Under oxic conditions (i.e., when the NADH/NAD+ ratio is low) it binds to its target sites to inhibit gene transcription but dissociates from these sites when the NADH/NAD+ ratio increases (Brekasis and Paget, 2003). Thus, it was expected that inactivation of Rex would lead to increased levels of those enzymes whose genes were under Rex-mediated control, leading to a consequent change in metabolic profiles. To predict such regulated genes we used the procedure outlined in Materials and Methods to search for potential Rex target sites in the genome. We identified a list of 47 putative Rex box sequences which potentially regulate the adjacent downstream genes (Supplementary Table S2) and defined a consensus sequence for C. pasteurianum, 5'- TTGTTAAWNNNTTAACAA. Of the non-fermentative genes previously reported, none were found using our approach (Ravcheev et al., 2012, Zhang et al., 2014). However, other notable putative Rex targets found include CLPA_c05450, a nitroreductase related to narAB or narK in C. carboxidivorans (Zhang et al., 2014), CLPA_c09900, a nicotinamidase family protein, CLPA_c17480 (spoVD2) encoding a stage V sporulation protein and CLPA_c39090, a NADPH dehydrogenase.
Given that a high ratio of NADH/NAD+ inhibits binding of Rex to DNA, it is expected that Rex-binding sites should be located upstream of genes encoding NADH-dependent enzymes. This is true for the alcohol dehydrogenases putatively involved in butanol and ethanol production (adhE2 and its two alleles adhE1 and adhE4), indirectly the thiolases that channel carbon into the reductive part of the fermentative pathway (thlA1 and thlA2), the crt-bcd-etfBA-hbd operon (crotonase-butyryl-CoA dehydrogenase-electron transfer flavoprotein B/A-hydroxybutyryl-CoA dehydrogenase) which uses 2 NADH to reduce acetyl-CoA to butyryl-CoA, and the reductive part of the dha (glycerol dehydratase) operon. In keeping with these observations, production of ethanol and n-butanol was elevated in the rex mutant when grown on either glycerol or glucose, as was demonstrated in C. acetobutylicum (Wietzke and Bahl, 2012).
By the same reasoning, and based on observations made in C. acetobutylicum (Ravcheev et al., 2012), we would have expected to find Rex sequences upstream of ldh (lactate dehydrogenase), hydA (hydrogenase) and ptb-buk (phosphotransbutyrylase-butyrate kinase), as was found in other species (Wietzke and Bahl, 2012, Hu et al., 2016), but none were found, even with relaxed algorithm parameters. Nevertheless, contrary to in silico expectations and the observations made in C. acetobutylicum, the C. pasteurianum rex mutant produced more lactate in glycerol fermentation compared to WT (Fig. 4). On the other hand, as expected, butyrate production was significantly reduced in the absence of Rex, and a clear butyrate reassimilation in glycerol fermentation was observed in the rex deletion mutant. Butyrate assimilation has been shown in C. pasteurianum when feeding external butyrate which led to increased butanol titres (Kao et al., 2013, Lin et al., 2015, Sabra et al., 2014) but reassimilation of butyrate in a standard fermentation was to our knowledge never observed before. Some of the higher n-butanol titres observed in the mutant strain could be explained by reassimilation and carbon recycling of butyrate.
Acetone formation was never observed in fermentations using the C. pasteurianum WT, in agreement with Biebl (2001), Sandoval et al. (2015) and Pyne et al. (2016). However, in the rex deletion mutant butyrate is visibly reassimilated which in the pathway proposed by Malaviya et al. (2012) is coupled to acetone formation via the acetoacetyl- CoA: butyrate:CoA transferase in a similar fashion to C. acetobutylicum (Jones and Woods, 1986). These findings suggest that C. pasteurianum may be able to reassimilate butyrate without the need of acetone production either through the reverse reaction of the butyrate kinase (buk) and the phosphotransbutyrylase (ptb), as previously suggested (Hüsemann and Papoutsakis, 1989, Millat et al., 2014), or via a third, as yet undescribed, mechanism proposed by Millat et al. (2014). Reassimilation of butyrate without acetone production has been described in several C. acetobutylicum inactivational mutants, most notably mutants of ctfA (Millat et al., 2014), ptb (phosphotransbutrylase) and adc (acetoacetate decarboxylase) (Lehmann et al., 2012) and butyrate kinase (buk) gene (Desai et al., 1999). The fact that C. pasteurianum does not produce acetone under any condition tested here, or elsewhere (Biebl, 2001, Pyne et al., 2016, Sandoval et al., 2015), makes this organism an ideal target for investigations of the butyrate and acetate uptake mechanisms not reliant on CtfAB.
