Abstract
Neural stem and progenitor cells (NSPCs) are heterogeneous populations of self-renewing stem cells and more committed progenitors that differentiate into neurons, astrocytes and oligodendrocytes. Accurately identifying and characterizing the different progenitor cells in this lineage has continued to be a challenge for the field. We found previously that populations of NSPCs with more neurogenic progenitors (NPs) can be distinguished from those with more astrogenic progenitors (APs) by their inherent biophysical properties, specifically the electrophysiological property of whole cell membrane capacitance, which we characterized with dielectrophoresis (DEP). Here we hypothesize that inherent electrophysiological properties are sufficient to define NPs and APs and test this by determining whether isolation of cells solely by these properties specifically separates NPs and APs. We found NPs and APs are enriched in distinct fractions after separation by electrophysiological properties using DEP. A single round of DEP isolation provided greater NP enrichment than sorting with PSA-NCAM, which is considered an NP marker. Additionally, cell surface N-linked glycosylation was found to significantly affect cell fate-specific electrophysiological properties, providing a molecular basis for the cell membrane characteristics. Inherent plasma membrane biophysical properties are thus sufficient to define progenitor cells of differing fate potential in the neural lineage, can be used to specifically isolate these cells, and are linked to patterns of glycosylation on the cell surface.
Keywords: Neural stem cell, neuron progenitor, astrocyte progenitor, biophysical properties, electrophysiological properties, dielectrophoresis, glycosylation
Introduction
NSPCs arise during early stages of CNS development to form the brain and spinal cord and have shown therapeutic potential for treating diverse conditions, such as spinal cord injury, Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, and stroke (1). NSPCs cultured for therapeutic purposes are heterogeneous, containing multipotent neural stem cells as well as specific progenitors giving rise to neurons, astrocytes, and oligodendrocytes. This heterogeneity makes it difficult to control the cellular composition of transplants and determining the ratios of the different cell types in the mixture has been complicated. Furthermore, the cell biological characteristics of specific progenitors such as NPs and APs have not been well defined.
Many cell surface biomarkers, such as PSA-NCAM, A2B5, CD133, CD15 (LeX), CD24, and CD184, have been used to assess neural cells (2, 3, 4, 5, 6). Yet, a clear set of markers for each progenitor is lacking since many markers are unable to distinguish progenitors from each other or from more or less differentiated cells in the lineage. For example, A2B5 has been characterized as a marker for both glial and neuronal progenitors and PSA-NCAM recognizes differentiated neurons as well as progenitors (7, 8). We developed an alternative approach to identify progenitors based on cells’ electrophysiological properties using dielectrophoresis (DEP), which induces motion of cells in non-uniform AC electric fields (9). Cells in DEP experience a frequency-dependent induced force that attracts them toward electrodes (positive DEP) or repels them away (negative DEP), depending on their inherent cellular traits. NPs and APs transition from negative to positive DEP at different frequencies in DEP, indicating these progenitors have distinct properties (10).
The response of cells to the electric fields in DEP is dominated by the plasma membrane at lower frequencies, within the range at which distinct differences exist for NPs and APs (10). A detailed analysis of cell behavior at these frequencies enabled calculation of both plasma membrane capacitance (ability to store charge) and conductance (ability to transmit charge). We found whole cell plasma membrane capacitance (Cm), but not conductance, distinguishes mouse and human NPs and APs (11). Cm dynamically and consistently changes with shifting fate potential of the cells, thereby serving as a predictive measure of fate (11). Thus, a non-marker based, quantitative electrophysiological measure reflects cell fate potential and indicates specific differences in the membrane compartments of distinct progenitors in the neural lineage.
Differentiation of adipose-derived stem cells along osteogenic and adipogenic lineages is detected by cell membrane capacitance measured by impedance spectroscopy, which, like DEP, employs a range of frequencies and measures cell electrophysiological properties (12). Furthermore, membrane capacitance measured by impedance analysis specifically identifies the six main leukocyte subpopulations in the hematopoietic lineage (13). Membrane capacitance can thus serve as a label-free quantitative indicator of stem cell fate potential in multiple lineages and could be exploited to investigate the biological differences between specific stem and progenitor cells. However, no studies to date have demonstrated separation of distinct progenitors in a lineage using inherent electrophysiological traits and no labels.
The consistent link between membrane capacitance and fate potential led to our hypothesis that inherent electrophysiological properties are sufficient to define progenitors in the neural lineage in the absence of markers or other cell lineage reporters. We tested this hypothesis by separating heterogeneous NSPCs, containing both NPs and APs, in DEP since this method can isolate cells with different electrophysiological properties. DEP has been used to sort a variety of cell types, including cancer cells from blood (14), NG2+ adipose stem cells from tissue (15), myotubes from C2C12 myoblasts (16), and neurons from NSPCs (17). DEP can separate cells differing in crossover frequency (fxo), which is the point at which there is no induced force in response to the DEP frequency and is strongly affected by membrane capacitance. In addition to their distinct membrane capacitance values, both human and mouse NPs and APs differ in fxo (11). Importantly, exposure to DEP at the time scales needed for sorting does not affect NSPC survival, proliferation or differentiation potential (18).
Despite the fact that membrane capacitance indicates cell phenotype in a variety of stem cell lineages, little is known about the underlying cell biological characteristics that contribute to this measure. We therefore investigated three potential contributors to the difference in membrane capacitance between NPs and APs: cell size, cell surface glycosylation, and resting membrane potential. Cell size is relevant since membrane capacitance as measured by DEP is a whole cell property and the total amount of plasma membrane can affect this measure. Thus, cells of different size but identical membrane composition will have different membrane capacitance values. Cell surface glycosylation is one way in which the membrane composition may affect membrane capacitance. N-linked glycosylation adds carbohydrates able to store charge to proteins expressed on the plasma membrane. Data from the literature suggest a significant role for glycosylation in neural development since N-glycosylation patterns change during developmental stages at which either NPs or APs are formed (19, 20). Lastly, neuron and astrocyte resting membrane potentials significantly differ, and their progenitors may begin to express channels resulting in distinct resting membrane potentials of NPs and APs. We tested whether cell size, cell surface glycosylation, or resting membrane potential contribute to fate-specific properties of progenitors in the neural lineage.
