Abstract
TBC1D15 belongs to the TBC (Tre‐2/Bub2/Cdc16) domain family and functions as a GTPase‐activating protein (GAP) for Rab GTPases. So far, the structure of TBC1D15 or the TBC1D15·Rab complex has not been determined, thus, its catalytic mechanism on Rab GTPases is still unclear. In this study, we solved the crystal structures of the Shark and Sus TBC1D15 GAP domains, to 2.8 Å and 2.5 Å resolution, respectively. Shark‐TBC1D15 and Sus‐TBC1D15 belong to the same subfamily of TBC domain‐containing proteins, and their GAP‐domain structures are highly similar. This demonstrates the evolutionary conservation of the TBC1D15 protein family. Meanwhile, the newly determined crystal structures display new variations compared to the structures of yeast Gyp1p Rab GAP domain and TBC1D1. GAP assays show that Shark and Sus GAPs both have higher catalytic activity on Rab11a·GTP than Rab7a·GTP, which differs from the previous study. We also demonstrated the importance of arginine and glutamine on the catalytic sites of Shark GAP and Sus GAP. When arginine and glutamine are changed to alanine or lysine, the activities of Shark GAP and Sus GAP are lost.
Keywords: shark, Sus, TBC1D15, GAP, crystal structure, GTPase activity assay
Introduction
Small GTPases regulate various cellular pathways and can be divided into five subfamilies: Ras, Rho, Rab, Arf/Sar and Ran.1, 2 Rab proteins belong to the Ras‐like superfamily and act as molecular switches by cycling between a GTP‐bound (active state) and GDP‐bound (inactive state), and they are regulated by the GTPase‐activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs).1, 3 Rab GEFs accelerate the exchange of GDP for GTP to activate Rabs. In contrast, Rab GAPs accelerate GTPase hydrolytic activity to inactivate Rabs.4, 5 GDI (GDP Dissociation Inhibitor) and GDF (GDI Displacement Factor) are another two substances involved in the cycling. GDI associates with inactive GDP‐bound geranyl‐geranylated Rab proteins, while GDF is required for GDI displacement and recruitment of Rabs to membranes.6, 7 According to previous research, there are more than 60 Rabs in mammalian systems,8, 9 and these Rab proteins exist in distinct intracellular compartments and serve as major coordinators of vesicle traffic.3
Rab7 is a small GTPase that regulates endo‐lysosomal trafficking.10 It can be divided into two forms: Rab7a and Rab7b. Rab7a primarily controls the transport from early endosomes to late endosomes and then from late endosomes to lysosomes.6 Rab7a can also regulate the secretion of endothelial microRNA through extracellular vesicles.11 Rab7b regulates the trafficking of endosomes‐lysosomes to the Golgi.12 Rab7 has different roles across many cellular functions, including phagocytosis,13 retrograde trafficking,6 the apoptotic response,14 autophagy, and mitophagy.15 Several studies have analyzed the interaction between Rab7a and disease. One example is that Rab7a could regulate peripherin assembly and its mutants influenced peripherin phosphorylation. Because peripherin is important for neurite outgrowth and axonal regeneration, the abnormal expression of peripherin would cause Charcot‐Marie‐Tooth type 2B (CMT2B) disease.16 Another example is that the depleted expression of Rab7a resulted in the increased accumulation of PrPC, as observed during a study on the relationship between Rab7a and Creutzfeldt‐Jakob disease (CJD)/Alzheimer's disease.17
Rab11 is another subfamily of small GTPases that regulates the recycling of endosomes to the plasma membrane.18 There are three subfamily members of Rab11: Rab11a, Rab11b and Rab25.19 Rab11a is mainly localized to the ERC/RE (endosomal recycling compartment/recycling endosome), while Rab11b is localized to the ERC and Rab25 is associated with the ARE (apical recycling endosome) in polarized epithelial cells.19 Rab11a is also an important protein that regulates many cellular pathways. Previous research showed that Rab11a is required for canalicular formation,20 and that Rab11a can also regulate the recycling of the human prostacyclin receptor (hlP).21 In addition, Rab11a can also mediate GLUT4 internalization together with its interacting proteins (FIPs).22 In microvillus inclusion disease (MVID), Rab11a interacts with MYO5B (mutations in the gene encoding myosin Vb) and Rab8a, which is required for the proper establishment of apical polarity.23
TBC1D15 is a protein that contains a conserved Tre2/Bub2/Cdc16 (TBC) domain. According to previous work, we found that TBC1D15 is an important protein that participates in several regulatory network. TBC1D15 can serve as a mitochondrial Rab GTPase‐activating protein (Rab‐GAP) and influence autophagosome biogenesis and morphology downstream of Parkin activation. TBC1D15 is recruited by mammalian Fist1, which is a mitochondrial receptor that functions in the regulation of mitochondrial morphology.24 TBC1D15 can also control stem cell self‐renewal when coupled with the Numb‐p53 complex.25 Moreover, TBC1D15 in Drosophila regulates synaptic growth by inhibiting Rab7 activity.26 TBC1D15 also has been identified as Rab7 GAP in vivo and in vitro, and can also modulate lysosomal morphology.27, 28 In addition to interacting with Rab7, TBC1D15 is required for the accumulation of RhoA during membrane blebbing and cytokinesis.29
TBC1D15 from Chiloscyllium plagiosum (Shark‐TBC1D15) is a 4213 bp gene that was first found in regenerated shark liver. Through in vitro GAP assays, we found that Shark‐TBC1D15 exhibited Rab‐GAP activity on Rab7.30 Shark‐TBC1D15 shows high sequence identity to those from other species, the identity between Shark‐ and Homo‐TBC1D15 is 58.8%, and the TBC domain is 78.1%, which indicate that they may share similar structures and function. At the N‐terminus of Shark‐TBC1D15 there is an APSL (Active Peptide from Shark Liver) domain, which is involved in liver regeneration in Chiloscyllium plagiosum.31 APSL also has many other functions, such as reducing the blood glucose level in mice with type 2 diabetes,32, 33 and is also noteworthy for its immunomodulatory and inhibitory activity on lipid peroxidation.34 Although we determined the GAP activity of Shark‐TBC1D15 in vitro, the catalytic mechanism remained not clear. Until now, the structure of TBC1D15 was not available, so it is desired for us to solve the structure of Shark‐TBC1D15 or Shark‐TBC1D15·Rab complex. Using these structures, we could elucidate the difference between Shark‐TBC1D15 and other TBC domain members and explain how Shark‐TBC1D15 acts on Rab7a to accelerate its hydrolysis.
