Abstract
Background
IRAK1 has been repoted to play an essential role in the development of multiple cancers. However, the clinical significance of IRAK1 in hepatocellular carcinoma (HCC) and the underlying molecular mechanism remain unclear. Therefore, we aimed to investigate the role of IRAK1 in the pathogenesis of HCC in this study.
Materials and methods
HCC tissues and para-carcinoma tissues were collected for immunohistochemistry (IHC) analysis to evaluate IRAK1 expression. Data of IRAK1 expression were downloaded from the cancer genome atlas (TCGA) for analyzing the clinical significance of IRAK1. Receiver operating characteristic (ROC) curve and survival analyses were carried out to assess the diagnostic and prognostic significance of IRAK1 in IHC and TCGA data. Additionally, we investigated the alteration of IRAK1 gene in HCC from cBioPortal to generate a network of the interaction between IRAK1 and the neighboring genes. The influence of IRAK1 gene alteration on the prognosis of HCC patients was evaluated by survival analysis.
Results
Analysis of both IHC and TCGA data revealed a significant upregulation of IRAK1 in HCC tissues. The IHC analysis revealed there was an increasing trend in IRAK1 expression among normal liver tissues, liver cirrhosis tissues, para-carcinoma tissues and HCC tissues. The ROC curves for IHC and TCGA data demonstrated that IRAK1 exhibited a significant diagnostic value for HCC. Moreover, IRAK1 expression was observed to be associated with tumor size, metastasis and T-stage. The survival analysis indicated that the upregulation of IRAK1 predicted a worse overall survival of HCC. Additionally, data from cBioPortal confirmed that 29% of HCC tissues possessed an alteration of the IRAK1 gene.
Conclusion
IRAK1 may act as an oncogene in the development of HCC with its overexpression in HCC. Moreover, IRAK1 might serve as a promising diagnostic and therapeutic target for HCC.
Keywords: IRAK1, hepatocellular carcinoma, immunohistochemistry, RNA-seq, diagnosis, prognosis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers and ranks second among the most fatal cancers.1,2 Approximately 250,000 new cases of HCC occur each year, with an estimated 600,000 HCC-related deaths.3,4 Surgery is currently the most common and effective therapy for HCC, although the prognosis of HCC patients remains unsatisfactory, with the 5-year survival rate ranging from 36% to 50%.5,6 Therefore, we aimed to determine an efficacious diagnostic and therapeutic target for HCC to improve the survival of HCC patients.
IRAK1, located on the X chromosome, belongs to the IRAK family.7,8 IRAK1 is a widely expressed serine/threonine kinase that plays a critical role in the TLR/IL1R signaling pathways.9 The activation of the downstream receptors could cause the phosphorylation of IRAK1, which subsequently binds to the E3 ubiquitin ligase and TRAF6, leading to the activation of the NF-κB and MAPK pathways.10–13 There is mounting evidence that IRAK1 promotes the initiation and progression of various types of cancers, including acute myeloid leukemia (AML), melanoma, lung cancer and breast cancer, by aberrantly stimulating the downstream signaling pathways.14–19
To date, only one study on the role of IRAK1 in HCC has reported that IRAK1 could accelerate cell growth and suppress cell apoptosis via its overexpression in HCC.8 However, that study did not provide information on the gene status of IRAK1 in HCC or analyze the significance of IRAK1 in the diagnosis and prognosis of HCC. Therefore, we carried out the present study to investigate the clinical significance of IRAK1, with a further investigation into the gene status of IRAK1 in the cBioPortal database of HCC, to shed light on the potential molecular mechanism of IRAK1 in the pathogenesis of HCC by analyzing the immunohistochemistry (IHC) and the cancer genome atlas (TCGA) data of IRAK1 expression in HCC.
Materials and methods
Patient samples
A total of 171 formalin-fixed and paraffin-embedded HCC tissues and 171 para-carcinoma tissues (153 males and 18 females) were obtained from the First Affiliated Hospital of Guangxi Medical University (Nanning, Guangxi, China) from March 2010 to December 2012. We also collected 37 cases of liver cirrhosis tissues and 33 normal tissues to compare the IRAK1 expression in different stages of HCC tissues. The research committee of Guangxi Medical University granted permission to conduct the study, and all participating patients signed the written informed consent for this study.
IHC
IHC analysis was performed on all formalin-fixed, paraffin-embedded tissue samples to identify the pattern of IRAK1 expression in HCC cells. The IRAK1 antibody (F-4, sc-5228, 1:50 dilution) provided by Santa Cruz Biotechnology (Heidelberg, Germany) was used for IHC detection. The detailed protocol of IHC was shown in our previous studies.20,21 The IRAK1 staining in HCC cells was analyzed through blind evaluation, which was performed by two pathologists who independently identified the intensity of staining and the positive staining ratio of IRAK1. The staining of IRAK1 was scored according to the following criteria: 1) scores of 0, 1, 2 and 3 indicated negative, weak, moderate and strong staining intensity of IRAK1, respectively, and 2) the proportion of cells positively stained for IRAK1 was graded as 0, 1, 2, 3 and 4 representing 0%, 1%–25%, 26%–50%, 51%–75% and 76%–100% positively stained cells, respectively. If the result of the two criteria was >2, the cells were considered to be positively immune reactive.
Extraction of TCGA data
TCGA data of IRAK1 expression in HCC and noncancerous tissues were downloaded to analyze IRAK1 expression in HCC and its correlations with the clinical parameters of HCC. Information about the alteration of the IRAK1 gene was available in cBioPortal OncoPrint (http://www.cBioPortal.org/index.do) and was analyzed to evaluate the status of IRAK1 gene in the development of HCC.22,23 Additionally, the genes associated with IRAK1 were accessed from cBioPortal OncoPrint to generate a network illustrating the interaction of the IRAK1-related genes. The Kaplan–Meier survival curves were drawn to examine the impact of the alteration of the IRAK1 gene on the prognosis of HCC patients.
