Abstract
Cystathionine β‐synthase (CBS) catalyzes the formation of l‐cystathionine from l‐serine and l‐homocysteine. The resulting l‐cystathionine is decomposed into l‐cysteine, ammonia, and α‐ketobutylic acid by cystathionine γ‐lyase (CGL). This reverse transsulfuration pathway, which is catalyzed by both enzymes, mainly occurs in eukaryotic cells. The eukaryotic CBS and CGL have recently been recognized as major physiological enzymes for the generation of hydrogen sulfide (H2S). In some bacteria, including the plant‐derived lactic acid bacterium Lactobacillus plantarum, the CBS‐ and CGL‐encoding genes form a cluster in their genomes. Inactivation of these enzymes has been reported to suppress H2S production in bacteria; interestingly, it has been shown that H2S suppression increases their susceptibility to various antibiotics. In the present study, we characterized the enzymatic properties of the L. plantarum CBS, whose amino acid sequence displays a similarity with those of O‐acetyl‐l‐serine sulfhydrylase (OASS) that catalyzes the generation of l‐cysteine from O‐acetyl‐l‐serine (l‐OAS) and H2S. The L. plantarum CBS shows l‐OAS‐ and l‐cysteine‐dependent CBS activities together with OASS activity. Especially, it catalyzes the formation of H2S in the presence of l‐cysteine and l‐homocysteine, together with the formation of l‐cystathionine. The high affinity toward l‐cysteine as a first substrate and tendency to use l‐homocysteine as a second substrate might be associated with its enzymatic ability to generate H2S. Crystallographic and mutational analyses of CBS indicate that the Ala70 and Glu223 residues at the substrate binding pocket are important for the H2S‐generating activity.
Keywords: amino acid, crystal structure, cystathionine β‐synthase, hydrogen sulfide, Lactobacillus plantarum
Short abstract
Abbreviations
- CBS
cystathionine β‐synthase
- CGL
cystathionine γ‐lyase
- HEPES
4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid
- HPLC
high performance liquid chromatography
- l‐OAS
O‐acetyl‐l‐serine
- NADH
reduced form of nicotinamide adenine dinucleotide
- OASS
O‐acetyl‐l‐serine sulfhydrylase
- PLP
pyridoxal 5′‐phosphate
- rms
root mean square
- TLC
thin layer chromatography.
Introduction
Cystathionine β‐synthase (CBS; EC 4.2.1.22), which is an enzyme requiring pyridoxal 5′‐phosphate (PLP) as a cofactor, works in a reverse transsulfuration pathway to generate l‐cysteine from l‐serine and l‐methionine.1, 2, 3 CBS catalyzes a β‐replacement reaction in which the hydroxyl group of l‐serine is replaced by l‐homocysteine to yield l‐cystathionine. The resulting l‐cystathionine is decomposed into l‐cysteine, ammonia, and α‐ketobutylic acid by another PLP‐dependent enzyme, cystathionine γ‐lyase (CGL; EC 4.4.1.1).3, 4, 5 This reverse transsulfuration pathway catalyzed by CBS and CGL mainly occurs in eukaryotic cells. In humans, l‐homocysteine, which is a nonessential amino acid synthesized from l‐methionine, is recognized as a toxic compound.6 Increased plasma level of l‐homocysteine, mainly caused by CBS deficiency, represents a risk indicator for thrombosis, atherosclerosis, and vascular disease, but it is not necessarily the causal factor. CBS‐deficient homocystinuria is an autosomal recessive inborn error of metabolism resulting from mutations in the CBS gene.
The catalytic mechanism of CBS is shown in Figure 1. PLP forms an internal aldimine bond with the amino group of a Lys residue in the active site of the enzyme. In the first step of the catalysis of CBS, an external aldimine linkage is formed between the amino group of l‐serine as the first substrate and the PLP at the active site. The Lys residue then acts as a base to catalyze an eliminative double bond formation, resulting in the release of a hydroxyl group and the generation of an α‐aminoacrylate intermediate. The subsequent step is the nucleophilic attack of the thiol group of l‐homocysteine on the β‐carbon of the intermediate. As a result, l‐cystathionine is generated in the form of an external aldimine adduct with PLP. When an amino group of the Lys residue again forms the internal aldimine bond, the product is released.
Figure 1.
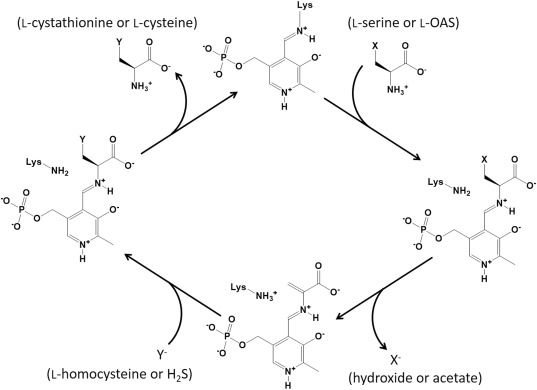
Catalytic mechanism of CBS and OASS for the syntheses of l‐cystathionine and l‐cysteine, respectively. X represents a hydroxyl group in l‐serine or an acetoxyl group in l‐OAS. The α‐aminoacrylate intermediate is formed after the generation of the external aldimine formed between the first substrate (l‐serine or l‐OAS) and PLP. Y represents the anionic form of the second substrate (l‐homocysteine or H2S), which nucleophilically attacks the β‐carbon of the α‐aminoacrylate intermediate. After the formation of an external aldimine between the product (l‐cystathionine or l‐cysteine) and PLP, the product is released concomitantly with the regeneration of an internal aldimine bond.
In addition to playing important roles in sulfur amino acid metabolism, human CBS and CGL have been recognized as major physiological resources of H2S, which is the third identified physiological gasotransmitter after nitric oxide and carbon monoxide.7 In general, H2S is an important signaling molecule for the cardiovascular and nervous systems. The molecule induces smooth muscle relaxation, and exhibits anti‐inflammatory and cytoprotective effects. With respect to the generation of H2S catalyzed by CBS,8, 9, 10 l‐cysteine is used as the first substrate instead of l‐serine. H2S is generated together with the formation of α‐aminoacrylate intermediate. The resulting intermediate may be degraded to ammonia and pyruvate, or may be converted to β‐substituted‐l‐alanine by a β‐replacement reaction with a nucleophilic group, such as hydroxide ion to form l‐serine, or thiol group of l‐cysteine or l‐homocysteine to form l‐lanthionine or l‐cystathionine, respectively.
In many Gram‐positive bacterial species, such as Bacillus subtilis, Bacillus anthracis, Staphylococcus aureus, and Lactobacillus plantarum, and in a few Gram‐negative bacterial species, such as Helicobacter pylori and Pseudomonas aeruginosa, the putative CBS‐ and CGL‐encoding genes are in a cluster in their genomes. In B. subtilis 11 and H. pylori,12 the CBS and CGL genes are clustered together with a putative S‐adenosyl‐l‐homocysteine nucleosidase gene, which might play a role in providing S‐ribosyl‐l‐homocysteine as a precursor for l‐homocysteine. In addition, the B. subtilis CBS, which is also referred to as YrhA or MccA, has been shown to be an O‐acetyl‐l‐serine (l‐OAS)‐dependent enzyme.11 Unlike its eukaryotic counterpart, the enzyme catalyzes the CBS reaction only when l‐serine is acetylated. In L. plantarum, the CBS and CGL genes are clustered together with a putative l‐serine O‐acetyltransferase gene,13 which might play a role in providing l‐OAS. Amino acid sequences of CBS display a similarity with those of l‐OAS sulfhydrylase (OASS; EC 2.5.1.47). OASS enzymes, which directly catalyze the synthesis of l‐cysteine from l‐OAS and H2S (Fig. 1), are found in many bacteria and plants.14, 15 The B. subtilis CBS has been reported to exhibit OASS activity at almost the same level as the l‐OAS‐dependent CBS activity.11 However, OASS activity of the B. subtilis CBS in the presence of high concentrations of the substrates l‐OAS and Na2S is about 100‐fold lower than that of its own OASS‐A (also called CysK). Considering gene organization and the low efficiency of the CBS enzyme to use H2S, cluster genes might play a role in the transfer of sulfur from l‐homocysteine to l‐cysteine, rather than in the fixation of sulfur.
It has been reported that deletion of the gene cluster consisting of the putative CBS and CGL genes in B. anthracis and P. aeruginosa suppresses H2S production.16 Moreover, addition of inhibitors against the human CBS and CGL reduces H2S production in B. anthracis, P. aeruginosa, and S. aureus.16 Like the eukaryotic counterparts, both the bacterial CBS and CGL enzymes may play roles in the generation of H2S, together with possible roles in the synthesis of l‐cysteine from l‐methionine. Surprisingly, deletion of the CBS and CGL genes and/or chemical inactivation of these enzymes render these pathogens highly sensitive to various antibiotics, although exogenous H2S suppresses this effect. The H2S‐mediated antibiotic resistance mechanism might involve a reduction in the oxidative stress imposed by the drugs. These findings indicate that compounds inhibiting the bacterial CBS or CGL can be useful in increasing the efficiency of antibiotics.
Similarity analysis of amino acid sequences of putative bacterial CBS enzymes, encoded by the cluster, suggests that such enzymes can be classified into two types, which we defined as A and B (Fig. 2). Similarly, OASS enzymes are classified into two types, A and B, based on their amino acid sequences.14, 15 In particular, OASS‐B isozymes are known to have a high ability to synthesize S‐sulfo‐l‐cysteine using l‐OAS with thiosulfate. Type‐A bacterial CBS enzymes, which include putative CBSs from B. subtilis, B. anthracis, H. pylori, S. aureus, and L. plantarum, are more similar to OASS‐A enzymes rather than to eukaryotic CBS enzymes. Eukaryotic CBSs have an N‐terminal heme‐binding domain and a C‐terminal S‐adenosyl‐l‐methionine‐binding regulatory domain, in addition to a catalytic domain.1, 17, 18, 19 On the other hand, bacterial CBS‐A enzymes, including the L. plantarum CBS, only have a catalytic domain, similar to the bacterial OASS‐A and OASS‐B enzymes. Given the enzymatic properties of the B. subtilis CBS,11 the enzymes classified into type A should have an l‐OAS‐dependent CBS activity. On the other hand, type‐B bacterial CBS enzymes, which include putative CBSs from P. aeruginosa, Mycobacterium tuberculosis, and Bifidobacterium longum, have a C‐terminal extension and sequence similarity with eukaryotic CBS enzymes. It is currently unknown whether bacterial CBS‐B enzymes need l‐OAS for their CBS activities and if the C‐terminal extension plays a role in the catalytic reaction.
Figure 2.
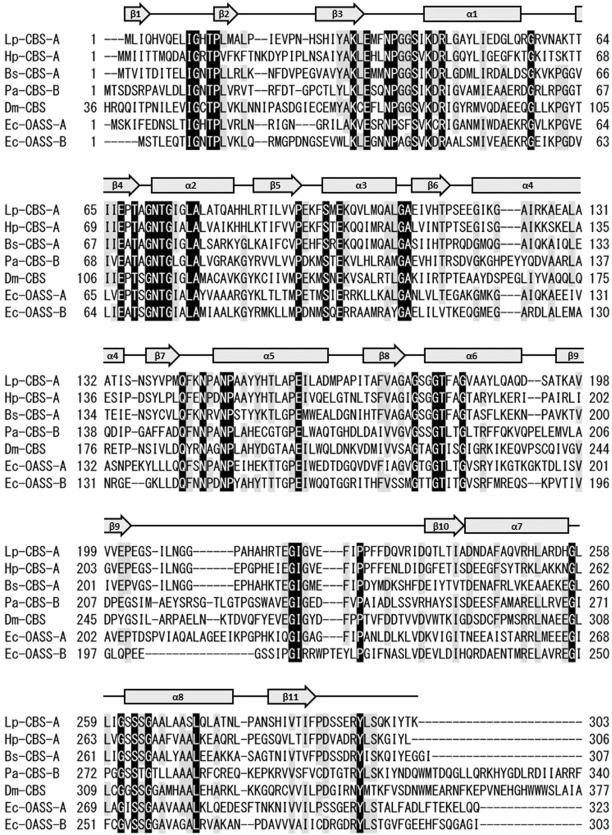
Sequence alignment of the L. plantarum CBS (Lp‐CBS‐A) with other CBS and OASS enzymes. Hp‐CBS‐A, H. pylori CBS‐A; Bs‐CBS‐A, B. subtilis CBS‐A; Pa‐CBS‐B, P. aeruginosa CBS‐B; Dm‐CBS, D. melanogaster CBS; Ec‐OASS‐A, E. coli OASS‐A; Ec‐OASS‐B, E. coli OASS‐B. The fully conserved residues in all sequences are marked with black shading. The residues conserved in more than five enzymes are marked with gray shading. In addition, secondary structures of the monomer in the l‐methionine‐bound L. plantarum CBS are shown above the sequence.