Results
Separation of cells by their inherent electrophysiological properties
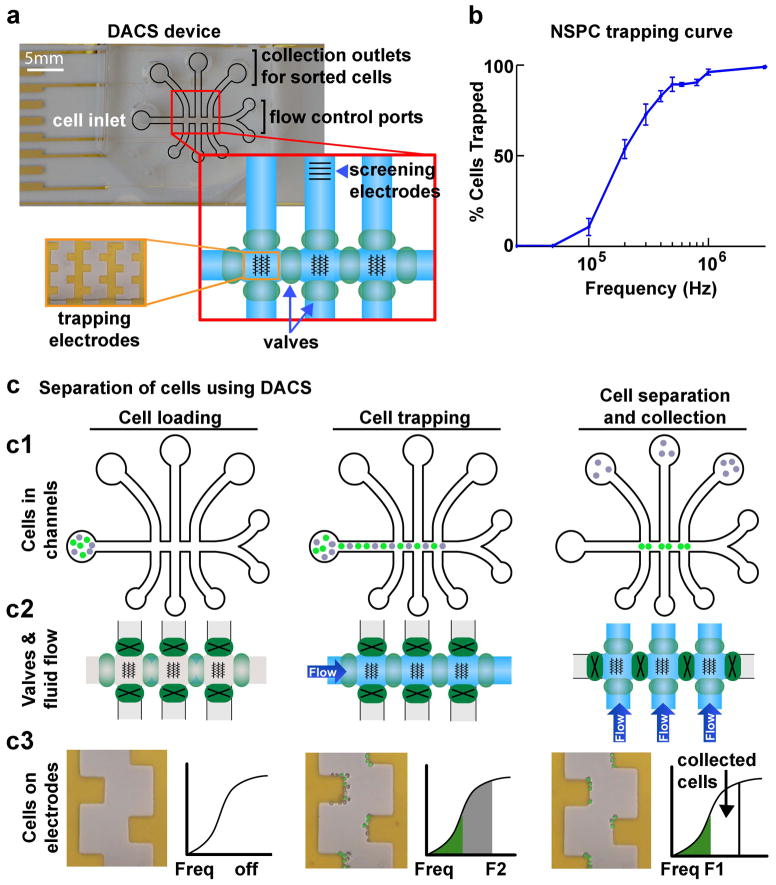
In order to test whether membrane electrophysiological properties are sufficient to define progenitors in the neural lineage we developed a microfluidic DEP-assisted cell sorting (DACS) device to isolate cells on the basis of their inherent properties (17). Cells can be separated by choosing a DEP frequency at which one cell type experiences positive DEP (frequency higher than that cell’s fxo) and the other negative DEP (frequency lower than that cell’s fxo) (Fig. S1). A heterogeneous cell population to be separated is loaded into the cell inlet of the DACS device, which has a series of microfluidic channels and DEP electrode arrays on the main channel floor (Fig. 1a). These electrode arrays are used to trap cells in positive DEP, while cells in negative DEP are removed. Pneumatic valves control the direction of fluid flow through the channels (Fig. 1a). Our system incorporates both DEP forces and hydrodynamic forces related to the fluid flow rate. Thus, positive DEP and cell trapping along the electrodes is dependent on inherent cellular properties (Cm and fxo) and the hydrodynamic force related to the fluid flow rate. We therefore define the threshold frequency (fth) as the frequency at which cells experience positive DEP in this system, and the fth reflects the combination of these two factors (17).
Figure 1. Microfluidic DACS separates cells at specific frequency bands.
a) DACS device for sorting cells. The DACS device is made up of microfluidic channels (outlined in black) with an inlet for loading cells, collection outlets for retrieval of sorted cells, and flow control ports for connection to pumps to regulate fluid flow. Direction of fluid flow in the channels is controlled by opening and closing pneumatic valves (green ovals in red enlarged box). DEP castellated trapping electrode arrays in the horizontal channel are shown by black lines in the schematic of the channels (red enlarged box) and in a magnified image of the gold electrodes in the device (orange enlarged box). A set of screening electrodes in the vertical channel of an outlet allows analysis of cells post-sorting.
b) The DEP trapping curve of E12.5 mNSPCs shows a gradual increase in the percentage of trapped cells until reaching ~100% and is used to set the frequency ranges for DACS separation of these cells.
c) Schematic describing the separation of cells using DACS. Three phases are illustrated - cell loading, cell trapping, and cell separation and collection. Panels depict the position of the cells in the channels (c1), valve operation and fluid flow in the channels (c2), and zoomed images of electrodes to show the separation of two cell populations (c3).
c1) Cells in the microfluidic channels. Cells are loaded into the inlet (left panel), flow through the horizontal channel and trap at electrodes (middle panel), and are sorted such that one population (gray cells) is directed to the collection outlets while the other (green cells) remain in the horizontal channel (right panel).
c2) Valve configuration and control of fluid flow. Pneumatic valves (green ovals) control fluid flow through the horizontal and vertical channels. Open valves (light green ovals) allow fluid flow, while closed valves (dark green ovals with a black X) restrict flow. Valves are open in the horizontal channel when cells are loaded (left panel) and flow in the horizontal channel carries cells to the trapping electrodes (middle panel). To collect separated cells, valves are closed in the horizontal channel and opened in the vertical channels to allow the separated cells to flow to the outlets (right panel).
c3) Images of cells on electrodes and trapping curve schematics describe cell separation. At the start of the cell loading phase there are no cells on the electrodes and the DEP frequency is off (left panel). During cell trapping, both populations of cells (gray and green pseudo-colored cells) are trapped along the electrodes since the DEP frequency is set to F2 and both cell populations have fth < F2 (middle panel). Green and gray colors in the trapping curve schematic correspond to the colors of cells on the electrodes. The gray cells are isolated from the green cells by setting the frequency to F1, allowing the gray cells having F1 < fth < F2 to leave the electrodes and flow to the collection outlets (right panel).
Embryonic day 12.5 mouse NSPCs (E12.5 mNSPCs) contain both NPs and APs so are an ideal population to test whether NP and AP inherent properties are sufficient to define and isolate these progenitors (11). Mouse NSPCs that differ in the numbers of NPs and APs have distinct electrophysiological properties (Cm: 8.2 mF/m2 NP-biased and 10.7 mF/m2 AP-biased; fxo: 48.0 kHz NP-biased and 35.1 kHz AP-biased) and based on profiles in DEP in the presence of fluid flow are likely to differ in fth (10, 11). To define appropriate frequencies for isolation of NPs and AP using the castellated electrode arrays in the DACS device, we generated a trapping curve for E12.5 mNSPCs by determining the percentage of cells experiencing positive DEP over a range of given frequencies (Fig. 1b). A homogenous population of cells would transition sharply to 100% trapped cells at the frequency at which those cells experience positive DEP (10, 17). In contrast, a heterogeneous population, such as E12.5 mNSPCs, transitions gradually to 100% over a range of frequencies (Fig. 1b). We identified frequencies ranging from 0 to 400 kHz to employ for isolation of NPs and APs from E12.5 mNSPCs since the heterogeneity of the population is observed in this range (Fig. 1b).
Figure 1c details the steps of cell separation in DACS and a video demonstrating cell separation in the DACS device is available in Supplemental Material (Supplemental Video 1). The heterogeneous cell population flows through the main, horizontal channel of the device while DEP electrodes are set at a specific frequency, F2 (Fig. 1c). All cells that have an fth < F2 are trapped along the electrodes (Fig. 1c, middle panels). Then, flow is stopped and a lower DEP frequency, F1, is set while valves are switched to redirect fluid flow to the perpendicular channels (Fig. 1c, right panels). Cells that do not experience positive DEP at F1 leave the electrodes and are directed to the collection wells (false gray cells in Fig. 1c), while cells that continue to experience positive DEP remain trapped and are not recovered (false green cells in Fig. 1c). Cells are trapped in discrete frequency bands and only those cells that have F1<fth<F2 are recovered, which is indicated on the trapping curve as the subset of collected cells (Fig. 1c, right lower panel).
Sorting enriches cells with similar electrophysiological properties
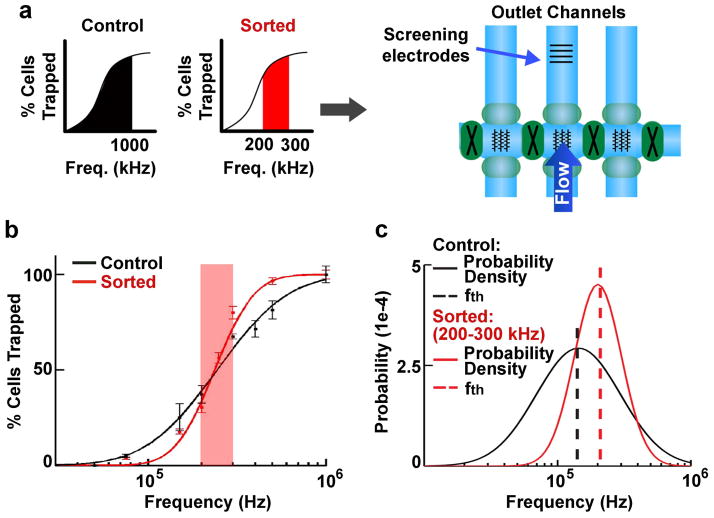
To determine whether isolated cells are more homogenous in electrophysiological properties, and thus were effectively sorted, a set of electrodes was included on a cell collection channel to allow immediate trapping curve analysis and calculation of the fth for the sorted cells (Fig. 2a). We analyzed cells sorted at 200–300 kHz because this band represents the middle of the frequency range of E12.5 mNSPCs (Fig. 1b). Controls for cells sorted by DACS included cells incubated in DEP buffer for the same amount of time as the cells that were sorted (buffer control) and cells that were in DEP buffer and went through the DACS device, but were collected by the frequency band 0–1000 kHz (unsorted control since this procedure collected all the cells in the population). The trapping curve of the 200–300 kHz sorted cells differs from that of unsorted control (0–1000 kHz) cells and is steeper in the 200–300 kHz frequency range (Fig. 2b), indicating successful sorting.