Statistical analysis
All statistical analyses were performed using SPSS 22.0 (IBM Corp., Armonk, NY, USA). The difference in the level of IRAK1 expression by IHC was calculated using the chi-square test. The quantitative data on IRAK1 obtained from TCGA were analyzed by Student’s t-test. With regard to the diagnostic value of IRAK1 expression in the IHC and TCGA data, the receiver operating characteristic (ROC) curve was drawn to evaluate the diagnostic capability of IRAK1 for HCC. A graded area under the curve (AUC) value for the ROC curves of 0.5–0.7, 0.7–0.9 or 0.9–1.0 represented a poor, moderate or high diagnostic value of IRAK1, respectively. The influence of IRAK1 on the prognosis of HCC patients was assessed by Kaplan–Meier analysis for the TCGA and IHC data. Moreover, the Kaplan–Meier analysis was applied to analyze the impact of IRAK1 alterations on the prognosis of HCC patients. Two-tailed value of P<0.05 was considered as statistically significant.
Results
Validation of the clinical significance of IRAK1 expression in HCC by IHC
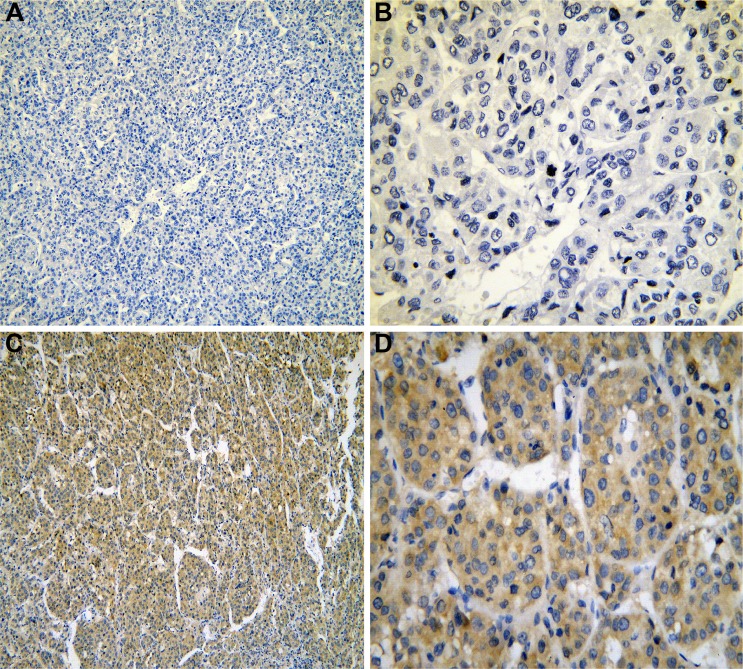
The IHC of normal liver, cirrhosis, normal liver para-HCC, cirrhosis para-HCC and HCC tissues is shown in Figures 1–5. To verify the aberrant expression of IRAK1 in HCC, we included a cohort of 171 samples of HCC. The result of the IHC showed the highest positive rate of IRAK1 in HCC tissues (48.5%) compared with para-HCC tissues (34.5%, P=0.008), liver cirrhosis tissues (24.3%, P=0.007) and normal liver tissues (15.2%, P<0.001; Table 1 and Figure 6A). Moreover, the IRAK1 expression increased noticeably in normal liver tissues, liver cirrhosis tissues, para-HCC tissues and HCC tissues (P<0.001; Table 1 and Figure 6A).
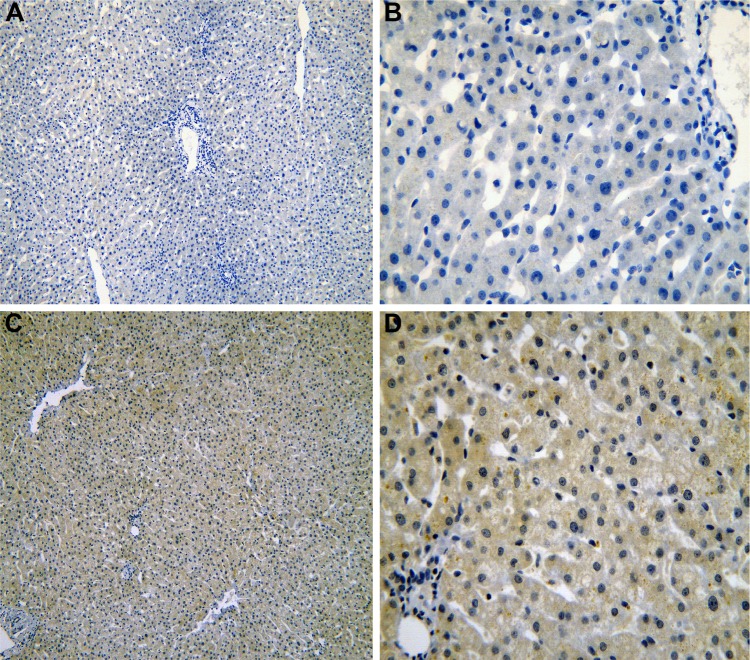
Figure 1.
The IHC of normal liver tissues.
Notes: IRAK1 negative staining in normal liver tissues (A: ×100 and B: ×400). IRAK1 positive staining in normal liver tissues (C: ×100 and D: ×400).
Abbreviation: IHC, immunohistochemistry.
Figure 2.
The IHC of liver tissues with cirrhosis.
Notes: IRAK1 negative staining in cirrhosis tissues (A: ×100 and B: ×400). IRAK1 positive staining in cirrhosis tissues (C: ×100 and D: ×400).
Abbreviation: IHC, immunohistochemistry.
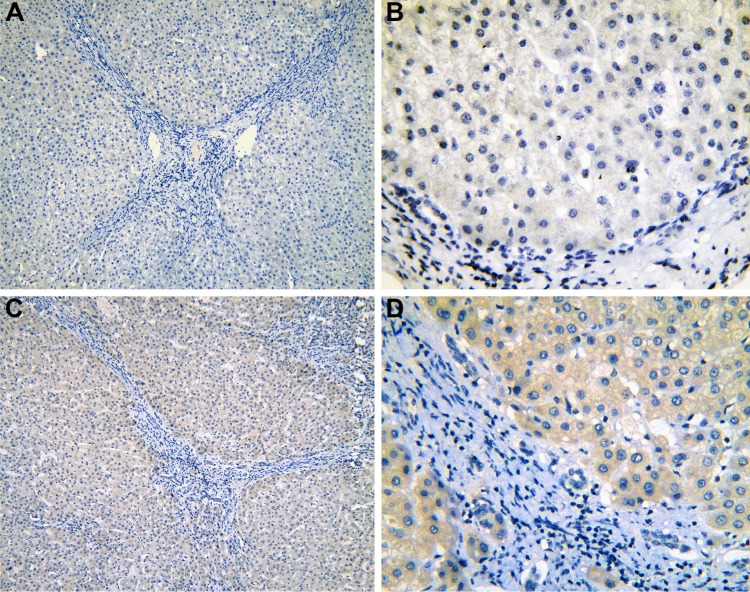
Figure 3.