To develop a drug inhibiting the H2S generation in bacteria, characterization of the H2S‐generating enzyme and the identification of the critical residues may be important. In the present study, we determined the detailed enzymatic properties and the crystal structure of the putative CBS enzyme from L. plantarum SN35N, which was previously isolated from pear in our laboratory,20, 21 as a representative of bacterial CBS‐A enzymes.
Results
Enzymatic characterization of the L. plantarum CBS
The L. plantarum CBS with C‐terminal His6‐tag, which was overexpressed in Escherichia coli and purified almost to homogeneity, displays OASS activity (Reaction 1 in Fig. 3 ) as well as l‐OAS‐dependent CBS activity (Reaction 2 in Fig. 3 ), but not a general CBS activity. In addition, the L. plantarum CBS has a low ability to synthesize S‐sulfo‐l‐cysteine.
Figure 3.
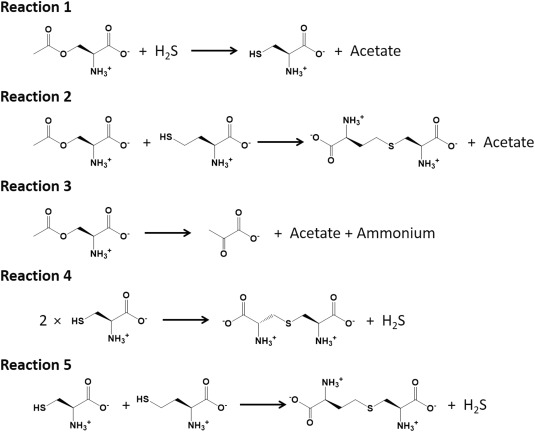
Reactions catalyzed by the L. plantarum CBS characterized in the present study.
Kinetic parameters for the OASS and the l‐OAS‐dependent CBS activities have been determined (Table 1). OASS activity of the L. plantarum CBS was significantly inhibited when l‐OAS was added at high concentrations. The substrate inhibition is frequently observed for the OASS enzymes. For example, the OASS‐A enzyme from Salmonella typhimurium (currently Salmonella enterica subsp. enterica serovar Typhimurium) showed the substrate inhibition by both substrates (l‐OAS and Na2S).22 Numerical analysis suggested that l‐OAS inhibits the catalytic activity of the L. plantarum CBS by the binding to the l‐OAS‐bound and the aminoacrylate‐bound forms with different binding affinities (Table 1). On the other hand, in the l‐OAS‐dependent CBS activity, the substrate inhibition was observed under the high concentration of l‐homocysteine. The l‐homocysteine may inhibit the activity mainly by the binding to the substrate‐free form, since the K I1 value was calculated to be much smaller than the K I2 value (Table 1). In the OASS and l‐OAS‐dependent CBS reactions, the K m value for l‐OAS and k cat are similar (Table 1). In addition, the K m for Na2S in l‐cysteine synthesis is similar to that for l‐homocysteine in l‐cystathionine synthesis (Table 1). Therefore, the k cat/K m values for l‐OAS and Na2S in l‐cysteine synthesis (1.4 ± 0.2 × 104 and 5.8 ± 0.8 × 104 M−1s−1, respectively) is similar to those for l‐OAS and l‐homocysteine in l‐cystathionine synthesis (1.9 ± 0.5 × 104 and 7.9 ± 1.3 × 104 M−1s−1, respectively).
Table 1.
Kinetic and Affinity Parameters
| Wild type | A70S | E223G | ||
|---|---|---|---|---|
| Reaction 1 | k cat (s−1) | 110 ± 9 | 3.9 ± 0.5 | 5.0 ± 0.7 |
| K m for l‐OAS (mM) | 8.0 ± 0.8 | 0.75 ± 0.15 | 28 ± 5 | |
| K I1 for l‐OAS (mM) | 43 ± 6 | – | 23 ± 7 | |
| K I2 for l‐OAS (mM) | 15 ± 2 | – | 71 ± 19 | |
| k cat/K m for l‐OAS (M−1s−1) | 1.4 ± 0.2 × 104 | 5.2 ± 1.2 × 103 | 1.8 ± 0.4 × 102 | |
| K m for Na2S (mM) | 1.9 ± 0.2 | 1.8 ± 0.3 | 0.28 ± 0.07 | |
| k cat/K m for Na2S (M−1s−1) | 5.8 ± 0.8 × 104 | 2.1 ± 0.3 × 103 | 1.8 ± 0.5 × 104 | |
| Reaction 2 | k cat (s−1) | 77 ± 7 | 3.5 ± 0.4 | 2.4 ± 0.3 |
| K m for l‐OAS (mM) | 4.2 ± 1.1 | 0.47 ± 0.17 | 19 ± 4 | |
| k cat/K m for l‐OAS (M−1s−1) | 1.9 ± 0.5 × 104 | 7.5 ± 2.9 × 103 | 1.2 ± 0.3 × 102 | |
| K m for l‐homocysteine (mM) | 1.0 ± 0.1 | 2.7 ± 0.5 | 0.26 ± 0.08 | |
| K I1 for l‐homocysteine (mM) | 1.2 ± 0.4 | 1.3 ± 0.5 | – | |
| K I2 for l‐homocysteine (mM) | 42 ± 36 | 36 ± 23 | – | |
| k cat/K m for l‐homocysteine (M−1s−1) | 7.9 ± 1.3 × 104 | 1.3 ± 0.3 × 103 | 9.2 ± 3.0 × 103 | |
| Reaction 3 | k cat (s−1) | 0.014 ± 0.001 | NDa | NDb |
| K m for l‐OAS (mM) | 0.034 ± 0.009 | NDa | NDb | |
| k cat/K m for l‐OAS (M−1s−1) | 4.3 ± 1.2 × 102 | NDa | NDb | |
| K d for l‐OAS (μM) | 5.1 ± 0.2 | < 0.1 | NDb | |
| Reaction 4 | k cat (s−1) | 1.1 ± 0.1 | NDc | 0.022 ± 0.003 |
| K m for l‐cysteine (mM) | 8.9 ± 1.1 | NDc | 37 ± 14 | |
| k cat/K m for l‐cysteine (M−1s−1) | 1.2 ± 0.1 × 102 | 2.0 ± 0.1 | 0.60 ± 0.24 | |
| Reaction 5 | k cat (s−1) | 7.6 ± 0.9 | 2.0 ± 0.3 | 0.33 ± 0.09 |
| K m for l‐cysteine (mM) | 5.9 ± 1.2 | 3.0 ± 0.2 | 37 ± 14 | |
| k cat/K m for l‐cysteine (M−1s−1) | 1.3 ± 0.3 × 103 | 1.5 ± 0.2 × 103 | 8.9 ± 4.2 | |
| K m for l‐homocysteine (mM) | 0.54 ± 0. 08 | 20 ± 2 | 0.40 ± 0.13 | |
| K I1 for l‐homocysteine (mM) | 1.2 ± 0.3 | – | – | |
| K I2 for l‐homocysteine (mM) | 8.0 ± 5.1 | – | – | |
| k cat/K m for l‐homocysteine (M−1s−1) | 1.4 ± 0.3 × 104 | 1.5 ± 0.2 × 102 | 8.3 ± 0.4 × 102 |
Parameters were unable to be determined due to the very low reactivity.
Due to protein aggregation, experiments could not be performed.
Since K m was too high, K m and k cat could not be determined.
The enzymatic ability of the L. plantarum CBS to generate H2S was also analyzed. When l‐cysteine was used as the only substrate for the L. plantarum CBS, H2S was generated with a low efficiency (the k cat/K m value was determined to be 1.2 ± 0.1 × 102 M−1s−1, Table 1). Namely, although the L. plantarum CBS can produce l‐cysteine from l‐OAS and H2S by the OASS activity, the enzyme can react with l‐cysteine to generate H2S. The possible byproducts formed from l‐cysteine, which are cogenerated together with H2S by CBS, are pyruvate, l‐serine, or l‐lanthionine (Reaction 4 in Fig. 3).10 After addition of l‐cysteine to the L. plantarum CBS, pyruvate was not detected in the reaction mixture where the coupling enzyme l‐lactate dehydrogenase and its cofactor NADH were present. On the other hand, HPLC analysis confirmed that lanthionine was formed in the reaction mixture, although l‐serine was not. Configuration of the product was not determined, because a mixture of l‐, d‐, and meso‐forms was used as an authentic sample. However, the product of the L. plantarum CBS is highly likely to be l‐lanthionine.
The generation of H2S by the L. plantarum CBS was greatly improved by using a mixture of l‐cysteine and l‐homocysteine, similar to the case of eukaryotic CBSs.10 Maximum velocity of the H2S synthesis in the presence of l‐homocysteine is 6.9‐fold higher than that in the absence of l‐homocysteine, although the K m value for l‐cysteine was reduced 1.5‐fold in the presence of l‐homocysteine (Table 1). Therefore, the k cat/K m value for l‐cysteine in the presence of l‐homocysteine (1.3 ± 0.3 × 103 M−1s−1) is one order higher than that in the absence of l‐homocysteine (1.2 ± 0.1 × 102 M−1s−1). HPLC analysis suggested that l‐cystathionine is a major byproduct generated from the equimolar mixture of l‐cysteine and l‐homocysteine (Reaction 5 in Fig. 3), while l‐lanthionine is a minor one. These results indicate that the L. plantarum CBS prefers l‐homocysteine to l‐cysteine as a second substrate.
Maximum velocities of the H2S synthesis using l‐cysteine as a first substrate (1.1 ± 0.1 and 7.6 ± 0.9 s−1 for Reactions 4 and 5, respectively, Table 1) are significantly lower than those of the reactions using l‐OAS as a first substrate (110 ± 9 and 77 ± 7 s−1 for Reactions 1 and 2, respectively, Table 1). However, the k cat/K m value for l‐homocysteine in Reaction 5 (1.4 ± 0.3 × 104 M−1s−1) is comparable with that for Na2S in Reaction 1 (5.8 ± 0.8 × 104 M−1s−1) or with that for l‐homocysteine in Reaction 2 (7.9 ± 1.3 × 104 M−1s−1). These data suggest that the enzyme can significantly generate H2S from l‐cysteine when the concentration of H2S is lower than that of l‐homocysteine.
Spectroscopic analysis of the L. plantarum CBS
When l‐OAS (1 mM) was added to the CBS solution (10 μM), the wavelength showing the maximum absorbance shifted from 412 to 470 nm, along with a concomitant increase in absorbance at 330 nm [Fig. 4(A)], indicating the formation of the aminoacrylate intermediate.22, 23, 24 In general, α‐aminoacrylate bound to PLP in the active site of OASS or CBS is slowly degraded to pyruvate or l‐serine in the absence of a second substrate.10, 23 HPLC analysis suggested that the L. plantarum CBS enzyme does not accelerate the conversion of l‐OAS to l‐serine. On the other hand, in contrast from the case of l‐cysteine addition to the L. plantarum CBS, the formation of pyruvate (Reaction 3 in Fig. 3) was increased in a manner dependent on l‐OAS concentration, suggesting that the aminoacrylate intermediate was converted to pyruvate in the absence of a second substrate. However, the k cat/K m value for l‐OAS in pyruvate synthesis (4.3 ± 1.2 × 102 M−1s−1, Table 1) is significantly lower than that in l‐cystathionine synthesis (1.9 ± 0.5 × 104 M−1s−1) or that in l‐cysteine synthesis (1.4 ± 0.2 × 104 M−1s−1).
Figure 4.