Figure 2. Increased purity of cells separated by DACS.
a) Cells were either unsorted control (entire frequency band 0–1000 kHz, black) or sorted (200–300 kHz frequency band, red) cells as depicted by graphs in the left panel. Both sets of cells were analyzed by screening electrodes in an outlet channel (right schematic). Cells were sorted in all 3 vertical channels at the same frequencies to improve yield and were screened in the middle channel.
b) Unsorted control (0–1000 kHz, black) and sorted cells (200–300 kHz, red) reveal distinct trapping curves (points are measured data and lines are fitted lognormal distributions)(17). The 200–300 kHz frequency range is indicated by red shading to highlight the steeper slope of the sorted cell trapping curve in this range than the control cell trapping curve, indicating greater purity of the sorted cells.
c) Log-normal distribution fitting of the trapping curves from (b) gives the resulting probability density functions (solid lines) for unsorted control (black) and sorted (red) cells. The most common trapping frequency for the population, the threshold frequency (fth, dashed lines) falls within the frequency band limits for the screened population (either 0–1000 kHz or 200–300 kHz) illustrating fidelity, while the peak height and shape (skewness) reflects the higher homogeneity of the sorted cells.
Trapping curves represent the behavior of all the cells in the population, each of which has an individual fth value, and the cumulative values of all the cells fit a log-normal distribution (17). The resulting log-normal distribution can be characterized by two values, the mode and the skewness of the distribution. The mode is the most probable fth for the cells in the population. The skewness is an indicator of the heterogeneity of the population with larger numbers being more heterogeneous and lower numbers being more homogenous (17).
We calculated the fth and skewness for sorted and unsorted control cells (Fig. 2c). Log-normal distributions of cells isolated in the 200–300 kHz band exhibit lower skewness (1.34) than the unsorted control (0–1000 kHz) population (3.09), indicating that cells isolated in this discrete band have more homogeneous electrophysiological properties. In addition, the calculated fth for the 200–300 kHz band (fth =200.9 kHz) falls in the frequency range of the band used for isolation and is different from the unsorted control population (fth = 143.6 kHz), demonstrating fidelity of the device and confirming that cells were sorted on the basis of their intrinsic electrophysiological properties.
Isolation of cells solely by electrophysiological properties specifically separates NPs and APs
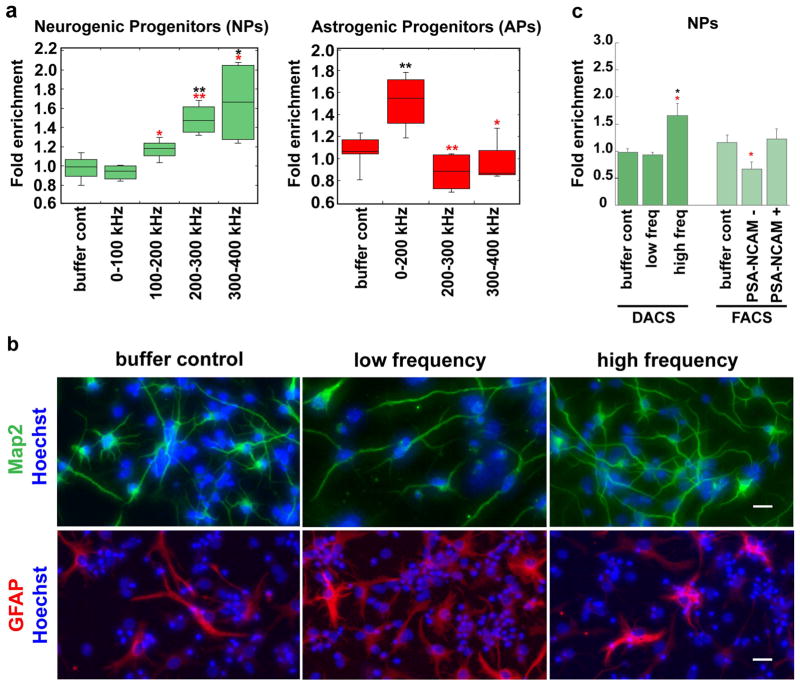
We hypothesized that inherent electrophysiological properties of NPs and APs would be sufficient to define these distinct progenitors. To test this hypothesis, we separated E12.5 mNSPCs by electrophysiological properties by isolating frequency bands of 100 kHz increments from 0–400 kHz since virtually all cells are trapped by positive DEP at 400 kHz (Fig. 1b). Reliable markers to identify APs and NPs are not available, so we examined fate potential by differentiating the sorted cells and determining their fate using immunocytochemistry to detect neurons and astrocytes (see Methods).
NPs and APs are enriched in distinct fractions after sorting on the basis of electrophysiological properties. NPs were elevated in higher frequency bands: 1.7 fold at 300–400 kHz and 1.5 fold at 200–300 kHz (Figs. 3a, left panel, 3b upper panels, S2). Unsorted controls had an average of 31% NPs, which means the maximum fold enrichment to achieve 100% NP purity is 3.2 fold. The cells isolated at 300–400 kHz had 52% NPs, and those isolated at 200–300 kHz had 45% NPs. APs were enriched at lower frequencies: 1.5 fold at 0–200 kHz over unsorted controls or cells isolated at higher frequency bands (Fig. 3a, right panel, 3b, lower panels). Unsorted controls had 19% APs cells and 0–200 kHz sorted cells had 30% APs. Isolation of NPs at higher frequencies and APs at lower frequencies was predicted by our previous findings that NSPCs with greater neurogenic potential respond to higher frequencies than those with greater astrogenic potential (10, 11). These data clarify that NPs and APs are specifically identified and isolated solely by their distinct inherent electrophysiological properties.
Figure 3. Prospective isolation of NPs and APs by electrophysiological properties.
a) Heterogeneous E12.5 mNSPCs were sorted into discrete frequency bands using DACS. Cells isolated at high frequency bands are significantly enriched for NPs as determined by ability to differentiate into Map2+/TuJ1+ neurons (left panel). In contrast, APs, determined by differentiation into GFAP+ astrocytes, are significantly enriched at lower frequencies (right panel). Sorted cells and DEP buffer control cells were normalized to unsorted control (0–1000 kHz) cells and data is presented as fold difference, n >/= 3 independent experiments. Percentages of cells are indicated in Results. Error bars show range, lines within box show average and data were analyzed with Student’s t-test. Black asterisks denote significance as compared to DEP buffer control (NPs: ** p= 0.0033 for 200–300k, * p=0.027, 300–400 kHz; APs: ** p=0.0069 for 0–200 kHz). Red asterisks denote significance as compared to low frequency sorted cells (0–100 kHz for NPs and 0–200 kHz for APs; NPs * p=0.032 for 100–200 kHz, ** p=0.0009 for 200–300K, * p=0.019 for 300–400kHz; APs ** p=0.0065 for 200–300 kHz, * p=0.04227 for 300–400 kHz).
b) Cells sorted by DACS were differentiated and immunostained to detect neurons or astrocytes. Representative images indicate greater numbers of Map2-positive neurons derived from cells sorted at high frequency (green, upper panels). Corresponding images showing Map2/TuJ1 costaining in neurons are in Fig. S2. More GFAP-positive astrocytes were formed from cells isolated at low frequency (red, lower panels). Images are of differentiated cells from unsorted DEP buffer control, low frequency band sort (0–100 kHz for Map2 staining and 0–200 kHz for GFAP staining) or high frequency band sort (300–400 kHz). Hoechst stained nuclei are blue and scale bars = 20 μm.
c) Graph indicates differences in NP enrichment by DACS and FACS with PSA-NCAM antibody. Significantly greater NP enrichment is found in high frequency bands than low frequency bands or DEP buffer control cells; each sample was normalized to unsorted control (0–1000 kHz) cells. In contrast, NPs were not enriched in PSA-NCAM (+) or (−) fractions isolated by FACS; each sample was normalized to FACS control, which was propidium iodide (PI) (−) live cells. PSA-NCAM (−) cells have significantly fewer NPs than PSA-NCAM (+) cells, but do not significantly differ from buffer control cells. Data is presented as fold difference, n>/=3 independent experiments and error bars show s.e.m. Black asterisks denote significance as compared to buffer control (DACS: * p=0.027). Red asterisks denote significance of low frequency sorted cells as compared to high frequency sorted cells (DACS: * p=0.018) or PSA-NCAM (−) as compared to PSA-NCAM (+) (FACS: * p=0.049).