The IHC of para-HCC tissues with cirrhosis.
Notes: IRAK1 negative staining in cirrhosis tissues (para-HCC; A: ×100 and B: ×400). IRAK1 positive staining in cirrhosis tissues (para-HCC; C: ×100 and D: ×400).
Abbreviations: IHC, immunohistochemistry; HCC, hepatocellular carcinoma.
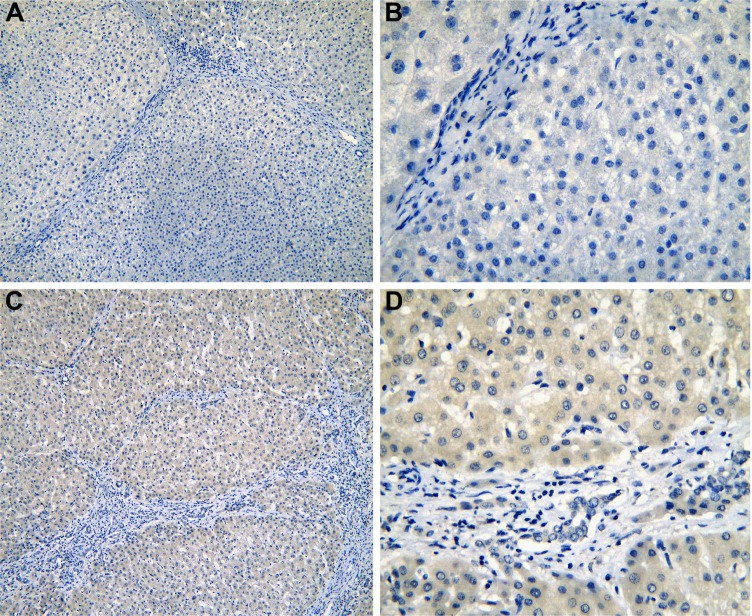
Figure 4.
The IHC of normal liver tissues (para-HCC).
Notes: IRAK1 negative staining in normal liver tissues (para-HCC; A: ×100 and B: ×400). IRAK1 positive staining in normal liver tissues (para-HCC; C: ×100 and D: ×400).
Abbreviations: IHC, immunohistochemistry; HCC, hepatocellular carcinoma.
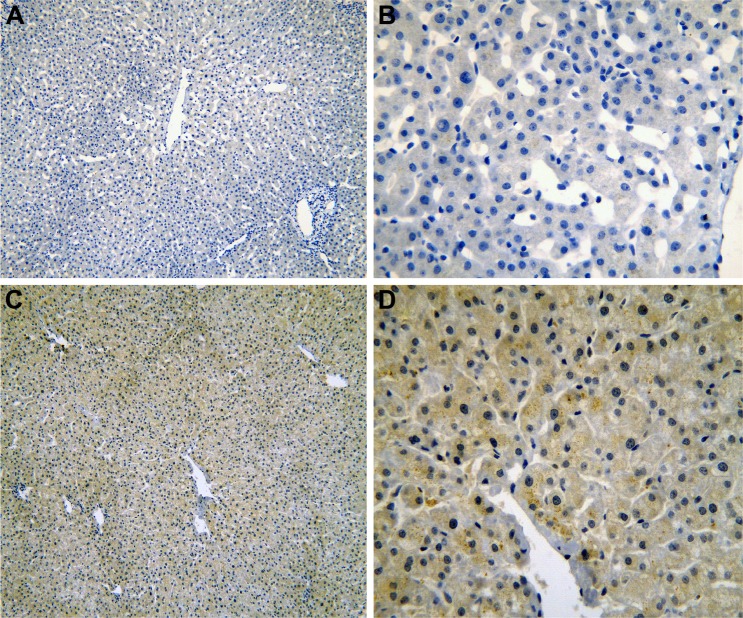
Figure 5.
The IHC of HCC tissues.
Notes: IRAK1 negative staining in HCC tissues (A: ×100 and B: ×400). IRAK1 positive staining in HCC tissues (C: ×100 and D: ×400).
Abbreviations: IHC, immunohistochemistry; HCC, hepatocellular carcinoma.
Table 1.
Expression of IRAK1 in different types of liver tissues
| Tissue | Patients (n) | IRAK1 negative, n (%) | IRAK1 positive, n (%) | χ2 | P-value |
|---|---|---|---|---|---|
| HCC vs | 171 | 88 (51.5) | 83 (48.5) | ||
| Para-carcinoma tissue | 171 | 112 (65.5) | 59 (34.5) | 6.936 | 0.008 |
| Liver cirrhosis | 37 | 28 (75.7) | 9 (24.3) | 7.230 | 0.007 |
| Normal | 33 | 28 (84.8) | 5 (15.2) | 12.570 | ,0.001 |
| Para-carcinoma tissue vs | 171 | 112 (65.5) | 59 (34.5) | ||
| Liver cirrhosis | 37 | 28 (75.7) | 9 (24.3) | 1.432 | 0.231 |
| Normal | 33 | 28 (84.8) | 5 (15.2) | 4.811 | 0.028 |
| Liver cirrhosis vs | 37 | 28 (75.7) | 9 (24.3) | ||
| Normal | 33 | 28 (84.8) | 5 (15.2) | 0.917 | 0.338 |
Note: Linear by linear association test (χ2=18.871, P<0.001) among normal, liver cirrhosis and para-carcinoma tissues, and HCC revealed an increasing trend of IRAK1 expression in these four tissues.
Abbreviation: HCC, hepatocellular carcinoma.
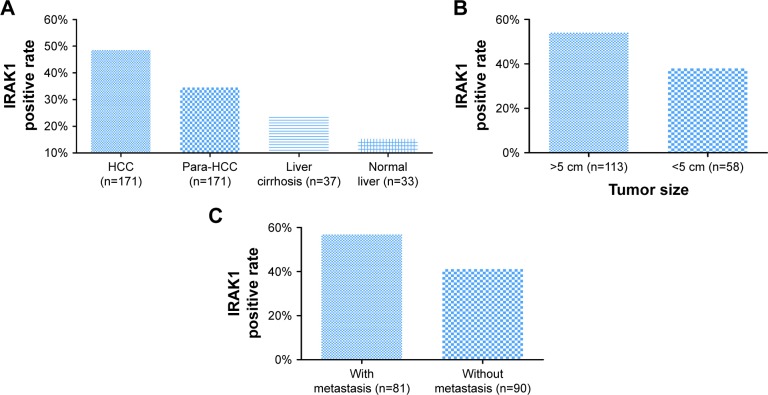
Figure 6.