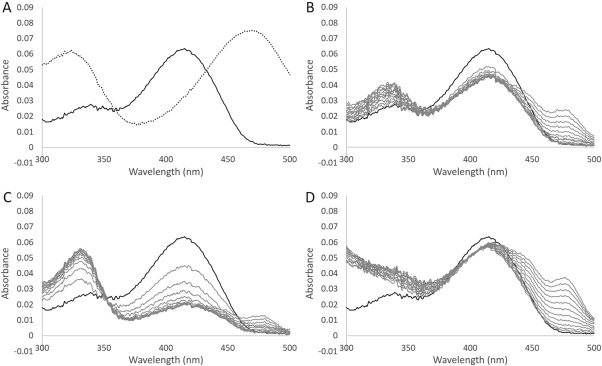
Absorption spectrum of the L. plantarum CBS. A: Absorption spectrum of solutions containing 10 μM CBS in the absence (solid line) or in the presence of 1 mM l‐OAS (dotted line). B, C: Changes in the spectrum after addition of 1 or 10 mM l‐cysteine to a solution containing CBS, respectively. D: Changes in the spectrum after addition of 1 mM lanthionine to a solution containing CBS. The bold line represents the spectrum before the addition of l‐cysteine or lanthionine. The spectrum was recorded every 3 min.
When l‐cysteine was added to the L. plantarum CBS solution, the wavelength displaying the maximum absorbance was immediately shifted from 412 to 420 nm, together with a decrease in absorbance and a concomitant increase in absorbance at 330 nm [Fig. 4(B,C)]. The change in the absorption spectrum after addition of l‐cysteine was dependent on concentration. The decrease in absorbance at 420 nm as well as the increase in absorbance at 330 nm was more pronounced when the concentration of l‐cysteine was higher. Furthermore, absorbance between 430 and 500 nm was increased by longer incubations [Fig. 4(B,C)]. In the case of lanthionine addition to the L. plantarum CBS solution, the wavelength showing the maximum absorbance immediately shifted from 412 to 420 nm, together with an increase in absorbance below 350 nm [Fig. 4(D)]. Moreover, the absorbance between 430 and 500 nm was increased by longer incubations. In particular, one absorption peak appeared at 480 nm.
Overall structure of the L. plantarum CBS
The structures of the wild‐type L. plantarum CBS were determined with the molecular replacement method, and refined at a 2.4 Å resolution (Table 1I). One asymmetric unit in the crystal contains four monomers. Like the majority of bacterial OASSs,14, 15 the L. plantarum CBS takes a dimeric structure in solution. In crystal, two monomers in the asymmetric unit form a dimer, while each of the other two monomers forms a dimer with a monomer in another asymmetric unit, which is related by a crystallographic twofold axis. The structural arrangement of the dimer subunits [Fig. 5(A)] is very similar to that of bacterial OASSs.
Figure 5.
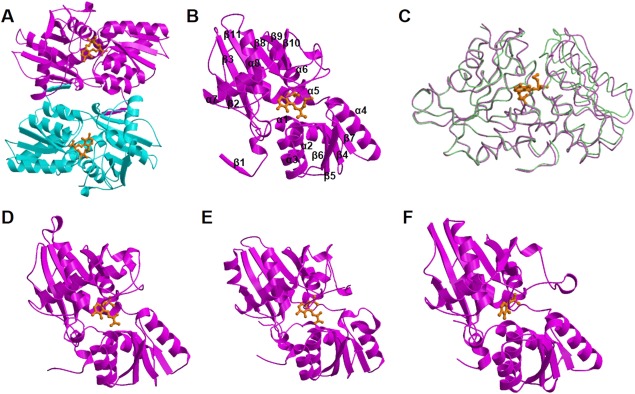
Ribbon diagrams of the crystal structures of the L. plantarum CBS. A: Dimeric structure of the K42A variant, which noncovalently binds to the external aldimine formed between PLP and l‐methionine. The two subunits are colored in magenta and cyan. B: Monomeric structure (subunit A) of the l‐methionine‐bound L. plantarum CBS. C: Superposition of the monomeric structures of the L. plantarum CBS. One of the l‐methionine‐unbound subunits and one of the l‐methionine‐bound subunits are colored in purple and light green, respectively. Superposition was calculated to maximally fit large domains. The external aldimine molecules formed between PLP and l‐methionine are also shown. D: Monomeric structure of M. tuberculosis OASS‐A complexed with aminoacrylate intermediate. E: Structure of catalytic core of Drosophila CBS complexed with aminoacrylate intermediate. F: Monomeric structure of the closed form of E. coli OASS‐B, in which PLP forms internal aldimine. View directions in D–F are the same with the direction in B. PLP or its amino acid complex was shown in the stick model colored in orange.
Each monomer of the L. plantarum CBS takes the typical fold of type II PLP enzymes,25 with the cofactor PLP covalently bound to an invariant Lys residue (Lys42) in the active site. The chain fold consists of eleven β‐strands, eight α‐helices, and three 310‐helices that are arranged in two domains, which are referred to as large and small domains [Fig. 5(B)]. The large domain consists of the residues Thr13–Met34 and Pro146–Lys303 and has a central five‐stranded parallel β‐sheet (β3, β11, β8, β9, β10), with β2 as an antiparallel addition to one of the edges (β3). This β‐sheet is flanked by helices α5 through α8. The small domain contains the residues His5–His12 and Met34–Asn145 with a central four‐stranded parallel β‐sheet (β7, β4, β5, β6) flanked by helices α1 through α4. N‐terminal residues containing the β1‐strand are positioned away from the main body of the subunit. The β1‐strand of one subunit interacts with the β2‐strand in the other subunit in a parallel fashion.
CBS and OASS enzymes are known to take two distinct conformations, open and closed.1, 2, 3, 14, 15 Binding of the first substrate into the active site of the enzyme induces a structural change from the open to the closed conformation, which is mediated by domain rotation. As a result, the binding pocket for the first and second substrates becomes smaller. In the crystal of the wild‐type L. plantarum CBS, which was grown in the absence of the substrate or its analogs, each of the four monomers in the asymmetric unit takes an open conformation [Figs. 5(C) and 6(A)]. The monomeric structures are very similar to each other, and the rms deviation values between pairs of two monomers were calculated between 0.15 and 0.39 Å for 303 Cα atoms. When compared among the four structures, the residues Pro202–Ile225 were shown to be more varied than the other residues. Specifically, the residues Pro202–Ile225 were poorly defined in the electron density map, and the B‐factors were calculated to be larger than the other residues.
In previous attempts to obtain structures of OASSs with bound substrates, active site variants carrying a substitution of the catalytic Lys residue with an Ala were designed. Structural analysis of such variants from S. typhimurium 26 and Arabidopsis thaliana 27 revealed that an external aldimine had formed at the active site via a reaction with l‐methionine during protein production. The S. typhimurium enzyme showed a dynamic conformational change to the closed form, although the l‐methionine complex of OASS from A. thaliana did not show any major structural rearrangements upon ligand binding. Another attempt to obtain the substrate‐bound structure was done with the OASS‐A (also referred to as CysK1) from M. tuberculosis.28 In this case, the α‐aminoacrylate intermediate was trapped by cryo‐freezing after soaking the crystal in a solution containing l‐OAS [Figs. 5(D) and 6(C)]. The trapped structure is similar to that of the l‐methionine‐bound S. typhimurium OASS. In addition, by using a similar method, carbanion (deprotonated l‐serine) and aminoacrylate intermediates were found in the crystal structure of CBS from Drosophila melanogaster 29 [Figs. 6(E) and 6(D)].
Figure 6.
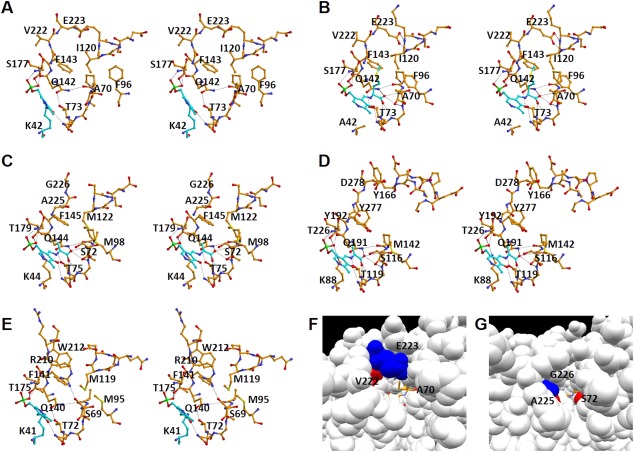
Active site structure. A: Active site of the wild‐type L. plantarum CBS in the open form in which the internal aldimine is formed. B: Active site of the K42A variant of the L. plantarum CBS in the closed form bound to the external aldimine. C: Active site of the M. tuberculosis OASS‐A in the closed form bound to the α‐aminoacrylate intermediate.28 D: Active site of the Drosophila CBS in the closed form bound to the α‐aminoacrylate intermediate.29 E: Active site of the E. coli OASS‐B in the closed form in which the internal aldimine is formed.33 Carbon atoms of PLP and the PLP‐attached residue (Lys residue in A and E) or PLP‐attached compounds (l‐methionine in B and α‐aminoacrylate in C and D) are shown in light sky blue. F: Substrate‐binding pocket of the L. plantarum CBS in the closed form. G: Substrate‐binding pocket of the M. tuberculosis OASS‐A in the closed form. In F & G, key amino acid residues are colored red or blue. External aldimine was shown in the stick model colored in orange.
In the present study, the K42A variant of the L. plantarum CBS, in which the catalytic Lys42 residue is replaced by Ala, was generated. The crystal structure of the L. plantarum CBS in a complex with the l‐methionine‐bound external aldimine was obtained from the crystal of the K42A variant grown in the presence of PLP and l‐methionine, and refined at a 3.3 Å resolution (Table 1I). One asymmetric unit in the crystal contains four dimers. Eight subunits in the asymmetric unit seem to take closed conformations upon binding of l‐methionine. The structures resemble one another, partly because the crystal structure was refined by imposing strong noncrystallographic symmetry restraints, due to the low quality of the crystal. One of four dimers was extraordinarily defined in the poor electron density map, indicating the imperfectness of the crystal.
The l‐methionine‐bound form of the L. plantarum CBS can be superimposed with the l‐methionine‐unbound form with an rms deviation between 0.77 and 1.00 Å for 303 Cα atoms. On the other hand, when calculated using the Cα atoms within each domain, rms deviation values decreased (between 0.55 and 0.65 Å in the case of small domains and between 0.39 and 0.52 Å in the case of large domains). The active site in each of the eight subunits determined seems to be closed by the rotation of the small domain [Fig. 5(C)]. As a result, the Pro93–Ser97 residues, which include the loop between the β5‐strand and the α3‐helix, and the Ser116–Lys121 residues, which include the loop between the β6‐strand and the α4‐helix and the first helical turn in the α4‐helix, located in the small domain move towards the residues Pro202–Ile225 in the large domain [Fig. 6(B)]. In contrast to the l‐methionine‐unbound form, the residues Pro202–Ile225 in the l‐methionine‐bound form were clearly defined in the electron density map.
The sequence homology search using the FASTA program30 indicates that the L. plantarum CBS is more similar to the bacterial OASS‐As than to the eukaryotic CBSs or to the bacterial OASS‐Bs (Fig. 2). For examples, the amino acid sequence of the L. plantarum CBS displays a 42% identity with that of the M. tuberculosis OASS‐A, a 38% identity with that of Drosophila CBS, and a 35% identity with that of the E. coli OASS‐B. The structural similarity search of proteins using the DALI program31 also suggests that the L. plantarum CBS is more similar to the bacterial OASS‐A than to the eukaryotic CBS or to the bacterial OASS‐B. A structure‐based sequence alignment of the L. plantarum CBS with CBS enzymes from other sources (H. pylori and D. melanogaster) and OASS enzymes (OASS‐A and OASS‐B from E. coli) is shown in Figure 2. The figure also includes the results of multiple sequence alignments with other bacterial CBSs, which was conducted with ClustalW.32
The l‐methionine‐bound form of the L. plantarum CBS can be well superimposed with the closed form of OASS and CBS enzymes. The best matched structure is the closed form (chain B) of the H. pylori CBS (unpublished data, PDB ID: 4R2V) with an rms deviation of 1.1 Å for 302 Cα atoms. The H. pylori CBS is annotated as cysteine synthase in PDB. However, considering gene organization12 and sequence similarity, the enzyme might have a CBS activity coupled with the generation of H2S, like the L. plantarum CBS. The second matched structure is the closed form of the M. tuberculosis OASS‐A28 (PDB ID: 2Q3D [Fig. 5(D)]) with an rms deviation of 1.5 Å for 301 Cα atoms. With respect to the similarity to other class enzymes, the L. plantarum CBS can be superimposed with the closed form of the Drosophila CBS29 (PDB ID: 3PC3, [Figs. 5(E)]) with an rms deviation of 1.8 Å for 300 Cα atoms, and with that of the E. coli OASS‐B33 (PDB ID: 2BHT, [Fig. 5(F)]) with an rms deviation of 1.6 Å for 288 Cα atoms. In particular, the active site architecture of the L. plantarum CBS is very similar to that of bacterial OASS‐As, but distinct from that of bacterial OASS‐Bs and eukaryotic CBSs (Fig. 6). This is consistent with the finding that the L. plantarum CBS cannot catalyze the general CBS reaction exhibited by eukaryotic CBS and has a low activity to synthesize S‐sulfo‐l‐cysteine characteristic to bacterial OASS‐B.
