Abstract
BACKGROUND & AIMS
We performed a genome-wide association study (GWAS) to identify genetic risk factors for drug-induced liver injury (DILI) from licensed drugs without previously reported genetic risk factors.
METHODS
We performed a GWAS of 862 persons with DILI and 10588 population-matched controls. The first set of cases was recruited prior to May 2009 in Europe (n=137) or the USA (n=274). The second set of cases were identified from May 2009 through May 2013 from international collaborative studies performed in Europe, the USA and South America. For the GWAS, we included only cases of European ancestry associated with a particular drug (but not flucloxacillin or amoxicillin-clavulanate). We used DNA samples from all subjects to analyze human leukocyte antigen (HLA) genes and single nucleotide polymorphisms (SNPs). After the discovery analysis was concluded, we validated our findings using data from 283 European patients with diagnosis of DILI associated with various drugs.
RESULTS
We associated DILI with rs114577328 (a proxy for A*33:01 a HLA class I allele; odds ratio [OR], 2.7; 95% CI, 1.9–3.8; P=2.4×10−8) and with rs72631567 on chromosome 2 (OR, 2.0; 95% CI, 1.6–2.5; P=9.7×10−9). The association with A*33:01 was mediated by large effects for terbinafine-, fenofibrate-, and ticlopidine-related DILI. The variant on chromosome 2 was associated with DILI from a variety of drugs. Further phenotypic analysis indicated that the association between DILI and A*33:01 was significant, genome wide, for cholestatic and mixed DILI, but not for hepatocellular DILI; the polymorphism on chromosome 2 associated with cholestatic and mixed DILI as well as hepatocellular DILI. We identified an association between rs28521457 (within the LRBA gene) and only hepatocellular DILI (OR, 2.1; 95% CI, 1.6–2.7; P=4.8×10−9). We did not associate any specific drug classes with genetic polymorphisms, except for statin-associated DILI, which was associated with rs116561224 on chromosome 18 (OR=5.4; 95% CI, 3.0–9.5; P=7.1×10−9). We validated the association between A*33:01 terbinafine- and sertraline-induced DILI. We could not validate the association between DILI and rs72631567, rs28521457, or rs116561224.
CONCLUSIONS
In a GWAS of persons of European descent with DILI, we associated HLA-A*33:01 with DILI due to terbinafine and possibly fenofibrate and ticlopidine. We identified polymorphisms that appear to be associated with DILI from statins, as well as 2 non–drug-specific risk factors.
Keywords: medication, liver damage, side effect, anti-fungal agent
Introduction
Hepatotoxicity is the second most common cause of drug attrition during development as well as for post-marketing withdrawal,1 and idiosyncratic drug-induced liver injury (DILI) accounts for 11%-17% of cases of acute liver failure in the United States and Europe.2, 3 The typical incidence of DILI varies from approximately 1% with the anti-tumor necrosis factor agents4 to 0.04% with some widely used antimicrobials such as amoxicillin-clavulanate.5 During the past 15 years, increasing progress on identifying genetic risk factors for DILI has been made. In particular, associations with HLA class I and II alleles have been reported for DILI caused by a range of drugs, though a particular HLA genotype does not appear to be relevant to all forms of idiosyncratic DILI.6
Previously, GWAS involving cohorts of DILI cases related to one particular drug only have resulted in identification of one or more drug-specific HLA risk alleles.7-11 A large study involving 783 DILI cases due to a range of different drugs also resulted in a genome-wide significant HLA signal, but this association was abolished once 296 cases of DILI due to flucloxacillin and amoxicillin-clavulanate were excluded.12 This partly reflects the fact that amoxicillin-clavulanate is a very common cause of DILI worldwide and flucloxacillin is an equally common cause in a number of Northern European countries.13 Therefore, DNA collections from DILI cases generally will be highly enriched in cases relating to these two drugs, making detection of associations related to other compounds more difficult.
We have expanded our previous study of DILI caused by a range of different drugs,12 and after excluding cases relating to amoxicillin-clavulanate and flucloxacillin, we have more than doubled the number of cases with additions from Europe, Australia, South America and the United States. We now report that HLA-A*33:01 is associated with risk of DILI, particularly due to terbinafine, fenofibrate and ticlopidine and especially with a cholestatic or mixed phenotype. We have also found novel non-major histocompatibility complex (MHC) related signals apparently shared across a range of different drugs; an intronic SNP, in the LPS-responsive vesicle trafficking, beach and anchor containing (LRBA) gene is associated with hepatocellular DILI and an intergenic SNP on chromosome 2, rs72631567, with DILI generally. An additional drug-specific genome-wide significant signal which could not be confirmed is also reported.
Materials and Methods
DILI discovery cohort
The cases in the study were from two separate recruitment phases. Phase I consists of 411 cases included in a previous study (from DILIN, DILIGEN and Eudragene)12 and phase II more recently recruited cases (n=451) of which a small subset was included in a recent report.14
Phase I cases
These cases included 413 DILI cases not due to amoxicillin-clavulanate or flucloxacillin, with a defined casual drug and with causality score greater than possible (RUCAM score≥3) recruited in Europe (n=137) or the USA (n=274) prior to 2009. Clinical characteristics of these cases and methods used for genotyping have been described in detail previously.12 Additional exome chip analysis (Illumina Infinium HumanCoreExome BeadChip) was performed on 150 of these 413 cases at the Broad Institute, Boston.
Phase II case recruitment-iDILIC
The iDILIC cases were recruited between May 2009 and May 2013 as part of an international collaborative study involving recruitment centers in the United Kingdom (Newcastle, Nottingham, Liverpool, London, Dundee), Sweden (Uppsala and Gothenburg), Spain (Malaga and Barcelona), France (Montpellier), the Netherlands (Utrecht), Germany (Kiel), Australia (Brisbane), Switzerland (Zurich), Finland (Helsinki), Argentina (Rosario), Uruguay (Montevideo) and Chile (Santiago). All participants provided written informed consent and each study had been approved by the appropriate national or institutional ethical review boards. For the GWAS, only cases of European ancestry where there were at least 2 cases due to a particular drug available (when phase I cases from Europe and the USA were also considered) and where the DILI was not due to either flucloxacillin or amoxicillin-clavulanate were included (n=339). Clinical inclusion criteria for all cases were those described by Aithal et al.15
Phase II case recruitment-DILIN
Details of the USA-based DILIN prospective study including IRB approval information have been described previously.16 A total of 112 eligible new cases of European ancestry and ≥ 18 years were included in the current GWAS. These new cases were selected from the larger DILIN sample collection such that only cases relating to drugs also included among the iDILIC cases were represented. Laboratory inclusion criteria were as described previously.16 Patients were excluded if there was known or suspected acetaminophen overdose, if there was a history of bone marrow or liver transplant prior to DILI onset or if there was a prior history of immune-related liver disease such as autoimmune hepatitis.
Additional cases used for confirmation of associations
After the discovery analysis was concluded, we enrolled an additional 283 European patients with diagnosis of DILI across multiple causal drugs (6 from iDILIC and 277 from DILIN networks recruited subsequent to the GWAS). The causal drug distribution is reported in Table S1A. An additional 12 statin DILI samples from the Spanish iDILIC network and 3 UK-DILIGEN cases were recruited later in the study to confirm the class specific association (Table S1B).
Out of the 283, we used 272 DILI cases to directly type SNPs associated across multiple drugs or specific for drugs/drug classes and 11 DILI cases for HLA typing to confirm HLA associations. An additional Chinese terbinafine DILI sample was also HLA typed.
Causality assessment
The iDILIC cases were evaluated by application of the Council for International Organizations of Medical Science (CIOMS) scale, also called the Roussel Uclaf Causality Assessment Method (RUCAM)15 and by expert review by a panel of three hepatologists. The pattern of liver injury was classified according to the International Consensus Meeting Criteria.17 Only cases having at least possible causality (score ≥3) were included in the study. For all cases in DILIN, causality assessment was by expert consensus as previously described.16
Controls
Since DILI has a very low prevalence, we used general population samples as study controls. We selected 10588 European ancestry controls from multiple available sources; Welcome Trust Case Control Consortium (WTCCC) (http://www.wtccc.org.uk), the population reference sample (POPRES)18, PGX4000119 and Spanish Bladder cancer cohort (phs000346.v1) from dbGAP.20 In order to increase the case/control ratio for Italian, Spanish and Swedish, we added samples from Hypergenes cohort (http://www.hypergenes.eu/dissemination.html#pub), the National Spanish DNA Bank (http://www.bancoadn.org/), Italian Penicillin Tolerant Controls (IPTC), and the Swedish Twin Registry (http://ki.se/en/research/the-swedish-twin-registry).
