Abstract
Purpose
Metastatic castration-resistant prostate cancer (mCRPC) is a heterogeneous disease for which better prognostic models for survival are needed. We examined the added value of circulating tumor cell (CTC) enumeration relative to common prognostic laboratory measures from CRPC patients.
Experimental Technique
Utility of CTC enumeration as a baseline and postbaseline prognostic biomarker was examined using data from two prospective randomized registration-directed trials (COU-AA-301 and ELM-PC4) within statistical models used to estimate risk for survival. Discrimination and calibration were used to measure model predictive accuracy and the added value for CTC enumeration in the context of a Cox model containing albumin, lactate dehydrogenase (LDH), prostate-specific antigen (PSA), hemoglobin, and alkaline phosphatase (ALK). Discrimination quantifies how accurately a risk model predicts short-term versus long-term survivors. Calibration measures the closeness of actual survival time to the predicted survival time.
Results
Adding CTC enumeration to a model containing albumin, LDH, PSA, hemoglobin, and ALK (“ALPHA”) improved its discriminatory power. The weighted c-index for ALPHA without CTCs was 0.72 (SE, 0.02) versus 0.75 (SE, 0.02) for ALPHA + CTCs. The increase in discrimination was restricted to the lower-risk cohort. In terms of calibration, adding CTCs produced a more accurate model-based prediction of patient survival. The absolute prediction error for ALPHA was 3.95 (SE, 0.28) versus 3.75 months (SE, 0.22) for ALPHA + CTCs.
Data Interpretation
Addition of CTC enumeration to standard measures provides more accurate assessment of patient risk in terms of baseline and postbaseline prognosis in the mCRPC population.
Keywords: biomarker, circulating tumor cells, incremental value, metastatic castration-resistant prostate cancer, predictive accuracy, risk model
Introduction
Studies in multiple tumor types have shown that circulating tumor cell (CTC) number measured with the CellSearch® assay is prognostic for survival before and after therapy (1-3). In 2004, the test received clearance from the US Food and Drug Administration as “an aid in the monitoring of patients with metastatic breast cancer” (4) and expanded to colorectal and prostate cancer in 2007 (5) and 2008 (6), respectively. Use in practice has been limited, in part because of variations in reimbursement by third party payers, the costs including the devices and reagents needed to perform and the technical personnel to run it, and uncertainties in how to use the test result in patient management. The central question is whether the use of the CTC enumeration result provides incremental information that improves the ability to assess the prognosis of the patient relative to an assessment without the result. If the prognosis of survival time incorporating CTC enumeration meaningfully impacts patient management, its diagnostic utility will be clear.
The conventional approach to determining whether a new biomarker adds value to current models is to establish its association with survival when combined with other known prognostic factors. The association analysis is often developed through a proportional hazards model, using the hazard ratio and P value affiliated with the new marker to establish its clinical importance. These association analyses alone, however, are not sufficient to assess the magnitude of the added value of a biomarker. Here, we go beyond the standard association analyses by assessing whether CTC enumeration before and after treatment improves risk classification and the prediction of survival time for patients with metastatic castration-resistant prostate cancer (mCRPC). To do so, we compared models that contained and excluded CTCs for their ability to discriminate and calibrate survival times.