Results
Expression and purification of proteins
The full length Shark TBC1D15 and Sus TBC1D15 contain 710 residues and 674 residues, respectively. The blue boxes show the predicted Rab·GAP‐TBC domains of Shark TBC1D15 and Sus TBC1D15 [Fig. 1(A)]. We tested four truncations and found that Shark GAP (residues: 307‐654) and Sus GAP (residues: 270‐617) are the best constructs for protein expression and purification.
Figure 1.
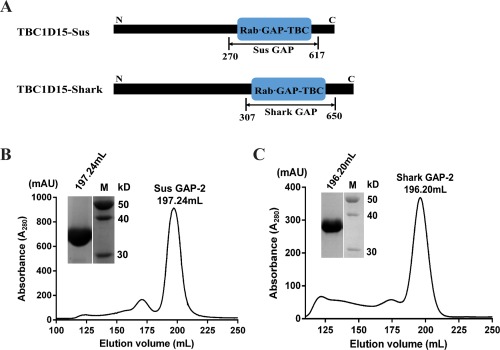
Expression and purification of Shark GAP and Sus GAP. (A) GAP domain truncations of TBC1D15‐Sus and TBC1D15‐Shark were determined based on structural prediction. The blue boxes indicate the RabGAP‐TBC domains by NCBI Blast (www.ncbi.nlm.nih.gov). Sus GAP contains the amino acid sequence from 270 to 617 while Shark GAP contains the sequence from 307 to 650. (B) Using Superdex 200 gel filtration chromatography (320 mL) to further purify Sus GAP, the elution fraction was analyzed by SDS‐PAGE. (C) Shark GAP was also further purified in the same manner as Sus GAP and the final purity of the protein was analyzed by SDS‐PAGE.
After expression and purification of the Shark and Sus GAPs, the proteins were further purified by Superdex 200 gel filtration chromatography (320 mL) to greater than 95% purity. As shown in [Fig. 1(B,C)], Sus GAP and Shark GAP were eluted at 196.20 mL and 197.24 mL, respectively. We collected the largest peak of the elution profile and conducted SDS‐PAGE. The bands from Shark GAP and Sus GAP correspond to molecular masses of 38 kDa.
The structures of the Rab GAP domains of shark TBC1D15 and Sus TBC1D15
Diffraction data for the Rab GAP domains of Shark TBC1D15 and Sus TBC1D15 were collected and scaled (Table 1). The Rab GAP domain of Sus TBC1D15 (Sus GAP) was solved at 2.5‐Å resolution using molecular replacement based on the TBC1D22A (PDB Code: 2QFZ) (there is no publication for 2QFZ) as initial model. The structure of the Shark TBC1D15 Rab GAP domain was subsequently solved at 2.8‐Å resolution using the Sus GAP structure as a research model.
Table 1.
Data Collection and Refinement Statistics
| Crystal statistics | Shark GAP | Sus GAP |
|---|---|---|
| Data collection statistics | ||
| Wavelength (Å) | 0.97872 | 0.9791 |
| Space group | C1 2 1 | P 63 2 2 |
| a, b, c, (Å) α, β, γ, (º) |
224.79 149.74 95.03 90 108.93 90 |
139.0 139.0 175.24 90 90 120 |
| Resolution range (Å) | 48.6–2.85 (2.91–2.85)a | 50–2.5 (2.60–2.50)a |
| No. of unique reflections | 69398 (4460) | 35232 (3896) |
| Completeness (%) | 100 (100) | 100 (100) |
| Redundancy | 7.7 (7.7) | 39.6 (36.6) |
| <I/σI> | 10.7 (1.7) | 22.2 (4.4) |
| Wilson B‐factor (Å2) | 66.0 | 36.1 |
| R merge(%) | 14.1 (116.5) | 17.3 (125.4) |
| CC1/2 | 0.994 (0.747) | 0.999 (0.915) |
| Refinement statistics | ||
| R work/R free (%) | 21.7/25.1 (34.1/36.3) | 22.0/26.3 (29.7/34.1) |
| No. of non‐hydrogen atoms | ||
| Macromolecules | 11173 | 5499 |
| Water | 72 | 160 |
| RMSD bond lengths (Å) | 0.005 | 0.004 |
| RMSD bond angles (º) | 0.987 | 0.918 |
| Ramachandran plot statistics | ||
| Most favored regions (%) | 98.5 | 97.4 |
| Additional allowed regions (%) | 1.5 | 2.6 |
| Outlier region (%) | 0 | 0 |
| Molprobity score | 1.3 | 1.5 |
Values in the highest resolution shell are shown in the parenthesis.
The crystals of Shark GAP have four protein molecules in the asymmetric unit, whereas those of Sus GAP have only two. There is 79.31% sequence identity between Shark GAP and Sus GAP (Fig. 2), so their structures are highly similar [Fig. 3(A)].
Figure 2.

Sequence alignment of the GAP domains of three species and Gyp1p. Sequence comparison was performed with the program CLUSTAL_W and Aline. Amino acid residues that were strictly conserved have an orange background. Three key motifs and conserved residues in red are shown above the aligned sequences. Secondary structural elements are shown above the aligned sequences. The pink spiral and blue helix indicate the Gyp1p and Shark GAP structural elements, respectively.
Figure 3.