Since alcohol production competes for electrons with the formation of molecular hydrogen, reducing the flux of electrons towards H2 synthesis might be expected to increase n-butanol titres. Various approaches have been reported that were designed to inhibit in vivo hydrogenase activity, for example, by the application of artificial electron carriers or sparging the culture with carbon monoxide (Datta and Zeikus, 1985, Hüsemann and Papoutsakis, 1989, Kim et al., 1984, Peguin et al., 1994;). The resultant increased NAD(P)H availability, led to more n-butanol and ethanol and less acetone being produced and a consequently improved alcohol: acetone ratio in C. acetobutylicum. However, previous attempts to inactivate the gene (hydA) encoding the major hydrogenase of C. acetobutylicum using the ClosTron were unsuccessful (Cooksley et al., 2012), suggesting that the gene is essential under the conditions tested. Whilst the C. pasteurianum hydA gene (CLPA_ c00280) sharing the closest similarity to the C. acetobutylicum hydA gene was recently knocked-down by Pyne et al. (2015) using an anti-sense approach, here we were able to generate a null mutant through the deletion of the entire CLPA_c00280 encoding gene.
In keeping with predictions, deletion of hydA led to increases in the products of NADH-consuming pathways, with the mutant producing 2.7-fold higher levels of ethanol and 5-fold higher levels of lactate (Fig. 4). In contrast, only a slight increase in n-butanol was observed with the mutant generating only 1.1-fold higher levels than the WT. Similar results were obtained with the knock-down mutant of Pyne et al. (2015). Although not a NADH-consuming pathway, a significant increase (1.8-fold) in acetate levels was also seen. Both the acetate and butyrate pathways generate ATP via substrate level phosphorylation. One consequence of which was an increase in biomass attained by the mutant (the mutant OD600 was 114% higher than the WT) and was characterised by a shorter fermentation time as adjudged by the earlier onset of increase in pH (after 18.7 h as opposed to 22.0 h in the WT). The same observation was made by Pyne et al. (2015). This increase in acetate production was not evident in the study of Dabrock et al. (1992), who used CO to inhibit hydrogenase activity on glucose grown C. pasteurianum. Here acetate production was reduced by almost 50%. In this instance, however, all hydrogenase activity would have been affected. The mutant strain utilised in our study, and that of Pyne et al. (2015), is only affected in the production of HydA (CLPA_c00280). Strain C. pasteurianum DSM 525 contains 3 more genes/ enzymes (CLPA_c07060-70, CLPA_c33960 and CLPA_c37830) (Poehlein et al., 2015) with homology to hydrogenase enzymes. With CLPA_c07060 and CLPA_c07070 C. pasteurianum seems to possess a rare [NiFe]-hydrogenase with the genes encoding the small and large subunit, respectively. The other gene encodes an iron only hydrogenase. All of these proteins might compensate for the loss of HydA (CLPA_ c00280).
Unexpectedly, PDO production was significantly reduced in the hydA mutant, by 19% compared to the WT (Fig. 4). This is counter intuitive as the PDO pathway is NADH-consuming. The study of Pyne et al. (2016) also reported such a reduction in levels of PDO (in this case 30%) in their knock-down mutant, while noting that PDO levels can vary widely between fermentations – an observation also made in other studies (Biebl, 2001, Taconi et al., 2009). Our data reinforces the view that the reduction in PDO production that results from ablation of hydA function is a real phenomenon. It should also be noted that the reduction in PDO and accompanying increase in biomass is contrary to suggestion of Biebl (2001) that the production of reducing equivalents by biomass build up has to be equalled by production of PDO to regenerate the pool of NAD+ to NADH.