The expression of IRAK1 in IHC.
Notes: (A) IRAK1 expression in different types of liver tissues. The highest IRAK1 expression was observed in HCC (48.5%) compared with para-HCC tissues (34.5%, P=0.008), liver cirrhosis tissues (24.3%, P=0.007) and normal liver tissues (15.2%, P<0.001). Moreover, the IRAK1 expression increased with the deterioration from noncancerous liver disease to HCC (P<0.001). (B) Tumor size and (C) tumor metastasis. IRAK1 expression was found to be higher in the group of patients with a large tumor size (54.0%) and metastasis (56.8%) than in the group of patients with a small tumor size (37.9%, P=0.047) and no metastasis (41.1%, P=0.041).
Abbreviations: IHC, immunohistochemistry; HCC, hepatocellular carcinoma.
Focusing on the relationships between IRAK1 and the clinicopathological features of HCC, we found a higher positive rate of IRAK1 expression in the groups of large tumor size (54.0%) and metastasis (56.8%) than that in the groups of small tumor size (37.9%, P=0.047; Figure 6B) and no metastasis (41.1%, P=0.041; Figure 6C). As shown in Table 2, the Spearman’s correlation test was utilized to evaluate the correlations between IRAK1 and the clinical features of HCC. The results indicated that IRAK1 expression was significantly correlated with tumor size (r=0.152, P=0.047) and metastasis (r=0.157, P=0.041). As for the diagnostic value of IRAK1 in HCC, the ROC curves indicated that IRAK1 showed a low diagnostic value for HCC (AUC =0.591, P=0.002; Figure 7).
Table 2.
Relationship between IRAK1 expression and clinicopathological features in HCC
| Clinicopathological parameters | n | IRAK1 negative, n (%) | IRAK1 positive, n (%) | χ2 | P-value | Correlation
|
|
|---|---|---|---|---|---|---|---|
| r | P-value | ||||||
| Age (years) | 0.431 | 0.432 | |||||
| <50 | 67 | 37 | 30 | −0.788 | 0.060 | ||
| $50 | 104 | 51 | 53 | ||||
| Gender | 0.713 | 0.715 | |||||
| Male | 153 | 78 (51.0) | 75 (49.0) | 0.135 | −0.028 | ||
| Female | 18 | 10 (55.6) | 8 (44.4) | ||||
| Edmondson grade | 0.199 | 0.173 | |||||
| High | 20 | 14 (70.0) | 6 (30.0) | 3.226 | 0.105 | ||
| Moderate | 98 | 49 (50.0) | 49 (50.0) | ||||
| Low | 53 | 25 (47.2) | 28 (52.8) | ||||
| Nodes | 0.679 | 0.700 | |||||
| Single | 68 | 30 (44.1) | 38 (55.9) | 0.152 | −0.034 | ||
| Multiple | 61 | 29 (47.5) | 32 (52.5) | ||||
| Tumor size (cm) | 0.047 | 0.047 | |||||
| <5 | 58 | 36 (62.1) | 22 (37.9) | 3.953 | 0.152 | ||
| ≥5 | 113 | 52 (46.0) | 61 (54.0) | ||||
| TNM stage | 0.071 | 0.072 | |||||
| I–II | 48 | 30 (62.5) | 18 (37.5) | 3.255 | 0.138 | ||
| III–IV | 123 | 58 (47.2) | 65 (52.8) | ||||
| Portal vein tumor thrombus | 0.829 | 0.831 | |||||
| Negative | 84 | 39 (46.4) | 45 (53.6) | 0.046 | 0.019 | ||
| Positive | 45 | 20 (44.4) | 25 (55.6) | ||||
| Vaso-invasion | 0.938 | 0.938 | |||||
| No | 77 | 35 (45.5) | 42 (54.5) | 0.006 | −0.007 | ||
| Yes | 52 | 24 (46.2) | 28 (53.8) | ||||
| Capsular | 0.972 | 0.972 | |||||
| Complete | 61 | 28 (45.9) | 33 (54.1) | 0.001 | 0.003 | ||
| No or infiltrate | 68 | 31 (45.6) | 37 (54.4) | ||||
| Metastasis | 0.041 | 0.041 | |||||
| No | 90 | 53 (58.9) | 37 (41.1) | 4.196 | 0.157 | ||
| Yes | 81 | 35 (43.2) | 46 (56.8) | ||||
| AFP | 0.453 | 0.457 | |||||
| Positive | 54 | 27 (50.0) | 27 (50.0) | 0.564 | 0.072 | ||
| Negative | 56 | 24 (42.9) | 32 (57.1) | ||||
Abbreviations: AFP, alpha fetoprotein; HCC, hepatocellular carcinoma; TNM, tumor–node–metastasis.
Figure 7.
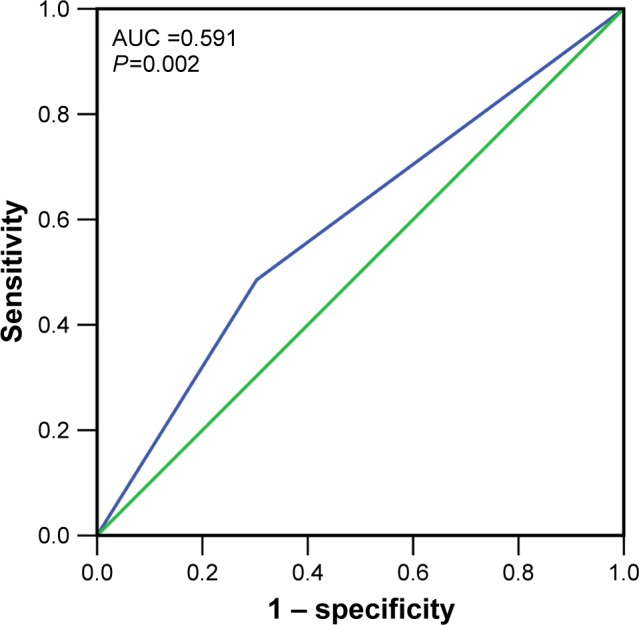
The ROC curves of IRAK1 expression by IHC in HCC.