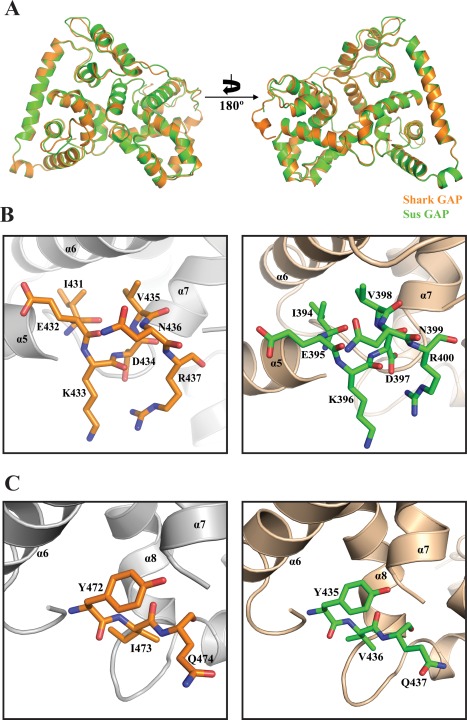
Crystal structures of the Shark GAP and Sus GAP. (A) Shark GAP (orange) is aligned with Sus GAP (green cyan) and the picture is rotated 180° and shown on the right. (B) The IxxDxxR motifs of Shark GAP (left) and Sus GAP (right) are shown as sticks. (C) The YxQ motifs of Shark GAP and Sus GAP are both shown as sticks, and the color corresponds to panel B.
Based on the structures of the Gyp1p Rab GAP domain,35 the Sus GAP and Shark GAP also have 16α‐helices but no β‐sheet elements, the helices of α1 to α16 are labeled on the structures and the different elements are labeled as affiliated helices [Fig. 4(A)]. Shark GAP and Sus GAP also have two important motifs: IxxDxxR and YxQ. IxxDxxR of the two proteins both lie on α5 and the residues are shown as sticks [Fig. 3(B)]. Similarly, the YxQ motif is also shown as sticks and lies between α6 and α7 [Fig. 3(C)].
Figure 4.
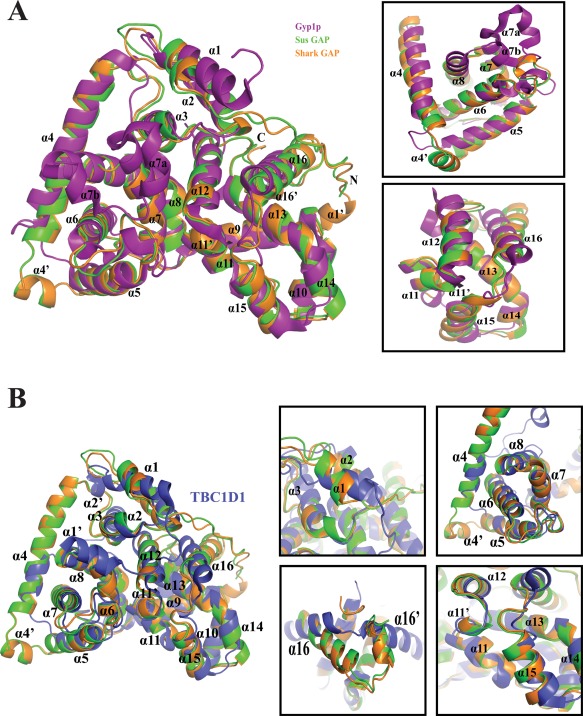
Structural comparisons of Shark GAP, Sus GAP, Gyp1p and TBC1D1. (A) Overall structure of Shark GAP (orange) compared to Sus GAP (green) and Gyp1p (purple) (PDB Code: 1FKM). The α‐helices are labeled in the cartoon. The significant differences are amplified (right). (B) The structural differences between Shark GAP, Sus GAP and TBC1D1 (blue) are obvious (left). The details (right) are amplified for further analysis.
Comparison of the structures of Sus GAP, shark GAP, yeast Gyp1p, and TBC1D1
The structures of Sus GAP and Shark GAP are compared with Yeast Gyp1p. There are several obvious structural differences between them. The amino‐terminus of Shark GAP has a short helix, termed α1′, which is not present in Sus GAP and Gyp1p. At the end of the carboxyl terminus of Shark GAP or Sus GAP, there is also a short helix α16′ that Gyp1p lacks. The α1 helices of Sus GAP and Shark GAP are shorter than that of Gyp1p. α1 contains seven residues in the Shark and Sus GAP domains, but in Gyp1p, it contains 15 residues that form a new configuration [Fig. 4(A)].
Between α4 and α5, there is an ancillary helix termed α4′ in the Shark and Sus GAP domains that is not present in Gyp1p. Another ancillary helix, α11′, lies between α11 and α12, while α12 occupies this position in Gyp1p. In the Gyp1p domain, α6 is much longer than that found in the Shark and Sus GAPs, which included the redundant residues of IPLYQFKS. Moreover, α7a and α7b are found in the Gyp1p domain,36 but in the Shark and Sus GAPs, a short loop replaces these two short helices [Fig. 4(A)]. α13 and α14 of Gyp1p connect continuously, while in the Shark and Sus GAPs, they are linked by a small loop.
TBC1D1 is a member of the TBC domain superfamily and the structure was solved in 2011.36 By comparing the structures of Sus GAP, Shark GAP and TBC1D1, we find that Sus GAP and Shark GAP show some notable differences although they have a similar overall structural frame [Fig. 4(B)]. TBC1D1 has α2′ between α1 and α2, which is absent in Sus and Shark GAPs. α4 in TBC1D1 is much shorter than that in the Sus and Shark GAPs. Moreover, the Sus and Shark GAPs also contain the ancillary helixes of α4′ and α11′. TBC1D1 has a new conformation about α16 and α16′ relative to Sus and Shark GAPs.
The catalytic mechanism of Sus GAP and shark GAP
The complex structure of Gyp1p·Rab33 provides extremely important information to analyze the catalytic mechanism.37 Using Gyp1p·Rab33 as the basic framework, we can mimic the complex structures of Sus GAP and Shark GAP separately with Rab7a and Rab11a [Fig. 5(A)]. These two members of Rab family, Rab7 (PDB Code:1T91) and Rab11a (PDB Code:1OIW), have been solved.8, 38 Rab7a substitutes the L67 to Q67 of Rab7 structure. Sequence alignment shows that Rab7a and Rab11a also have switch I region and switch II region, similar to Rab33, which are crucial for nucleotide binding and hydrolysis [Fig. 5(B)].
Figure 5.