5. Conclusion
In the current report we have developed and exemplified a robust tool kit for the metabolic engineering of C. pasteurianum based on the use of pyrE alleles. To maximise gene transfer we deployed a novel approach based on the isolation of a highly transformable variant within the host population. In keeping with previous work on other Gram-positive chassis, we once again showed the advantage of using a pyrE minus strain as the host through the rapid complementation of the deletion mutants by the integration of the complementing gene into the genome at the pyrE locus concomitant with ACE-mediated restoration of prototrophy. The system was used to make in-frame deletion mutants of pivotal genes involved in solvent production, namely hydA (hydrogenase), rex (Rex response regulator) and dhaBCE (glycerol dehydratase). We were, for the first time in C. pasteurianum, able to eliminate PDO synthesis and demonstrate its production was essential for growth on glycerol as a sole carbon source. Inactivation of both rex and hydA resulted in increase in n-butanol titres, representing the first steps towards improving the utilisation of C. pasteurianum as a chassis for the industrial production of this important chemical.
Competing interests
All of the authors declare that they have no competing interests.
Acknowledgements
This work was supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC) [grant number BB/L004356/1] as part of the TSB/BBSRC “Advancing the Industrial Application of Synthetic Biology Feasibility Study Competition”. CR acknowledges the financial support of the European Community's Seventh Framework Program “CLOSTNET” [Grant number PEOPLE-ITN-2008–237942] while AGH acknowledges the support of the Forman Hardy Trust and the University of Nottingham, Vice-Chancellors Scholarship. The authors thank Klaus Winzer and Wouter Kuit for fruitful discussions and Matthew Abbott for his help with the HPLC methodology and analysis.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ymben.2017.01.009.
Appendix A. Supplementary material
Supplementary material
.
References
- Barbirato F., Grivet J.P., Soucaille P., Bories A. 3-Hydroxypropionaldehyde, an inhibitory metabolite of glycerol fermentation to 1,3-propanediol by enterobacterial species. Appl. Environ. Microbiol. 1996;62:1448–1451. doi: 10.1128/aem.62.4.1448-1451.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebl H. Fermentation of glycerol by Clostridium pasteurianum—batch and continuous culture studies. J. Indust. Microbiol. Biotechnol. 2001;27:18–26. doi: 10.1038/sj.jim.7000155. [DOI] [PubMed] [Google Scholar]
- Brekasis D., Paget M.S. A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3. EMBO J. 2003;22(2):4856–4865. doi: 10.1093/emboj/cdg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartman S.T., Minton N.P. A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl. Environ. Microbiol. 2010;76:1103–1109. doi: 10.1128/AEM.02525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartman S.T., Kelly M.L., Heeg D., Heap J.T., Minton N.P. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl. Environ. Microbiol. 2012;78:4683–4690. doi: 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.K., Blaschek H.P. Acetate enhances solvent production and prevents degeneration in Clostridium beijerinckii BA101. Appl. Microbiol. Biotechnol. 1999;52(2):170–173. doi: 10.1007/s002530051504. [DOI] [PubMed] [Google Scholar]
- Cherubini, F., Jungmeier, G., Mandl, M., Philips, C., Wellisch, M., Jørgensen, H., Skiadas, I., Boniface, L., Dohy, M., Pouet, J.C. and Willke, T., 2009. IEA Bioenergy Task 42: Report on Participating Countries. Document of IEA Bioenergy Task 42 ‘Biorefineries’.