The analysis was performed using data from patients enrolled in the phase III registration trial of abiraterone acetate plus prednisone (COU-AA-301; NCT00638690) (7, 8) and independently validated using the data from patients in a second registration trial of similar design evaluating orteronel plus prednisone (ELM-PC4; NCT01193244) (9). The analysis included patients from both the treatment and control arms of these studies with the purpose of determining whether the predictive accuracy of the baseline and postbaseline CTC biomarker was agnostic to treatment. Also noteworthy is that the COU-AA-301 trial population was docetaxel refractory and the ELM-PC4 population was chemotherapy naïve (7-9), which enabled the examination of the CTC signal in multiple mCRPC populations
Methods
Study design and participants
A proportional hazards model for survival was developed using baseline and posttreatment data from patients enrolled in the completed phase III registration trials of abiraterone acetate plus prednisone (COU-AA-301) and orteronel plus prednisone (ELM-PC4). The results of the trials have been reported previously (7-9). The biomarkers used for clinical prognostication were albumin, lactate dehydrogenase (LDH), prostate-specific antigen (PSA), hemoglobin, and alkaline phosphatase (ALK), which have previously been shown to be prognostic for survival in multivariate analysis and are components of several prediction models (nomograms) that estimate survival times in men with mCRPC (10-14). The baseline values of albumin, LDH, PSA, hemoglobin, and ALK, along with the increase or decrease in PSA at week 13 relative to baseline, represent our submodel (called “ALPHA” in this article). For the ELM-PC4 analysis, a weighted proportional hazards model was used to account for the nonproportionality in the model. The weights enable the interpretation of the model coefficients as the average hazard ratio over time. All analyses were based on a landmark time of 12 weeks.
The objective of this study was to determine the incremental information provided by early post-treatment CTC measures in predicting patient survival. For each patient, a risk score was computed from both the ALPHA model and the model developed by adding baseline CTC enumeration and the increase or decrease in CTCs at week 13 relative to baseline. This model is referred to as ALPHA + CTC. Adding CTCs to the submodel indicates that both the baseline CTC values and the relative change in CTCs were included. For the CTC and PSA markers, the relative change was defined as:
Because patients may not have detectable CTCs at baseline, CTC baseline values equal to 0 were recorded as 1 for the CTC relative change variable. The risk score for each patient is a weighted sum of his biomarkers in the proportional hazards model, where the weights are the regression coefficients derived from the model.
Statistical analysis methods
Discrimination represents the model’s strength in differentiating long-term survivors from short-term survivors. It is illustrated graphically using negative and positive predictive value statistics (15). The negative predictive value is depicted with a Kaplan-Meier curve estimating the probability of survival using the cohort of patients with low-risk scores derived from the proportional hazards models. The positive predictive value is defined as the probability of dying (one minus Kaplan-Meier) for patients with model-based high-risk scores. For the current analysis, the negative predictive value is computed using patients with the lowest 25% of the risk scores (presumed best prognosis), and the positive predictive value is calculated using patients with the highest 25% of the risk scores (presumed worst prognosis). To assess the added value of CTC enumeration, the negative and positive predictive values were computed for both models. An enhanced negative predictive value with CTCs would be indicated by a Kaplan-Meier survival curve that is above the survival curve derived from the model developed without the CTC marker, while an improvement in the positive predictive value is represented by a higher one minus Kaplan-Meier curve for the model that included CTCs.
In addition to a graphical analysis, the weighted concordance index (c-index) was computed to summarize the discriminatory power of each model to evaluate the added value of CTCs (16). The concordance index is the proportion of all pairs of patients, where the patient with a longer survival time also has a smaller risk score. The weighted c-index ranges between 0.5 and 1.0, where the value 0.5 indicates that the model cannot discriminate between long-term survivors and short-term survivors and a value of 1.0 indicates perfect discrimination. A 10-fold cross-validation procedure was used to compute the weighted c-indices and bootstrap 95% confidence intervals were computed for the associated parameter.
A calibration metric is used to compare the model-based predicted survival time with the actual survival time of a patient (17). To gauge calibration for each model, the relationship between the observed time to death (for patients who died) and the median predicted survival time is illustrated graphically. Perfect calibration is represented by a 45° line through the origin showing that the observed times and the predicted median survival times were equal for all patients.
A summary measure of calibration, termed the absolute prediction error (APE), is computed (18). The APE computes a weighted difference between the actual survival time and a predicted median survival time (19). The patient’s predicted survival time is computed from his model-based risk score. An APE equal to zero indicates that the model-based predicted median survival is exactly equal to the true survival for all patients, which equates to the data lining up in a 45° line in the calibration plot described above. As the disparity between the observed and predicted survival increases, the APE increases.