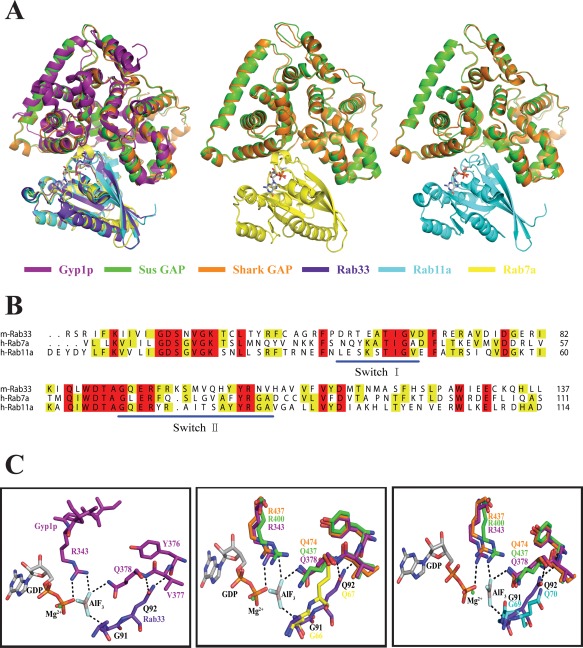
The catalytic mechanism of Shark GAP and Sus GAP mimics the Gyp1p with Rab33 complex. (A) Overall structure of the Gyp1p‐Rab33‐GDP‐AlF3 complex (PDB Code: 2G77) as a basic framework to mimic the complexes of Shark/Sus GAP with Rab7a/Rab11a (PDB Code: 1OIW). Rab7a is based on Rab7 (PDB Code: 1T91) and was changed L67 to Q67. The overall structures are shown in cartoons with the corresponding colors indicated underneath of the pictures. The GDP in the complexes is shown in stick models. (B) Sequence alignment of Rab33, Rab7a and Rab11a. The key motifs of switch I and switch II are indicated with blue lines. (C) Polar interactions in the Gyp1p‐Rab33‐GDP‐AlF3 complex are shown on the left. Shark GAP and Sus GAP are aligned with Gyp1p, while Rab7a (middle) and Rab11a (right) are aligned with Rab33. The color is the same as in panel A.
In the Gyp1p‐Rab33‐GDP‐AlF3 complex, the Arg343 in the IxxDxxR motif forms two hydrogen bonds with the oxygen from the β‐phosphate of GDP and a fluoride ion [Fig. 5(C)]. The same fluoride ion forms another hydrogen bond with Gyp1p‐Gln378 in the YxQ motif. Rab33 provides Gln92 in the DxxGQ motif to adjust a bipartite polar interaction with the Gyp1p‐Tyr376 and Gyp1p‐Gln378. Rab33‐Gly91 in the DxxGQ motif mediates the interaction with the fluoride ion of AlF3. Similar to Gyp1p, Shark GAP and Sus GAP offer the same “arginine finger” from the IxxDxxR motif and the same “glutamine finger” from the YxQ motif. Hence, the catalytic mechanism for Shark GAP and Sus GAP may be similar to Gyp1p. Shark GAP possibly provides Arg437 and Gln474 to mediate the interaction with GDP and Rab7a through the corresponding hydrogen bond. Because Rab11a contains the key residues of the Gly69 and Gln70 in switch II, which correspond to Gly66 and Gln67 in Rab7a, when the substrate is changed to Rab11a, the functional residues of Shark GAP are not altered. Sus GAP may act on the substrates of Rab7a and Rab11a using the same bonds [Fig. 5(C)].
GAP assay of shark GAP and Sus GAP
The Rab GAP activities were determined for the Shark and Sus GAP domains using human Rab7a⋅GTP and Rab11a⋅GTP complexes as substrates. Rab7⋅GTP has been shown to be an in vitro substrate for Human TBC1D1528 or Shark TBC1D15.30 Rab11a⋅GTP is another specific substrate for TBC1D15,39 but the interaction between TB1D15 and Rab11a⋅GTP has not been tested.
In the GAP assay, Rab7a⋅GTP and Rab11a⋅GTP were provided in excess for Shark GAP and Sus GAP for catalysis. Free phosphate generated by Shark and Sus GAPs‐catalyzed GTP hydrolysis were detected at 360 nm absorbance for 600 s. Shark GAP and Sus GAP can obviously catalyze Rab7a⋅GTP [Fig. 6(B,C)] and Rab11a⋅GTP [Fig. 6(D,E)] hydrolysis, as free phosphate is subsequently produced. The reaction rate is increased when the concentration of Shark GAP and Sus GAP are increased from 5 to 10 μM.
Figure 6.
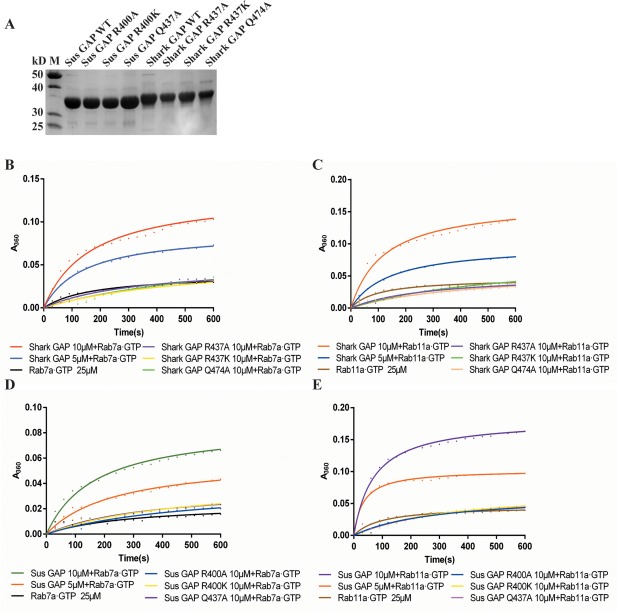
Shark and Sus GAPs compared with their variants with regards to GTP hydrolysis on Rab7a·GTP and Rab11a·GTP. (A) Protein purification of Shark and Sus GAPs with their variants; the purity is identified by SDS‐PAGE. (B) GAP assay of Shark GAP and its arginine (R437) and glutamine (Q474) variants using Rab7a·GTP as the substrate. (C) Rab11a·GTP as the substrate to determine the GAP activity of Shark GAP and variants. (D) GAP assay of Sus GAP and its arginine (R400) and glutamine (Q437) variants using Rab7a·GTP as the substrate. (E) GAP activity of Sus GAP and variants on Rab11a·GTP.