To determine the robustness of inherent electrophysiological properties as defining traits of progenitors in the neural lineage, we compared the isolation of NPs using DACS to that using PSA-NCAM antibody in FACS, which has been reported to isolate NPs from mouse, rat and human CNS (8, 21, 22). FACS analysis of E12.5 NSPCs indicated an average of 79.8 % (+/− 3.3 s.e.m., n=4) PSA-NCAM (+) cells in the population. The list of all the controls for cells sorted by FACS is in Methods, but shown in Figure 3 are cells incubated in FACS buffer for the same amount of time as the sorted cells (buffer control) and samples were normalized to live cells sorted by FACS for PI staining (FACS control). We find sorting mNSPCs for PSA-NCAM (+) cells does not significantly enrich NPs (Fig. 3c). PSA-NCAM (−) cells do not have significantly reduced numbers of NPs as compared to unsorted cells; however, there are significantly fewer NPs compared to PSA-NCAM (+) cells (Fig. 3c). The fact that PSA-NCAM sorting does not enrich NPs is not surprising since approximately 30% of the starting E12.5 mNSPCs become neurons, yet 79.8 % are PSA-NCAM (+), indicating PSA-NCAM is not strictly an NP marker. In contrast, cells sorted by high frequency DACS show a significant increase in NPs (Fig. 3c). These results demonstrate that inherent electrophysiological properties are a more robust indicator of fate potential than the marker PSA-NCAM.
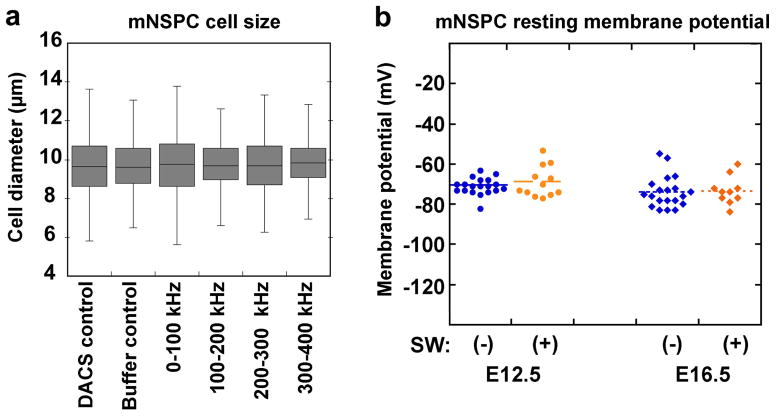
NPs and APs do not differ in cell size or resting membrane potential
The distinct fth of NPs and APs and their sorting in DACS could reflect differences in cell size and the total amount of plasma membrane (23). To test whether cell size differences that would increase the amount of membrane could account for the distinct properties of NPs and APs, we measured the diameters of cells sorted by DACS. Unsorted mNSPCs are a mixture of cells ranging in diameter from 7.5–12 μm when in suspension, allowing for the possibility that cell subpopulations of different size will be sorted at different DEP frequencies. However, measuring cells immediately after sorting reveals no difference in the average size of cells sorted at any frequency (Fig. 4a). The average diameters of sorted cells also do not differ from those of unsorted control (0–1000 kHz) or buffer control cells (Fig. 4a). This is consistent with previous data showing no difference in size between cell populations with more NPs (mouse E12.5 or human SC27 NSPCs) and those with more APs (mouse E16.5 or human SC23 NSPCs) (11). We conclude that distinct electrophysiological properties of NPs and APs are not due to size differences of the cells and NPs and APs do not significantly differ in size.
Figure 4. NSPCs isolated into discrete frequency bands do not differ in cell size and resting membrane potential does not differ with mNSPC fate potential or glycosylation state.
a) No difference was observed in the diameters of control cells (unsorted DACS control 0–1000 kHz and DEP buffer control) and cells sorted at distinct frequency bands (0–100 kHz, 100–200 kHz, 200–300 kHz and 300–400 kHz). Error bars show range, lines within box show average. Data were analyzed with Student’s t-test, p>0.05 all comparisons, n=3 independent experiments, n>500 cells per condition.
b) Resting membrane potential was measured using patch-clamp analysis. No significant difference in resting membrane potential was detected between cells isolated at E12.5 (blue circles) or E16.5 (blue diamonds) although these cells differ in fate potential and membrane capacitance (11) (E12.5 (−): −71.0 mV +/− 4.2 s.d., n=19 cells; E16.5 (−): −73.9 mV +/− 7.8 s.d., n=20 cells; p=0.16). Furthermore, treatment of E12.5 (orange circles) or E16.5 (orange diamonds) mNSPCs with the glycosylation inhibitor Swainsonine (+) did not alter their resting membrane potential (E12.5 (+): −68.7 +/− 2.2; n=12 cells, p=0.29 compared to untreated E12.5)(E16.5 (+): −73.3 mV +/− 7.0 s.d.; n=10 cells, p=0.84 compared to untreated E16.5). Each point on the graph represents an individual cell.
We also examined whether resting membrane potential (RMP) differs between NSPCs with distinct fate potentials since this electrophysiological property can correlate with differentiation state (24). Patch clamp analysis indicated no significant differences in RMP between E12.5 and E16.5 mouse NSPCs (E12.5 −71.0 mV +/− 4.2 s.d., E16.5 −73.9 mV +/−7.8 s.d., n> 18 cells, p=0.16) although these cells differ in fate potential (Fig. 4b) (11). Consistent with this finding, we previously showed membrane conductance, or the ability of the membrane to transmit charge, did not reflect the fate potential of these cells (11). These data suggest ion channel activity likely does not underlie the fate-specific progenitor signatures.
Cell surface glycosylation significantly affects cell fate-specific electrophysiological properties
Since size and RMP do not explain the distinct electrophysiological properties of NPs and APs, we considered other plasma membrane components that could contribute to these cells’ Cm and fth values (25). We chose to focus on cell surface N-glycans since almost all membrane proteins are glycosylated, making N-glycans a major component of membranes (26). In addition, N-glycans could affect Cm through the sugar moieties’ large size, ability to store charge or their role in membrane clustering and receptor retention at the plasma membrane (Fig. S3) (27). N-glycans have a significant role in cell-cell and cell-matrix interactions in the developing brain when NPs and APs are formed (28). Finally, glycosylation enzyme levels and their products found on membranes increase during mouse cortical development in whole brain analysis (19, 20), leading us to propose that mNSPC glycosylation might also differ during development and affect progenitor cell inherent electrophysiological properties.