Graphs and summary statistics of discrimination and calibration were computed. We evaluated the added value of CTC enumeration as a clinical predictor of survival. The results were validated using a patient cohort from the independent randomized clinical ELM-PC4 trial, which was designed to test the efficacy of orteronel plus prednisone in men with mCRPC.
Results
The randomized clinical trial comparing abiraterone acetate plus prednisone versus prednisone alone for patients with mCRPC enrolled 1195 patients. Of these, 949 had all baseline markers drawn and 648 patients were alive with CTC and PSA markers recorded at week 13 and were eligible for the landmark analysis. In addition, four patients had data values with extreme leverage points that had an overly influential effect on the analysis. One patient had a PSA relative change (from baseline to week 13) approximately equal to 100, two patients had CTC relative changes equal to 99, and the final patient had a CTC relative change approximately equal to 23. With these additional four patients excluded, data from 644 patients were used in the analysis (CONSORT diagram, Fig. 1A). The median survival time for the 644 patients under study was 16.8 months (95% CI, 15.6–18.0). A summary of the marker values for these 644 patients is provided in Table 1.
Figure 1.
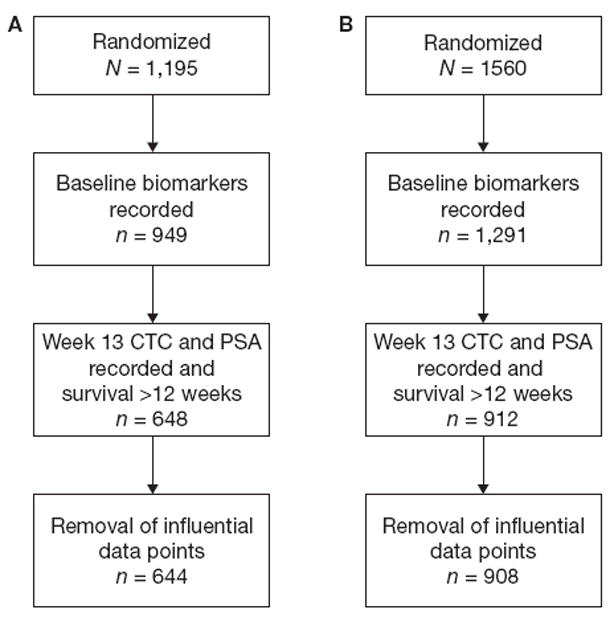
Consort diagrams for the COU-AA-301 (A) and ELM-PC4 (B) studies.
Table 1.
Summary of biomarker measures.
| Marker | COU-AA-301 study (N = 644) | ELM-PC4 study (N = 908) | ||
|---|---|---|---|---|
|
| ||||
| Median | (Range) | Median | (Range) | |
| Baseline albumin | 4.1 | (2.9–4.9) | 4.4 | (2.5–5.4) |
| Baseline ALK | 114.5 | (33–4617) | 96.0 | (35–3,281) |
| Baseline CTC | 4.0 | (0–100) | 2.0 | (0–5,153) |
| Baseline hemoglobin | 12.0 | (7.3–16.5) | 12.9 | (4.7–17.5) |
| Baseline LDH | 215.0 | (84–3,373) | 186.0 | (67–1,868) |
| Baseline PSA | 119.2 | (0.4–10,110) | 49.7 | (1.7–3,906) |
| Week 13 CTC | 1.0 | (0–100) | 0 | (0–1,635) |
| Week 13 PSA | 97.5 | (0.1–8,582) | 31.6 | (0–7682) |
| Age | 70 | (42–95) | 71 | (49–89) |
Abbreviations: ALK, alkaline phosphatase; CTC, circulating tumor cells; LDH, lactate dehydrogenase; PSA, prostate-specific antigen.
Table 2 provides a summary of the log relative risk coefficients from the proportional hazards model and their attendant standard errors and P values. In addition to the significance of many of the factors, the baseline and relative change in CTC enumeration demonstrated a strong association with survival time. However, association alone is not a sufficient measure of the prognostic utility of individual markers.