With Rab7a⋅GTP as the substrate, the catalytic efficiency parameters kcat/Km for Shark GAP and Sus GAP are (85.43 ± 1.77) × 103 M−1s−1 and (22.42 ± 2.39) × 103 M−1s−1, respectively. Using Rab11a⋅GTP as the substrate, the corresponding kcat/Km for Shark GAP and Sus GAP are (119.36 ± 23.70) × 103 M−1s−1 and (186.33 ± 9.65) × 103 M−1s−1, respectively. Both Shark GAP and Sus GAP show higher catalytic ability on Rab11a⋅GTP than Rab7a⋅GTP (1.4‐fold and 8.3‐fold respectively), Sus GAP shows higher catalytic activity on Rab11a⋅GTP and lower catalytic activity on Rab7a⋅GTP than Shark GAP.
Mutational analysis of conserved arginine and glutamine in the catalytic domain of shark GAP and Sus GAP
Point mutations were generated to analyze the catalytic function of Shark GAP and Sus GAP. Shark GAP and Sus GAP contain two critical motifs, IxxDxxR and YxQ. Although earlier studies have determined that the arginine in IxxDxxR and the glutamine in YxQ are key residues for GAP catalytic function, no such studies have been performed for TBC1D15. Therefore, we designed a series of protein variants to analyze this activity, including Arg437 (Shark GAP) and Arg400 (Sus GAP) to Ala or Lys, and Gln474 (Shark GAP) and Gln437 (Sus GAP) to Ala. The protein variants display high expression levels compared with the wild type [Fig. 6(A)].
When those two crucial residues Arg437 (Shark GAP) and Arg400 (Sus GAP) are mutated to Ala or Lys, the proteins lose catalytic activity on Rab7a⋅GTP or Rab11a⋅GTP [Fig. 6(B–E)]. The catalytic efficiency parameters kcat/Km for these variants are extremely lower compared to the wild‐type proteins. For Shark GAP, when the arginine is mutated to alanine (Shark GAP R437A), the kcat/Km values on Rab7a⋅GTP and Rab11a⋅GTP are (5.35 ± 4.88) × 103 M−1s−1 and (2.72 ± 0.50) × 103 M−1s−1, respectively. For Sus GAP R437A, the corresponding kcat/Km values are (2.27 ± 2.03) × 103 M−1s−1 on Rab7a⋅GTP and (5.90 ± 0.78) × 103 M−1s−1 on Rab11a⋅GTP.
Moreover, for Shark GAP R437K, the kcat/Km values are (1.84 ± 1.08) × 103 M−1s−1 on Rab7a⋅GTP and (5.07 ± 0.71) × 103 M−1s−1 on Rab11a⋅GTP; for Shark GAP Q474A, the kcat/Km values are (5.62 ± 3.37) × 103 M−1s−1 on Rab7a⋅GTP and (4.53 ± 1.26) × 103 M−1s−1 on Rab11a⋅GTP. In addition, for Sus GAP R437K, the kcat/Km values are (4.10 ± 0.06) × 103 M−1s−1 on Rab7a⋅GTP and (6.62 ± 1.90) × 103 M−1s−1 on Rab11a⋅GTP. For Sus GAP Q474A, the kcat/Km values are (2.68 ± 1.35) × 103 M−1s−1 on Rab7a⋅GTP and (6.42 ± 0.66) × 103 M−1s−1 on Rab11a⋅GTP. Therefore, the Shark GAP variants decrease the catalytic activity by ∼20‐fold on Rab7a⋅GTP and ∼29‐fold on Rab11a⋅GTP. For Sus GAP, the protein variants decrease the activity by ∼7‐fold on Rab7a⋅GTP and ∼30‐fold on Rab11a⋅GTP. Thus, the protein variants of Shark GAP and Sus GAP abolish the catalytic activity on Rab7a⋅GTP and Rab11a⋅GTP.
Discussion
In this study, we solved the structures of the GAP domains of Shark and Sus TBC1D15. Although Shark and Sus belong to different classes and exist in different environments, their TBC1D15 Rab GAP domains are extremely similar. This may suggest the conservation of TBC1D15 proteins among different species. The GAP domain of Homo‐TBC1D15 shares 97.99% sequence identity with Sus GAP and 78.16% with Shark GAP (Fig. 2) using ClustalW and BioEdit. Thus, we can predict that the structure of the Homo‐TBC1D15 GAP domain may be similar to Sus GAP. So far, the full‐length structure of Homo‐TBC1D15 or even the GAP domain have not been solved, although Homo‐TBC1D15 is an important protein involved in physiological metabolism. These two solved structures can help us better understand the functions of Homo‐TBC1D15 and analyze how it regulate the functions of the small GTPase Rabs.
The newly determined crystal structures of Shark GAP and Sus GAP display new variations compared to the structures of yeast Gyp1p Rab GAP domain and TBC1D1. For instance, yeast Gyp1p has ancillary helices of α7a and α7b which are not present in Shark and Sus GAPs or TBC1D1.35, 36 Shark GAP and Sus GAP have two special helices (α4′ and α11′) among α4/α5 and α11/α12, but the function of these two regions is unknown. We predict that α4′ may be involved in substrate recognition and that α11′ may influence the substrate binding capacity of the protein. Gyp1p does not contain short ancillary helices (α1′and α16′), while Shark and Sus GAPs as well as TBC1D1 all have these motifs. In addition, α16 of Shark GAP and Sus GAP show a new orientation that differs from Gyp1p and TBC1D1. The similarity of the overall framework may determine the major function of the GAP domain, because TBC1D15,28, 40 TBC1D141 and Gyp1p all have binding affinity to GTPase (Rab). On the other hand, the different regions may be required for the substrate binding specificity and other functions, but these hypotheses should be further explored.