Active site of the L. plantarum CBS
In the crystal structure of the L. plantarum CBS, as in the structures of the OASS and CBS enzymes reported previously,1, 2, 3, 14, 15 the internal aldimine bond is formed between the amino group of an invariant Lys residue (Lys42) and PLP [Fig. 6(A)]. The pyridine ring of PLP forms hydrogen bonds with invariant Asn72 and Ser263 residues (Figs. 2 and 7). On the other hand, its phosphate group forms hydrogen bonds with the well‐conserved Gly176, Ser177, Gly178, and Thr180 residues (Fig. 7). Gly176 and Thr180 are almost invariant among CBS and OASS enzymes (Fig. 2).
Figure 7.
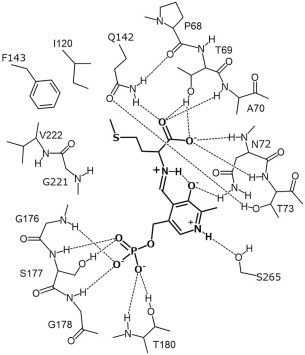
Schematic representation of the interactions between the residues in the L. plantarum CBS and the external aldimine molecule formed between PLP and l‐methionine.
To compare the substrate‐binding pocket among bacterial CBS‐A, OASS‐A, OASS‐B, and eukaryotic CBS, the pocket structures in the closed forms of L. plantarum CBS, M. tuberculosis OASS‐A,28 Drosophila CBS,29 and E. coli OASS‐B33 are depicted in Figure 6(B–E), respectively. The substrate‐binding pocket of the L. plantarum CBS was clarified by the crystal structure of the l‐methionine‐bound form of the K42A variant [Figs. 6(B) and 7]. The carboxyl group of the external aldimine adduct forms hydrogen bonds with the hydroxyl oxygen atom of Thr69, the Nε2 atom of Gln142, and backbone nitrogen atoms of Ala70, Asn72, and Thr73 [Figs. 6(B) and 7]. In addition, the Oε1 atom of the Gln142 residue forms a hydrogen bond with the hydroxyl group of a conserved Thr73 residue. Thr69, Asn72, Thr73, and Gln142 of the L. plantarum CBS are highly conserved among OASS and CBS enzymes (Fig. 2), and the structures of the carboxylate‐binding site are similar [Fig. 6(B–E)]. However, although Ala70 of the L. plantarum CBS is conserved among bacterial CBS‐As, it is replaced by Ser in eukaryotic CBSs and bacterial OASSs (Fig. 2).
The side chain of PLP‐bound l‐methionine is surrounded by main‐chain portions of Gly176 and Gly219–Gly221 in the large domain and hydrophobic side chains of Ala70, Phe96, Ile120, and Phe143 in the small domain [Figs. 6(B) and 7]. The Glu223 residue is positioned at the upper part of the l‐methionine side chain, as shown in Figure 6B. The Ile120 residue in the L. plantarum CBS, which is located at the opposite side of the Glu223 residue in the substrate‐binding pocket, is positioned in the N‐terminal part of the α4‐helix [Fig. 6(A,B)]. Especially, the Glu223 residue in the l‐methionine‐bound form interacts with the N‐terminal part of the α4‐helix, which has a positive potential due to the helix dipole effect [Fig. 6(B)].
When compared with the amino acid sequence close to the Glu223 residue in the L. plantarum CBS, a three‐residue‐insertion is observed in the sequence of bacterial OSAA‐Bs (Fig. 2). As a result, the structure of this region in OASS‐B, which may be important for the binding of thiosulfate as the second substrate, is very different from that of the corresponding regions in the L. plantarum CBS [Fig. 6(B,E)]. On the other hand, eukaryotic CBSs have a three‐residue‐insertion in the sequence close to a residue corresponding to Ile120 in the L. plantarum CBS (Fig. 2). As a result, the structure of this region in eukaryotic CBSs is very different from that of the corresponding regions in the L. plantarum CBS [Fig. 6(B,D)], and the size of the second substrate‐binding pocket in eukaryotic CBSs is larger than that in the L. plantarum CBS. Amino acid sequence comparison suggests that bacterial CBS‐Bs might have similar structures with eukaryotic ones in this region (Fig. 2).
When compared with the amino acid sequences close to the Ile120 and Glu223 residues in the L. plantarum CBS, bacterial OASS‐As display no insertion or deletion (Fig. 2). The structure around the Ile120 residue in the L. plantarum CBS, which interacts with the Glu223 residue in the closed form, is similar to that of the corresponding part in the bacterial OASS‐A enzymes [Fig. 6(B,C)]. However, the Glu223 residue, which is conserved among bacterial CBS‐As, is replaced by Gly in bacterial OASS‐As (Fig. 2). Therefore, the inter‐domain interaction is not formed in the bacterial OASS‐As due to the absence of a side chain in the corresponding Gly residue [Fig. 6(B,C)]. The Glu223 residue in the L. plantarum CBS seems to restrict the closure of the two domains, thereby determining the lower limit of the size of the substrate‐binding pocket. In addition, the side chain of the adjacent Val222 residue in the L. plantarum CBS protrudes inside of the large domain [Fig. 6(B)], whereas that of the corresponding Ala residues in the bacterial OASS‐A protrude towards the substrate‐binding pocket [Fig. 6(C)]. As a result, the size of the second substrate‐binding pocket in the closed form of the L. plantarum CBS is much larger than that of the bacterial OASS‐As [Fig. 6(F,G)].
Mutational analysis
Amino acid sequence and crystal structure of the L. plantarum CBS are similar to those of bacterial OASS‐As, but the enzymatic properties of the L. plantarum CBS are distinct from those of bacterial OASS‐As. The Ala70 and Glu223 residues, which are present in the second substrate‐binding pocket in the L. plantarum CBS, are well conserved among bacterial CBS‐As, whereas they are replaced by the Ser and Gly residues in the bacterial OASS‐As, respectively. To clarify the roles of the Ala70 and Glu223 residues in the L. plantarum CBS, we investigated the kinetic and spectroscopic properties of the A70S and E223G variants of the enzyme (Table 1).
Similar to the wild type, when l‐OAS (1 mM) was added to the A70S variant (10 μM), the wavelength showing the maximum absorbance shifted from 412 to 470 nm, with a concomitant increase in absorbance at 330 nm [Fig. 8(A)]. However, the titration analysis showed that the absorbance at 470 nm of the variant was simply proportional to the concentrations of added l‐OAS, until the concentrations of l‐OAS reached the concentration of the protein. Therefore, although the apparent K d value of this variant towards l‐OAS could not be determined, it was estimated to be lower than that of the wild type (5.1 ± 0.2 μM, Table 1). In addition, the formation of pyruvate was not detected after addition of l‐OAS to the variant, in contrast to the result obtained from the wild type. The K m value of the variant for l‐OAS was decreased 11‐ and 8.9‐fold in the OASS reaction and l‐OAS‐dependent CBS reaction, respectively (Table 1). On the other hand, the k cat value of the variant was also decreased 28‐ and 22‐fold in the OASS and l‐OAS‐dependent CBS reactions, respectively (Table 1).
Figure 8.
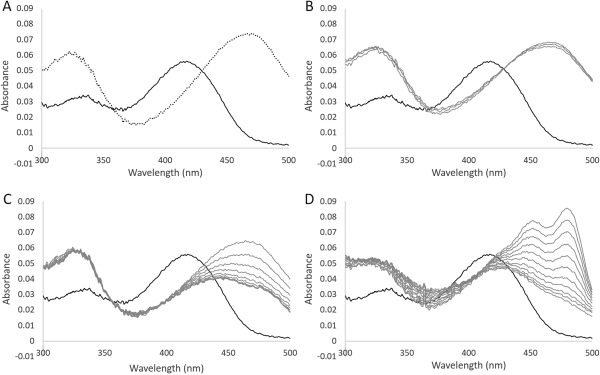
Absorption spectrum of the A70S variant of the L. plantarum CBS. A: Absorption spectrum of solutions containing 10 μM A70S variant in the absence (solid line) or in the presence of 1 mM l‐OAS (dotted line). B, C: Changes in the spectrum after addition of 1 or 10 mM l‐cysteine to a solution containing the A70S variant, respectively. D: Changes in the spectrum after addition of 1 mM lanthionine to a solution containing the A70S variant. The bold line represents the spectrum before the addition of l‐cysteine or lanthionine. The spectrum was recorded every 3 min.
In contrast to the wild type, when l‐cysteine (1 or 10 mM) was added to the variant, the wavelength showing the maximum absorbance immediately shifted from 412 to 470 nm, with a concomitant increase in absorbance at 330 nm, suggesting the accumulation of the aminoacrylate intermediate [Fig. 8(B,C)]. Subsequently, the peak at 470 nm decreased, together with a broadening of the peak. The decrease in absorbance at 470 nm and the broadening of the peak were accelerated by increasing l‐cysteine concentrations. When 1 mM lanthionine was added to the variant, the wavelength displaying the maximum absorbance immediately shifted from 412 to 425 nm, together with a decrease in absorbance and a broadening of the peak, while absorbance at 325 nm was increased. Subsequently, two absorption peaks at 455 and 480 nm gradually appeared [Fig. 8(D)].
With respect to the H2S generation activity from l‐cysteine, the k cat/K m value of the A70S variant was 60‐fold lower than that of the wild type (Table 1). On the other hand, with respect to the H2S generation activity from a mixture of l‐cysteine and l‐homocysteine, the K m value of the A70S variant for the first substrate displayed a 2.0‐fold decrease, whereas the K m value for the second substrate displayed a 37‐fold increment (Table 1). As a result, the k cat/K m value of the A70S variant for l‐cysteine is similar with that of the wild type, whereas the k cat/K m value for l‐homocysteine is 93‐fold decreased (Table 1). Evaluating the k cat/K m values, the enzymatic ability to utilize l‐cysteine or l‐homocysteine as a second substrate is commonly lowered. Similarly, the k cat/K m value of the variant for Na2S and l‐homocysteine was decreased 28‐ and 61‐fold in the OASS and l‐OAS‐dependent CBS reactions, respectively, mainly due to the decrease in the k cat values (Table 1). In summary, the A70S variant exhibits an increased affinity towards the first substrate, l‐OAS or l‐cysteine; however, its reactivity towards H2S, l‐cysteine or l‐homocysteine as second substrates, is decreased.
Concerning the E223G variant, the addition of l‐OAS was found to induce aggregation of the protein. Therefore, the apparent K d value towards l‐OAS and the kinetic parameters for the pyruvate formation activity from l‐OAS could not be determined. Similar to the case of the A70S variant, the k cat value of the E223G variant was decreased 22‐ and 32‐fold in the OASS and l‐OAS‐dependent CBS reactions, respectively. On the other hand, the K m value of the variant for l‐OAS was increased to 3.5‐ and 4.5‐fold in the OASS and l‐OAS‐dependent CBS reactions, respectively. Therefore, as shown in Table 1, the k cat/K m value of the variant for l‐OAS (1.8 ± 0.4 × 102 and 1.2 ± 0.3 × 102 M−1s−1 for Reactions 1 and 2, respectively) was decreased by two orders of magnitude when compared to the wild type. On the other hand, the K m value of the E223G variant for Na2S in the OASS reaction and the value for l‐homocysteine in the l‐OAS‐dependent CBS reaction were decreased than those of the wild type. As a result, the k cat/K m value for Na2S in the OASS reaction and the value for l‐homocysteine in the l‐OAS‐dependent CBS reaction were decreased only 3.2‐ and 8.5‐fold, respectively.