Notes: The ROC curves indicated that IRAK1 (blue line) showed a weak diagnostic ability for HCC (AUC =0.591, P=0.002). Green line denotes reference line.
Abbreviations: ROC, receiver operating characteristic; IHC, immunohistochemistry; HCC, hepatocellular carcinoma; AUC, area under the curve.
IRAK1 expression detected by RNA-sequencing (RNA-seq) data in TCGA database
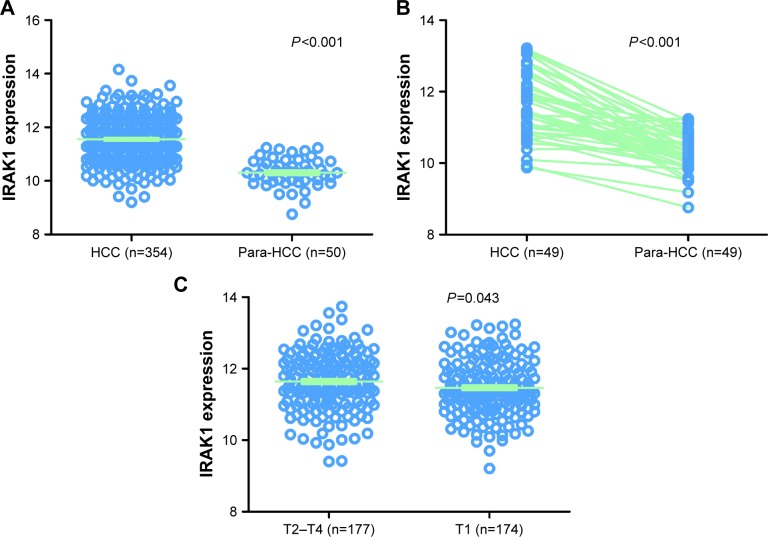
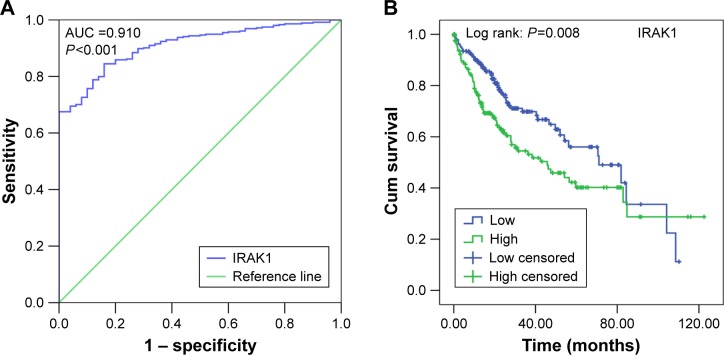
The clinical significance of IRAK1 expression in HCC based on TCGA data was investigated. The results revealed that an upregulation of IRAK1 was detected in 354 cases of HCC (10.3058±0.5212) compared with 50 adjacent normal liver tissues (11.5503±0.7946, P<0.001; Figure 8). Furthermore, a paired test of 49 pairs of HCC tissues and matched adjacent normal liver tissues showed a similar trend of IRAK1 expression between HCC and normal liver tissues (11.6174±0.8979 vs 10.2921±0.5174, respectively, P<0.001; Figure 8B). We also used the ROC curves to estimate the diagnostic value of IRAK1 in HCC. An AUC value of 0.910 indicated a high diagnostic value (95% CI: 0.878–0.942, P<0.001; Figure 9A).
Figure 8.
The expression of IRAK1in HCC in the TCGA data.
Notes: (A) The expression of IRAK1 was upregulated in 354 cases of HCC tissues (11.5503±0.7946) compared with 50 adjacent normal liver tissues (10.3058±0.5212, P<0.001). (B) The paired t-test on 49 pairs of HCC tissues and the corresponding adjacent normal liver tissues confirmed this finding (11.6174±0.8979 vs 10.2921±0.5174, P<0.001). (C) Moreover, this significant difference in IRAK1 expression was observed only in the advanced tumor stage (T2–T4: 11.6363±0.8153 vs T1: 11.4647±0.7677, P=0.043).
Abbreviations: HCC, hepatocellular carcinoma; TCGA, the cancer genome atlas.
Figure 9.
The ROC curve and Kaplan–Meier survival curve of IRAK1 expression for HCC from TCGA data.
Notes: (A) ROC curve. An AUC value of 0.910 indicated a strong diagnostic value (95% CI: 0.878–0.942, P<0.001). (B) Kaplan–Meier survival curve. The median survival time was 45.7 months for the IRAK1 high-expression group and 71.0 months for the IRAK low-expression group (log rank: P=0.008).
Abbreviations: ROC, receiver operating characteristic; HCC, hepatocellular carcinoma; TCGA, the cancer genome atlas; AUC, area under the curve; Cum, cumulative.
With regard to the relationships between IRAK1 expression and clinicopathological characteristics, the expression of IRAK1 significantly increased only in the advanced tumor stage (T2–T4: 11.6363±0.8153 vs T1: 11.4647±0.7677, P=0.043; Table 3 and Figure 8C).
Table 3.
Expression of IRAK1 detected by RNA sequencing and clinicopathological parameters in HCC in TCGA
| Clinicopathological parameters | n | IRAK1 expression
|
||
|---|---|---|---|---|
| Mean ± SD | t | P-value | ||
| Tissue (unmatched) | <0.001 | |||
| Adjacent noncancerous liver | 50 | 10.3058±0.5212 | 14.649 | |
| HCC | 354 | 11.5503±0.7946 | ||
| Tissue (matched) | <0.001 | |||
| Adjacent noncancerous liver | 49 | 10.2921±0.5174 | 10.703 | |
| HCC | 49 | 11.6174±0.8979 | ||
| Age (years) | 0.497 | |||
| <60 | 163 | 11.5168±0.7733 | −0.680 | |
| ≥60 | 190 | 11.5746±0.8132 | ||
| Gender | 0.116 | |||
| Male | 241 | 11.5958±0.8123 | 1.575 | |
| Female | 113 | 11.4534±0.7499 | ||
| AJCC pathologic T | 0.043 | |||
| T1 | 174 | 11.4647±0.7677 | −2.029 | |
| T2–T4 | 177 | 11.6363±0.8153 | ||
| AJCC pathologic N | 0.225 | |||
| N0 | 241 | 11.5107±0.7965 | F=1.498* | |
| N1 | 3 | 11.1300±0.4606 | ||
| NX | 109 | 11.6453±0.7940 | ||
| AJCC pathologic M | 0.149 | |||
| M0 | 256 | 11.5036±0.7794 | F=1.912* | |
| M1 | 4 | 11.3702±0.6400 | ||
| MX | 94 | 11.6853±0.8317 | ||
| Pathologic stage | 0.135 | |||
| I–II | 245 | 11.4982±0.7800 | −1.499 | |
| III–IV | 83 | 11.6407±0.8398 | ||
| Vaso-invasion | 0.704 | |||
| − | 196 | 11.4945±0.7900 | −0.380 | |
| + | 105 | 11.5311±0.7961 | ||
Note:
One-way ANOVA was performed.