Table 2.
Summary of association analysis in the COU-AA-301 and ELM-PC4a proportional hazards analysis.
| Factor | Coefficient | SE (coefficient) | P value |
|---|---|---|---|
| COU-AA-301 | |||
| Baseline albumin | − 0.678 | 0.171 | <0.001 |
| Baseline log (ALK) | − 0.031 | 0.076 | 0.681 |
| Baseline log (CTC) | 0.253 | 0.043 | <0.001 |
| Baseline haemoglobin | − 0.008 | 0.041 | 0.842 |
| Baseline log (LDH) | 1.161 | 0.135 | <0.001 |
| Baseline log (PSA) | 0.050 | 0.037 | 0.173 |
| Relative change in CTC | 0.098 | 0.014 | <0.001 |
| Relative change in PSA | 0.222 | 0.029 | <0.001 |
|
| |||
| ELM-PC4 | |||
|
| |||
| Baseline albumin | − 0.037 | 0.021 | 0.084 |
| Baseline log (ALK) | 0.200 | 0.105 | 0.056 |
| Baseline log (CTC) | 0.327 | 0.044 | <0.001 |
| Baseline hemoglobin | − 0.013 | 0.005 | 0.007 |
| Baseline log (LDH) | 0.047 | 0.175 | 0.787 |
| Baseline log (PSA) | 0.067 | 0.047 | 0.153 |
| Relative change in CTC | 0.089 | 0.026 | <0.001 |
| Relative change in PSA | 0.292 | 0.053 | <0.001 |
Abbreviations: ALK, alkaline phosphatase; CTC, circulating tumor cells; LDH, lactate dehydrogenase; PSA, prostate-specific antigen.
A weighted proportional hazards model was used to account for the nonproportionality in the ELM-PC4 risk model.
We examined the weighted c-index to numerically evaluate the value of adding CTCs to the model in terms of discrimination. The discriminatory power of ALPHA + CTC was 0.75 (SE, 0.02). The weighted c-index for the model containing ALPHA (without CTCs) was 0.72 (SE, 0.02) (Table 3). A 95% bootstrap confidence interval for the difference in these two measures, indicating the magnitude of the improvement in discrimination due to the addition of CTCs, is (0.02, 0.05)
Table 3.
Predictive accuracy summary measures from the COU-AA-301 and ELM-PC4 studies.
| Weighted c-index | APE | |||
|---|---|---|---|---|
|
| ||||
| COU-AA-301 | ELM-PC4 | COU-AA-301 | ELM-PC4 | |
| ALPHA | 0.72 | 0.71 | 3.95 | 3.83 |
| ALPHA + CTC | 0.75 | 0.75 | 3.75 | 3.56 |
Abbreviations: APE, absolute prediction error; ALPHA, alubmin, lactate dehydrogenase, prostate-specific antigen, hemoglobin, and alkaline phosphatase; CTC, circulating tumor cells.
A deeper analysis, presented visually, demonstrated where CTC enumeration provides added value as a prognostic marker of survival. A visual assessment of the improvement in discrimination, based on the addition of CTC, was obtained by graphing the negative and positive predictive values. For the negative predictive value (Fig. 2A), patients with low risk scores are expected to have prolonged survival times relative to the entire cohort. This improvement in survival is magnified in the model that includes CTCs. For the positive predictive value, which reflects patients with the poorest prognosis, there is no benefit to adding CTCs to the prognostic model (Fig. 2B). Thus, in terms of discrimination, the prognostic utility of CTC enumeration is manifested in the lower-risk cohort; the existing markers are sufficient for prognosis with the high-risk cohort.
Figure 2.