With respect to the H2S‐generating activity from l‐cysteine, the k cat/K m value of the E223G variant was 200‐fold lower than that of the wild type due to the decrease in the k cat value and the increase in the K m value (Table 1). Similarly, the E223G variant has a decreased ability to generate H2S from a mixture of l‐cysteine and l‐homocysteine, although the K m value for l‐homocysteine is slightly lower than that of the wild type. The k cat/K m values for both l‐cysteine and l‐homocysteine were decreased 150‐fold and 17‐fold, respectively (Table 1). In summary, the E223G variant exhibits an increased affinity towards the second substrate, Na2S or l‐homocysteine, whereas it exhibits a decreased affinity towards the first substrate, l‐OAS or l‐cysteine. In addition, the maximum velocity of the variant was decreased in all of the β‐replacement reactions.
Discussion
H2S‐generating ability of the L. plantarum CBS
Based on their amino acid sequences, bacterial CBS‐A enzymes are similar to OASS‐A enzymes, which directly generate l‐cysteine by using l‐OAS and H2S, while bacterial CBS‐B enzymes are similar to eukaryotic CBS enzymes. In humans, CBS is known as one of the major physiological sources to generate H2S.7 In the present study, we focused on the enzymatic properties and structure of the L. plantarum CBS‐A as an H2S‐generating enzyme.
The L. plantarum CBS was not able to catalyze the general CBS reaction exhibited by eukaryotic forms. However, similar to the CBS enzyme from B. subtilis,11 the L. plantarum CBS exhibits an l‐OAS‐dependent CBS activity and an OASS activity (Fig. 3). Furthermore, although the enzymatic ability of the L. plantarum CBS to generate H2S by condensation of two l‐cysteine molecules is low, the enzyme efficiently generates H2S from a mixture of l‐cysteine and l‐homocysteine, together with the formation of l‐cystathionine (Table 1).
The CBS and OASS enzymes have H2S‐consuming and H2S‐generating activities in varying degrees. For examples, eukaryotic CBSs can generate H2S from l‐cysteine especially in the presence of l‐homocysteine,10 whereas they can generate l‐cysteine from l‐serine and H2S with low catalytic efficiency.34 On the other hand, although the main activity of the OASS enzymes is the synthesis of l‐cysteine from l‐OAS and H2S, they can generate H2S from l‐cysteine via the same mechanism with eukaryotic CBSs.35
The k cat value in the H2S‐consumig OASS reaction of the L. plantarum CBS is significantly higher than that in the H2S‐generating reactions (Table 1). The reason should be that acetoxyl group in l‐OAS can be easily liberated from the active site than the thiol group in l‐cysteine due to the absence of the reverse reaction. However, the k cat/K m value of the enzyme for Na2S in the H2S‐consumig l‐cysteine synthesis is comparable to that for l‐homocysteine in the H2S‐generating cystathionine synthesis. It should be noticed that the k cat/K m value of the L. plantarum CBS for Na2S in l‐cysteine synthesis is much lower than that of general OASS enzymes (e.g., 3.4 × 108 and 2.4 × 107 M−1s−1 for the E. coli OASS‐A36 and the S. typhimurium OASS‐A37, respectively). These results indicate that the L. plantarum CBS is an H2S‐generating enzyme, similar to eukaryotic CBSs, rather than an H2S‐consuming enzyme. The preference of the L. plantarum CBS towards l‐homocysteine as a second substrate, which is comparable with that towards H2S, may be a factor associated with its ability to generate H2S.
Characteristic residues in the substrate‐binding pocket of the L. plantarum CBS
To clarify the structural factors determining the substrate preference of the L. plantarum CBS, its crystal structures were determined. One is in an open conformation without the substrate or its analogs, while the other is in a closed conformation bound with external aldimine formed between PLP and l‐methionine [Fig. 5(C)]. The overall structure of the L. plantarum CBS is highly similar to that of bacterial OASS‐A enzymes [Fig. 5(B,D)]. However, even in the closed form, the substrate‐binding pocket of the L. plantarum CBS has a larger size compared to bacterial OASS‐As [Fig. 6(B,C,F,G)] due to the following reasons: the lack of the hydroxyl group of the Ala70 residue, which is replaced by Ser in OASS‐A enzymes (Fig. 2), the orientation of the Val222 residue [Fig. 6(B)], which is replaced by Ala in OASS‐A enzymes (Fig. 2), and the inter‐domain interaction via the Glu223 residue [Fig. 6(B)], which is replaced by Gly in OASS‐A enzymes (Fig. 2).
The hydroxyl group of the Ser residue in eukaryotic CBSs and bacterial OASSs, which is a counterpart of Ala70 in the L. plantarum CBS, might assist the binding of the first substrate (l‐serine, l‐OAS or l‐cysteine) to form the external aldimine with PLP, given that the group is expected to be within hydrogen‐bonding distance from the γ‐position atom of the substrate amino acid (oxygen in the case of l‐serine or l‐OAS, sulfur in the case of l‐cysteine). In fact, the corresponding hydrogen bond is found in the crystal structure of Drosophila CBS complexed with l‐serine.29 In addition, the substitution of the Ser residue to Ala has been reported to significantly diminish the catalytic efficiency of yeast CBS.38 Similarly, the hydroxyl group of the Ser residue might help the binding of the second substrate (H2S, l‐cysteine or l‐homocysteine), given that the hydroxyl group is expected to interact with a sulfur atom of the second substrate. The increased hydrophobicity caused by the substitution to Ala in the L. plantarum CBS seems to be a disadvantage for the enzymatic function.
The orientation of the Ala residue in the bacterial OASS‐A, which is a counterpart of Val222 in the L. plantarum CBS, may be determined by the particular backbone conformation of this region, since the Ala residue is sandwiched by invariant Gly residues having large freedom in backbone torsion angles (Fig. 2). In the L. plantarum CBS, the orientation of the side chain of the Val222 residue seems to be differently fixed, since the conformation of the next Glu223 residue is restricted. In addition, the Glu223 residue itself seems to restrict the closure of the two domains by forming the inter‐domain interactions [Fig. 6(B,F)]. We assume that the pocket size of the L. plantarum CBS is largely adjusted by the Glu223 residue conserved among bacterial CBS‐As.
Interpretation of the spectroscopic study
Spectroscopic analyses of the bacterial OASS and eukaryotic CBS enzymes demonstrate that the β‐replacement reaction progresses through the formation of the aminoacrylate intermediate.8, 9, 10, 22, 23, 24, 35 In the case of OASS, the intermediate was clearly observed when mixed with l‐OAS. On the other hand, in the case of eukaryotic CBS, the intermediate was generated by the addition of l‐serine, and immediately after the addition of l‐cysteine or l‐cystathionine. As found in the OASS enzymes, addition of l‐OAS to the L. plantarum CBS generates the aminoacrylate intermediate [Fig. 4(A)], although the intermediate was significantly degraded to pyruvate (Reaction 3 in Table 1). On the other hand, the spectra characteristic to the aminoacrylate intermediate was not observed after the addition of l‐cysteine or lanthionine to the L. plantarum CBS [Fig. 4(B–D)], at least under the current condition using the ordinary spectrometer.
Figure 9 shows possible structures formed after the addition of l‐cysteine or l‐lanthionine to the L. plantarum CBS. Addition of each amino acid is expected to generate the corresponding external aldimine immediately. The l‐cysteine‐ and l‐lanthionine‐bound external aldimine would be further converted to the aminoacrylate intermediate, together with the release of H2S and l‐cysteine, respectively.
Figure 9.
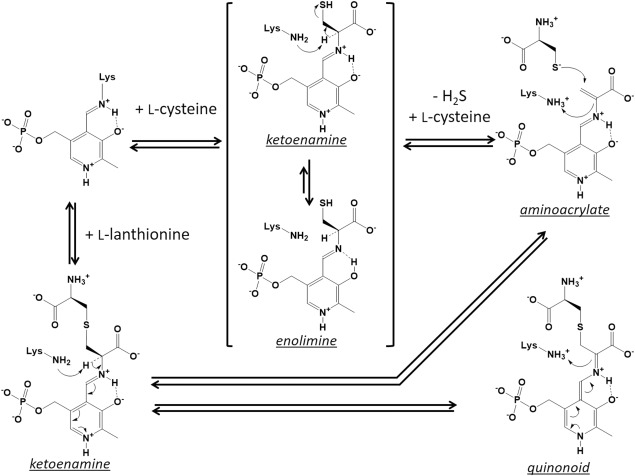
Possible structures formed after addition of l‐cysteine or l‐lanthionine to the L. plantarum CBS. Concerning the external aldimine formed between PLP and l‐cysteine, the enolimine tautomer might be contained in a higher ratio than the ketoenamine tautomer. Condensation of two l‐cysteine molecules to generate l‐lanthionine progresses through the formation of the aminoacrylate intermediate. With regard to the reaction with l‐lanthionine, external aldimine is likely to be converted to the quinonoid intermediate by deprotonation.
Spectroscopic analysis indicated that the aminoacrylate intermediate did not accumulate after addition of l‐cysteine [Fig. 4(B,C)]. The main species observed after the addition of l‐cysteine may be l‐lanthionine‐bound aldimine. That is, prior to the degradation of the intermediate to pyruvate, a thiol group of the second l‐cysteine would quickly attack to the β‐carbon of the intermediate to generate it. Alternatively, it is also possible that the released H2S reacts with the intermediate to regenerate l‐cysteine‐bound aldimine. Therefore, in the reaction mixture, l‐cysteine‐ and l‐lanthionine‐bound external aldimines might be present.
During the late incubation period after addition of lanthionine, the absorbance between 430 and 500 nm increased [Fig. 4(D)]. The quinonoid form of the external aldimine, which is considered to be an intermediate in the reaction of some PLP‐requiring enzymes, such as tryptophan synthase, are known to have an absorption peak at long wavelengths (about 500 nm).39 Although the actual species generated by the enzyme reaction is currently unknown, the species with absorption peak at 480 nm might be a quinonoid form of the l‐lanthionine‐bound external aldimine, which corresponds to one of the deprotonated forms (Fig. 9). If so, l‐lanthionine‐bound external aldimine and its quinonoid form might be present together with the aminoacrylate intermediate in the reaction mixture. The quinonoid‐like peak was also found during the late incubation period after the addition of l‐cysteine [Fig. 4(B,C)], although the molar ratio would be low.
When an l‐amino acid‐bound external aldimine is generated within OASS, two tautomers are known to be formed. It has been suggested that the ketoenamine tautomer has a maximum absorbance at approximately 420 nm, while the enolimine tautomer has a maximum absorbance at approximately 330 nm.40 The main species, which was quickly generated after addition of l‐cysteine or lanthionine, is likely to be the ketoenamine tautomer of the l‐lanthionine‐bound external aldimine (Fig. 9 ). The species formed at the stationary phase are dependent on the concentrations of l‐cysteine, l‐lanthionine, and H2S present in the reaction mixture. Based on kinetic parameters (Table 1), H2S is more reactive to the aminoacrylate intermediate than l‐cysteine. If the concentration of the added l‐cysteine is high enough, the ratio of l‐cysteine‐bound external aldimine is expected to be increased in the stationary phase. This is because l‐lanthionine, which forms an external aldimine with PLP, would be replaced by a high concentration of l‐cysteine, and the generated l‐cysteine‐bound aldimine is likely unreactive due to the high concentration of H2S present in solution. Based on the results obtained with different concentrations of l‐cysteine [Fig. 4(B,C)], the ratio of the enolimine tautomer is likely high for the l‐cysteine‐bound external aldimine generated within the L. plantarum CBS (Fig. 9).
Importance of Ala70 and Glu223 on the H2S‐generatinig activity
The L. plantarum CBS, which generates H2S from l‐cysteine especially in the presence of l‐homocysteine, is structurally similar with bacterial OASS‐As, which utilizes H2S to synthesize l‐cysteine. However, the Ala70 and Glu223 residues existing in the substrate‐binding pocket of the L. plantarum CBS are replaced by Ser and Gly in the bacterial OASS‐As, respectively (Fig. 2).