The catalytic activities of Shark GAP and Sus GAP were measured using Rab7a⋅GTP and Rab11a⋅GTP as substrates. When Rab7a⋅GTP was used as the substrate, for Shark GAP, the kcat/Km was 85430 M−1s−1, whereas for Sus GAP, the kcat/Km was 22420 M−1s−1. However, when the substrate was changed to Rab11a⋅GTP, for Shark GAP, the kcat/Km was 119360 M−1s−1, whereas for Sus GAP, the kcat/Km was 186330 M−1s−1. From those results, we found that Shark GAP and Sus GAP also show GAP activity, although they are the C‐terminal part of the whole TBC1D15 proteins. Furthermore, for the substrates of Rab7a⋅GTP and Rab11a⋅GTP, Shark GAP and Sus GAP also have different catalytic activities between them. The kcat/Km of Shark GAP acts on Rab11a⋅GTP by approximately 1.4‐fold greater than on Rab7a⋅GTP, whereas for Sus GAP, the ratio (kcat/KmRab11a/ kcat/KmRab7a) is improved to ∼8‐fold. This indicated that the GAP activity of shark‐TBC1D15 is largely due to the GAP domain on the C‐terminus of the whole protein.30 We determined that Rab11a is the best substrate for Shark GAP and Sus GAP, which is in contrast to previous research.27, 28
Our research also identified arginine and glutamine in the catalytic site of Shark GAP and Sus GAP to be indispensable for their GAP activity. We used the structure of the yeast Gyp1p‐Rab33‐GDP‐AlF3 complex as the model to mimic the catalytic mechanism of our proteins. Through structural alignment, we found that the arginine (IxxDxxR) and glutamine (YxQ) of Shark GAP and Sus GAP lie in critical catalytic sites, thus, we assume that they can form similar hydrogen bonds to interact with substrates. This explains why all the protein variants reduced the GAP activity. Thus, the double‐finger mechanism is applicable for Shark GAP and Sus GAP. We speculate that the glutamine may stabilize the conformation with Rabs through hydrogen bonding, but we do not know the mechanism underlying the preference of Shark GAP and Sus GAP for Rab11a. One possible reason is that the distance between glutamine (YxQ) and Rabs may have an effect on this preference. Thus, further research is required. Finally, the phenomenon that Shark GAP and Sus GAP are both GAP proteins for Rab11a may help us to understand the function of Rab11a.
In conclusion, we have solved the structures of the GAP domains of Shark‐TBC1D15 and Sus‐TBC1D15. Sequence alignment showed high identity between Sus GAP and Homo GAP. Thus, Sus GAP provides structural information to analyze the GAP domain of Homo‐TBC1D15. Although Shark and Sus belong to different classes, their GAP domain show a high level of similarity regarding to structure and function. Our research demonstrates the highly conserved nature of TBC1D15. Through analysis of the molecular interface of yeast Gyp1p·Rab33, the arginine and glutamine figures are also involved in Shark GAP and Sus GAP. The double‐finger mechanism may be common in proteins which contain a GAP domain. However, we cannot use the model of Gyp1p·Rab33 to explain the preference of Shark GAP and Sus GAP for Rab11a. Although we had hoped to crystallize Shark GAP/Sus GAP in complex with Rab7a/Rab11a, we have not obtained these crystals.
Material and Methods
Cloning
The plasmids of the pETDuet His‐SUMO‐Shark TBC1D15 and pETDuet His‐SUMO ‐Sus TBC1D15 were obtained from our laboratory.30 The GAP domain sequences of the two species (amino acid residues 270‐617 and 307‐654 of Sus TBC1D15 and Shark TBC1D15, respectively) were amplified from original plasmids and cloned into the pETDuet vector for recombinant expression in E. coli.
Point mutations were made by the recombination method using ClonExpress Entry One Step Cloning Kit (Vazyme). The amplification products from the forward and reverse reactions were digested for 4 h at 37°C by Dpn I. Then, at the same temperature, the digested products were recombined for 1 h. The next steps were common transformation and identification.
Protein expression and purification
The constructs were transformed into the Escherichia coli BL21 (DE3) strain (Novagen) and the cells were grown at 37°C in LB media. Protein expression was induced overnight at 16°C with 0.1 mM isopropyl‐β‐D‐thiogalactopyranoside (IPTG) after the OD600 reached 0.8‐1.0.
The cells were disrupted with a high pressure cracker (ATS Engineering Inc. AH‐2010). The lysate was clarified by centrifugation at 16,000g for 45 min and the supernatant was loaded onto a 50 mL gravity column (GR Healthcare) loaded with 8 mL Ni2+ beads. The column was washed with 100 mL washing buffer (20 mM Tris pH 8.0, 200 mM NaCl, 48.75 mM imidzaole, 10% glycerol) and the proteins were eluted with eluting buffer (20 mM Tris pH 8.0, 200 mM NaCl, 262.5 mM imidzaole, 10% glycerol). Purification of diverse Rabs should have 10 mM MgCl2 in the above buffers. The SUMO‐fusion parts were removed by Ulp1 protease digestion. For crystallization, Sus GAP and Shark GAP were further purified using Superdex 200 gel filtration chromatography (320 mL) containing size buffer (10 mM Tris pH 7.5, 200 mM ammonium acetate, 1 mM EDTA, 1 mM DTT) and then concentrated to ∼15 mg/mL. The purity of the proteins was established by SDS‐PAGE gel and the concentrations were determined using the Qubit ® Protein Assay Kit.
GAP assay
The influence of GAPs on Rab hydrolytic activity was determined using the EnzChek Phosphate Assay Kit (Invitrogen). Full‐length Rab7a and Rab11a were purified and incubated with a 25‐fold molar excess of GTP on ice for 3 h in 20 mM Tris pH 7.5, 200 mM NaCl, 10% glycerol. After incubation, the excess of GTP was removed by a Desalting column (HiPrepTM 26/10 Desalting, GE Healthcare Life Science) and Rab‐GTP complexes were concentrated to 5 mg/mL. For the enzymatic kinetics experiment, 25 μM GTP‐loaded Rab GTPases were mixed with various concentrations of GAPs. The release of inorganic phosphates was monitored with a Multiscan spectrometer (SpectraMax 190, Molecular Devices) at 360 nm absorbance. The data were analyzed following the method to calculate the catalytic efficiency (kcat/Km).37
Crystallization and data collection
The Sus GAP and Shark GAP crystals were grown using sitting‐drop vapor diffusion at 22°C by mixing the protein and the reservoir solution at a 1:1 ratio for the first screening using Hampton Research crystallization kits®. The hanging‐drop vapor diffusion method was used for Shark GAP crystal‐optimization. Diffraction‐quality crystals of Sus GAP appeared at the condition of 13 mg/mL with a reservoir solution containing 50 mM MES sodium Salt (pH 6.5), 20% (w/v) PEG1000, 100 mM sodium chloride and 200 mM magnesium chloride. Shark GAP crystals were obtained at the reservoir solution containing 1.35 M ammonium sulfate, 0.09 M BIS‐TRIS propane (pH 7.0) and 0.01 M calcium chloride dehydrate. Crystals were soaked with 10% ethylene glycol in reservoir solution and then flash‐cooled in liquid nitrogen. X‐ray diffraction data of Sus GAP were collected at beamline BL17U1 of the SSRF (Shanghai Synchrotron Radiation Facility) and the data of Shark GAP were collected at LS‐CAT, at Advanced Photon Source in Chicago, IL, USA.