Genotyping
DNA preparation from Phase II cases
For iDILIC cases, DNA was prepared as described previously.8 DILIN DNA was extracted from lymphocytes and stored at the NIDDK biosample repository at Rutgers University, Piscataway, NJ.
Genome-wide analysis
Genome-wide genotyping of the phase II and 150 phase I cases was performed by the Broad Institute, Boston by Illumina Infinium HumanCoreExome BeadChip. iDILIC and DILIN cases were genotyped in two separate batches. A total of 505740 markers shared across the batches passed quality control (QC) and no samples were excluded for low quality profile. (see Supplemental Materials and Methods). Details on the genotype data available for each control collection are reported in Table S2.
Imputation
SNP imputation was performed in batches dividing the cohorts according to genotyping platforms. Imputation methods are described in detail in the Supplementary Appendix. For HLA genotypes, four digit HLA alleles were inferred using HIBAG.21
SNP genotyping
The top associated imputed SNPs were validated by SNP genotyping in subsets of iDILIC cases and in the overall DILIN cohort (see Supplementary Appendix). The SNPs were further confirmed in additional cases using TaqMan® predesigned and custom SNP genotyping assays (ThermoFisher Scientific, Waltham, MA) in accordance with the manufacturer's recommendations.
HLA genotyping
High resolution genotyping of HLA-A, B, C, DRB1, DQA1 and DQB1 was performed on selected cases by Histogenetics (Ossining, New York). Sequencing data files were analyzed using Histogenetics’ proprietary analysis software (Histomatcher and HistoMagic) for HLA genotype calling. Allele assignments are based on IMGT/HLA Database release version 2.21.0, dated April 2008 (http://www.ebi.ac.uk/imgt/hla/).
Statistical analysis
The effect of population structure was assessed through principal components analysis (PCA) using the smartPCA program from the EIGENSTRAT package (version 3.0).22 Single marker and haplotype association analyses and heterogeneity test analyses were carried out by PLINK.23 The statistical association of each marker, HLA alleles and SNPs, was determined in a logistic regression framework with scores for the first seven principal components as covariates under an additive model using PLINK. We used the same statistical test for sub-population analyses, using two, seven and ten most significant principal components as covariates in Italian, Spanish and North European populations, respectively. We set the genome-wide traditional significance p-value threshold to 5.0×10−8 to correct for multiple testing.24 When we obtained genome-wide significant signals, we tested for independent effects from the neighboring variants by including the most associated variants as a covariate and then testing the significance of others in the region. We also tested interaction effects among them by including interaction terms in the logistic regression. Differences in clinical characteristics among sample groups were tested by Fisher's exact test. All detailed analyses and Manhattan plots were performed with R (Version 3.0.2).25 Regional plots were drawn by LocusZoom.26
Results
Clinical characteristics of the cases
Clinical details of the DILI cases included in the main GWAS are summarized in Table 1. A variety of different causative drugs were represented but the most common was diclofenac with 67 cases, followed by nitrofurantoin with 64 cases. A few drugs, including azathioprine, isoniazid, fenofibrate, and diclofenac had significantly disproportionate number of cases in one of the two recruitment phases. Details of all the causative drugs are shown in Table S3.
Table 1.
Clinical details of the DILI cases included in the GWAS
| CHARACTERISTICS | Phase 1 (n=411) |
Phase 2 (n=451) |
Combined (n=862) |
|---|---|---|---|
| Clinical information | |||
| Age (mean years) | 51 | 54 | 53 |
| % Female | 63.0% | 60.5% | 61.8% |
| ALT (mean IU/L) | 895.1 | 757.7 | 822.2 |
| ALP (mean IU/L) | 388.2 | 282.5 | 330.6 |
| Latency (mean days) | 201.7 | 177.4 | 188.2 |
| Injury type | |||
| Cholestatic | 76 | 87 | 163 |
| Hepatocellular | 202 | 272 | 474 |
| Mixed | 69 | 91 | 160 |
| Not available* | 64 | 1 | 65 |
| Genotype chip | |||
| Illumina 1 M | 261 | 261 | |
| Illumina 1M/Illumina Infinium
HumanCoreExome BeadChip |
150 | 150 | |
| Illumina Infinium HumanCoreExome BeadChip | 447 | 447 | |
| Illumina HumanOmniExpress BeadChip | 4 | 4 | |
| Country of birth | |||
| USA | 274 | 112 | 386 |
| UK | 71 | 79 | 150 |
| Spain | 16 | 95 | 111 |
| Sweden | 81 | 81 | |
| France | 30 | 7 | 37 |
| Germany | 20 | 20 | |
| Italy | 16 | 1 | 17 |
| Others | 4 | 56 | 60 |
Because of the retrospective nature of the phase I study, minimal clinical information needed to establish the type of injury were not available for a subset of initial NSAID DILI cases from DILIGEN because of missing ALP and upper limit of normal values.
Overall analysis
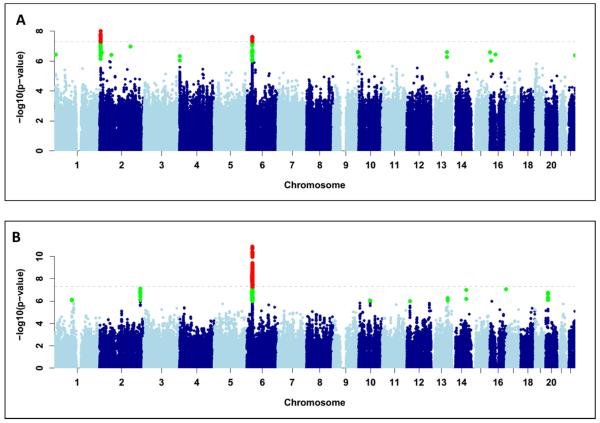
The discovery cohort included 862 European ancestry DILI cases (411 from phase I12 and 451 from Phase II) and 10,588 controls. PCA showed that all cases (including those from South America) clustered within three major groups (Italian, Spanish and Northern European) and matched with the population controls (Figure S1A). Consistent with the previous study,12 phase I cases were predominantly Northwest European. The most significant genome-wide associated SNPs were rs72631567 on chromosome 2 (OR=2.0, 95% CI =1.6-2.5, p-value=9.7×10−9) and rs114577328 in the MHC region of chromosome 6 (OR=2.7, 95% CI=1.9-3.8, p-value=2.4×10−8)(See Figure 1A, Table 2 and Figures S2 and S3). Data for both SNPs had been obtained by imputation in cases and controls and subsequently validated by SNP typing (see Supplementary Methods). The associations were consistent among geographic clusters and study phases (Table S4) and not due to artefact/s of population structure, missing genotypes rate (Table S5) or variability in imputation quality among populations or genotyping platforms (see Supplementary Methods).
Figure 1.
Manhattan plots displaying the association results of (A) the overall analysis (n=864); (B) terbinafine only cases (n=14 cases). SNPs in green have a significance level less than 5×10−6 and red have a significance level less than 5×10−8.
Table 2.
Association effect size of rs72631567, A*33:01 and rs28521457 across different liver injury patterns
| COHORTs | Variant | OR | 95% CI | P | AF Cases |
AF Controls |
|---|---|---|---|---|---|---|
|
Entire
DILI cohort |
rs72631567 | 2.0 | 1.6-2.5 | 9.7×10−9 | 0.05 | 0.03 |
| A*33:01 | 2.6 | 1.8-3.7 | 7.0×10−8 | 0.02 | 0.01 | |
| rs28521457 | 1.5 | 1.3-1.9 | 5.1×10−5 | 0.06 | 0.04 | |
|
| ||||||
|
Cholestatic
and Mixed DILI cohort |
rs72631567 | 2.4 | 1.7-3.4 | 9.5×10−7 | 0.06 | 0.03 |
| A*33:01 | 5.0 | 3.3-7.9 | 4.2×10−13 | 0.04 | 0.01 | |
| rs28521457 | 1.0 | 0.7-1.5 | 0.9 | 0.04 | 0.04 | |
|
| ||||||
|
Hepatocellular
DILI cohort |
rs72631567 | 1.6 | 1.2-2.3 | 2.5×10−3 | 0.04 | 0.03 |
| A*33:01 | 1.5 | 0.8-2.6 | 0.19 | 0.01 | 0.011 | |
| rs28521457 | 2.1 | 1.6-2.7 | 4.8×10−9 | 0.08 | 0.040 | |
COHORTs = type of comparison; Variant = associated variant; OR=Odds Ratio; 95% CI = 95% Confidence Interval of the odds ratio; P=logistic p-value; AF = allele frequency
For the chromosome 2 SNP rs72631567, breakdown by drug showed that 10 unrelated drug causes had an OR greater than 2.0 with at least two carriers (Table S6). Ciprofloxacin-related cases showed the strongest association (n=21, OR=7.4, 95% CI=17.3-161, p-value = 4.0×10−6).