Spectroscopic analysis of the A70S variant of the L. plantarum CBS demonstrated that, differently from the case of wild type, aminoacrylate intermediate was accumulated by the addition of l‐cysteine [Fig. 8(B,C)]. However, if the concentration of l‐cysteine is high enough, a part of the intermediate would be converted to l‐lanthionine‐bound external aldimine by reaction with the second l‐cysteine molecule. Based on the profile of the spectrum, enolimine tautomer of the l‐cysteine‐bound external aldimine might coexist with aminoacrylate intermediate at the stationary phase after the addition of high concentration of l‐cysteine to the A70S variant [Fig. 8(C)]. The l‐cysteine‐bound external aldimine would be increased in a similar way for the wild type. It should be noticed that the A70S substitution elongates the life of the aminoacrylate intermediate probably due to a lowered reactivity with H2S or another l‐cysteine as a second substrate (Table 1).
Kinetic and affinity analyses of the A70S variant clearly indicate that the substitution increases its affinity towards l‐OAS and l‐cysteine as first substrates (Table 1). Moreover, the variant lost the activity to form pyruvate from l‐OAS, indicating that the substitution increased the stability of the aminoacrylate intermediate. In fact, the substitution of the corresponding Ser residue to Ala in the yeast CBS has been reported to induce the α,β‐elimination activity to synthesize pyruvate in the reaction mixture.38 The similar substitution on the Salmonella OASS‐A was shown to reduce the rate constant for the aminoacrylate formation from l‐OAS.41 The introduced hydroxyl group in the A70S variant is likely to increase the affinities towards the first substrates (l‐OAS and l‐cysteine) by forming a hydrogen bond with the atom in the γ‐position. Additionally, the introduced hydroxyl group might prevent the invasion of a water molecule into the active site, leading to the stabilization of the aminoacrylate intermediate.
On the other hand, the substitution decreases the reactivity towards second substrates (H2S, l‐cysteine, and l‐homocysteine). Although the reason is currently unknown, we presumed that the hydrogen‐bonding interaction, which should be formed between the nucleophilic atom of the second substrate and the introduced hydroxyl group in the A70S variant, inhibits the movement of the nucleophilic atom towards the position suitable for the β‐replacement reaction, which may lead to the reduction of the k cat values (Reactions 1, 2, and 5 in Table 1). These propositions are also supported by the spectroscopic results that indicate the great stability of the aminoacrylate intermediate formed after the addition of l‐cysteine [Fig. 8(B,C)]. In addition, in the reactions using l‐cysteine as a first substrate (Reactions 4 and 5 in Table 1), H2S molecule derived from l‐cysteine would remain at the substrate‐binding site due to the interaction with the introduced hydroxyl group, thereby increasing the K m values for the second substrates (l‐cysteine or l‐homocysteine).
With respect to the reactivity of the E223G variant, the k cat values in all of the β‐replacement reactions were decreased (Table 1). In this variant, the substrate‐binding pocket is expected to be narrower than that of the wild type, which might inhibit the release of the products. On the other hand, although the K m values for the second substrate (Na2S or l‐homocysteine) were decreased, those for the first substrate (l‐OAS or l‐cysteine) were increased (Table 1). To accommodate the first substrate in the narrow substrate‐binding pocket, the hydrogen‐bonding interactions would be required to offset the steric disadvantage. One of the hydrogen bonds might be formed by using the Ser residue conserved among OASS enzymes, however, the residue corresponds to Ala70 in the L. plantarum CBS. On the other hand, the narrow substrate‐binding pocket in this variant assists the binding of the second substrate to the position in which the nucleophilic atom can attack the aminoacrylate intermediate.
Currently, the residues in the L. plantarum CBS that interact with l‐cysteine or l‐homocysteine as a second substrate are unknown. Considering the architecture of the substrate‐binding pocket [Fig. 6(B)], a possible interaction might occur between the amino group of l‐cysteine or l‐homocysteine and the carboxyl group of the Glu223 residue, although its substitution to Gly did not increase the K m value for l‐homocysteine (Table 1). It has been reported that the substitution of an Asp residue to Ala in the yeast CBS, which is correspondent with Glu223 of the L. plantarum CBS, increased K m for l‐homocysteine. This suggests that the carboxyl group at this position interacts with the amino group of l‐homocysteine.42 In addition, given that an imidazole group of the His215 residue, which is conserved among the bacterial CBS‐A enzymes, is positioned near the Glu223 residue, the positive charge on the His215 imidazole might help the interaction with the carboxyl group of the second substrate.
Conclusion
In summary, the catalytic site of the L. plantarum CBS is larger and more hydrophobic than that of bacterial OASS‐A enzymes [Fig. 6(B,C,F,G)]. Although the hydrophobic nature of the catalytic site pocket is a disadvantage for the binding of l‐cysteine as a first substrate, the large size characteristic might allow the binding of l‐cysteine. On the other hand, the hydrophobic characteristic of the pocket seems to allow the productive binding of l‐homocysteine as a second substrate. In addition, the large size and the hydrophobic characteristics may be helpful for the release of H2S and l‐cystathionine from the active site. Therefore, the L. plantarum CBS can efficiently generate H2S from a mixture of l‐cysteine and l‐homocysteine.
Another research group has reported that the suppression of H2S generation in bacteria made them more sensitive to various antibiotics.16 The compounds, which specifically inhibit the L. plantarum CBS, are expected to suppress H2S generation in the pathogen possessing CBS‐A enzymes, and might become drugs to enhance the effect of antibiotics against bacteria. A screening of substances inhibitory to the L. plantarum CBS is in progress.
Materials and Methods
Preparation of the L. plantarum CBS
A gene encoding CBS from L. plantarum SN35N was amplified by PCR with the KOD DNA polymerase (TOYOBO) using a sense primer, 5′‐ATATCATATGCTCATTCAGCACGTTCAAGAACTTATC‐3′ (NdeI site underlined), and an antisense primer, 5′‐ATATCTCGAGTTTCGTATAGATTTTTTGGCTCAGA‐3′ (XhoI site underlined). The amplified DNA fragment was digested with NdeI and XhoI and inserted into the pET‐21a(+) vector (Novagen) to generate an expression plasmid for CBS. E. coli BL21(DE3) cells harboring the expression vector were grown at 28°C in Overnight Express Autoinduction System 1 (Novagen). Cells were harvested by centrifugation and disrupted by sonication. C‐terminal His6‐tagged CBS was purified by Ni(II) affinity chromatography using the His‐Bind resin (Novagen) according to the supplier's instructions. The fractions containing CBS were dialyzed against a 20 mM Tris–HCl buffer (pH 7.5) containing 0.2 M NaCl, 1 mM EDTA, and 0.2 mM PLP.
By using the pET‐28a(+) vector (Novagen) and the amplified CBS gene containing stop codon, we tried to obtain the construct to express the N‐terminal His6‐tagged CBS, whose tag sequence can be removed by thrombin cleavage. However, since the recombinant protein was found in the insolubilized fraction, we used the C‐terminal His6‐tagged CBS for all the experiments. The C‐terminal region in the M. tuberculosis OASS‐B (also called CysM), which has a long C‐terminal extension sequence, has been reported to regulate the catalytic activity by inserting to the active site.43 In the current crystal structures, the C‐terminal His6‐tag sequence is invisible in the electron density map, and the visible C‐terminal residues are found at the protein surface.
Mutagenesis
The QuikChange Site‐Directed Mutagenesis Kit (Stratagene) was used to generate the CBS variants K42A and A70S according to the supplier's instructions. The mutagenic primers containing the desired mutations (underlined) were as follows (sense only): 5′‐CGGCGGCAGTATTGCGGACCGGCTGGGAG‐3′ (K42A) and 5′‐CAACGATTATTGAACCAACTTCTGGTAACACCGG‐3′ (A70S). On the other hand, the KOD ‐Plus‐ Mutagenesis Kit (TOYOBO) was used to generate the E223G variant according to the supplier's instructions. The mutagenic primers containing the desired mutations (underlined) were as follows: 5′‐GGATTCATCCCACCATTTTTCGATCAGG‐3′ (sense), 5′‐GACGCCGATTCCCTCGGTACG‐3′ (antisense). To generate the expression vector for the CBS variant, the vector for wild‐type CBS was amplified using sense and antisense primers. The mutation was confirmed by DNA sequencing. Expression and purification of substituted proteins were conducted using the same method employed for the wild type. For the K42A variant, 1 mM PLP and 10 mM l‐methionine were added to all the buffers used for the purification to avoid precipitation.
Analysis of molecular mass
molecular mass of the purified enzymes was estimated by HPLC using a Superdex 200 10/300 GL column (GE Healthcare) at 0.40 mL min−1 with 50 mM sodium phosphate buffer (pH 7.0) containing 0.15 M NaCl. The proteins used for calibration were as follows: ferritin (440 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), and RNase A (13.7 kDa). Proteins were detected at 280 nm using a multiwavelength detector (MD‐2010, JASCO).
Absorption spectroscopy
Prior to spectroscopic analysis, CBS was dialyzed against 100 mM potassium phosphate buffer (pH 7.5) containing 1 mM EDTA, 0.2 M NaCl, and 1 μM PLP. The absorption spectrum of CBS (10 μM) in the absence or presence of given concentrations of l‐OAS, l‐cysteine, or lanthionine was recorded between 250 and 500 nm using a V‐550 spectrophotometer (JASCO). In all cases, tris(2‐carboxyethyl)phosphine was added to the buffer at a final concentration of 10 mM before the measurement. The mixture was measured in a 1‐cm pathlength cuvette at 20°C.
The binding affinities of the wild‐type CBS or its variants for l‐OAS were measured by monitoring changes in the absorbance of PLP. The starting volume of the reaction mixture was 400 μL, and 3–6 μL of the l‐OAS solution were added continuously. The titration range of substrate concentration was 0.1–370 μM. The apparent K d for l‐OAS was calculated by fitting data to the following equation.
| (1) |
In Eq. (1), Abs is the absorbance at 470 nm in the presence of CBS and l‐OAS at the concentrations of E t and L t, respectively. A max is the maximum difference of absorbance at 470 nm of the unitary concentration of CBS, caused by the complete binding of l‐OAS to the enzyme and the following conversion into the aminoacrylate intermediate. L is the concentration of unbound l‐OAS, which is calculated as follows:
| (2) |
A max and K d were determined with the least square method.
Product analysis
Before measurement of the enzymatic activity of CBS, stock solutions of l‐OAS (HCl form), Na2S, sodium thiosulfate, l‐cysteine, and l‐homocysteine were freshly prepared, and their pHs were adjusted to about 7 using HCl or NaOH.
The OASS activity of the L. plantarum CBS was measured under the various pH conditions (pH 4.0–9.15 in broad‐range buffer containing 3,3‐dimethyl‐glutaric acid, Tris, and 2‐amino‐2‐methyl‐1,3‐propanediol, pH 6.0–8.0 in HEPES–KOH buffer). The pH of 7.5 in the HEPES–KOH buffer was found to maximize the activity. Formation of the enzymatic product was conveniently analyzed by TLC using silica gel 60 F254 (Merck). The reaction mixture, consisting of 100 mM HEPES–KOH (pH 7.5), 10 mM tris(2‐carboxyethyl)phosphine, 10 mM first substrate (l‐OAS, l‐serine, or l‐cysteine) and 10 mM second substrate (Na2S, sodium thiosulfate, or l‐homocysteine), and 10–500 μg mL−1 CBS, was incubated at 37°C. As a control, the same reaction mixture without CBS was used. After an overnight incubation, the enzymatic reaction was stopped by incubating at 100°C for 5 min, and the precipitate was removed by centrifugation. The supernatant was used for product analysis. To detect l‐cysteine and l‐homocysteine, the supernatant was mixed with an equivalent volume of a 10 mM N‐ethylmaleimide solution and incubated at room temperature for 20 min before TLC analysis. The solvent used for the chromatogram was 2‐butanone‐pyridine‐water (18:5:10). Amino acids were visualized using ninhydrin. When l‐OAS was used as a first substrate, l‐serine was detected in the reaction mixture. We found that boiling treatment of l‐OAS stimulated the generation of l‐serine.