Data processing, structure determination and refinement
Diffraction data for the Rab GAP domains of Shark TBC1D15 and Sus TBC1D15 were reduced and scaled using MOSFLM42 and SCALA43 (Table 1). The Rab GAP domain of Sus TBC1D15 (Sus GAP) was solved by molecular replacement using Phaser44 with the structural model of TBC1D22A (PDB Code: 2QFZ) as the search model. The structure of the Shark TBC1D15 Rab GAP domain was subsequently solved using the Sus GAP structure as a research model. Both structures were manually rebuilt in COOT45 and refined using the Phenix46 software package.
Conflict of Interest
The authors disclose that they have no conflicts of interest with regards to content of this article.
Author Contributions
Yanna Chen, Weidong Wang, H. Eric Xu and Zhengbing Lv designed the study. Yanna Chen, Xin Gu, Weidong Wang, Dandan Cheng and Yinghua Ge performed the experiments. Xin Gu and X. Edward Zhou collected the diffraction data of Shark GAP and Sus GAP, and also solved and analyzed the structures. Yanna Chen wrote the manuscript. Yanna Chen, Weidong Wang, Dandan Cheng, Yinghua Ge, Fei Ye, H. Eric Xu and Zhengbing Lv discussed the results. H. Eric Xu and Zhengbing Lv revised and commented on the manuscript.
Acknowledgments
We thank the staff at SSRF (Shanghai Synchrotron Radiation Facility) and APS (Advanced Photon Source) for their assistance in data collection.
Accession number: The atomic coordinates and structure factors of the Shark GAP and Sus GAP have been deposited in the PDB (http://www.rcsb.org) (PDB ID codes 5TUB and 5TUC).
Contributor Information
H. Eric Xu, Email: Eric.Xu@vai.org.
Zhengbing Lv, Email: zhengbingl@zstu.edu.cn.
References
- 1. Reiner DJ, Lundquist EA (2016) Small GTPases. WormBook, ed. The C. elegans Research Community, WormBook, doi:10.1895/wormbook.1.67.2. [Google Scholar]
- 2. Kawauchi T (2011) Regulation of cell adhesion and migration in cortical neurons. Small GTPases 2:36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stenmark H (2009) Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10:513–525. [DOI] [PubMed] [Google Scholar]
- 4. Pfeffer SR (2001) Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol 11:487–491. [DOI] [PubMed] [Google Scholar]
- 5. Bos JL, Rehmann H, Wittinghofer A (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129:865–877. [DOI] [PubMed] [Google Scholar]
- 6. Guerra F, Bucci C (2016) Multiple Roles of the Small GTPase Rab7. Cells 5:PMID: 27548222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cherfils J, Zeghouf M (2013) Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 93:269–309. [DOI] [PubMed] [Google Scholar]
- 8. Wu M, Wang T, Loh E, Hong W (2005) Structural basis for recruitment of RILP by small GTPase Rab7. EMBO J 24:1491–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tan SX, Ng Y, Burchfield JG, Ramm G, Lambright DG, Stockli J, James DE (2012) The Rab GTPase‐activating protein TBC1D4/AS160 contains an atypical phosphotyrosine‐binding domain that interacts with plasma membrane phospholipids to facilitate GLUT4 trafficking in adipocytes. Mol Cell Biol 32:4946–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang T, Ming Z, Xiaochun W, Hong W (2011) Rab7: role of its protein interaction cascades in endo‐lysosomal traffic. Cell Signal 23:516–521. [DOI] [PubMed] [Google Scholar]
- 11. Jae N, McEwan DG, Manavski Y, Boon RA, Dimmeler S (2015) Rab7a and Rab27b control secretion of endothelial microRNA through extracellular vesicles. FEBS Lett 589:3182–3188. [DOI] [PubMed] [Google Scholar]
- 12. Progida C, Nielsen MS, Koster G, Bucci C, Bakke O (2012) Dynamics of Rab7b‐dependent transport of sorting receptors. Traffic 13:1273–1285. [DOI] [PubMed] [Google Scholar]
- 13. Harrison RE, Bucci C, Vieira OV, Schroer TA, Grinstein S (2003) Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol Cell Biol 23:6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edinger AL, Cinalli RM, Thompson CB (2003) Rab7 prevents growth factor‐independent survival by inhibiting cell‐autonomous nutrient transporter expression. Dev Cell 5:571–582. [DOI] [PubMed] [Google Scholar]
- 15. Yamano K, Fogel AI, Wang C, Bliek AM, Youle RJ (2014) Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. elife 3:e01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cogli L, Progida C, Thomas CL, Spencer‐Dene B, Donno C, Schiavo G, Bucci C (2013) Charcot‐Marie‐Tooth type 2B disease‐causing RAB7A mutant proteins show altered interaction with the neuronal intermediate filament peripherin. Acta Neuropathol 125:257–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zafar S, Younas N, Correia S, Shafiq M, Tahir W, Schmitz M, Ferrer I, Andréoletti O, Zerr I (2017) Strain specific altered regulatory response of Rab7a and Tau in Creutzfeldt‐Jakob disease and Alzheimer's disease. Mol Neurobiol 54:697–709. [DOI] [PubMed] [Google Scholar]
- 18. Jagoe WN, Lindsay AJ, Read RJ, McCoy AJ, McCaffrey MW, Khan AR (2006) Crystal structure of rab11 in complex with rab11 family interacting protein 2. Structure 14:1273–1283. [DOI] [PubMed] [Google Scholar]
- 19. Kelly EE, Horgan CP, McCaffrey MW (2012) Rab11 proteins in health and disease. BioChem Soc Trans 40:1360–1367. [DOI] [PubMed] [Google Scholar]