The chromosome 6 SNP rs114577328 is the SNP proxy of an uncommon HLA class I allele, HLA-A*33:01. Indeed this SNP was in near-perfect LD with A*33:01 (r2=0.98). From the imputed HLA allele assignments, a strong association with DILI for this allele is confirmed (OR=2.6; 95% CI=1.8-3.7, p-value=8.0×10−8, Figure S4). Including rs114577328 or A*33:01 as a covariate removed any association in the MHC region, indicating that there is only one MHC association signal (Figure S5). The A*33:01 association appears independent of the chromosome 2 signal, since rs72631567 when conditioned on A*33:01 showed an almost unchanged effect size (ORrs72631567 = 1.7, 95% CI= 1.25-2.2, p-value = 0.0006). There was no statistically significant interaction effect between the two signals (p-value = 0.5).
Breakdown by drug showed DILI due to terbinafine was most strongly associated with the HLA-A*33:01 signal (OR=40.5, 95% CI=12.5-131.4, p-value=6.7×10−10) and a similarly strong association was seen with rs114577328 (OR=58.7, 95% CI=18.31-188.2, p-value=7.3×10−12, Figure 1B and Figure S6). As summarized in Table 3, in addition to terbinafine cases, cases due to six additional drugs showed an association with A*33:01 with p-values lower than 0.01. The largest case subset related to terbinafine but we found that A*33:01 was also a risk factor for ticlopidine (OR=163.1, 95% CI=16.2-1642.0, p-value=0.00002), methyldopa (OR=97.8, 95% CI=12.3-743.0, p-value=0.00001) and fenofibrate DILI (OR=58.7; 95% CI=12.3-279.8; p-value=3.2×10−7). Indeed, although fewer positive carriers were observed, A*33:01 also seems to be a common risk factor for enalapril (OR=34.8), sertraline (OR=29) and erythromycin DILI (OR=10.2). An erythromycin case was positive for A*33:03, an allele rare in European population controls (AF=0.002) which belongs to the A*33 group. Overall we found that 87% (n=36) of the A*33:01 positive carriers were also positive for HLA-B*14:02 and HLA-C*08:02. The haplotype showed a larger OR than A*33:01 as single marker in terbinafine (ORhaplotype =49.2, p-value=9.54×10−11), ticlopidine (ORhaplotype =201; p-value= 7.2×10−6), fenofibrate (ORhaplotype 68.5; p-value=1.1×10−7) and erythromycin (ORhaplotype = 13.1; p-value=0.002) DILI but not with DILI as a phenotype (ORhaplotype =2.7; p-value= 1.6×10−7, Table S7).
Table 3.
Association effect size of A*33:01 signal for the causal drugs enriched in A*33:01 positive carriers
| DRUGs | Number of cases tested |
Number of A*33:01 alleles in cases |
OR | 95% CI | P | CF |
|---|---|---|---|---|---|---|
| TICLOPIDINE | 5 | 4 | 163.1 | 16.2- 1642 |
0.00002 | 0.8 |
| METHYLDOPA | 4 | 2 | 97.8 | 12.8- 743.8 |
0.00001 | 0.5 |
| FENOFIBRATE | 7 | 4 | 58.7 | 12.3- 279.8 |
3.2*10−7 | 0.43 |
| TERBINAFINE | 14 | 6 | 40.5 | 12.5- 131.4 |
6.7*10−10 | 0.43 |
| ENALAPRIL | 4 | 2 | 34.8 | 3.9-302.9 | 0.001 | 0.5 |
| SERTRALINE | 5 | 2 | 29 | 4-207.2 | 0.0008 | 0.4 |
| ERYTHROMYCIN | 10 | 2 | 10.2 | 2-51.7 | 0.005 | 0.2 |
DRUGs=Causal drug involved; OR=Odds Ratio; 95% CI = 95% Confidence Interval of the odds ratio; P=logistic p-value; CF= Carrier Frequency
We verified the imputed A*33:01 genotype by sequence-based HLA typing in 35 cases related to the main A*33:01-associated drugs (Table S8). The A*33:01 predictions were confirmed in all cases except that one methyldopa case was negative for this allele (false positive) and an additional terbinafine case was a carrier (false negative). This validation result suggests that methyldopa might not share the HLA risk factor. The validation confirmed that all the A*33:01-positive terbinafine cases carried the complete HLA A*33:01-B*14:02-C*08:02 haplotype, increasing the strength of the haplotype association in the terbinafine DILI cases (ORhaplotype=70; p-value=8.7×10−13) and in the overall analyses (ORhaplotype=2.8; p-value=5.1×10−8). We also typed the A*33:01 proxy SNP across DILIN cases to confirm imputed genotypes. We found only one new carrier of the minor allele, not related to the major-A*33:01 associated drugs.
Analysis by type of injury and causative drugs
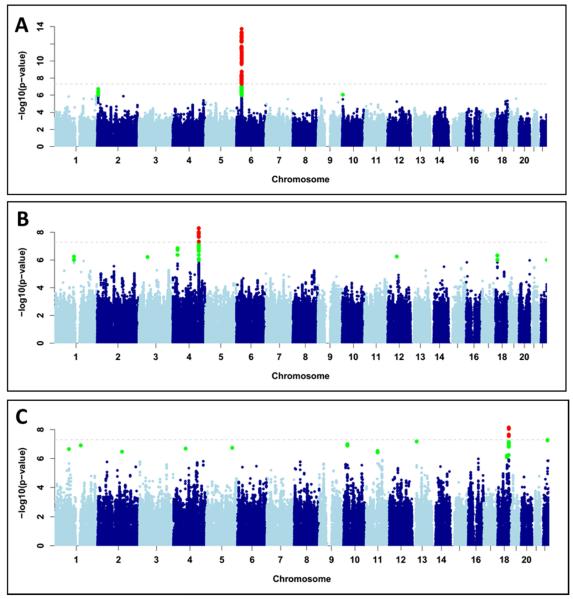
We further investigated the association of genotypes with particular patterns of DILI by grouping the cases into hepatocellular (HC) and cholestatic/mixed (CM) pattern. The chromosome 2 association described above was similar in the two phenotypic categories (direct comparison between CM cases vs HC cases, logistic p-value=0.5), although the effect was marginally stronger in the CM cases (Table 2). The association with rs114577328 was genome-wide significant only in the CM cases (n=323, OR=5.3, 95% CI=3.4-8.2, p-value=4.5×10−14, Figure 2B and Figure S6) and similarly with A*33:01 (OR=5.1; 95% CI=3.3-7.9, p-value=4.2×10−13, Figure 2B and Figure S6). Conditional analysis on the variant and HLA allele indicated only one genetic association was present in the region, as shown for the main analysis (Figure S7). There was no association between the proxy SNP or A*33:01 in the HC cases (n=474, OR for A*33:01=1.5, 95% CI=0.82-2.6, p-value=0.19, Table 2). The A*33:01-B*14:02-C*08:02 haplotype showed an ORhaplotype = 5.6; p-value = 2.5×10−13 in CM cases.
Figure 2.
Manhattan plot displaying the association results for (A) Cholestatic/Mixed only cases (n=323); (B) Hepatocellular only cases (n=474 cases); (C) Statin cases (n=59). SNPs in green have a significance level less than 5×10−6 and red have a significance level less than 5×10−8.