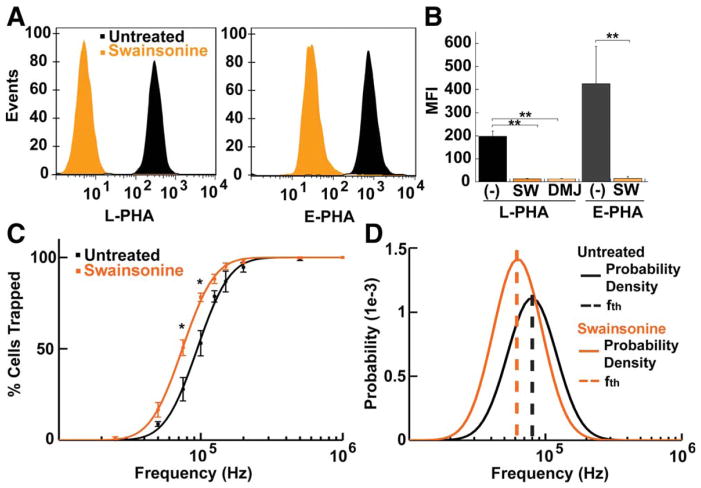
We used two different inhibitors that reduce the amount of complex N-glycans on the cell surface to test whether altering cell surface glycosylation affects NSPC inherent electrophysiological properties. These inhibitors, swainsonine (SW) affecting mannosidase II and deoxymannojirimycin (DMJ) affecting mannosidase I, significantly reduce the levels of complex N-glycans on mNSPCs as detected by flow cytometry with lectins L-PHA and E-PHA (Fig. 5a, b). The significant reduction of these complex N-glycans after inhibitor treatment allowed us to examine the effect of different glycosylation states on intrinsic electrophysiological properties.
Figure 5. Glycosylation inhibitors reduce plasma membrane complex N-glycans, revealing contributions of glycosylation to mNSPC electrophysiological properties.
a) Lectin binding analysis of cell surface N-glycosylation. Representative flow cytometry profiles demonstrate decreased binding of complex N-glycan specific lectins L-PHA-FITC (left panel) or E-PHA-FITC (right panel) to E16.5 mNSPCs treated with the glycosylation inhibitor swainsonine (SW). Histograms depict the distribution of untreated (black) or SW-treated (orange) cells (events, y axis) that stained for lectin (fluorescence intensity, x-axis).
b) Mean fluorescence intensity (MFI) of L-PHA and E-PHA lectin binding demonstrates that glycosylation inhibitors SW and deoxymannojirimycin (DMJ) reduce complex N-glycans by >93%. Cells were untreated (−) or treated with SW or DMJ for 2 days prior to lectin binding. Error bars show s.e.m., n=3 independent experiments, ** p=0.0001.
c) Untreated and SW-treated E16.5 mNSPCs have distinct DEP trapping curves (points are measured data of the percentage of cells trapped at each frequency and lines are fitted lognormal distributions). Error bars show s.e.m. Data points at each frequency were analyzed with Student’s t-test, * p= 0.032 for 75 kHz, p=0.018 for 100 kHz, n=4 independent experiments.
d) The lognormal distribution of trapping curves from (c) give the probability distribution functions (solid lines) for untreated (black) and SW-treated (orange) cells to reveal distinct fth for each set of cells (dashed lines).
Analysis of mNSPCs by DEP trapping curves and calculation of fth after treatment with glycosylation inhibitors indicated significant shifts in cell electrophysiological properties. The trapping curve of SW-treated cells is significantly shifted to lower frequencies (Fig. 5c) and the fth for cells treated with glycosylation inhibitors are lower than those of untreated cells (SW-treated 62.6 kHz, DMJ-treated 55.4 kHz, untreated 79.6 kHz) (Fig. 5d). The skewness of the trapping curves was comparable among all samples, suggesting similar heterogeneity of the cell populations (Fig. 5d). Although glycosylation inhibition affects cell electrophysiological properties, both cell size (cell diameters: untreated 10.7 +/− 0.2 s.d., SW-treated 10.7 +/− 0.2 s.d., DMJ-treated, 10.8 +/−0.3 s.d., n>300, p>0.55 for all comparisons) and RMP (Fig. 4) remain unchanged. These studies indicate N-glycans are a cell surface molecular component contributing to inherent electrophysiological properties.
Discussion
Prospective enrichment of NPs and APs from a heterogeneous population using inherent cell membrane electrophysiological properties demonstrates that these properties are sufficient to identify distinct progenitors in the neural lineage. These findings suggest a new concept in developmental biology - that inherent membrane electrophysiological properties define specific cell populations during differentiation. Evidence for this concept is now available for multiple lineages. In the hematopoietic stem cell lineage, the electrophysiological properties of human CD34+ stem cells distinguish these cells from more differentiated blood cells and the six main leukocyte subpopulations in the human hematopoietic lineage have distinct membrane capacitance values (13, 29, 30). Progression of human adipose-derived stem cells along either adipogenic or osteogenic lineages is dynamically detected by changes in membrane capacitance during differentiation and nestin- and NG2-positive progenitor cells are isolated from adipose tissue by DEP (12, 15). Furthermore, inherent electrophysiological properties of mouse myoblast progenitors shift as these cells differentiate into myotubes (16). We previously found the potential of mouse cortical NSPCs to form either neurons or astrocytes is indicated by the response of the cells in DEP and the fate potential of both human and mouse cortical NSPCs is predicted by the cells’ membrane capacitance values (10, 11). We show for the first time here that electrophysiological properties are sufficient to prospectively isolate two distinct progenitors on the basis of their fate potential from a heterogeneous cell population, further indicating how intricately progenitor cell electrophysiological properties are linked to their fate potential.
Separation of distinct cell types in DEP is often most robust for cells that are significantly different from each other, particularly in cell size. Cell types prospectively separated by DEP include leukemia cells from blood (31), human breast cancer cells from blood (32), human neuroblastoma and glioma cells (33), human peripheral blood mononuclear cells from a human monocyte cell line (33), isolation of human leukocyte subtypes (34), and neurons from NSPCs (17). In many of these cases the cells differed in size or membrane morphology. In contrast, we show that progenitors in the neural lineage are prospectively isolated from a naturally occurring heterogeneous population of cells on the basis of electrophysiological properties independent of size, known surface marker expression (PSA-NCAM), and resting membrane potential. Thus, properties of the membrane itself, and not a difference in the amount of membrane due to variations in cell size, define progenitors in the neural lineage and are sufficient for their isolation.
We are the first to identify complex N-glycans on the cell surface as significant contributors to cell fate specific electrophysiological properties since changing N-glycan expression with glycosylation inhibitors alters NSPC electrophysiological properties. In fact, glycosylation differences alone could be sufficient to explain the distinct electrophysiological properties of NPs and APs since treatment of cells with glycosylation inhibitors causes a shift in DEP trapping curves equal to the difference in NP and AP trapping curves (10). N-glycans have functional roles that can contribute to cell fate and function (Fig. S3). For example, complex N-glycans that are highly branched and recognized by L-PHA are important for cell surface residency and clustering of receptors such as EGFR (27), regulation of T cells and autoimmune diseases (35, 36), and tumor growth and metastasis (37). In addition, complex bisecting N-glycans recognized by E-PHA affect functionality of receptors such as E-cadherin (38) and inhibit proliferation of hepatocytes (39). N-glycans regulate tyrosine kinase receptors, TGF beta family receptors, and cell-cell and cell-substrate adhesion receptors (Fig. S3). These receptors and their associated N-glycans may not only contribute to cell electrophysiological properties, but directly regulate cell fate potential as well.
In addition to N-glycans, other glycosylated membrane components may contribute to distinct NP and AP electrophysiological properties. For example, glycosphingolipids (GSLs) modulate receptor clustering in lipid rafts (40), which may influence both membrane electrophysiological properties and receptor function (Fig. S3). Moreover, GSL composition changes during brain development (41), and brain gangliosides are GSLs with sialic acid residues that contribute negative charge to the outer membrane. Additionally, a recent study suggests the proportion of saturated lipids may affect inherent electrophysiological properties (16). Future studies will focus on defining the aggregate molecular characteristics underlying fate-specific membrane electrophysiological properties.