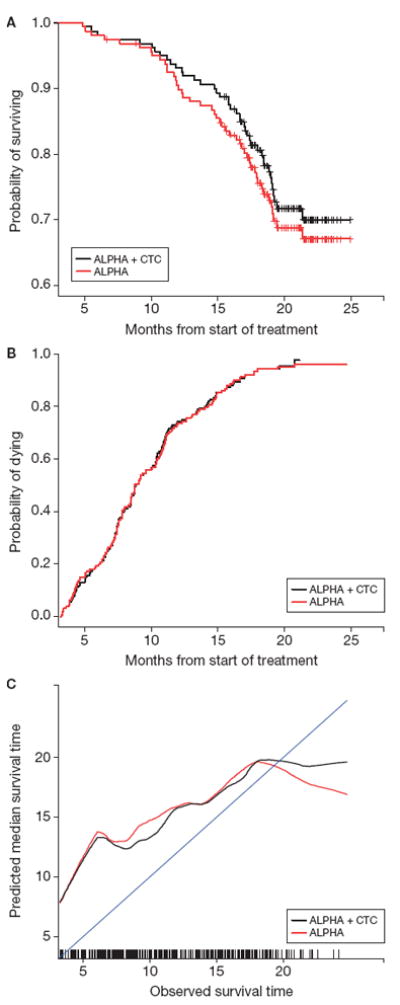
COU-AA-301 negative (A) and positive (B) predictive value and calibration (C) curves. Note: Negative predictive value includes patients in the lowest quartile and positive predictive value those in the highest of risk scores within each model. An improvement in the negative predictive value among these low-risk patients would be shown by an increase in the Kaplan-Meier estimate. An improvement in the positive predictive value among these high-risk patients would be demonstrated by an increase in the one minus Kaplan-Meier estimate. Calibration represents the relationship between observed survival time and the median predicted survival time. A model with good calibration would closely approximate the 45° line through the origin.
Taken together, the graphical and numerical discrimination analyses show that the addition of CTC enumeration improves the discriminatory power of the risk model relative to standard prognostic factors in this patient population and that the discrimination benefit is found in the lower-risk cohort.
Calibration measures the closeness of the actual survival time to the model-based predicted survival. The APE is recorded to summarize the distance between the observed survival times and predicted median survival times. The APE for ALPHA + CTC is 3.75 months (SE, 0.22). Removing CTCs from this model (ALPHA) increases the APE to 3.95 (SE, 0.28). Therefore, using APE, the inclusion of CTCs to the model provides a modest improvement in the accuracy of the predicted survival time (Table 3). The reduction in the APE due to CTC is 0.20 and the 95% confidence for this difference is (-0.02, 0.51).
The graphical calibration analysis compared the estimated median survival times to the observed survival times using smoothed curves for an uncluttered visual of this relationship (Fig. 2c). The frequencies for the observed survival times are illustrated by the vertical lines at the bottom of the plot. As shown, the calibration curve for ALPHA + CTC is closer to the 45° line early in the follow-up period, but there remains significant distance between this curve and exact calibration in the early time period.
Data from 908 patients enrolled on the ELM-PC4 study were used to validate the discrimination and calibration analyses (Fig. 1B, Table 2). In this patient population, the discrimination metric for ALPHA + CTC produced a weighted c-index of 0.75 (SE, 0.01). When removing CTCs from the model, the weighted c-index decreased to 0.71 (SE, 0.01) (Table 3). A 95% confidence for the difference in the two measures is (0.02, 0.64). A similar pattern occurred with the calibration metric. The APE for ALPHA + CTC was 3.56 months (SE, 0.24) and increased to 3.83 months (SE, 0.22) when CTCs were omitted from the model. The 95% confidence for this difference is (0.02, 0.66). The negative (Fig. 3A) and positive (Fig. 3B) predictive value curves and the calibration curves (Fig. 3C) produce parallel information to these summary measures. These results validate the discrimination and calibration analyses obtained from the COU-AA-301 set.
Figure 3.