The CM only terbinafine-specific OR increased two fold compared with the value for all terbinafine cases (OR=88.1, 95% CI = 19.3-402.4, p-value = 7.5×10−9) since all the A*33:01 carriers belonged to this injury type. Following the injury correlation pattern established for terbinafine, A*33:01 appeared to be a stronger risk factor for CM injury than for HC injury also for fenofibrate, ticlopidine, enalapril and erythromycin-related DILI. This was not the case for injury due to sertraline and methyldopa. These top seven drugs account for 51% (n=21) of all A*33:01 positive cases (Table S9). Sixteen other drugs account for the rest of the carriers showing slight enrichment in CM phenotypes, which showed a marginal association with A*33:01 (OR = 2.6, 95% CI = 1.4-4.9, p-value = 0.003, Table S9 and Table S10).
We detected a new HC-specific genome-wide significant signal on chromosome 4 (Figure 2B). The signal lies within the LRBA (LPS-responsive vesicle trafficking, beach and anchor containing) gene with the imputed variant rs28521457, located in an intronic region, the most significant SNP (OR=2.1, 95% CI=1.6-2.7, p-value=4.8×10−9)(Table 2 and Figure 2B). The allele frequency for this SNP in the CM cases (0.04) was comparable to that in controls with no evidence of association with this phenotype. The risk allele was carried by more than 4% of the HC cases in cases due to a total of 45 drugs but in general, there were no drug-specific signals (Table S11).
We also investigated associations with particular causative drugs or specific therapeutic classes where a group including more than 40 samples was available. Detail on the groups studied is summarized in Table S12. Genome-wide significance was detected only for one group examined, the statins, with no significant signals for the other drug classes (Figure S8 and Table S13). In the case of the statins, rs116561224, a common intergenic SNP on chromosome 18, was genome-wide significant (OR=5.4, 95% CI=3.0-9.5, p-value=7.1×10−9, Figure 2C and Figure S9) with the signal mainly driven by simvastatin (Table S14).
Confirmation of associations
The European cohort used to confirm the associations (n=283) had a wider range of causal drugs, mostly different from the discovery cohort (Table S1). Later in time, we had access to 15 additional cases relating specifically to the statin cohort.
The A*33:01 association was further investigated in the additional cases by directly genotyping rs114577328 in 272 cases and by direct HLA typing on 11 additional samples who developed DILI due to drugs for which we had detected an enrichment in A*33:01 alleles in the discovery cases. Overall, the rs114577328 carriers were enriched in cases from drugs previously associated with the allele (Table S15). Eight out of all 23 additional cases relating to drugs previously associated with A*33:01 were shown to carry this allele or the proxy SNP (allele frequency 0.17) compared with an expected population frequency of 0.01. We specifically confirmed the association of A*33:01/rs114577328 with terbinafine having a carrier frequency of 0.63 (5 out 8 terbinafine-related cases across both the injury types) and with sertraline at a carrier frequency of 0.75 (3 out 4 sertraline-related cases) (Table S15). Although fenofibrate had a high carriage rate for A*33:01 in the discovery cohort, none of the 7 additional cases carried this allele or the proxy SNP. Few additional cases were available for other A*33:01-related drugs to confirm the association.
Interestingly, a terbinafine DILI case from Finland was positive for A*33:05, a very rare allele in the general European population (AF = 0.0001, USA NMDP European Caucasian in http://www.allelefrequencies.net/, n=1,242,890) and Finnish27 populations. An additional terbinafine DILI case of Chinese origin was positive for A*33:03. In total, 10 of the 24 additional cases (23 European cases and one single Chinese case) were carriers of an A*33 allele, in line with expectations based on the effects observed in the discovery sample.
We further genotyped rs72631567 and rs28521457 in 272 additional European cases. The rs72631567 and rs28521457 variants were found at AFs comparable to those for controls (AFrs72631567 = 0.022 and AFrs28521457 in HC only = 0.025) and so the association was not confirmed. However, rs72631567 carriers were slightly enriched in ciprofloxacin, atorvastatin and mercaptopurine-induced DILI cases, as in the discovery cohort, with ORs in the same direction in both cohorts (Table S16). Similarly, rs28521457 carriers seemed to be more common in the same subgroup of causal drugs in both cohorts (Table S17). This suggested a limited replication of the signal for these drugs.
We also attempted to confirm the rs116561224 signal for statins. The number of additional cases available for this purpose was small (n=29, Table S1b) with only four simvastatin cases. None of the statin cases were positive for rs116561224 so the signal could not be confirmed.
Discussion
Our previous studies have been successful in identifying genetic risk factors for both flucloxacillin and amoxicillin-clavulanate DILI.4, 6 However, our most recent GWAS did not identify any risk factors that were common for DILI in general or specific genetic risk factors for DILI due to individual drugs which accounted for a smaller number of cases of DILI.12 The current study included 451 additional cases of DILI due to a wide variety of causative drugs, including at least 10 DILI cases relating to each of 22 different drugs. This increase in numbers and the exclusion of the amoxicillin-clavulanate and flucloxacillin cases together with use of improved imputation methods has enabled the detection and confirmation of a novel genome-wide significant signal relating to a relatively rare HLA class I allele A*33:01. Though three other interesting signals were detected in the course of the study, an intergenic signal on chromosome 2, an intronic SNP in LRBA in HC cases only and a signal on chromosome 18 for statins, the failure to confirm these signals is a limitation. There are some indications that, as observed for HLA-A*33:01, the chromosome 2 and LRBA signals are shared across multiple unrelated drugs instead of being non-drug-specific risk variants. As supporting evidence, the chromosome 2 signal has been consistently associated in both replication and discovery cohorts with DILI due to ciprofloxacin, atorvastatin and mercaptopurine. There remains a possibility that replication could be achieved in a larger study involving a different mix of causative drugs but the degree of heterogeneity in drugs originally associated with the signals also increases the risk that these were chance observations.
Interestingly, unlike previously recognized HLA associations for DILI, A*33:01 also appears to be a risk factor for DILI due to several, structurally unrelated drugs. Our results also suggest that a haplotype comprising A*33:01, B*14:02, and C*08:02 may participate in concert to confer risk for DILI, as opposed to A*33:01 alone. However, because these alleles are so highly correlated, our current sample size does not allow us to distinguish between these possible explanations by genetic association evidence alone. This conceivable hypothesis could be further verified in a larger study, or by experiments with recombinant HLA proteins.28
In the case of terbinafine where the A*33:01 association showed genome-wide significance for cases relating to this drug only, information on the underlying mechanism for hepatotoxicity is limited, N-dealkylation leads to the formation of an aldehyde metabolite, TBF-A, and this metabolite shows reactivity with glutathione.29 It has been proposed that the GSH-adduct is transported across the canalicular membrane and concentrated in the bile where it may cause damage to biliary epithelial cells. There is limited data from the various case reports on an underlying inflammatory mechanism but it has been demonstrated that treatment of monocytes with terbinafine results in the release of the proinflammatory cytokines IL-8 and TNF-alpha.30 Metabolism of terbinafine is complex involving several different cytochromes P450.31 However, there was no evidence from the GWAS for a role for either CYP genes or innate immunity genes in the terbinafine DILI cases studied.
The other drugs showing the most convincing associations with A*33:01 were fenofibrate, ticlopidine and sertraline. Failure to see individual genome-wide significant associations with these drugs is likely to be due to fewer cases being available than for terbinafine. The A*33:01 association was seen for 3 of 7 cases due to fenofibrate, all with CM DILI. The literature on fenofibrate DILI is quite limited, but it appears that this drug is extensively metabolized, mainly by CYP3A4,32 and there is a report of a drug interaction resulting in cholestatic injury33 together with other isolated reports of idiosyncratic cholestatic DILI.34
Ticlopidine-related DILI has been well studied previously, including two studies investigating genetic risk factors in Japanese individuals. Cholestatic liver injury also predominates in this form of DILI.35, 36 Ticlopidine is subject to extensive metabolism by several cytochrome P450 isoforms and carboxyesterase.37 A study in rats suggests that adducts are formed following metabolism by cytochrome P450 with evidence for toxicity after biliary excretion of glutathione-conjugated metabolites via MRP2-facilitated transport.38 In previous studies, 22 Japanese patients with ticlopidine DILI showed an association with an HLA haplotype including A*33:03 (odds ratio 13).39 In line with current observations, the association was strongest with cholestatic cases with 12 out of 14 cases positive for A*33:03. It should be noted that A*33:03 is relatively common in Japan with approximately 10 to 15% of individuals carrying this allele.