In summary, inherent electrophysiological properties provide a unique quantitative measure of stem and progenitor fate potential and are sufficient to define and isolate progenitors of the neural lineage. Cell surface glycosylation contributes to these plasma membrane properties and likely plays a role in directing fate potential by modulating cell surface receptors.
Materials and Methods
DACS and trapping curves
The DACS device was fabricated as previously described (17) except screening electrodes were introduced in the outlet channel. We designed the DACS device with castellated electrode arrays that have the advantage of providing greater cell sorting fidelity since a symmetrical DEP electric field is generated whether the fluid flow is parallel, when cells are loaded in the channel, or perpendicular, when cells are collected (17). For DACs experiments, cells were washed twice with DEP buffer (8.5% sucrose, 0.3% glucose, adjusted to 110 μS/cm conductivity with RPMI). Cells were loaded into the DACS device at 1–2 × 106 cells/ml in DEP buffer at the inlet. Fluid flow was controlled by pulling with a syringe pump connected to the outlet at a flow rate of 1.0 to 1.5 μl/min. Cells were trapped along all three electrode arrays to increase throughput. The frequency was set at F2 for 30 seconds to trap cells then fluid flow was reversed to remove non-trapped cells for an additional 30 seconds. Valves were switched to collection mode and F1 was applied while buffer flowed through the perpendicular channels at 1.5 μl/min to the collection wells. To screen collected cells, three 5-second videos were recorded with a Canon EOS Rebel T2i of cells at the screening electrodes for each screening frequency. Cells in the videos were counted to quantify the percentage trapped cells at each frequency. Cell diameters of sorted cells and controls were obtained by imaging cells with an EVOS microscope immediately after isolation and analyzing images with ImageJ software to measure cell diameters.
Controls for cells sorted by DACS included cells incubated in DEP buffer for the same amount of time as the cells that were sorted (buffer control) and cells that were in DEP buffer and went through the DACS device, but were collected by the frequency band 0–1000 kHz (unsorted control since this procedure collected all the cells in the population). Sorted cells and buffer control cells were normalized to unsorted control (0–1000 kHz) cells.
The trapping curve for E12.5 mNSPCs was determined by analyzing the percentage of cells that trap on the castellated electrodes in the DACs device at different frequencies. Three 5-second videos were recorded with a Canon EOS Rebel T2i of cells for each frequency. Cells in the videos were counted to quantify the percentage trapped cells at each frequency.
Trapping curves for glycosylation inhibitor studies were performed in a single channel microfluidic device as previously described (10) except DEP buffer was adjusted to 110 μS/cm with KCl and the applied field was at 16 Vpp with a Tektronix AFG3101 function generator. An Olympus BX41 microscope, Olympus TH4-100 light source and Canon EOS Rebel T2i camera were used to perform the trapping experiments and videos were captured with Camstudio screen capture software.
Cell culture
NSPCs from cortices of CD1 mouse embryo day 12.5 (E12.5) or E16.5 were cultured as neurospheres as described previously (10, 11, 18). Mouse NSPC growth medium consisted of: DMEM, 1x B27, 1x N2, 1 mM sodium pyruvate, 2 mM glutamine, 1 mM N-acetylcysteine (Sigma Aldrich, St. Louis, MO), 20 ng/mL EGF (BD Biosciences, Bedford, MA), 10 ng/mL FGF (BD Biosciences, Bedford, MA), and 2 μg/mL heparin (Sigma, St. Louis, MO). For all experiments requiring single cell suspensions (DACS, trapping curves, FACS, flow cytometry), cells were dissociated with Neurocult Chemical Dissociation Kit (Stem Cell Technologies). Cells were differentiated for 3 days to analyze neuronal differentiation or for 5–7 days to analyze astrocyte differentiation in media lacking EGF, FGF and heparin. For differentiation, cells were plated in 3 mm diameter PDMS microwells placed on glass coverslips (Carolina Biological Supply) coated with 10 μg/ml poly-D-lysine (MP Biomedicals) and 20 μg/ml laminin (Invitrogen). For glycosylation inhibitor studies, dissociated E16.5 mNSPCs were seeded at 150,000 cells/ml in regular growth media and neurosphere culture conditions in the presence or absence of 500 nM swainsonine (Sigma) or 1 μm deoxymannojirimycin (Calbiochem) for 2 days.
Immunostaining and Cell Quantitation
Cells were fixed with 4% paraformaldehyde for 10 min as described previously (10). Cells were immunostained overnight at 4° C with mouse anti-Map2 (HM2) (Sigma) at 1:200 and rabbit anti-TuJ1 (Fitzgerald Laboratories) at 1:100 or mouse anti-GFAP (Sigma) at 1:200. Secondary antibodies (Vector Labs, Alexa 488 and Alexa 555) were used at 1:200 at room temperature in the dark for 2 hrs. All antibodies were diluted in 1% bovine serum albumin (IgG-free, Jackson Labs) in PBS. Cell nuclei were counterstained with Hoechst 33342 at 1:500 (2 μg/ml in phosphate buffered saline, Invitrogen). Coverslips with cells were mounted with Vectashield (Vector Labs) and imaged with a Nikon Eclipse Ti microscope with a 20x objective and NIS element AR3.10 software.
Percentages of cells that differentiated into neurons or astrocytes were calculated from at least 3 separate experiments with images of at least 10 randomly selected fields and more than 600 total cells analyzed for each cell population. Hoechst-stained nuclei were used to determine the total cell number. Stringent criteria were applied to assess generation of neurons and astrocytes from NSPCs and we restricted cells in our analyses to those that met the following criteria: 1) cells counted as neurons co-expressed the neuronal markers MAP2 and TuJ1 and had neurites at least two times the length of the cell body (Fig. S2) and 2) cells counted as astrocytes were GFAP-positive cells exhibiting a filamentous, cytoskeletal pattern of GFAP reactivity in the cytoplasm. Undifferentiated NSPCs from E12.5 cortices do not express GFAP and thus would not be confused with astrocytes in our analyses (10). Positively stained cells were counted in ImageJ and each cell was marked to prevent double counting.
Flow cytometry and FACS
For flow cytometry analysis of lectin binding, dissociated E16.5 mNSPCs were washed with 5% BSA in PBS and incubated for 30 min on ice in the dark with 20 μg/ml FITC-L-PHA or FITC-E-PHA (Vector Labs). Cells were washed twice, passed through a 35 μm mesh (Falcon 352235) and incubated with 2 μg/ml propidium iodide to distinguish live and dead cells. Cells were subjected to flow cytometry (BD LSR II) and data was analyzed with FlowJo software. For FACS isolation, dissociated E12.5 mNSPCs were incubated for 30 min on ice with mouse anti-PSA-NCAM (Millipore) at 1:50 in FACS buffer (PBS without calcium or magnesium (Mediatech), 0.1% IgG-free BSA (Jackson ImmunoResearch), and 1.55 g/L glucose (Fisher)). Washed cells were incubated 30 min on ice with goat anti-mouse Alexa Fluor 488 secondary antibody (Invitrogen) (1:50) in FACS buffer. Samples were passed through 35μm mesh and sorted with the FACS Aria II system (Becton Dickinson) after an appropriate gating design was created based on controls and optimized machine settings. To sort, cells were passed through a 70-μm nozzle at a sheath pressure of 25 PSI with an events rate of 1000–3000 events/second. Cells were sorted into collection tubes with 0.5 ml differentiation medium supplemented with 1x antibiotic-antimycotic (Invitrogen) prior to plating on pDL-laminin coated coverslips for differentiation. All FACS sorting was performed by a dedicated, BD certified FACS operator in the UCI Stem Cell Core Facility. FACS samples included: unstained buffer control, a propidium iodide- (PI) stained control, a secondary antibody only control, a secondary and PI only control (FACS control), PSA-NCAM and secondary antibody stained with and without PI.