Abbreviations: HCC, hepatocellular carcinoma; TCGA, the cancer genome atlas; AJCC, American Joint Committee on Cancer; ANOVA, analysis of variance; SD, standard deviation.
As for the prognostic value of IRAK1 for HCC, the cohort of 354 HCC patients was separated into two groups according to the cutoff value of the mean level of IRAK1 expression. The median survival time was 45.7 months in the IRAK1 high-expression group and 71.0 months in the IRAK1 low-expression group (log rank: P=0.008; Figure 9B).
IRAK1 alteration analyzed in cBioPortal database
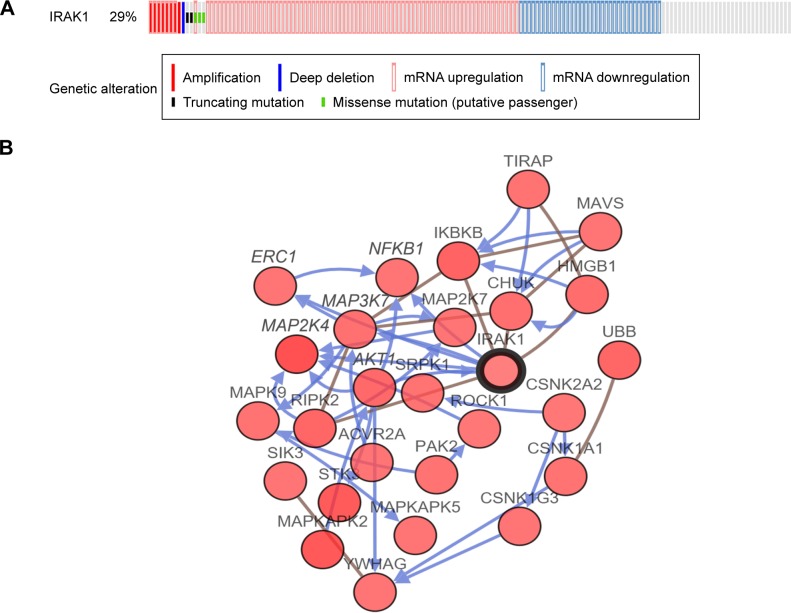
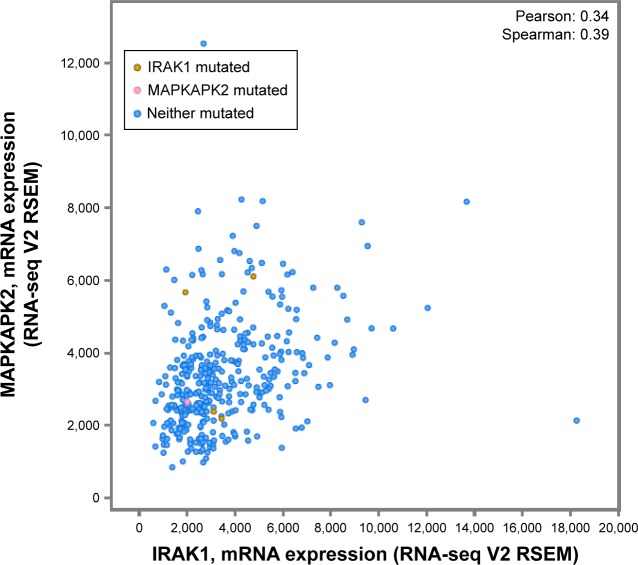
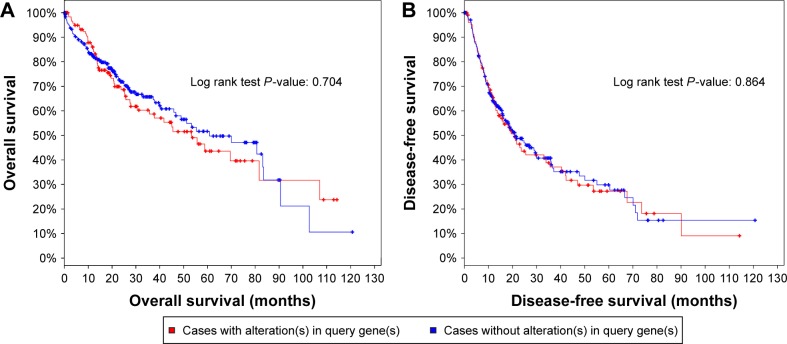
We investigated the genetic alteration of IRAK1 with a cohort of 440 cases of HCC available in the cBioPortal database. The result showed that 29% (126/440) cases of HCC in this cohort exhibited IRAK1 alteration, including 85 cases of mRNA upregulation, 35 cases of mRNA downregulation, eight cases of amplification, three cases of missense mutation, two cases of truncating mutation and one case of deep deletion. Among them, eight genes had the mixed type of alteration (Figure 10A). Moreover, a network of the interaction of IRAK1 with several frequently altered neighboring genes was drawn (Figure 10B). The network reflected clear interaction between IRAK1 and several genes, including MAP3K7, MAP2K4, NFKB1, AKT1 and ERC1. Moreover, Pearson correlation analysis was performed to investigate the correlations between IRAK1 expression and its potential targets in the network. The results revealed that the expressions of several target genes significantly correlated with IRAK1 expression (HMGB1: r=−0.13, MAP3K7: r=−0.15, ROCK1: r=−0.13, CSNK1A1: r=−0.15, CSNK1G3: r=−0.13, MAPKAPK5: r=0.18, ACVR2A: r=−0.19, MAPKAPK2: r=0.34, all P<0.05). Remarkably, MAPKAPK2 showed relatively a good correlation to IRAK1 (Figure 11). The survival analysis (overall survival and disease-free survival) was performed to compare the prognosis of the patient group with and without IRAK1 alteration. However, the results did not show statistical significance (Figure 12).