The observations on the HLA association for Japanese ticlopidine DILI cases were followed up by a report that those carrying a −2320T>C polymorphism in CYP2B6 were more susceptible to ticlopidine DILI due to high CYP2B6 expression (OR 2, p-value=0.04).40 The CYP2B6 polymorphism (rs7254579)41 is less frequent in Europeans than in Asians (MAFcau = 0.29, MAFasian = 0.45) and its low effect size limited our ability to replicate the association in this small European ticlopidine DILI population, but the non-significant effect is in the same direction as the previous study (OR=2.8, 95% CI=0.7-10.18, p-value=0.11).
In contrast with the three drug examples above, sertraline is associated predominantly with HC DILI.42-44 In line with this phenotypic association, there is evidence that sertraline can cause mitochondrial damage28 and induce endoplasmic reticulum stress45 in liver cells. There are parallels with a previous example of a HLA risk factor (DRB1*15:01) which is associated with predominantly CM DILI with amoxicillin-clavulanate but HC DILI with lumiracoxib.9, 10
In line with the Japanese report of a role for A*33:03 in ticlopidine DILI39 and a case report showing an association between A*33 and tiopronin-induced cholestasis in a Chinese patient,46 we found two DILI cases positive for A*33:03 after direct HLA typing. One of these was a Chinese terbinafine DILI case and the second a European-American with erythromycin DILI. In our imputed GWAS dataset, A*33:03 was carried by eight DILI cases due to a range of drugs in our cohort and showed an apparent risk effect for CM DILI but this was not genome-wide significant (OR=2.1, 95% CI=1.02-4.6, p-value=0.04). Another A*33 allele, A*33:05, was also represented in a terbinafine case from Finland. There is very strong homology between these three HLA-A*33 alleles at the protein sequence level with A*33:03 differing at only two positions from A*33:01 (Tyr instead of His at position 171 and Lys instead of Arg at position 186) and A*33:05 differing at only one position (Arg at 54 in place of Gln). In particular, all three alleles conserve the key residues for specific peptide binding within the B and F pockets.47 This is in contrast to a related HLA allele A*31:01, which is associated with carbamazepine-induced skin rash,48 but does not appear to be a risk factor for DILI, where the B pocket sequence, though homologous, is not conserved.47
As mentioned above, the association of a common HLA allele with DILI due to chemically-unrelated compounds had been observed previously for DRB1*15:0110 with amoxicillin-clavulanate and lumiracoxib and for DRB1*07:0149 with DILI from lapatinib and ximelagatran. The association of A*33:01 with DILI in general and secondary to a number of structurally dissimilar compounds is consistent with these observations. Together with recent findings from in vitro studies on T-cell responses to flucloxacillin and amoxicillin-clavulanate,50, 51 these observations support the hypothesis that either the parent drug or metabolites bind covalently to cellular or circulating proteins to form adducts in a mechanism that is probably slightly different to the direct drug effect seen with hypersensitivity reactions to abacavir.52 Adduct formation may then allow binding to the peptide binding groove of HLA molecules leading to activation and differentiation of T-cells with a consequent adaptive immune response-mediated liver injury. Evidence that the majority of the drugs showing the A*33:01 association undergo hepatic metabolism and biliary excretion may explain the stronger association of A*33:01 with CM DILI and could indicate that, unlike in the case of flucloxacillin and amoxicillin-clavulanate, metabolites contribute to the toxicity mechanism. Further investigation of potential interaction of both the various drugs and their metabolites with the A*33:01 gene product by molecular modelling and in vitro studies on T cells as previously undertaken for flucloxacillin50 would be of interest.
The novel association of HC DILI with LRBA is interesting because this gene is a biologically plausible DILI candidate. LRBA deficiency due to rare mutations is associated with primary immunodeficiency of variable severity with a particular feature of decreased regulatory T cell (Treg) levels, other immunodeficiencies and inherited autoimmune disease.53-56 Patients with mutations in LRBA leading to immunodeficiency have been demonstrated to show loss of cytotoxic T lymphocyte antigen-4 (CTLA4).57 Studies in a mouse model suggest that low CTLA4 is a risk factor for DILI.58 Unlike the HLA-A*33:01 association, no genome-wide significant associations for single drugs were detected with the LRBA SNP and there were no obvious features in common between cases positive for the variant other than the HC phenotype. It remains possible that this association could be replicated if a larger cohort were available.
The intergenic signal on chromosome 2 is from a region 800 kb upstream from SOX11, is independent of A*33:01 and associated with an almost two-fold risk of DILI with the top SNP showing a frequency of 0.02 in Europeans. This risk factor seems to be shared across unrelated drugs among which ciprofloxacin showed the strongest association. The ENCODE project suggests there are no regulatory elements in this region so the basis for the signal is unclear. Neither rs72631567 nor any of its LD SNPs (r2>0.5) are known eQTL variants (http://www.gtexportal.org/). The failure to confirm this association and the absence of any apparent biological basis suggests the observed significance could have been a chance finding.
Most data for individual drug classes that were comparatively well represented in our cohort were entirely negative but the finding of a signal for statins which was driven by several class members was entirely novel. Similar to the more general signal seen on chromosome 2, the chromosome 18 is intergenic with the closest known gene, cadherin 19 located approx. 300000 bp downstream. Although functionally such a protein could be of relevance to the liver injury process,59 any biological significance seems tenuous. The failure to confirm the signal in additional cases could be due to the availability of only a small cohort of additional cases which reflects the rarity of this form of DILI.60
In conclusion, this study has detected a novel HLA association (HLA-A*33:01) in cases of DILI due to a number of different drugs, together with several novel non-HLA signals. Overall sensitivity and specificity of the A*33:01 allele as a predictor of DILI is low but our findings may be important for future drug treatment in cases of DILI due to one of the drugs for which the A*33:01 association is relevant. Follow-up studies are required to further explore the intergenic signal on chromosome 2, the biologically interesting signal in LRBA and the rs116561224 signal for statins in larger cohorts.
Supplementary Material
Figure S1. Scatterplots representing the first two principal components of the current study cohort. The homogenous distribution between cases and controls across the three major European clusters is shown. In panel (a) cases from phase II are highlighted in red and the cases from phase I in orange. In panel (b) the cholestatic/mixed cases are highlighted in red and the hepatocellular cases in blue.
Figure S2. Regional Manhattan plots for chromosome 2 and 4 in the region of the rs72631567 and rs28521457 signals. (A) Chromosome 2 for the overall cohort, (B) Chromosome 2 for the overall cohort conditioned on rs72631567. (C) Chromosome 4 for the HC only cohort, (D) Chromosome 4 for the HC only cohort conditioned on rs28521457.
Figure S3. QQ plots. (a) for the overall original analysis (b) after eliminating variants in MHC region. The QQ plot in the (b) panel highlights the signal on chromosome 2. The inflation factor is 1.05 after correction and 4.65 before correction.
Figure S4. Scatterplot representing the first two principal components of the current study cohort. The A*33:01-positive cases and their injury type are highlighted. A*33:01-positive cases are homogenously distributed in all the major population clusters.
Figure S5. MHC Regional Manhattan plots for (a) the overall cohort (b) the same cohort conditioned on A*33:01 (purple dot) and (c) the same cohort conditioned on rs114577328 (purple dot).
Figure S6. QQ plot for (a) Terbinafine cases only, (b) Cholestatic/Mixed cases only, (c) Hepatocellular cases only.
Figure S7. MHC Region Manhattan plots for (a) all cholestatic/mixed cases (b) the same cases conditioned on rs114577328 (purple dot), the top SNP (c) the same cases conditioned on A*33:01 (purple dot), the top HLA allele
Figure S8. Manhattan plots for DILI due to (A) Anti-TB drugs, (B) Fluoroquinolones, (C) NSAIDs, (D) Diclofenac (E) Nitrofurantoin.
Figure S9. Regional Manhattan plots for chromosome 18 in the area of the rs116561224 signal for (a) statin-induced DILI cases and (b) the same cases with conditioning on rs116561224
Table S1. Causal drugs in the replication cohort.