Resting membrane potential measurements
Cells were grown as neurospheres and plated on laminin-coated coverslips after mechanical trituration. Resting membrane potential measurements were made 8–36 hours after plating at room temperature in whole-cell patch clamp configuration with an Axopatch 200B amplifier controlled by pClamp 10 and Digidata 1440A (Molecular Devices). The bath solution consisted of 145 mM NaCl, 3 mM KCl, 3 mM CaCl2, 2 mM MgCl2, 8 mM glucose, 10 mM HEPES, pH 7.3 and osmolality of 313 mOsm/kg. Patch pipettes were pulled from standard-walled borosilicate glass and had an access resistance of 2 – 4 MΩ when filled with a solution composed of 137 mM KCl, 9 mM NaCl, 1 mM MgCl2, 5 mM EGTA, 5 mM ATP, 0.4 mM GTP, 10 mM HEPES pH 7.3, and osmolality of 313 mOsm/kg. The membrane potential was measured immediately after establishing whole-cell configuration by setting the current clamp mode to zero (I=0 mode).
Statistics
Data are shown as the average of at least 3 separate experiments. The data were subject to Student’s t-test with significance p<0.05. The n for each experiment is listed in the figure legends.
Supplementary Material
Schematic depicts two hypothetical cells with distinct biophysical properties and responses to DEP frequencies (schematic adapted from (1)). The fxo of each cell is the point at which the line (blue or orange) intersects the x-axis. At the frequency denoted by the dashed green line, the blue cell experiences positive DEP (frequency higher than fxo) and is trapped along the electrode while the orange cell experiences negative DEP (frequency lower than fxo) and is not trapped. The different frequency responses of the cells can be used to separate them along DACS trapping electrode arrays.
Cells sorted by DACS were differentiated and immunostained with Map2 and TuJ1 (which detects class III beta tubulin) to detect neurons. Cells were considered neurons if they costained with the two markers and had neurites greater than 3 times the length of the cell body (see Methods). Greater numbers of neurons were derived from cells sorted at high frequency. Images are of differentiated cells from unsorted DEP buffer control, low frequency band sort (0–100 kHz) or high frequency band sort (300–400 kHz). Hoechst stained nuclei are blue and scale bar = 20 μm.
Schematic of mNSPC plasma membrane components that are heavily glycosylated and may contribute to progenitor cell differences in membrane capacitance. The phospholipid bilayer is estimated to be 7–10 nm thick. Complex N-glycans on growth factor receptors affect their residence time on the cell surface and ligand binding. Membrane proteoglycans have large repeating sugar structures such as heparan sulfate, whose sugar components contribute to growth factor presentation and clustering of receptors. Glycosylation also contributes to clustering of cell-matrix receptors depicted by glycosylated alpha and beta integrin heterodimers in glycosphingolipid (GSL) rich lipid rafts. Additionally, many cell-cell receptors such as NCAM are heavily glycosylated (as is the case for PSA-NCAM as depicted). Difference in glycosylation between cell types could contribute to variations in electrophysiological properties.
Acknowledgments
We thank M. Arredondo for help with cell diameter measurements, V. Scarfone and C. Tu for technical assistance with FACS, and E. Monuki for critical reading of the manuscript. We gratefully acknowledge the support of the Sue & Bill Gross Stem Cell Research Center and a gift by Pearl Tze Hosfiel and Keith Hosfiel. This work was supported in part by the California Institute for Regenerative Medicine (CIRM) RT1-01074 and TG2-01152, NIH AG23583, and UL1 TR000153 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and the NIH Roadmap for Medical Research
Footnotes
Jamison L. Nourse: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing
Javier Lopez Prieto: Conception and design, collection and/or assembly of data, device fabrication, data analysis and interpretation, manuscript writing
Amanda R. Dickson: collection and/or assembly of data
Jente Lu: Conception and design
Medha M. Pathak: collection and/or assembly of data, data analysis and interpretation
Francesco Tombola: Conception and design
Michael Demetriou: Conception and design
Abraham P. Lee: Conception and design, manuscript editing, financial support
Lisa A. Flanagan: Conception and design, data interpretation, manuscript writing, financial support, final approval of manuscript
References
- 1.Kim SU, de Vellis J. Stem cell-based cell therapy in neurological diseases: a review. Journal of neuroscience research. 2009;87:2183–2200. doi: 10.1002/jnr.22054. [DOI] [PubMed] [Google Scholar]
- 2.Pruszak J, Ludwig W, Blak A, et al. CD15, CD24 and CD29 Define a Surface Biomarker Code for Neural Lineage Differentiation of Stem Cells. Stem Cells. 2009;27:2928–2940. doi: 10.1002/stem.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao MS, Noble M, Mayer-Proschel M. A tripotential glial precursor cell is present in the developing spinal cord. Proc Natl Acad Sci U S A. 1998;95:3996–4001. doi: 10.1073/pnas.95.7.3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panchision DM, Chen HL, Pistollato F, et al. Optimized flow cytometric analysis of central nervous system tissue reveals novel functional relationships among cells expressing CD133, CD15, and CD24. Stem Cells. 2007;25:1560–1570. doi: 10.1634/stemcells.2006-0260. [DOI] [PubMed] [Google Scholar]
- 5.Pfenninger CV, Roschupkina T, Hertwig F, et al. CD133 is not present on neurogenic astrocytes in the adult subventricular zone, but on embryonic neural stem cells, ependymal cells, and glioblastoma cells. Cancer Res. 2007;67:5727–5736. doi: 10.1158/0008-5472.CAN-07-0183. [DOI] [PubMed] [Google Scholar]
- 6.Yuan SH, Martin J, Elia J, et al. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS One. 2011;6:e17540. doi: 10.1371/journal.pone.0017540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maric D, Maric I, Chang YH, et al. Prospective cell sorting of embryonic rat neural stem cells and neuronal and glial progenitors reveals selective effects of basic fibroblast growth factor and epidermal growth factor on self-renewal and differentiation. J Neurosci. 2003;23:240–251. doi: 10.1523/JNEUROSCI.23-01-00240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mujtaba T, Piper DR, Kalyani A, et al. Lineage-restricted neural precursors can be isolated from both the mouse neural tube and cultured ES cells. Dev Biol. 1999;214:113–127. doi: 10.1006/dbio.1999.9418. [DOI] [PubMed] [Google Scholar]
- 9.Pethig R, Markx GH. Applications of dielectrophoresis in biotechnology. Trends Biotechnol. 1997;15:426–432. doi: 10.1016/S0167-7799(97)01096-2. [DOI] [PubMed] [Google Scholar]
- 10.Flanagan LA, Lu J, Wang L, et al. Unique dielectric properties distinguish stem cells and their differentiated progeny. Stem Cells. 2008;26:656–665. doi: 10.1634/stemcells.2007-0810. [DOI] [PubMed] [Google Scholar]
- 11.Labeed FH, Lu J, Mulhall HJ, et al. Biophysical characteristics reveal neural stem cell differentiation potential. PLoS One. 2011;6:e25458. doi: 10.1371/journal.pone.0025458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagnaninchi PO, Drummond N. Real-time label-free monitoring of adipose-derived stem cell differentiation with electric cell-substrate impedance sensing. Proc Natl Acad Sci U S A. 2011;108:6462–6467. doi: 10.1073/pnas.1018260108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vykoukal DM, Gascoyne PR, Vykoukal J. Dielectric characterization of complete mononuclear and polymorphonuclear blood cell subpopulations for label-free discrimination. Integr Biol (Camb) 2009;1:477–484. doi: 10.1039/b906137a. [DOI] [PubMed] [Google Scholar]