Figure 10.
The alteration of IRAK1 and its interaction in altered neighboring genes in 440 HCC cases from the cBioPortal database.
Notes: (A) IRAK1 alteration. In this cohort, 29% (126/440) of HCC cases exhibited IRAK1 alteration, including 85 cases of mRNA upregulation, 35 cases of mRNA downregulation, eight cases of amplification, three cases of missense mutation, two cases of truncating mutation and one case of deep deletion. Among them, eight cases possessed a mixed type of alteration. (B) Network of IRAK1 and altered neighboring genes. The network reflected easily identified interactions between IRAK1 and several genes, including MAP3K7, MAP2K4, NFKB1, AKT1 and ERC1.
Abbreviation: HCC, hepatocellular carcinoma.
Figure 11.
Correlation between IRAK1 and MAPKAPK2.
Note: Pearson correlation analysis revealed that IRAK1 expression was significantly correlated with MAPKAPK2 expression (r=0.34, P<0.05).
Abbreviation: RNA-seq, RNA sequencing.
Figure 12.
Analysis of the overall survival (A) and disease-free survival (B) of HCC patients with and without IRAK1 alteration.
Note: No statistically significant difference was identified.
Abbreviation: HCC, hepatocellular carcinoma.
Discussion
Using reverse transcription polymerase chain reaction (RT-PCR), Western blot and immunohistochemical staining, Li et al8 confirmed that IRAK1 exhibited a higher expression in 33 HCC cases and noted that IRAK1 may augment the malignant potential of HCC by stimulating the proliferation of cells and suppressing the apoptosis of cells. However, no study has elaborated on the diagnostic and prognostic value of IRAK1 for HCC or the potential molecular mechanisms of IRAK1 in the pathogenesis of HCC. To date, this study is the first one to comprehensively investigate the clinical significance of IRAK1 in HCC with an analysis of IHC, TCGA and gene alteration data from cBioPortal.
In agreement with the study by Li et al,8 the results of our study indicated that IRAK1 had a significantly higher expression in HCC tissues than in noncancer tissues of both the IHC and TCGA data. The large sample size of the current study may provide more powerful evidence to verify the upregulation of IRAK1 in HCC. We also observed an increasing trend of IRAK1 expression in normal liver tissues, liver cirrhosis tissues, para-HCC tissues and HCC tissues, which suggests that a higher expression of IRAK1 was closely associated with the malignant evolution of liver tissues. Thus, our results implied that IRAK1 may act as an oncogene in the tumorigenesis of HCC.
Consistent with an oncogenic effect of IRAK1 in HCC, the results obtained from the IHC and TCGA data in our study revealed that IRAK1 exhibited a significant diagnostic value for HCC, especially in TCGA data (AUC >0.9, P<0.001), which has not been reported in previous studies. Significantly, IRAK1 correlated well with the prognosis of HCC patients. Therefore, we anticipated that IRAK1 may serve as a potential diagnostic and prognostic tool for HCC. Further prospective study would be necessary to confirm the diagnostic and prognostic value of IRAK1 in HCC, especially in Asia.
To further explore the clinical significance of IRAK1 in HCC, we examined the relationships between IRAK1 expression and the clinicopathological variables of HCC. Analysis of our IHC and TCGA data revealed that IRAK1 expression was related to the malignant phenotype of HCC, including larger tumor sizes, tumor metastasis and advanced tumor stage, which was consistent with the previous results indicating that IRAK1 could strengthen the proliferative ability and inhibit the apoptosis of HCC cells.8 It can be deduced from our result that IRAK1 may promote the progression of HCC by enhancing the growth, invasion and metastasis of cancer cells.
The oncogenic roles of IRAK1 in other cancers have been widely studied. In T-cell acute lymphoblastic leukemia (T-ALL), IRAK1/4 signaling promoted the progression of T-ALL by activating TRAF6, which subsequently strengthened MCL1, a protein with antiapoptotic effect.24 In breast cancer, IRAK1 was reported to facilitate the growth of triple-negative breast cancer (TNBC) by inducing the expression of NF-κB-related cytokine.19 Toll-like receptors (TLRs) are important molecules that regulate the innate immunity and participate in tissue repair, autoimmune disease and tumor microenvironment.25,26 The study by Fonte et al indicated that TLR1/2, TLR2/6 and TLR9 stimulated the proliferation of cancer cells in splenic marginal zone lymphoma (SMZL). Moreover, the inhibition of IRAK1/4, upstream kinases of the TLR signaling pathway, reduced the cell viability induced by TLRs.27 Additionally, miR-146, which belongs to the miRNA family, has been shown to suppress the development of cancers, including gastric cancer, pancreatic cancer and breast cancer, which was implemented by targeting IRAK1.28–30 Thus, we assumed that IRAK1 may play an oncogenic role in HCC by regulating the expression of downstream molecules, such as MCL1 and NF-κB-related cytokine, and modulating the TLR signaling pathways. The aberrant expression of miR-146a may also lead to the carcinogenic function of IRAK1 in HCC. Further in vivo and in vitro studies on the molecular mechanisms of IRAK1 in HCC are needed to decipher the pathogenesis of HCC.
To facilitate the understanding of the function of IRAK1 in HCC, we further investigated the alteration of the IRAK1 gene in HCC. The results indicated that approximately one-third of the genes exhibited IRAK1 alterations, among which the mRNA upregulation was the predominant type of alteration, followed by mRNA downregulation, amplification and missense mutation. The upregulation of IRAK1 may cause an increased level of IRAK1 expression in HCC, as confirmed by our results. As for the missense mutation of IRAK1, a Phe196Ser change in IRAK1 was detected in primary effusion lymphoma (PEL),31 which caused the constitutive phosphorylation of IRAK1 in PEL and promoted the progression of PEL. We postulated that the specific missense mutation of IRAK1 existed in HCC to facilitate the development of IRAK1 via the phosphorylation of IRAK1. Then, we attempted to trace the causes of the IRAK1 alteration by investigating the interaction between the IRAK1 gene and the neighboring frequently altered genes. From the network, we found that several genes, including MAP3K7, MAP2K4, NFKB1, AKT1 and ERC1, interact directly with IRAK1. Moreover, MAPKAPK2 showed relatively good correlation to IRAK1 and is worth be validated in future work. Thus, we speculated that these genes may influence the alteration of IRAK1 gene and exert a carcinogenic effect on IRAK1 to promote the occurrence and development of HCC. However, the bioinformatics analysis may produce a result with weak evidence. Thus, whether these genes were involved in the alteration of IRAK1 gene needs further in vitro and in vivo validation in future studies.