Table S2. Genotyping details for the DILI control cohorts
Table S3. Causative drugs across the overall DILI cohort
Table S4. Effect of the A*33:01, rs72631567 and rs28521457 signals across populations and recruitment phases
Table S5. Missing genotypes rate for the most associated SNPs within case and control groups in the comparisons where SNPs were significant
Table S6. Causative drugs across the rs72631567 signal on chromosome 2
Table S7. Summary of A*33:01-B*14:02-C*08:02 haplotype specific analysis across study cohorts
Table S8. Summary of the validation by direct HLA typing
Table S9. The A*33:01 signal across the main six A*33:01-associated drugs by type of injury
Table S10. List of all causal drugs where at least one case carries a A*33:01 allele
Table S11. The most represented causative drugs across the rs28521457 signal on chromosome 4
Table S12. Summary of drug and class comparisons with more than 40 samples
Table S13. The most associated variants for each drug in the class-specific analysis
Table S14. Causative drugs associated with rs116561224 signal in the statin cohort
Table S15. Causative drugs associated with the rs114577328/A*33:01 signal in the additional case set
Table S16. Causative drugs associated with the chromosome 2 rs72631567 signal in the additional case set
Table S17. Causative drugs associated with the chromosome 4 rs28521457 signal in the additional case set
Acknowledgments
Significant case enrollment, sample ascertainment and analytical/data coordinating support on the DILIGEN, Spanish DILI registry, Eudragene and iDILIC studies was provided by the International Serious Adverse Events Consortium (iSAEC). We are extremely grateful to Arthur Holden of iSAEC for his continuing support. We are grateful to Julia Patch and Julian Arbuckle for technical, study management and assistance with recruitment on the iDILIC study and to Daniele Cusi (Hypergenes), Patrik K. Magnusson (Swedish Twin Registry) and Javier Martin (Spanish DNA bank) for provision of control data. Contributors to sample collection via iDILIC, the Spanish DILI registry, EUDRAGENE, DILIN, DILIGEN and iSAEC are listed in the Appendix.
Funding
The genome-wide association study, HLA genotyping and iDILIC case enrollment and sample collection was funded by the International Serious Adverse Events Consortium with (Phase 2) membership support from Abbott, Amgen, Daiichi-Sankyo, GlaxoSmithKline,Merck, Novartis, Pfizer, Roche, Sanofi-Aventis, Takeda, and the Wellcome Trust. This is a summary of independent research partly (the DILIGEN and iDILIC sample collection) funded by the National Institute for Health Research (NIHR) Nottingham Digestive Diseases Biomedical Research Unit at the Nottingham University Hospitals NHS Trust and University of Nottingham. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. The DILIN (https://dilin.dcri.duke.edu/) is supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health (NIH) as a Cooperative Agreement (U01s) under grants: U01-DK065176 (Duke), U01-DK065201 (UNC), U01-DK065184 (Michigan), U01-DK065211 (Indiana), U01DK065193 (UConn), U01-DK065238 (UCSF/CPMC), U01-DK083023 (UTSW), U01-DK083027 (TJH/UPenn), U01-DK082992 (Mayo), U01-DK083020 (USC), U01-DK100928 (Icahn). Additional funding is provided by CTSA grants UL1 RR025761 (Indiana), UL1 TR001111 (UNC), and UL1 UL1 RR024986 (UMich). The EUDRAGENE collaboration received support from the EC 5th Framework program (QLRI-CT-2002-02757). The Spanish DILI Registry is partly funded by the Spanish Medicine Agency, Fondo Europeo de Desarrollo Regional - FEDER (P10-CTS-6470, FIS PI12/00378). CIBERehd is funded by Instituto de Salud Carlos III. The Swedish case collection (SWEDEGENE) has received support from the Swedish Medical Products Agency, the Swedish Society of Medicine (2008-21619), Swedish Research Council (Medicine 521-2011-2440), and Swedish Heart and Lung Foundation (20120557). MM was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at Guy's and St Thomas' NHS Foundation Trust and King's College London. LI was supported by Instituto de Salud Carlos III (EC08/00250). MA was supported by CONICYT PIA/Basal PFB12.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- DILI
drug-induced liver injury
- GWAS
genome wide association study
- OR
Odd Ratio
- RUCAM
Roussel Uclaf Causality Assessment Method
- AF
Allele Frequency
- HC
Hepatocellular
- CM
Cholestatic-Mixed
- eQTL
Expression quantitative trait loci
- MHC
Major Histocompatibility Complex
- MAF
Minor allele frequency
- LD
Linkage Disequilibrium
- HLA
Human leukocyte antigen
- SNP
Single Nucleotide Polymorphism
Footnotes
Conflicts of interest: The authors disclose the following: Dr Nelson is an employee of GlaxoSmithKline. Drs Chalasani, Fontana, and Watkins report consulting agreements and research grants with several pharmaceutical companies but none represent as potential conflicts for this paper. The DILIN causality committee considers potential conflicts while assigning cases for adjudication to individual investigators. The remaining authors disclose no conflicts.
Author contribution:
Study concept and design: Guruprasad P. Aithal, Paul B. Watkins, Matthew R. Nelson, Thomas J. Urban and Ann K. Daly
Case recruitment and data acquisition: Guruprasad P. Aithal, Marco Arrese, Einar Bjornsson, Raul J. Andrade, Pär Hallberg, Mia Wadelius, M. Isabel Lucena, Camilla Stephens, Sally A. Coulthard, Anke H. Maitland-van der Zee, Jennifer H. Martin, Ingolf Cascorbi, Christopher P. Day, John F. Dillon, Tarja Laitinen, Dominique Larrey, Mariam Molokhia, Gerd A. Kullak-Ublick, Luisa Ibáñez, Munir Pirmohamed, Shengying Qin, Fernando Bessone, Marco Arrese, Elizabeth Powell, Anita Conforti, Emmanuelle Bondon-Guitton, Alfonso Carvajal, Robert J. Fontana, Nelia Hernández, Jane I. Grove, Jose Serrano, Andrew Stolz, Victor Navarro, Herbert L. Bonkovsky, Huiman X. Barnhart, Naga Chalasani, Paul B. Watkins and Ann K. Daly
Case adjudication: Guruprasad P. Aithal, Einar Bjornsson and Raul J. Andrade for iDILIC; Robert J. Fontana, Paul H. Hayashi, Andrew Stolz, Jose Serrano and Paul B. Watkins for DILIN
Sample preparation and laboratory analysis: Camilla Stephens, Sally A. Coulthard Mark J. Daly and Jane Grove
Data analysis and interpretation: Paola Nicoletti, Ashley Sawle, Yufeng Shen, Aris Floratos, Guruprasad P. Aithal, Mark J. Daly, Elizabeth T. Cirulli, Thomas J. Urban, Matthew R. Nelson, Paul B. Watkins and Ann K. Daly
Writing the manuscript: Paola Nicoletti, Guruprasad P. Aithal, Thomas J. Urban, Paul B. Watkins and Ann K. Daly
Critical revision of manuscript for important intellectual content particularly in statistical analysis: Matthew R. Nelson and Yufeng Shen
Drs Nicoletti, Aithal, Urban and Daly contributed equally to this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stevens JL, Baker TK. The future of drug safety testing: expanding the view and narrowing the focus. Drug Discov Today. 2009;14:162–7. doi: 10.1016/j.drudis.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Gulmez SE, Larrey D, Pageaux GP, et al. Transplantation for acute liver failure in patients exposed to NSAIDs or paracetamol (acetaminophen): the multinational case-population SALT study. Drug Saf. 2013;36:135–44. doi: 10.1007/s40264-012-0013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reuben A, Koch DG, Lee WM. Drug-induced acute liver failure: results of a U.S. multicenter, prospective study. Hepatology. 2010;52:2065–76. doi: 10.1002/hep.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjornsson ES, Gunnarsson BI, Grondal G, et al. Risk of drug-induced liver injury from tumor necrosis factor antagonists. Clin Gastroenterol Hepatol. 2015;13:602–8. doi: 10.1016/j.cgh.2014.07.062. [DOI] [PubMed] [Google Scholar]