- Clomburg J.M., Gonzalez R. Anaerobic fermentation of glycerol: a platform for renewable fuels and chemicals. Trends Biotechnol. 2013;31(1):20–28. doi: 10.1016/j.tibtech.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Cooksley C.M., Davis I.J., Winzer K., Chan W.C., Peck M.W., Minton N.P. Regulation of neurotoxin production and sporulation by a putative agrBD signaling system in proteolytic Clostridium botulinum. Appl. Environ. Microb. 2010;76(13):4448–4460. doi: 10.1128/AEM.03038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksley C.M., Zhang Y., Wang H., Redl S., Winzer K., Minton N.P. Targeted mutagenesis of the Clostridium acetobutylicum acetone–butanol–ethanol fermentation pathway. Metab. Eng. 2012;14(6):630–641. doi: 10.1016/j.ymben.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Dabrock B., Bahl H., Gottschalk G. Parameters affecting solvent production by Clostridium pasteurianum. Appl. Environ. Microbiol. 1992;58:1233–1239. doi: 10.1128/aem.58.4.1233-1239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta R., Zeikus J.G. Modulation of acetone-butanol-ethanol fermentation by carbon monoxide and organic acids. Appl. Environ. Microbiol. 1985;49:522–529. doi: 10.1128/aem.49.3.522-529.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R.P., Harris L.M., Welker N.E., Papoutsakis E.T. Metabolic flux analysis elucidates the importance of the acid-formation pathways in regulating solvent production by Clostridium acetobutylicum. Metab. Eng. 1999;1:206–213. doi: 10.1006/mben.1999.0118. [DOI] [PubMed] [Google Scholar]
- Dobson R., Gray V., Rumbold K. Microbial utilization of crude glycerol for the production of value-added products. J. Indust. Microbiol. Biotechnol. 2012;39:217–226. doi: 10.1007/s10295-011-1038-0. [DOI] [PubMed] [Google Scholar]
- Dürre P. Biobutanol: an attractive biofuel. Biotechnol. J. 2007;2:1525–1534. doi: 10.1002/biot.200700168. [DOI] [PubMed] [Google Scholar]
- Ehsaan M., Kuit W., Zhang Y., Cartman S.T., Heap J.T., Winzer K., Minton N.P. Mutant generation by allelic exchange and genome resequencing of the biobutanol organism Clostridium acetobutylicum ATCC 824. Biotechnol. Biofuels. 2016;9:4. doi: 10.1186/s13068-015-0410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap J.T., Ehsaan M., Cooksley C.M., Ng Y.K., Cartman S.T., Winzer K., Minton N.P. Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker. Nucl. Acids Res. 2012;40:e59. doi: 10.1093/nar/gkr1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap J.T., Pennington O.J., Cartman S.T., Minton N.P. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods. 2009;78:79–85. doi: 10.1016/j.mimet.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Hu L., Huang H., Yuan H., Tao F., Xie H., Wang S. Rex in Clostridium kluyveri is a global redox-sensing transcriptional regulator. J. Biotechnol. 2016;233:17–25. doi: 10.1016/j.jbiotec.2016.06.024. [DOI] [PubMed] [Google Scholar]
- Hüsemann M.H., Papoutsakis E.T. Comparison between in vivo and in vitro enzyme activities in continuous and batch fermentations of Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 1989;30:585–595. [Google Scholar]
- Jang Y.S., Lee J.Y., Lee J., Park J.H., Im J.A., Eom M.H., Lee J., Lee S.H., Song H., Cho J.H., Lee S.Y. Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum. MBio. 2012;3:e00314–12. doi: 10.1128/mBio.00314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen T.Ø., Kvist T., Mikkelsen M.J., Christensen P.V., Westermann P. Fermentation of crude glycerol from biodiesel production by Clostridium pasteurianum. J. Indust. Microbiol. Biotechnol. 2012;39:709–717. doi: 10.1007/s10295-011-1077-6. [DOI] [PubMed] [Google Scholar]
- Johnson D.T., Taconi K.A. The glycerin glut: options for the value‐added conversion of crude glycerol resulting from biodiesel production. Environ. Prog. 2007;26:338–348. [Google Scholar]
- Johnson E.E., Rehmann L. The role of 1,3-propanediol production in fermentation of glycerol by Clostridium pasteurianum. Bioresour. Technol. 2016;209:1–7. doi: 10.1016/j.biortech.2016.02.088. [DOI] [PubMed] [Google Scholar]
- Jones D.T., Woods D.R. Acetone-butanol fermentation revisited. Microbiol. Rev. 1986;50:484. doi: 10.1128/mr.50.4.484-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao W.C., Lin D.S., Cheng C.L., Chen B.Y., Lin C.Y., Chang J.S. Enhancing butanol production with Clostridium pasteurianum CH4 using sequential glucose–glycerol addition and simultaneous dual-substrate cultivation strategies. Bioresour. Technol. 2013;135:324–330. doi: 10.1016/j.biortech.2012.09.108. [DOI] [PubMed] [Google Scholar]
- Kerr, B.J., Shurson, G.C., Johnston, L.J., Dozier, W.A., 2011. Utilization of crude glycerin in nonruminants. INTECH Open Access Publisher.