- 20. Wakabayashi Y, Dutt P, Lippincott‐Schwartz J, Arias IM (2005) Rab11a and myosin Vb are required for bile canalicular formation in WIF‐B9 cells. Proc Natl Acad Sci 102:15087–15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wikstrom K, Reid HM, Hill M, English KA, O'Keeffe MB, Kimbembe CC, Kinsella BT (2008) Recycling of the human prostacyclin receptor is regulated through a direct interaction with Rab11a GTPase. Cell Signal 20:2332–2346. [DOI] [PubMed] [Google Scholar]
- 22. Schwenk RW, Luiken JJ, Eckel J (2007) FIP2 and Rip11 specify Rab11a‐mediated cellular distribution of GLUT4 and FAT/CD36 in H9c2‐hIR cells. Biochem Biophys Res Commun 363:119–125. [DOI] [PubMed] [Google Scholar]
- 23. Knowles BC, Roland JT, Krishnan M, Tyska MJ, Lapierre LA, Dickman PS, Goldenring JR, Shub MD (2014) Myosin Vb uncoupling from RAB8A and RAB11A elicits microvillus inclusion disease. J Clin Invest 124:2947–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Onoue K, Jofuku A, Ban‐Ishihara R, Ishihara T, Maeda M, Koshiba T, Itoh T, Fukuda M, Otera H, Oka T, Takano H, Mizushima N, Mihara K, Ishihara N (2013) Fis1 acts as a mitochondrial recruitment factor for TBC1D15 that is involved in regulation of mitochondrial morphology. J Cell Sci 126:176–185. [DOI] [PubMed] [Google Scholar]
- 25. Feldman DE, Chen C, Punj V, Machida K (2013) The TBC1D15 oncoprotein controls stem cell self‐renewal through destabilization of the Numb‐p53 complex. PloS One 8:e57312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee MJ, Jang S, Nahm M, Yoon JH, Lee S (2013) Tbc1d15‐17 regulates synaptic development at the Drosophila neuromuscular junction. Mol Cells 36:163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peralta ER, Martin BC, Edinger AL (2010) Differential effects of TBC1D15 and mammalian Vps39 on Rab7 activation state, lysosomal morphology, and growth factor dependence. J Biol Chem 285:16814–16821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang X‐M, Walsh B, Mitchell CA, Rowe T (2005) TBC domain family, member 15 is a novel mammalian Rab GTPase‐activating protein with substrate preference for Rab7. Biochem Biophys Res Commun 335:154–161. [DOI] [PubMed] [Google Scholar]
- 29. Takahara Y, Maeda M, Hasegawa H, Ito S, Hyodo T, Asano E, Takahashi M, Hamaguchi M, Senga T (2014) Silencing of TBC1D15 promotes RhoA activation and membrane blebbing. Mol Cell Biochem 389:9–16. [DOI] [PubMed] [Google Scholar]
- 30. Li Y, Wang W, Cheng D, Wang T, Lu C, Chen J, Nie Z, Zhang W, Lv Z, Wu W, Shu J (2015) A New member of the TBC1D15 family from Chiloscyllium plagiosum: Rab GTPase‐activating protein based on Rab7 as a substrate. Mar Drugs 13:2955–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lv Z, Ou Y, Li Q, Zhang W, Ye B, Wu W (2009) Expression, purification and bioactivities analysis of recombinant active peptide from shark liver. Mar Drugs 7:258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Y, Chen Y, Chen J, Zhang W, Sheng Q, Chen J, Yu W, Nie Z, Zhang Y, Wu W, Wang L, Indran IR, Li J, Qian L, Lv Z (2013) A shark liver gene‐derived active peptide expressed in the silkworm, Bombyx mori: preliminary studies for oral administration of the recombinant protein. Mar Drugs 11:1492–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang F, Wu W (2005) Antidiabetic effect of a new peptide from Squalus mitsukurii liver (S‐8300) in streptozocin‐induced diabetic mice. J Pharm Pharmacol 57:1575–1580. [DOI] [PubMed] [Google Scholar]
- 34. Huang F, Lv Z, Li Q, Wei L, Zhang L, Wu W (2005) Study on hepatoprotective effect of peptide S‐8300 from shark liver. World J Gastroenterol 11:1809–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rak A, Fedorov R, Alexandrov K, Albert S, Goody RS, Gallwitz D, Scheidig AJ (2000) Crystal structure of the GAP domain of Gyp1p: first insight into interaction with Ypt/Rab proteins. EMBO J 19:5105–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park SY, Jin W, Woo JR, Shoelson SE (2011) Crystal structures of human TBC1D1 and TBC1D4 (AS160) RabGTPase‐activating protein (RabGAP) domains reveal critical elements for GLUT4 translocation. J Biol Chem 286:18130–18138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pan X, Eathiraj S, Munson M, Lambright DG (2006) TBC‐domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual‐finger mechanism. Nature 442:303–306. [DOI] [PubMed] [Google Scholar]
- 38. Pasqualato S, Senic‐Matuglia F, Renault L, Goud B, Salamero J, Cherfils J (2004) The structural GDP/GTP cycle of Rab11 reveals a novel interface involved in the dynamics of recycling endosomes. J Biol Chem 279:11480–11488. [DOI] [PubMed] [Google Scholar]
- 39. Fukuda M (2011) TBC proteins: GAPs for mammalian small GTPase Rab? Biosci Rep 31:159–168. [DOI] [PubMed] [Google Scholar]
- 40. Frasa MA, Koessmeier KT, Ahmadian MR, Braga VM (2012) Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat Rev Mol Cell Biol 13:67–73. [DOI] [PubMed] [Google Scholar]
- 41. Roach WG, Chavez JA, Miinea CP, Lienhard GE (2007) Substrate specificity and effect on GLUT4 translocation of the Rab GTPase‐activating protein Tbc1d1. Biochem J 403:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG (2011) iMOSFLM: a new graphical interface for diffraction‐image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Evans P (2006) Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. [DOI] [PubMed] [Google Scholar]
- 44. McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]