- 14.Gascoyne PR, Noshari J, Anderson TJ, et al. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis. 2009;30:1388–1398. doi: 10.1002/elps.200800373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vykoukal J, Vykoukal DM, Freyberg S, et al. Enrichment of putative stem cells from adipose tissue using dielectrophoretic field-flow fractionation. Lab Chip. 2008;8:1386–1393. doi: 10.1039/b717043b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muratore M, Srsen V, Waterfall M, et al. Biomarker-free dielectrophoretic sorting of differentiating myoblast multipotent progenitor cells and their membrane analysis by Raman spectroscopy. Biomicrofluidics. 2012:6. doi: 10.1063/1.4746252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prieto JL, Lu J, Nourse JL, et al. Frequency discretization in dielectrophoretic assisted cell sorting arrays to isolate neural cells. Lab Chip. 2012;12:2182–2189. doi: 10.1039/c2lc21184j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu J, Barrios C, Dickson AR, et al. Advancing practical usage of microtechnology: a study of the functional consequences of dielectrophoresis on neural stem cells. Integr Biol. 2012;4:1223–1236. doi: 10.1039/c2ib20171b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flaris NA, Shindler KS, Kotzbauer PT, et al. Developmentally-regulated lectin binding in the embryonic mouse telencephalon. Brain Res. 1995;678:99–109. doi: 10.1016/0006-8993(95)00173-n. [DOI] [PubMed] [Google Scholar]
- 20.Ishii A, Ikeda T, Hitoshi S, et al. Developmental changes in the expression of glycogenes and the content of N-glycans in the mouse cerebral cortex. Glycobiology. 2007;17:261–276. doi: 10.1093/glycob/cwl076. [DOI] [PubMed] [Google Scholar]
- 21.Piper DR, Mujtaba T, Keyoung H, et al. Identification and characterization of neuronal precursors and their progeny from human fetal tissue. Journal of neuroscience research. 2001;66:356–368. doi: 10.1002/jnr.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer-Proschel M, Kalyani AJ, Mujtaba T, et al. Isolation of lineage-restricted neuronal precursors from multipotent neuroepithelial stem cells. Neuron. 1997;19:773–785. doi: 10.1016/s0896-6273(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 23.Voldman J. Electrical forces for microscale cell manipulation. Annu Rev Biomed Eng. 2006;8:425–454. doi: 10.1146/annurev.bioeng.8.061505.095739. [DOI] [PubMed] [Google Scholar]
- 24.Sundelacruz S, Levin M, Kaplan DL. Role of membrane potential in the regulation of cell proliferation and differentiation. Stem Cell Rev. 2009;5:231–246. doi: 10.1007/s12015-009-9080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desai SP, Vahey MD, Voldman J. Electrically Addressable Vesicles: Tools for Dielectrophoresis Metrology. Langmuir. 2009;25:3867–3875. doi: 10.1021/la803330c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi M, Kuroki Y, Ohtsubo K, et al. Core fucose and bisecting GlcNAc, the direct modifiers of the N-glycan core: their functions and target proteins. Carbohydr Res. 2009;344:1387–1390. doi: 10.1016/j.carres.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 27.Lau KS, Partridge EA, Grigorian A, et al. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 28.Yanagisawa M, Yu RK. The expression and functions of glycoconjugates in neural stem cells. Glycobiology. 2007;17:57–74. doi: 10.1093/glycob/cwm018. [DOI] [PubMed] [Google Scholar]
- 29.Talary MS, Mills KI, Hoy T, et al. Dielectrophoretic separation and enrichment of CD34+ cell subpopulation from bone marrow and peripheral blood stem cells. Med Biol Eng Comput. 1995;33:235–237. doi: 10.1007/BF02523050. [DOI] [PubMed] [Google Scholar]
- 30.Stephens M, Talary MS, Pethig R, et al. The dielectrophoresis enrichment of CD34+ cells from peripheral blood stem cell harvests. Bone Marrow Transplant. 1996;18:777–782. [PubMed] [Google Scholar]
- 31.Becker FF, Wang XB, Huang Y, et al. The removal of human leukaemia cells from blood using interdigitated microelectrodes. J Phys D: Appl Phys. 1994;27:2659–2662. [Google Scholar]
- 32.Becker FF, Wang XB, Huang Y, et al. Separation of human breast cancer cells from blood by differential dielectric affinity. Proc Natl Acad Sci U S A. 1995;92:860–864. doi: 10.1073/pnas.92.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Joo S, Duhon M, et al. Dielectrophoretic cell separation and gene expression profiling on microelectronic chip arrays. Anal Chem. 2002;74:3362–3371. doi: 10.1021/ac011273v. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Huang Y, Wang XB, et al. Differential analysis of human leukocytes by dielectrophoretic field-flow-fractionation. Biophys J. 2000;78:2680–2689. doi: 10.1016/S0006-3495(00)76812-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Demetriou M, Granovsky M, Quaggin S, et al. Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature. 2001;409:733–739. doi: 10.1038/35055582. [DOI] [PubMed] [Google Scholar]
- 36.Mkhikian H, Grigorian A, Li CF, et al. Genetics and the environment converge to dysregulate N-glycosylation in multiple sclerosis. Nat Commun. 2011;2:334. doi: 10.1038/ncomms1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Granovsky M, Fata J, Pawling J, et al. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat Med. 2000;6:306–312. doi: 10.1038/73163. [DOI] [PubMed] [Google Scholar]
- 38.Liwosz A, Lei T, Kukuruzinska MA. N-glycosylation affects the molecular organization and stability of E-cadherin junctions. J Biol Chem. 2006;281:23138–23149. doi: 10.1074/jbc.M512621200. [DOI] [PubMed] [Google Scholar]
- 39.Yang X, Tang J, Rogler CE, et al. Reduced hepatocyte proliferation is the basis of retarded liver tumor progression and liver regeneration in mice lacking N-acetylglucosaminyltransferase III. Cancer Res. 2003;63:7753–7759. [PubMed] [Google Scholar]
- 40.Simons K, Toomre D. Lipid rafts and signal transduction. Nature reviews Molecular cell biology. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 41.Ngamukote S, Yanagisawa M, Ariga T, et al. Developmental changes of glycosphingolipids and expression of glycogenes in mouse brains. Journal of Neurochemistry. 2007;103:2327–2341. doi: 10.1111/j.1471-4159.2007.04910.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic depicts two hypothetical cells with distinct biophysical properties and responses to DEP frequencies (schematic adapted from (1)). The fxo of each cell is the point at which the line (blue or orange) intersects the x-axis. At the frequency denoted by the dashed green line, the blue cell experiences positive DEP (frequency higher than fxo) and is trapped along the electrode while the orange cell experiences negative DEP (frequency lower than fxo) and is not trapped. The different frequency responses of the cells can be used to separate them along DACS trapping electrode arrays.
Cells sorted by DACS were differentiated and immunostained with Map2 and TuJ1 (which detects class III beta tubulin) to detect neurons. Cells were considered neurons if they costained with the two markers and had neurites greater than 3 times the length of the cell body (see Methods). Greater numbers of neurons were derived from cells sorted at high frequency. Images are of differentiated cells from unsorted DEP buffer control, low frequency band sort (0–100 kHz) or high frequency band sort (300–400 kHz). Hoechst stained nuclei are blue and scale bar = 20 μm.
Schematic of mNSPC plasma membrane components that are heavily glycosylated and may contribute to progenitor cell differences in membrane capacitance. The phospholipid bilayer is estimated to be 7–10 nm thick. Complex N-glycans on growth factor receptors affect their residence time on the cell surface and ligand binding. Membrane proteoglycans have large repeating sugar structures such as heparan sulfate, whose sugar components contribute to growth factor presentation and clustering of receptors. Glycosylation also contributes to clustering of cell-matrix receptors depicted by glycosylated alpha and beta integrin heterodimers in glycosphingolipid (GSL) rich lipid rafts. Additionally, many cell-cell receptors such as NCAM are heavily glycosylated (as is the case for PSA-NCAM as depicted). Difference in glycosylation between cell types could contribute to variations in electrophysiological properties.