Conclusion
IRAK1 was significantly overexpressed in HCC and showed promise as a diagnostic and therapeutic target for HCC with its oncogenic function in HCC.
Acknowledgments
The study was supported by the fund of Youth Science Foundation of Guangxi Medical University (GXMUYSF201624). The authors thank the American Journal Experts (AJE) for editing the manuscript.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist. 2010;15(suppl 4):5–13. doi: 10.1634/theoncologist.2010-S4-05. [DOI] [PubMed] [Google Scholar]
- 4.White DL, Thrift AP, Kanwal F, Davila J, El-Serag HB. Incidence of hepatocellular carcinoma in all 50 United States, from 2000 through 2012. Gastroenterology. 2016 Nov 23; doi: 10.1053/j.gastro.2016.11.020. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balogh J, Victor D, 3rd, Asham EH, et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41–53. doi: 10.2147/JHC.S61146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colagrande S, Inghilesi AL, Aburas S, Taliani GG, Nardi C, Marra F. Challenges of advanced hepatocellular carcinoma. World J Gastroenterol. 2016;22(34):7645–7659. doi: 10.3748/wjg.v22.i34.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janssens S, Beyaert R. Functional diversity and regulation of different interleukin-1 receptor-associated kinase (IRAK) family members. Mol Cell. 2003;11(2):293–302. doi: 10.1016/s1097-2765(03)00053-4. [DOI] [PubMed] [Google Scholar]
- 8.Li N, Jiang J, Fu J, et al. Targeting interleukin-1 receptor-associated kinase 1 for human hepatocellular carcinoma. J Exp Clin Cancer Res. 2016;35(1):140. doi: 10.1186/s13046-016-0413-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao Z, Henzel WJ, Gao X. IRAK: a kinase associated with the interleukin-1 receptor. Science. 1996;271(5252):1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 10.Flannery S, Bowie AG. The interleukin-1 receptor-associated kinases: critical regulators of innate immune signalling. Biochem Pharmacol. 2010;80(12):1981–1991. doi: 10.1016/j.bcp.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 11.Wesche H, Henzel WJ, Shillinglaw W, Li S, Cao Z. Pillars article: MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity. 1997;7:837–847. [PubMed] [Google Scholar]; J Immunol. 2013;190(1):5–15. [PubMed] [Google Scholar]
- 12.Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13(2):85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 13.Zhang G, Ghosh S. Toll-like receptor-mediated NF-kappaB activation: a phylogenetically conserved paradigm in innate immunity. J Clin Invest. 2001;107(1):13–19. doi: 10.1172/JCI11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhyasen GW, Bolanos L, Fang J, et al. Targeting IRAK1 as a therapeutic approach for myelodysplastic syndrome. Cancer Cell. 2013;24(1):90–104. doi: 10.1016/j.ccr.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivastava R, Geng D, Liu Y, et al. Augmentation of therapeutic responses in melanoma by inhibition of IRAK-1,-4. Cancer Res. 2012;72(23):6209–6216. doi: 10.1158/0008-5472.CAN-12-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behrens C, Feng L, Kadara H, et al. Expression of interleukin-1 receptor-associated kinase-1 in non-small cell lung carcinoma and preneoplastic lesions. Clin Cancer Res. 2010;16(1):34–44. doi: 10.1158/1078-0432.CCR-09-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Dang Y, Li P, Rong M, Chen G. Expression of IRAK1 in lung cancer tissues and its clinicopathological significance: a microarray study. Int J Clin Exp Pathol. 2014;7(11):8096–8104. [PMC free article] [PubMed] [Google Scholar]
- 18.Scheeren FA, Kuo AH, van Weele LJ, et al. A cell-intrinsic role for TLR2-MYD88 in intestinal and breast epithelia and oncogenesis. Nat Cell Biol. 2014;16(12):1238–1248. doi: 10.1038/ncb3058. [DOI] [PubMed] [Google Scholar]
- 19.Wee ZN, Yatim SM, Kohlbauer VK, et al. IRAK1 is a therapeutic target that drives breast cancer metastasis and resistance to paclitaxel. Nat Commun. 2015;6:8746. doi: 10.1038/ncomms9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li JJ, Luo J, Lu JN, et al. Relationship between TRAF6 and deterioration of HCC: an immunohistochemical and in vitro study. Cancer Cell Int. 2016;16:76. doi: 10.1186/s12935-016-0352-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Liang L, Liu Y, et al. Clinicopathological significance of STAT4 in hepatocellular carcinoma and its effect on cell growth and apoptosis. Onco Targets Ther. 2016;9:1721–1734. doi: 10.2147/OTT.S100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269) doi: 10.1126/scisignal.2004088. pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Younger K, Gartenhaus R, et al. Inhibition of IRAK1/4 sensitizes T cell acute lymphoblastic leukemia to chemotherapies. J Clin Invest. 2015;125(3):1081–1097. doi: 10.1172/JCI75821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Neill LA, Golenbock D, Bowie AG. The history of toll-like receptors – redefining innate immunity. Nat Rev Immunol. 2013;13(6):453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 26.Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nat Rev Cancer. 2009;9(1):57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- 27.Fonte E, Agathangelidis A, Reverberi D, et al. Toll-like receptor stimulation in splenic marginal zone lymphoma can modulate cell signaling, activation and proliferation. Haematologica. 2015;100(11):1460–1468. doi: 10.3324/haematol.2014.119933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kogo R, Mimori K, Tanaka F, Komune S, Mori M. Clinical significance of miR-146a in gastric cancer cases. Clin Cancer Res. 2011;17(13):4277–4284. doi: 10.1158/1078-0432.CCR-10-2866. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Vandenboom TG, 2nd, Wang Z, et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70(4):1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27(42):5643–5647. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang D, Chen W, Xiong J, Sherrod CJ, Henry DH, Dittmer DP. Interleukin 1 receptor-associated kinase 1 (IRAK1) mutation is a common, essential driver for Kaposi sarcoma herpesvirus lymphoma. Proc Natl Acad Sci U S A. 2014;111(44):E4762–E4768. doi: 10.1073/pnas.1405423111. [DOI] [PMC free article] [PubMed] [Google Scholar]