Generation of l‐serine, l‐lanthionine, or l‐cystathionine in the reaction mixture was also analyzed by HPLC after derivatization with O‐phthaldialdehyde and mercaptoethanol.44 After a reaction mixture (100 μL) consisting of 100 mM HEPES‐KOH (pH 7.5), 10 mM l‐cysteine or 10 mM l‐OAS, and 10 μg mL−1 CBS was incubated at 37°C for 30 min in the presence or absence of 10 mM l‐homocysteine, the reaction was stopped by addition of 10 μL of 50% trichloroacetic acid, and the precipitate was removed by centrifugation. The pH of the supernatant was neutralized to about 7 with a small amount of NaOH, and 440 μL of 0.2 M borate buffer (pH 9.6) was added. A 50 μL aliquot of each sample was derivatized with 25 μL of 0.2 M sodium borate buffer (pH 9.6) containing 90 mM O‐phthaldialdehyde, 180 mM 2‐mercaptoethanol, and 10% (v/v) methanol for 1 min at room temperature. A 10 μL aliquot of the derivatized sample was injected into an ODS‐A column (YMC, 100 × 4.6 mM) equilibrated with 80% solvent A (100 mM sodium acetate at pH 4.75) and 20% solvent B (methanol). The ratio of solvent B linearly increased from 20 to 50% in 10 min, and from 50 to 100% in the next 10 min. The flow rate was set to 1.0 mL min−1. A derivative from the product was detected measuring the absorbance at 340 nm with a multi‐wavelength detector. We found that the amount of l‐serine formed from l‐OAS with CBS was almost the same as that formed in the absence of CBS.
Kinetic analysis
In general, the continuous assay is a superior method for the kinetic analysis than the end product assay, since the former method is not influenced by the product inhibition. However, the method convenient to measure the concentration of the synthesized l‐cysteine (in the presence of Na2S) and l‐cystathionine is absent. Therefore, we have employed the end product assay method for the measurement of the OASS and l‐OAS dependent CBS reactions. The concentration range of substrate was set to cover the K m value. However, we did not verify the kinetic response under the conditions where the substrate concentration exceeds the upper limits (100 mM for l‐OAS, 40 mM for l‐homocysteine, 4 mM for Na2S, and 150 mM for l‐cysteine).
The ability of CBS to synthesize l‐cysteine was determined using a colorimetric method.45 To determine the kinetic parameters, a reaction mixture (100 μL), consisting of 100 mM HEPES‐KOH (pH 7.5), 1 mM tris(2‐carboxyethyl)phosphine, 0.2–100 mM l‐OAS, 0.1–4 mM Na2S, and CBS at the appropriate concentration was incubated at 37°C for 10 min. The amount of enzyme added to the reaction mixture was adjusted so that the amount of the enzymatic products is less than 30% of the maximum amount which is estimated from the initial concentrations of two substrates (1, 20, and 5 μg mL−1 in the case of wild type, A70S variant, and E223G variant, respectively). The enzyme reaction was stopped by addition of 250 μL of a solution containing 0.2 M H2SO4 and 1 mM NaNO2. The amount of l‐cysteine formed by CBS was determined with the procedure described previously.45
The ability of CBS to synthesize l‐cystathionine was also determined using the colorimetric method.46 To determine the kinetic parameters, the reaction mixture (100 μL), consisting of 100 mM HEPES–KOH (pH 7.5), 1 mM tris(2‐carboxyethyl)phosphine, 0.2–100 mM l‐OAS, 0.1–20 mM l‐homocysteine, and appropriate amount of enzyme (1, 20, and 10 μg mL−1 in the case of wild type, A70S variant, and E223G variant, respectively), was incubated at 37°C for 10 min. Enzyme activity was stopped by addition of 10 μL of 50% trichloroacetic acid. The amount of l‐cystathionine formed by CBS was determined with the method described previously.46
The ability of CBS to generate H2S from l‐cysteine or from a mixture of l‐cysteine and l‐homocysteine was measured in the presence of lead(II) acetate.47 Formation of PbS was continuously monitored measuring the absorbance at 390 nm. To determine the kinetic parameters, the reaction mixture (0.6 mL), consisting of 100 mM HEPES–KOH (pH 7.5), 1 mM tris(2‐carboxyethyl)phosphine, 1–150 mM l‐cysteine, and appropriate concentrations of CBS (7.5, 75, and 75 μg mL−1 in the case of wild type, A70S variant, and E223G variant, respectively), was incubated at 37°C. In addition, the reaction mixture (0.6 mL), consisting of 100 mM HEPES–KOH (pH 7.5), 1 mM tris(2‐carboxyethyl)phosphine, 1–150 mM l‐cysteine, 0.1–40 mM l‐homocysteine, and appropriate concentrations of CBS (5, 10, and 25 μg mL−1 in the case of wild type, A70S variant, and E223G variant, respectively), was incubated at 37°C. A molar extinction coefficient of 5,500 M– 1 cm−1 was used for lead sulfide. Lead acetate (0.4 mM) did not inhibit l‐OAS‐dependent CBS activity measured in the above assay.
The ability of CBS to generate pyruvate from l‐OAS was measured in the presence of l‐lactate dehydrogenase and NADH. Formation of pyruvate was detected by continuous monitoring of the absorbance at 340 nm. To determine the kinetic parameters for l‐OAS, the reaction mixture (0.6 mL), consisting of 100 mM HEPES–KOH (pH 7.5), 0.2 mM NADH, 20 U mL−1 l‐lactate dehydrogenase, 0.01–1 mM l‐OAS, and 500 μg mL−1 CBS, was incubated at 37°C.
Each measurement was done in duplicate. CBS generally catalyzes a two‐substrate reaction with a ping‐pong mechanism.22 In this case, initial reaction rates at different substrate concentrations are expressed by Eq. (3).
| (3) |
In Eq. (3), v is the initial velocity, E t, S A, and S B are the concentrations of the enzyme, the first substrate, and the second substrate, respectively, and k cat and K m are the catalytic and the Michaelis–Menten constants, respectively. The k cat and two K m values were evaluated using the nonlinear least square method by fitting to Eq. (3). For H2S generation from a mixture of l‐cysteine and l‐homocysteine, the generation should be coupled with the formation of l‐cystathionine and l‐lanthionine. Kinetic parameters were determined after subtracting the estimate of the H2S generation accompanied with the l‐lanthionine synthesis.
When using single substrate (Reactions 3 and 4 in Fig. 3), k cat and K m values were evaluated using the nonlinear least square method by fitting to Eq. (4).
| (4) |
For H2S generation from l‐cysteine, the first and the second substrate concentrations are identical. The K m value determined means the sum of two Michaelis–Menten constants. In the case of Reaction 4 of A70S mutant (Table 1), k cat/K m value for l‐cysteine was calculated from the slope of the straight line in the kinetic graph.
In some cases, substrate inhibition was strongly suggested. In this case, data were fitted to (5).
| (5) |
The K I values are the constants for substrate inhibition. In the H2S‐generation activity from the mixture of l‐cysteine and l‐homocysteine, substrate inhibition was observed for the both substrates. In the presence of high concentration of l‐cysteine, l‐lanthionine synthesis is stimulated instead of the l‐cystathionine synthesis, which may lead to the decrease in the H2S generation. Therefore, kinetic parameters were determined using data obtained in the presence of low concentrations of l‐cysteine where the substrate inhibition does not occur.
Crystallography
Prior to crystallization, the CBS solution was concentrated to 10 mg mL−1 using Amicon Ultra filters (Millipore). CBS crystals were grown using the sitting‐drop vapor‐diffusion method, with a 1:1 (v/v) ratio of protein solution to precipitant solution. Stick‐like crystals were formed within 3 days by using a 0.1 M HEPES–NaOH buffer (pH 8.0) containing 0.7 M Li2SO4 as a precipitant solution.
Crystals were flash‐frozen before data collection with a cryoprotectant containing 30% (v/v) glycerol. Diffraction intensities of the crystals were collected using synchrotron radiation from BL38B1 at SPring‐8 (Hyogo, Japan). X‐ray diffraction was measured with a CCD camera at the station, and intensities were integrated and scaled using the combination of the Mosflm and Scala programs in the CCP4 program suite.48 The tertiary structure of CBS was revealed by the molecular replacement method using the atomic coordinates of the M. tuberculosis OASS‐A28 (PDB code: 2Q3D), as a search model and the program Molrep in the CCP4 program suite.48 The model was refined by simulated annealing and conventional restrained refinement methods using the CNS program.49 A subset of 5% of the reflections was used to monitor the free R factor (R free).50 Each refinement cycle includes the refinement of the positional parameters and individual isotropic B‐factors, and the revision of the model, which was visualized by the program Xfit from the XtalView software package.51
Like the wild‐type CBS solution, the solution of the K42A variant was concentrated to 10 mg mL−1 using Amicon Ultra filters. The substituted protein was crystallized within 1 month using a 0.1 M sodium citrate buffer (pH 5.6) containing 0.5 M (NH4)2SO4 and 1.2 M Li2SO4 as a precipitant solution. X‐ray diffraction intensities of the crystal were collected using synchrotron radiation from BL38B1 at SPring‐8. Diffraction intensities were integrated and scaled using the HKL2000 program.52 The structure was solved by molecular replacement using Molrep, and refined by CNS. Details of data collection and refinement statistics are shown in Table 2.
Table 2.
Data Collection and Refinement Statistics
| Data set | Wild type | K42A |
|---|---|---|
| Data collection | ||
| Space group | C2 | P43212 |
| Cell dimensions | ||
| a (Å) | 139.89 | 165.94 |
| b (Å) | 146.34 | 165.94 |
| c (Å) | 82.79 | 259.59 |
| β (º) | 90.00 | 90 |
| Wavelength (Å) | 1.00000 | 1.00000 |
| Resolution (Å) | 100–2.40 | 100–3.30 |
| Unique reflections | 62,935 | 55,046 |
| Redundancya | 3.4 (2.5) | 5.8 (6.0) |
| Completeness (%)a | 97.6 (85.0) | 99.9 (100) |
| R merge (%)a, b | 6.4 (41.2) | 7.2 (49.8) |
| I/σa | 19.8 (2.0) | 26.2 (3.8) |
| Refinement | ||
| Resolution (Å) | 30–2.40 | 30–3.30 |
| Used reflections | 62,494 | 53,762 |
| No. of atoms | ||
| Protein | 9,076 | 18,120 |
| Ligand | 204 | 272 |
| Water | 406 | 0 |
| R (%) | 18.9 | 23.8 |
| R free (%) | 22.1 | 28.3 |
| Rms deviationsc | ||
| Bond length (Å) | 0.006 | 0.009 |
| Bond angle (º) | 1.2 | 1.3 |
| Mean B‐factor (Å2) | ||
| Protein | 52.0 | 45.5 |
| Ligand | 74.3 | 53.6 |
| Water | 53.9 | – |
| Ramachandran plot (%) | ||
| Favored | 96.0 | 91.2 |
| Allowed | 3.4 | 8.2 |
| Outliers | 0.7 | 0.6 |
Values in parentheses are for the highest resolution bin.
R merge = Σ|I −〈I〉 |/ΣI, where 〈I〉 is the observed intensity and is the mean value of I.
Rms deviations are calculated by CNS.49
ACCESSION NUMBERS
Atomic coordinates and structure factors of wild‐type CBS from Lactobacillus plantarum and the l‐methionine‐bound K42A variant have been deposited in the Protein Data Bank with the accession codes 5B1H and 5B1I, respectively.
CONFLICT OF INTEREST
All the authors have declared no conflict of interest.
ACKNOWLEDGMENTS
DNA sequence determination was carried out at the Analysis Center of Life Science, Hiroshima University. We are grateful to the beamline staff at SPring‐8, Japan, for their kind help with the X‐ray data collection. We also thank to Ms. C. Yasutake for the assistance in the protein purification.
Contributor Information
Yasuyuki Matoba, Email: ymatoba@hiroshima-u.ac.jp.
Masanori Sugiyama, Email: sugi@hiroshima-u.ac.jp.