- Kim B.H., Bellows P., Datta R., Zeikus J.G. Control of carbon and electron flow in Clostridium acetobutylicum fermentations: utilization of carbon monoxide to inhibit hydrogen production and to enhance butanol yields. Appl. Environ. Microbiol. 1984;48:764–770. doi: 10.1128/aem.48.4.764-770.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann D., Hönicke D., Ehrenreich A., Schmidt M., Weuster-Botz D., Bahl H., Lütke-Eversloh T. Modifying the product pattern of Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 2012;94:743–754. doi: 10.1007/s00253-011-3852-8. [DOI] [PubMed] [Google Scholar]
- Liew F., Henstra A.M., Kӧpke M., Winzer K., Simpson S.D., Minton N.P. Metabolic engineering of Clostridium autoethanogenum for selective alcohol production. Metab. Eng. 2017 doi: 10.1016/j.ymben.2017.01.007. (in press), pii: S1096-7176(17)30031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D.S., Yen H.W., Kao W.C., Cheng C.L., Chen W.M., Huang C.C., Chang J.S. Bio-butanol production from glycerol with Clostridium pasteurianum CH4: the effects of butyrate addition and in situ butanol removal via membrane distillation. Biotechnol. Biofuels. 2015;8:1. doi: 10.1186/s13068-015-0352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Xu Y., Zheng Z., Liu D. 1, 3‐Propanediol and its copolymers: research, development and industrialization. Biotechnol. J. 2010;5:1137–1148. doi: 10.1002/biot.201000140. [DOI] [PubMed] [Google Scholar]
- Lunnen K.D., Barsomian J.M., Camp R.R., Card C.O., Chen S.Z., Croft R., Looney M.C., Meda M.M., Moran L.S., Nwankwo D.O., Slatko B.E. Cloning type-II restriction and modification genes. Gene. 1988;74:25–32. doi: 10.1016/0378-1119(88)90242-9. [DOI] [PubMed] [Google Scholar]
- Maervoet V.E., De Maeseneire S.L., Avci F.G., Beauprez J., Soetaert W.K., De Mey M. 1,3-propanediol production with Citrobacter werkmanii DSM17579: effect of a dhaD knock-out. Microb. Cell. Fact. 2014;13:1. doi: 10.1186/1475-2859-13-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaviya A., Jang Y.S., Lee S.Y. Continuous butanol production with reduced byproducts formation from glycerol by a hyper producing mutant of Clostridium pasteurianum. Appl. Microbiol. Biotechnol. 2012;93:1485–1494. doi: 10.1007/s00253-011-3629-0. [DOI] [PubMed] [Google Scholar]
- Market and Market, 2015. 1,3-Propanediol (PDO) Market by Applications (PTT, Polyurethane, Cosmetic, Personal Care & Home Cleaning & Others) & Geography - Global Market Trends & Forecasts to 2021, Report code: CH3091, available online 〈http://www.marketsandmarkets.com/Market-Reports/1-3-propanediol-pdo-market-760.html〉.