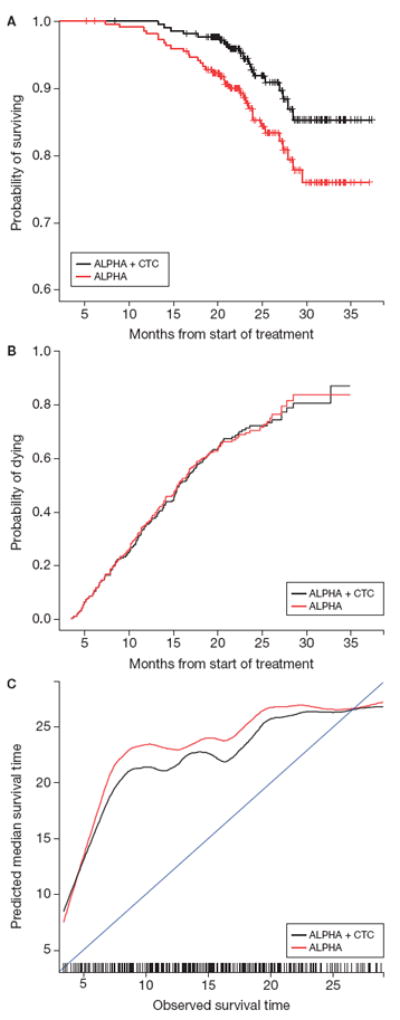
ELM-PC4 negative (A) and positive (B) predictive value and calibration (c) curves. Note: Negative predictive value includes patients in the lowest quartile of risk scores within each model; positive predictive value includes patients in the highest quartile of risk scores within each model; calibration represents the relationship between observed survival time and the median predicted survival time.
Discussion
Understanding the clinical importance of a post-therapy measure to assess long-term trial endpoints is an essential step in establishing outcome measures as indicators of clinical benefit that can be used to support drug approval. To do so requires a determination of the prognosis of the patient cohort using risk factors measured at baseline and postbaseline. The present study shows that the incorporation of the CTC number at baseline and the relative change in CTC number from baseline to week 13 provides an improvement in the predictive accuracy of a prognostic model for low-risk patients with respect to survival time. The results were validated using patient data from a comparable mCRPC trial, but with chemotherapy-naïve patients, in contrast to the initial study based on patients with prior docetaxel treatment.
The discrimination analysis in the COU-AA-301 marker data demonstrated that, for defining a low-risk cohort, the addition of CTC number to the risk model produced higher survival rates relative to a risk model developed without CTC enumeration. Thus, the addition of CTC enumeration to the submodel improved the negative predictive value of the risk classification model. The finding was validated using an independent cohort of patients treated in the ELM-PC4 trial, where the addition to CTCs to the submodel showed even greater separation in the survival curves among the low-risk cohort developed with and without CTCs.
The calibration analysis established that the CTC-based model provided only a modest improvement in predicting survival time. Neither curve approaches the 45° line of equivalence between the model and actual survival time. There are two components that impact the accuracy of a calibration analysis. First, point prediction of survival time is complicated by many factors not related to disease, such as age or comorbidities. Second, there are disease-related factors, such as number of bone metastases, performance status, and Brief Pain Inventory-Short Form score, to name a few, that are not included in the model. In our analysis, this is evident by the calibration curves in Fig. 3: the prediction of early deaths is poor, as shown by the distance or separation from the 45° line.
Complete biomarker data was not available for all patients. For patients who are alive at 13 weeks and missing either the week 13 CTC or PSA recording, the data are not missing data at random. For the COU-AA-301 study, the median survival time for these patients is 13.4 months compared to a median survival time of 16.8 for the analysis cohort. However, the potential biases in the discrimination or calibration comparisons should approximately cancel because the same patient cohort was used for both risk models (ALPHA and ALPHA+CTC).
The use of biomarkers during follow-up to accurately determine prognosis is essential for disease management. To this end, serial biologic profiling of the disease before treatment, during treatment, and at progression, and determining the association of the profiles with later events such as radiographic progression-free survival and/or overall survival, was included in the updated recommendations from the Prostate Cancer Clinical Trials Working Group 3 (20). If applied early in the posttreatment period, the biomarker results may be useful for informing the decision to continue treatment if the patient-specific risk score is favorable or to discontinue treatment if the computed risk score is not.