REFERENCES
- 1. Miles EW, Kraus JP (2004) Cystathionine β‐synthase: structure, function, regulation, and location of homocystinuria‐causing mutations. J Biol Chem 279:29871–29874. [DOI] [PubMed] [Google Scholar]
- 2. Aitken SM, Kirsch JF (2005) The enzymology of cystathionine biosynthesis: strategies for the control of substrate and reaction specificity. Arch Biochem Biophys 433:166–175. [DOI] [PubMed] [Google Scholar]
- 3. Aitken SM, Lodha PH, Morneau DJ (2011) The enzymes of the transsulfuration pathways: active‐site characterizations. Biochim Biophys Acta 1814:1511–1517. [DOI] [PubMed] [Google Scholar]
- 4. Messerschmidt A, Worbs M, Steegborn C, Wahl MC, Huber R, Laber B, Clausen T (2003) Determinants of enzymatic specificity in the Cys‐Met‐metabolism PLP‐dependent enzymes family: crystal structure of cystathionine γ‐lyase from yeast and intrafamiliar structure comparison. Biol Chem 384:373–386. [DOI] [PubMed] [Google Scholar]
- 5. Sun Q, Collins R, Huang S, Holmberg‐Schiavone L, Anand GS, Tan CH, van den Berg S, Deng LW, Moore PK, Karlberg T, Sivaraman J (2009) Structural basis for the inhibition mechanism of human cystathionine γ‐lyase, an enzyme responsible for the production of H2S. J Biol Chem 284:3076–3085. [DOI] [PubMed] [Google Scholar]
- 6. Castro R, Rivera I, Blom HJ, Jakobs C, Tavares de Almeida I (2006) Homocysteine metabolism, hyperhomocysteinaemia and vascular disease: an overview. J Inherit Metab Dis 29:3–20. [DOI] [PubMed] [Google Scholar]
- 7. Szabó C (2007) Hydrogen sulphide and its therapeutic potential. Nat Rev Drug Discov 6:917–935. [DOI] [PubMed] [Google Scholar]
- 8. Singh S, Ballou DP, Banerjee R (2011) Pre‐steady‐state kinetic analysis of enzyme‐monitored turnover during cystathionine β‐synthase‐catalyzed H2S generation. Biochemistry 50:419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yadav PK, Banerjee R (2012) Detection of reaction intermediates during human cystathionine β‐synthase‐monitored turnover and H2S production. J Biol Chem 287:43464–43471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Singh S, Padovani D, Leslie RA, Chiku T, Banerjee R (2009) Relative contributions of cystathionine β‐synthase and γ‐cystathionase to H2S biogenesis via alternative trans‐sulfuration reactions. J Biol Chem 284:22457–22466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hullo MF, Auger S, Soutourina O, Barzu O, Yvon M, Danchin A, Martin‐Verstraete I (2007) Conversion of methionine to cysteine in Bacillus subtilis and its regulation. J Bacteriol 189:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doherty NC, Shen F, Halliday NM, Barrett DA, Hardie KR, Winzer K, Atherton JC (2010) Helicobacter pylori, LuxS is a key enzyme in cysteine provision through a reverse transsulfuration pathway. J Bacteriol 192:1184–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu M, Nauta A, Francke C, Siezen RJ (2008) Comparative genomics of enzymes in flavor‐forming pathways from amino acids in lactic acid bacteria. Appl Environ Microbiol 74:4590–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rabeh WM, Cook PF (2004) Structure and mechanism of O‐acetylserine sulfhydrylase. J Biol Chem 279:26803–26806. [DOI] [PubMed] [Google Scholar]
- 15. Mozzarelli A, Bettati S, Campanini B, Salsi E, Raboni S, Singh R, Spyrakis F, Kumar VP, Cook PF (2011) The multifaceted pyridoxal 5′‐phosphate‐dependent O‐acetylserine sulfhydrylase. Biochim Biophys Acta 1814:1497–1510. [DOI] [PubMed] [Google Scholar]
- 16. Shatalin K, Shatalina E, Mironov A, Nudler E (2011) H2S: a universal defense against antibiotics in bacteria. Science 334:986–990. [DOI] [PubMed] [Google Scholar]
- 17. Ereño‐Orbea J, Majtan T, Oyenarte I, Kraus JP, Martínez‐Cruz LA (2013) Structural basis of regulation and oligomerization of human cystathionine β‐synthase, the central enzyme of transsulfuration. Proc Natl Acad Sci USA 110:E3790–E3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ereño‐Orbea J, Majtan T, Oyenarte I, Kraus JP, Martínez‐Cruz LA (2014) Structural insight into the molecular mechanism of allosteric activation of human cystathionine β‐synthase by S‐adenosylmethionine. Proc Natl Acad Sci USA 111:E3845–E3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McCorvie TJ, Kopec J, Hyung SJ, Fitzpatrick F, Feng X, Termine D, Strain‐Damerell C, Vollmar M, Fleming J, Janz JM, Bulawa C, Yue WW (2014) Inter‐domain communication of human cystathionine β‐synthase: structural basis of S‐adenosyl‐l‐methionine activation. J Biol Chem 289:36018–36030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jin H, Higashikawa F, Noda M, Zhao X, Matoba Y, Kumagai T, Sugiyama M (2010) Establishment of an in vitro Peyer's patch cell culture system correlative to in vivo study using intestine and screening of lactic acid bacteria enhancing intestinal immunity. Biol Pharm Bull 33:289–293. [DOI] [PubMed] [Google Scholar]
- 21. Zhao X, Higashikawa F, Noda M, Kawamura Y, Matoba Y, Kumagai T, Sugiyama M (2012) The obesity and fatty liver are reduced by plant‐derived Pediococcus pentosaceus LP28 in high fat diet‐induced obese mice. PLoS One 7:e30696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cook PF, Wedding RT (1976) A reaction mechanism from steady state kinetic studies for O‐acetylserine sulfhydrylase from Salmonella typhimurium LT‐2. J Biol Chem 251:2023–2029. [PubMed] [Google Scholar]
- 23. Cook PF, Hara S, Nalabolu S, Schnackerz KD (1992) pH dependence of the absorbance and 31P NMR spectra of O‐acetylserine sulfhydrylase in the absence and presence of O‐acetyl‐l‐serine. Biochemistry 31:2298–2303. [DOI] [PubMed] [Google Scholar]
- 24. Jhee KH, McPhie P, Miles EW (2000) Yeast cystathionine β‐synthase is a pyridoxal phosphate enzyme but, unlike the human enzyme, is not a heme protein. J Biol Chem 275:11541–11544. [DOI] [PubMed] [Google Scholar]
- 25. Schneider G, Kack H, Lindqvist Y (2000) The manifold of vitamin B6 dependent enzymes. Structure 8:R1–R6. [DOI] [PubMed] [Google Scholar]
- 26. Burkhard P, Tai CH, Ristroph CM, Cook PF, Jansonius JN (1999) Ligand binding induces a large conformational change in O‐acetylserine sulfhydrylase from Salmonella typhimurium . J Mol Biol 291:941–953. [DOI] [PubMed] [Google Scholar]
- 27. Bonner ER, Cahoon RE, Knapke SM, Jez JM (2005) Molecular basis of cysteine biosynthesis in plants. Structural and functional analysis of O‐acetylserine sulfhydrylase from Arabidopsis thaliana . J Biol Chem 280:38803–38813. [DOI] [PubMed] [Google Scholar]
- 28. Schnell R, Oehlmann W, Singh M, Schneider G (2007) Structural insights into catalysis and inhibition of O‐acetylserine sulfhydrylase from Mycobacterium tuberculosis. Crystal structures of the enzyme α‐aminoacrylate intermediate and an enzyme–inhibitor complex. J Biol Chem 282:23473–27481. [DOI] [PubMed] [Google Scholar]
- 29. Koutmos M, Kabil O, Smith JL, Banerjee R (2010) Structural basis for substrate activation and regulation by cystathionine β‐synthase (CBS) domains in cystathionine β‐synthase. Proc Natl Acad Sci USA 107:20958–20963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holm L, Sander CJ (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233:123–138. [DOI] [PubMed] [Google Scholar]
- 32. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Claus MT, Zocher GE, Maier THP, Schulz GE (2005) Structure of the O‐acetylserine sulfhydrylase isoenzyme CysM from Escherichia coli . Biochemistry 44:8620–8626. [DOI] [PubMed] [Google Scholar]
- 34. Ono BI, Kijima K, Inoue T, Miyoshi SI, Matsuda A, Shinoda S (1994) Purification of properties of Saccharomyces cerevisiae cystathionine β‐synthase. Yeast 10:333–339. [DOI] [PubMed] [Google Scholar]
- 35. Woehl EU, Tai CH, Dunn MF, Cook PF (1996) Formation of the alpha‐aminoacrylate immediate limits the overall reaction catalyzed by O‐acetylserine sulfhydrylase. Biochemistry 35:4776–4783. [DOI] [PubMed] [Google Scholar]
- 36. Mino K, Yamanoue T, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K (2000) Effects of bienzyme complex formation of cysteine synthetase from Escherichia coli on some properties and kinetics. Biosci Biotechnol Biochem 264:1628–1640. [DOI] [PubMed] [Google Scholar]
- 37. Tai CH, Nalabolu SR, Jacobson TM, Minter DE, Cook PF (1993) Kinetic mechanisms of the A and B isozymes of O‐acetylserine sulfhydrylase from Salmonella typhimurium LT‐2 using the natural and alternative reactants. Biochemistry 32:6433–6442. [DOI] [PubMed] [Google Scholar]
- 38. Aitken SM, Kirsch JF (2004) Role of active‐site residues Thr81, Ser82, Thr85, Gln157, and Tyr158 in yeast cystathionine β‐synthase catalysis and reaction specificity. Biochemistry 43:1963–1971. [DOI] [PubMed] [Google Scholar]
- 39. Toney MD (2005) Reaction specificity in pyridoxal phosphate enzymes. Arch Biochem Biophys 433:279–287. [DOI] [PubMed] [Google Scholar]
- 40. Schnackerz KD, Tai CH, Simmons JW, 3rd , Jacobson TM, Rao GS, Cook PF (1995) Identification and spectral characterization of the external aldimine of the O‐acetylserine sulfhydrylase reaction. Biochemistry 34:12152–12160. [DOI] [PubMed] [Google Scholar]
- 41. Tian H, Guan R, Salsi E, Campanini B, Bettati S, Kumar VP, Karsten WE, Mozzarelli A, Cook PF (2010) Identification of the structural determinants for the stability of substrate and aminoacrylate external Schiff bases in O‐acetylserine sulfhydrylase‐A. Biochemistry 49:6093–6103. [DOI] [PubMed] [Google Scholar]
- 42. Lodha PH, Shadnia H, Woodhouse CM, Wright JS, Aitken SM (2009) Investigation of residues Lys112, Glu136, His138, Gly247, Tyr248, and Asp249 in the active site of yeast cystathionine β‐synthase. Biochem Cell Biol 87:531–540. [DOI] [PubMed] [Google Scholar]
- 43. Agren D, Schnell R, Schneider G (2009) The C‐terminal of CysM from Mycobacterium tuberculosis protects the aminoacrylate intermediate and is involved in sulfur donor selectivity. FEBS Lett 583:330–336. [DOI] [PubMed] [Google Scholar]
- 44. Canevari L, Vieira R, Aldegunde M, Dagani F (1992) High‐performance liquid chromatographic separation with electrochemical detection of amino acids focusing on neurochemical application. Anal Biochem 205:137–142. [DOI] [PubMed] [Google Scholar]
- 45. Kredich NM, Tomkins GM (1966) The enzymatic synthesis of l‐cysteine in Escherichia coli and Salmonella typhimurium . J Biol Chem 241:4955–4965. [PubMed] [Google Scholar]
- 46. Kashiwamata S, Greenberg DM (1970) Studies on cystathionine synthase of rat liver: properties of the highly purified enzyme. Biochim Biophys Acta 212:488–500. [DOI] [PubMed] [Google Scholar]
- 47. Willhardt I, Wiederanders B (1975) Activity staining of cystathionine‐β‐synthetase and related enzymes. Anal Biochem 63:263–266. [DOI] [PubMed] [Google Scholar]
- 48. Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr Sect D 50:760–763. [DOI] [PubMed] [Google Scholar]
- 49. Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse‐Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr Sect D Biol Crystallogr 54:905–921. [DOI] [PubMed] [Google Scholar]
- 50. Brünger AT (1992) The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472–475. [DOI] [PubMed] [Google Scholar]
- 51. McRee DE (1992) A visual protein crystallographic software system for X11/XView. J Mol Graphics 10:44–46. [Google Scholar]
- 52. Otwinowski Z, Minor W (1997) Processing of X‐ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. [DOI] [PubMed] [Google Scholar]