- 5.Bjornsson ES, Bergmann OM, Bjornsson HK, et al. Incidence, presentation, and outcomes in patients with drug-induced liver injury in the general population of Iceland. Gastroenterology. 2013;144:1419–25. 1425, e1–3. doi: 10.1053/j.gastro.2013.02.006. quiz e19-20. [DOI] [PubMed] [Google Scholar]
- 6.Urban TJ, Daly AK, Aithal GR. Genetic Basis of Drug-Induced Liver Injury: Present and Future. Semin Liver Dis. 2014;34:123–133. doi: 10.1055/s-0034-1375954. [DOI] [PubMed] [Google Scholar]
- 7.Kindmark A, Jawaid A, Harbron CG, et al. Genome-wide pharmacogenetic investigation of a hepatic adverse event without clinical signs of immunopathology suggests an underlying immune pathogenesis. Pharmacogenomics J. 2008;8:186–95. doi: 10.1038/sj.tpj.6500458. [DOI] [PubMed] [Google Scholar]
- 8.Daly AK, Donaldson PT, Bhatnagar P, et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet. 2009;41:816–819. doi: 10.1038/ng.379. [DOI] [PubMed] [Google Scholar]
- 9.Singer JB, Lewitzky S, Leroy E, et al. A genome-wide study identifies HLA alleles associated with lumiracoxib-related liver injury. Nat Genet. 2010;42:711–4. doi: 10.1038/ng.632. [DOI] [PubMed] [Google Scholar]
- 10.Lucena MI, Molokhia M, Shen Y, et al. Susceptibility to Amoxicillin-Clavulanate-Induced Liver Injury Is Influenced by Multiple HLA Class I and II Alleles. Gastroenterology. 2011;141:338–47. doi: 10.1053/j.gastro.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spraggs CF, Budde LR, Briley LP, et al. HLA-DQA1*02:01 Is a Major Risk Factor for Lapatinib-Induced Hepatotoxicity in Women With Advanced Breast Cancer. J Clin Oncol. 2011;29:667–73. doi: 10.1200/JCO.2010.31.3197. [DOI] [PubMed] [Google Scholar]
- 12.Urban TJ, Shen Y, Stolz A, et al. Limited contribution of common genetic variants to risk for liver injury due to a variety of drugs. Pharmacogenet Genomics. 2012;22:784–95. doi: 10.1097/FPC.0b013e3283589a76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bjornsson ES. Drug-induced liver injury: an overview over the most critical compounds. Arch Toxicol. 2015;89:327–34. doi: 10.1007/s00204-015-1456-2. [DOI] [PubMed] [Google Scholar]
- 14.Nicoletti P, Werk AN, Sawle A, et al. HLA-DRB1*16: 01-DQB1*05: 02 is a novel genetic risk factor for flupirtine-induced liver injury. Pharmacogenet Genomics. 2016;26:218–24. doi: 10.1097/FPC.0000000000000209. [DOI] [PubMed] [Google Scholar]
- 15.Aithal GP, Watkins PB, Andrade RJ, et al. Case Definition and Phenotype Standardization in Drug-Induced Liver Injury. Clin Pharmacol Ther. 2011;89:806–815. doi: 10.1038/clpt.2011.58. [DOI] [PubMed] [Google Scholar]
- 16.Fontana RJ, Watkins PB, Bonkovsky HL, et al. Drug-Induced Liver Injury Network (DILIN) prospective study: rationale, design and conduct. Drug Saf. 2009;32:55–68. doi: 10.2165/00002018-200932010-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benichou C, Danan G, Flahault A. Causality assessment of adverse reactions to drugs--II. An original model for validation of drug causality assessment methods: case reports with positive rechallenge. J Clin Epidemiol. 1993;46:1331–6. doi: 10.1016/0895-4356(93)90102-7. [DOI] [PubMed] [Google Scholar]
- 18.Nelson MR, Bryc K, King KS, et al. The Population Reference Sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am J Hum Genet. 2008;83:347–58. doi: 10.1016/j.ajhg.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Y, Nicoletti P, Floratos A, et al. Genome-wide association study of serious blistering skin rash caused by drugs. Pharmacogenomics J. 2012;12:96–104. doi: 10.1038/tpj.2010.84. [DOI] [PubMed] [Google Scholar]
- 20.Tryka KA, Hao L, Sturcke A, et al. NCBI's Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–9. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng X, Shen J, Cox C, et al. HIBAG--HLA genotype imputation with attribute bagging. Pharmacogenomics J. 2014;14:192–200. doi: 10.1038/tpj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy MI. Casting a wider net for diabetes susceptibility genes. Nat Genet. 2008;40:1039–40. doi: 10.1038/ng0908-1039. [DOI] [PubMed] [Google Scholar]
- 25.The R Project for Statistical Computing. http://wwwr-projectorg.
- 26.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haimila K, Perasaari J, Linjama T, et al. HLA antigen, allele and haplotype frequencies and their use in virtual panel reactive antigen calculations in the Finnish population. Tissue Antigens. 2013;81:35–43. doi: 10.1111/tan.12036. [DOI] [PubMed] [Google Scholar]
- 28.Ostrov DA, Grant BJ, Pompeu YA, et al. Drug hypersensitivity caused by alteration of the MHC-presented self-peptide repertoire. Proc Natl Acad Sci U S A. 2012;109:9959–64. doi: 10.1073/pnas.1207934109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iverson SL, Uetrecht JP. Identification of a reactive metabolite of terbinafine: insights into terbinafine-induced hepatotoxicity. Chem Res Toxicol. 2001;14:175–81. doi: 10.1021/tx0002029. [DOI] [PubMed] [Google Scholar]
- 30.Mizuno K, Fukami T, Toyoda Y, et al. Terbinafine stimulates the proinflammatory responses in human monocytic THP-1 cells through an ERK signaling pathway. Life Sci. 2010;87:537–44. doi: 10.1016/j.lfs.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Vickers AE, Sinclair JR, Zollinger M, et al. Multiple cytochrome P-450s involved in the metabolism of terbinafine suggest a limited potential for drug-drug interactions. Drug Metabolism and Disposition. 1999;27:1029–38. [PubMed] [Google Scholar]
- 32.Miller DB, Spence JD. Clinical pharmacokinetics of fibric acid derivatives (fibrates) Clin Pharmacokinet. 1998;34:155–62. doi: 10.2165/00003088-199834020-00003. [DOI] [PubMed] [Google Scholar]
- 33.Lucena MI, Andrade RJ, Vicioso L, et al. Prolonged cholestasis after raloxifene and fenofibrate interaction: A case report. World J Gastroenterol. 2006;12:5244–6. doi: 10.3748/wjg.v12.i32.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hajdu D, Aiglova K, Vinklerova I, et al. Acute cholestatic hepatitis induced by fenofibrate. J Clin Pharm Ther. 2009;34:599–602. doi: 10.1111/j.1365-2710.2009.01029.x. [DOI] [PubMed] [Google Scholar]
- 35.Skurnik YD, Tcherniak A, Edlan K, et al. Ticlopidine-induced cholestatic hepatitis. Ann Pharmacother. 2003;37:371–5. doi: 10.1345/aph.1A406. [DOI] [PubMed] [Google Scholar]
- 36.Pizarro AE, Andrade RJ, Garcia-Cortes M, et al. [Acute hepatitis due to ticlopidine. A report of 12 cases and review of the literature] Rev Neurol. 2001;33:1014–20. [PubMed] [Google Scholar]
- 37.Kim MJ, Jeong ES, Park JS, et al. Multiple cytochrome P450 isoforms are involved in the generation of a pharmacologically active thiol metabolite, whereas paraoxonase 1 and carboxylesterase 1 catalyze the formation of a thiol metabolite isomer from ticlopidine. Drug Metabolism and Disposition. 2014;42:141–52. doi: 10.1124/dmd.113.053017. [DOI] [PubMed] [Google Scholar]
- 38.Yoshikado T, Takada T, Yamamoto H, et al. Ticlopidine, a cholestatic liver injury-inducible drug, causes dysfunction of bile formation via diminished biliary secretion of phospholipids: involvement of biliary-excreted glutathione-conjugated ticlopidine metabolites. Mol Pharmacol. 2013;83:552–62. doi: 10.1124/mol.112.081752. [DOI] [PubMed] [Google Scholar]
- 39.Hirata K, Takagi H, Yamamoto M, et al. Ticlopidine-induced hepatotoxicity is associated with specific human leukocyte antigen genomic subtypes in Japanese patients: a preliminary case-control study. Pharmacogenomics J. 2008;8:29–33. doi: 10.1038/sj.tpj.6500442. [DOI] [PubMed] [Google Scholar]
- 40.Ariyoshi N, Iga Y, Hirata K, et al. Enhanced susceptibility of HLA-mediated ticlopidine-induced idiosyncratic hepatotoxicity by CYP2B6 polymorphism in Japanese. Drug Metab Pharmacokinet. 2010;25:298–306. doi: 10.2133/dmpk.25.298. [DOI] [PubMed] [Google Scholar]