In this study, we found that CTC enumeration measured at baseline and early in the treatment phase, regardless of treatment received, provided incremental value to the clinical factors and laboratory test results acquired in the course of routine clinical practice. A limitation of the CTC CellSearch assay is that it only defines one circulating tumor cell type. Whether non-selection based assays that enable the identification and enumeration of multiple cell types is more informative is unknown. At this point, however, the results support a role for postbaseline CTC containing biomarkers as an indicator of prognosis. More research is needed to go beyond its prognostic utility and examine its clinical utility as an intermediate response variable.
Supplementary Material
Translational Relevance.
Previous studies in multiple tumor types have shown that pre- and posttreatment circulating tumor cell (CTC) number measured with the CellSearch® assay is prognostic for survival. Use of CTC enumeration in practice has been limited, in part due to costs, variations in reimbursement by third-party payers, and questions about the prognostic utility. In this study, we explored in greater depth the magnitude of the added value of CTCs in terms of its predictive accuracy for survival time. We found that CTC enumeration measured at baseline and early in the treatment phase provided incremental value in predictive accuracy relative to known biomarkers acquired in the course of routine clinical practice. These findings and those in previous studies suggest a role for posttreatment CTC containing biomarkers as an indicator of patient risk.
Acknowledgments
Editorial assistance was provided by Lashon Pringle, PhD, of PAREXEL, and was funded by Janssen Global Services, LLC.
Financial support: These analyses were funded by the following sources: NIH/NCI P50-CA92629 SPORE in Prostate Cancer, NIH/NCI Cancer Center Support Grant P30 CA008748, Department of Defense Prostate Cancer Research Program (PC071610 and PC121111), Prostate Cancer Foundation Challenge Award, David H. Koch Fund for Prostate Cancer Research.
Footnotes
Disclosure of Potential Conflicts of Interest: G. Heller reports research funding to Memorial Sloan Kettering Cancer Center from Janssen Research & Development, and he has served as an uncompensated consultant to Takeda Millennium. K. Fizazi has nothing to disclose. R. McCormack is an employee of Janssen Oncology Research & Development, LLC. A. Molina was an employee of Janssen Research & Development at the time of the analysis, and owns stock in Johnson & Johnson. D. MacLean and I.J. Webb are employees of Takeda Pharmaceuticals, which sponsored the C21004 and C21005 studies in men with metastatic CRPC. F. Saad been involved in advisory boards, received honoraria and served as an investigator for research funded by Astellas, Janssen, Takeda Millennium. J.S. de Bono has nothing to disclose. H.I. Scher reports research funding to Memorial Sloan Kettering Cancer Center from Janssen Research & Development, BIND Therapeutics, Medivation, Innocrin Pharma, and Exelixis; has served as an uncompensated consultant to AstraZeneca, BIND Pharmaceuticals, Blue Earth Diagnostics, Bristol Myers Squibb, Celegene, Genentech, Medivation, Novartis, Pfizer, Takeda Millennium, and Endocyte; has served as a paid consultant to Astellas, Elseiver's Practice Up-date, Ferring Pharmaceuticals, MED IQ, Merck, Oncology STAT, Roche, Sanofi Aventis, WCG Oncology, and Chugai Academy for Advanced Oncology; has received honoraria from BIND Pharmaceuticals and Blue Earth Diagnostics.
References
- 1.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–21. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 2.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 3.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–9. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 4.US Food and Drug Administration. [21 December, 2015];CellSearch™ Epithelial Cell Kit/CellSpotter™ Analyzer - K031588. Available from US FDA Web site [Internet]. Available from: http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/DeviceApprovalsandClearances/Recently-ApprovedDevices/ucm081239.htm.
- 5.US Food and Drug Administration. [21 December, 2015];CellSearch™ Epithelial Cell Kit/CellSpotter™ Analyzer - K071729 510(k) Summary. US FDA Web site [Internet]. Available from: http://www.accessdata.fda.gov/cdrh_docs/reviews/K071729.pdf.