- Millat T., Voigt C., Janssen H., Cooksley C.M., Winzer K., Minton N.P., Bahl H., Fischer R.J., Wolkenhauer O. Coenzyme A-transferase-independent butyrate re-assimilation in Clostridium acetobutylicum—evidence from a mathematical model. Appl. Microbiol. Biotechnol. 2014;98:9059–9072. doi: 10.1007/s00253-014-5987-x. [DOI] [PubMed] [Google Scholar]
- Minton N.P., Ehsaan M., Humphreys C.M., Little G.T., Baker J., Henstra A.M., Liew F., Kelly M.L., Sheng L., Schwarz K., Zhang Y. A roadmap for gene system development in Clostridium. Anaerobe. 2016;41:104–112. doi: 10.1016/j.anaerobe.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon C., Lee C.H., Sang B.I., Um Y. Optimization of medium compositions favoring butanol and 1,3-propanediol production from glycerol by Clostridium pasteurianum. Bioresour. Technol. 2011;102:10561–10568. doi: 10.1016/j.biortech.2011.08.094. [DOI] [PubMed] [Google Scholar]
- Münch R., Hiller K., Barg H., Heldt D., Linz S., Wingender E., Jahn D. PRODORIC: prokaryotic database of gene regulation. Nucl. Acids Res. 2003;31:266–269. doi: 10.1093/nar/gkg037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch R., Hiller K., Grote A., Scheer M., Klein J., Schobert M., Jahn D. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics. 2005;21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- Ng Y.K., Ehsaan M., Philip S., Collery M.M., Janoir C., Collignon A., Cartman S.T., Minton N.P. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS One. 2013;8:e56051. doi: 10.1371/journal.pone.0056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien R.W., Morris J.G. Oxygen and the growth and metabolism of Clostridium acetobutylicum. Microbiol. 1971;68:307–318. doi: 10.1099/00221287-68-3-307. [DOI] [PubMed] [Google Scholar]
- Papoutsakis E.T., Meyer C.L. Fermentation equations for propionic‐acid bacteria and production of assorted oxychemicals from various sugars. Biotechnol. Bioeng. 1985;27(1):67–80. doi: 10.1002/bit.260270109. [DOI] [PubMed] [Google Scholar]
- Peguin S., Goma G., Delorme P., Soucaille P. Metabolic flexibility of Clostridium acetobutylicum in response to methyl viologen addition. Appl. Microbiol. Biotechnol. 1994;42:611–616. [Google Scholar]
- Percheron G., Brechignac F., Soucaille P., Condoret J.S. Carbon dioxide desorption from fermentation broth by use of oxygen vectors. Bioprocess Eng. 1995;12(1–2):11–16. [Google Scholar]
- Poehlein A., Grosse-Honebrink A., Zhang Y., Minton N.P., Daniel R. Complete genome sequence of the nitrogen-fixing and solvent-producing Clostridium pasteurianum DSM 525. Genome Announc. 2015;3 doi: 10.1128/genomeA.01591-14. (e01591-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne M.E., Moo-Young M., Chung D.A., Chou C.P. Development of an electrotransformation protocol for genetic manipulation of Clostridium pasteurianum. Biotechnol. Biofuels. 2013;6:1. doi: 10.1186/1754-6834-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne M.E., Moo-Young M., Chung D.A., Chou C.P. Antisense-RNA-mediated gene downregulation in Clostridium pasteurianum. Fermentation. 2015;1(1):113–126. [Google Scholar]
- Pyne M.E., Sokolenko S., Liu X., Srirangan K., Bruder M.R., Aucoin M.G., Moo-Young M., Chung D.A., Chou C.P. Disruption of the reductive 1,3-propanediol pathway triggers production of 1, 2-propanediol for sustained glycerol fermentation by Clostridium pasteurianum. Appl. Environ. Microbiol. 2016;82:5375–5388. doi: 10.1128/AEM.01354-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quispe C.A., Coronado C.J., Carvalho J.A., Jr Glycerol: production, consumption, prices, characterization and new trends in combustion. Renew. Sustain. Energy Rev. 2013;27:475–493. [Google Scholar]
- Ravcheev D.A., Li X., Latif H., Zengler K., Leyn S.A., Korostelev Y.D., Kazakov A.E., Novichkov P.S., Osterman A.L., Rodionov D.A. Transcriptional regulation of central carbon and energy metabolism in bacteria by redox-responsive repressor Rex. J. Bacteriol. 2012;194:1145–1157. doi: 10.1128/JB.06412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REN21, 2015. Renewables Global Status Report, REN21Secretariat, Editor. 2015. p. ISBN 978-3-9815934-6-4.