- 41.Raccor BS, Claessens AJ, Dinh JC, et al. Potential contribution of cytochrome P450 2B6 to hepatic 4-hydroxycyclophosphamide formation in vitro and in vivo. Drug Metab Dispos. 2012;40:54–63. doi: 10.1124/dmd.111.039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fartoux-Heymann L, Hezode C, Zafrani ES, et al. Acute fatal hepatitis related to sertraline. J Hepatol. 2001;35:683–4. doi: 10.1016/s0168-8278(01)00159-3. [DOI] [PubMed] [Google Scholar]
- 43.Tabak F, Gunduz F, Tahan V, et al. Sertraline hepatotoxicity: report of a case and review of the literature. Dig Dis Sci. 2009;54:1589–91. doi: 10.1007/s10620-008-0524-3. [DOI] [PubMed] [Google Scholar]
- 44.Suen CF, Boyapati R, Simpson I, et al. Acute liver injury secondary to sertraline. BMJ Case Rep. 2013;2013 doi: 10.1136/bcr-2013-201022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S, Xuan J, Couch L, et al. Sertraline induces endoplasmic reticulum stress in hepatic cells. Toxicology. 2014;322:78–88. doi: 10.1016/j.tox.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurosaki M, Takagi H, Mori M. HLA-A33/B44/DR6 is highly related to intrahepatic cholestasis induced by tiopronin. Dig Dis Sci. 2000;45:1103–8. doi: 10.1023/a:1005585515826. [DOI] [PubMed] [Google Scholar]
- 47.Sidney J, Peters B, Frahm N, et al. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCormack M, Alfirevic A, Bourgeois S, et al. HLA-A*3101 and carbamazepine-induced hypersensitivity reactions in Europeans. N Engl J Med. 2011;364:1134–43. doi: 10.1056/NEJMoa1013297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parham LR, Briley LP, Li L, et al. Comprehensive genome-wide evaluation of lapatinib-induced liver injury yields a single genetic signal centered on known risk allele HLA-DRB1*07:01. Pharmacogenomics J. 2016;16:180–5. doi: 10.1038/tpj.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monshi MM, Faulkner L, Gibson A, et al. Human leukocyte antigen (HLA)-B*57:01-restricted activation of drug-specific T cells provides the immunological basis for flucloxacillin-induced liver injury. Hepatology. 2013;57:727–39. doi: 10.1002/hep.26077. [DOI] [PubMed] [Google Scholar]
- 51.Kim SH, Saide K, Farrell J, et al. Characterization of amoxicillin- and clavulanic acid-specific T cells in patients with amoxicillin-clavulanate-induced liver injury. Hepatology. 2015;62:887–99. doi: 10.1002/hep.27912. [DOI] [PubMed] [Google Scholar]
- 52.Illing PT, Vivian JP, Dudek NL, et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature. 2012;486:554–8. doi: 10.1038/nature11147. [DOI] [PubMed] [Google Scholar]
- 53.Gamez-Diaz L, August D, Stepensky P, et al. The extended phenotype of LPS-responsive beige-like anchor protein (LRBA) deficiency. J Allergy Clin Immunol. 2016;137:223–30. doi: 10.1016/j.jaci.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 54.Cohen JI. Primary Immunodeficiencies Associated with EBV Disease. Curr Top Microbiol Immunol. 2015;390:241–65. doi: 10.1007/978-3-319-22822-8_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bogaert DJ, Dullaers M, Lambrecht BN, et al. Genes associated with common variable immunodeficiency: one diagnosis to rule them all? J Med Genet. 2016 doi: 10.1136/jmedgenet-2015-103690. [DOI] [PubMed] [Google Scholar]
- 56.Levy E, Stolzenberg MC, Bruneau J, et al. LRBA deficiency with autoimmunity and early onset chronic erosive polyarthritis. Clin Immunol. 2016 doi: 10.1016/j.clim.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 57.Lo B, Zhang K, Lu W, et al. Autoimmune disease. Patients with LRBA deficiency show CTLA4 loss and immune dysregulation responsive to abatacept therapy. Science. 2015;349:436–40. doi: 10.1126/science.aaa1663. [DOI] [PubMed] [Google Scholar]
- 58.Metushi IG, Hayes MA, Uetrecht J. Treatment of PD-1(-/-) mice with amodiaquine and anti-CTLA4 leads to liver injury similar to idiosyncratic liver injury in patients. Hepatology. 2015;61:1332–42. doi: 10.1002/hep.27549. [DOI] [PubMed] [Google Scholar]
- 59.Cadwell CM, Su W, Kowalczyk AP. Cadherin tales: Regulation of cadherin function by endocytic membrane trafficking. Traffic. 2016 doi: 10.1111/tra.12448. [DOI] [PubMed] [Google Scholar]
- 60.Bjornsson E, Jacobsen EI, Kalaitzakis E. Hepatotoxicity associated with statins: reports of idiosyncratic liver injury post-marketing. J Hepatol. 2012;56:374–80. doi: 10.1016/j.jhep.2011.07.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Scatterplots representing the first two principal components of the current study cohort. The homogenous distribution between cases and controls across the three major European clusters is shown. In panel (a) cases from phase II are highlighted in red and the cases from phase I in orange. In panel (b) the cholestatic/mixed cases are highlighted in red and the hepatocellular cases in blue.
Figure S2. Regional Manhattan plots for chromosome 2 and 4 in the region of the rs72631567 and rs28521457 signals. (A) Chromosome 2 for the overall cohort, (B) Chromosome 2 for the overall cohort conditioned on rs72631567. (C) Chromosome 4 for the HC only cohort, (D) Chromosome 4 for the HC only cohort conditioned on rs28521457.
Figure S3. QQ plots. (a) for the overall original analysis (b) after eliminating variants in MHC region. The QQ plot in the (b) panel highlights the signal on chromosome 2. The inflation factor is 1.05 after correction and 4.65 before correction.
Figure S4. Scatterplot representing the first two principal components of the current study cohort. The A*33:01-positive cases and their injury type are highlighted. A*33:01-positive cases are homogenously distributed in all the major population clusters.
Figure S5. MHC Regional Manhattan plots for (a) the overall cohort (b) the same cohort conditioned on A*33:01 (purple dot) and (c) the same cohort conditioned on rs114577328 (purple dot).
Figure S6. QQ plot for (a) Terbinafine cases only, (b) Cholestatic/Mixed cases only, (c) Hepatocellular cases only.
Figure S7. MHC Region Manhattan plots for (a) all cholestatic/mixed cases (b) the same cases conditioned on rs114577328 (purple dot), the top SNP (c) the same cases conditioned on A*33:01 (purple dot), the top HLA allele
Figure S8. Manhattan plots for DILI due to (A) Anti-TB drugs, (B) Fluoroquinolones, (C) NSAIDs, (D) Diclofenac (E) Nitrofurantoin.
Figure S9. Regional Manhattan plots for chromosome 18 in the area of the rs116561224 signal for (a) statin-induced DILI cases and (b) the same cases with conditioning on rs116561224
Table S1. Causal drugs in the replication cohort.
Table S2. Genotyping details for the DILI control cohorts
Table S3. Causative drugs across the overall DILI cohort
Table S4. Effect of the A*33:01, rs72631567 and rs28521457 signals across populations and recruitment phases
Table S5. Missing genotypes rate for the most associated SNPs within case and control groups in the comparisons where SNPs were significant
Table S6. Causative drugs across the rs72631567 signal on chromosome 2
Table S7. Summary of A*33:01-B*14:02-C*08:02 haplotype specific analysis across study cohorts
Table S8. Summary of the validation by direct HLA typing
Table S9. The A*33:01 signal across the main six A*33:01-associated drugs by type of injury
Table S10. List of all causal drugs where at least one case carries a A*33:01 allele
Table S11. The most represented causative drugs across the rs28521457 signal on chromosome 4
Table S12. Summary of drug and class comparisons with more than 40 samples
Table S13. The most associated variants for each drug in the class-specific analysis
Table S14. Causative drugs associated with rs116561224 signal in the statin cohort
Table S15. Causative drugs associated with the rs114577328/A*33:01 signal in the additional case set
Table S16. Causative drugs associated with the chromosome 2 rs72631567 signal in the additional case set
Table S17. Causative drugs associated with the chromosome 4 rs28521457 signal in the additional case set