- 6.US Food and Drug Administration. [21 December, 2015];CellSearch™ Circulating Tumor Cell Kit - K073338. Premarket notification - expanded indications for use - metastatic prostate cancer. Available from US FDA Web site [Internet]. Available from: http://www.accessdata.fda.gov/cdrh_docs/pdf7/K073338.pdf.
- 7.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fizazi K, Scher HI, Molina A, Logothetis CJ, Chi KN, Jones RJ, et al. Abiraterone acetate for treatment of metastatic castration-resistant prostate cancer: final overall survival analysis of the COU-AA-301 randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2012;13:983–92. doi: 10.1016/S1470-2045(12)70379-0. [DOI] [PubMed] [Google Scholar]
- 9.Saad F, Fizazi K, Jinga V, Efstathiou E, Fong PC, Hart LL, et al. Orteronel plus prednisone in patients with chemotherapy-naive metastatic castration-resistant prostate cancer (ELM-PC 4): a double-blind, multicentre, phase 3, randomised, placebo-controlled trial. Lancet Oncol. 2015;16:338–48. doi: 10.1016/S1470-2045(15)70027-6. [DOI] [PubMed] [Google Scholar]
- 10.Armstrong AJ, Garrett-Mayer ES, Yang YC, de WR, Tannock IF, Eisenberger M. A contemporary prognostic nomogram for men with hormone-refractory metastatic prostate cancer: a TAX327 study analysis. Clin Cancer Res. 2007;13:6396–403. doi: 10.1158/1078-0432.CCR-07-1036. [DOI] [PubMed] [Google Scholar]
- 11.Fizazi K, Massard C, Smith M, Rader M, Brown J, Milecki P, et al. Bone-related parameters are the main prognostic factors for overall survival in men with bone metastases from castration-resistant prostate cancer. Eur Urol. 2015;68:42–50. doi: 10.1016/j.eururo.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Halabi S, Small EJ, Kantoff PW, Kattan MW, Kaplan EB, Dawson NA, et al. Prognostic model for predicting survival in men with hormone-refractory metastatic prostate cancer. J Clin Oncol. 2003;21:1232–7. doi: 10.1200/JCO.2003.06.100. [DOI] [PubMed] [Google Scholar]
- 13.Halabi S, Lin CY, Kelly WK, Fizazi KS, Moul JW, Kaplan EB, et al. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J Clin Oncol. 2014;32:671–7. doi: 10.1200/JCO.2013.52.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smaletz O, Scher HI, Small EJ, Verbel DA, McMillan A, Regan K, et al. Nomogram for overall survival of patients with progressive metastatic prostate cancer after castration. J Clin Oncol. 2002;20:3972–82. doi: 10.1200/JCO.2002.11.021. [DOI] [PubMed] [Google Scholar]
- 15.Altman DG. Practical Statistics for Medical Research. 1. New York: Chapman and Hall; 1991. [Google Scholar]
- 16.Uno H, Cai T, Pencina MJ, D’Agostino RB, Wei LJ. On the c-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Statistics in Medicine. 2011;30:1105–1117. doi: 10.1002/sim.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Royston P, Altman DG. External validation of a Cox prognostic model: principles and methods. BMC Med Res Methodol. 2013;13:33. doi: 10.1186/1471-2288-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawless JF, Yuan Y. Estimation of prediction error for survival models. Statistics in Medicine. 2010;29:262–74. doi: 10.1002/sim.3758. [DOI] [PubMed] [Google Scholar]
- 19.Heller G, Simonoff JS. Prediction in censored survival data: a comparison of the proportional hazards and linear regression models. Biometrics. 1992;48:101–115. [PubMed] [Google Scholar]
- 20.Scher HI, Morris MJ, Stadler WM, Higano C, Basch E, Fizazi K, et al. Trial design and objectives for castration-resistant prostate cancer: Updated recommendations from the Prostate Cancer Clinical Trials Working Group 3. J Clin Oncol. 2016 doi: 10.1200/JCO.2015.64.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