- Renbaum P., Abrahamove D., Fainsod A., Wilson G.G., Rottem S., Razin A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1 (M Sssl) Nucl. Acids Res. 1990;18:1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards D.F., Linnett P.E., Oultram J.D., Young M. Restriction endonucleases in Clostridium pasteurianum ATCC 6013 and C. thermohydrosulfuricum DSM 568. Microbiol. 1988;134:3151–3157. doi: 10.1099/00221287-134-12-3151. [DOI] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sabra W., Groeger C., Sharma P.N., Zeng A.P. Improved n-butanol production by a non-acetone producing Clostridium pasteurianum DSMZ 525 in mixed substrate fermentation. Appl. Microbiol. Biotechnol. 2014;98:4267–4276. doi: 10.1007/s00253-014-5588-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarchami T., Johnson E., Rehmann L. Optimization of fermentation condition favoring butanol production from glycerol by Clostridium pasteurianum DSM 525. Bioresour. Technol. 2016;208:73–80. doi: 10.1016/j.biortech.2016.02.062. [DOI] [PubMed] [Google Scholar]
- Sandoval N.R., Venkataramanan K.P., Groth T.S., Papoutsakis E.T. Whole-genome sequence of an evolved Clostridium pasteurianum strain reveals Spo0A deficiency responsible for increased butanol production and superior growth. Biotechnol. Biofuels. 2015;8:1. doi: 10.1186/s13068-015-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo J.D., Dürre P., Woods D.R. Characterization and expression of the hydrogenase-encoding gene from Clostridium acetobutylicum P262. Microbiol. 1995;141:171–180. doi: 10.1099/00221287-141-1-171. [DOI] [PubMed] [Google Scholar]
- Sauer C., Syvertsson S., Bohorquez L., Cruz R., Harwood C., Van Rij T., Hamoen L.W. Effect of genome position on heterologous gene expression in Bacillus subtilis; an unbiased analysis. ACS Synth. Biol. 2016;5:942–947. doi: 10.1021/acssynbio.6b00065. [DOI] [PubMed] [Google Scholar]
- Sheng L., Kovács K., Winzer K., Zhang Y., Minton N.P. Development and implementation of rapid metabolic engineering tools for chemical and fuel production in Geobacillus thermoglucosidasius NCIMB 11955. Biotechnol. Biofuels. 2017;10:5. doi: 10.1186/s13068-016-0692-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som T., Tomizawa J.I. Origin of replication of Escherichia coli plasmid RSF 1030. Mol. Gen. Genet. 1982;187:375–383. doi: 10.1007/BF00332615. [DOI] [PubMed] [Google Scholar]
- Steiner E., Scott J., Minton N.P., Winzer K. An agr quorum sensing system that regulates granulose formation and sporulation in Clostridium acetobutylicum. Appl. Environ. Microbiol. 2012;78:1113–1122. doi: 10.1128/AEM.06376-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taconi K.A., Venkataramanan K.P., Johnson D.T. Growth and solvent production by Clostridium pasteurianum ATCC® 6013™ utilizing biodiesel‐derived crude glycerol as the sole carbon source. Environ. Prog. Sust. Energ. 2009;28(1):100–110. [Google Scholar]
- Wang E., Bauer M.C., Rogstam A., Linse S., Logan D.T., Von Wachenfeldt C. Structure and functional properties of the Bacillus subtilis transcriptional repressor rex. Mol. Microbiol. 2008;69:466–478. doi: 10.1111/j.1365-2958.2008.06295.x. [DOI] [PubMed] [Google Scholar]
- Werpy Todd, Petersen Gene, Aden A., Bozell J., Holladay J., White J., Manheim Amy, Eliot D., Lasure L., Jones S. Top value added chemicals from biomass. volume 1-results of screening for potential candidates from sugars and synthesis gas. No. DOE/GO-102004-1992. Dep. Energy Wash. DC. 2004 [Google Scholar]
- Wietzke M., Bahl H. The redox-sensing protein rex, a transcriptional regulator of solventogenesis in Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 2012;96:749–761. doi: 10.1007/s00253-012-4112-2. [DOI] [PubMed] [Google Scholar]
- Yang F., Hanna M.A., Sun R. Value-added uses for crude glycerol--a byproduct of biodiesel production. Biotechnol. Biofuels. 2012;5:1. doi: 10.1186/1754-6834-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani S.S., Gonzalez R. Anaerobic fermentation of glycerol: a path to economic viability for the biofuels industry. Curr. Opin. Biotechnol. 2007;18:213–219. doi: 10.1016/j.copbio.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Zhang J. Study of poly (trimethylene terephthalate) as an engineering thermoplastics material. J. Appl. Polym. Sci. 2004;91:1657–1666. [Google Scholar]
- Zhang L., Nie X., Ravcheev D.A., Rodionov D.A., Sheng J., Gu Y., Yang S., Jiang W., Yang C. Redox-responsive repressor rex modulates alcohol production and oxidative stress tolerance in Clostridium acetobutylicum. J. Bact. 2014;196:3949–3963. doi: 10.1128/JB.02037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material