Abstract
We aimed to analyze the antimicrobial resistance phenotypes and to elucidate the molecular mechanisms underlying resistance to cephalosporins, ciprofloxacin, and azithromycin in Salmonella enterica serovar Typhimurium isolates identified from patients with diarrhea in Shanghai. The isolates showed high rates of resistance to traditional antimicrobials, and 20.6, 12.7, and 5.5% of them exhibited decreased susceptibility to cephalosporins, ciprofloxacin, and azithromycin, respectively. Notably, 473 (84.6%) isolates exhibited multidrug resistance (MDR), including 161 (28.8%) isolates that showed an ACSSuT profile. Twenty-two MDR isolates concurrently exhibited decreased susceptibility to cephalosporins and ciprofloxacin, and six of them were co-resistant to azithromycin. Of all the 71 isolates with decreased susceptibility to ciprofloxacin, 65 showed at least one mutation (D87Y, D87N, or D87G) in gyrA, among which seven isolates simultaneously had mutations of parC (S80R) (n = 6) or parC (T57S/S80R) (n = 1), while 49 isolates with either zero or one mutation in gyrA contained plasmid-mediated quinolone resistance (PMQR) genes including qnrB, qnrS, and aac(6′)-Ib-cr. Among the 115 cephalosporin-resistant isolates, the most common ESBL gene was blaCTX-M, followed by blaTEM-1, blaOXA-1, and blaSHV -12. Eight subtypes of blaCTX-M were identified and blaCTX-M-14 (n = 22) and blaCTX-M-55 (n = 31) were found to be dominant. To the best of our knowledge, this is the first report of the presence of blaCTX-M-123 and blaCTX-M-125 in S. Typhimurium. Besides, mphA gene was identified in 15 of the 31 azithromycin-resistant isolates. Among the 22 isolates with reduced susceptibility to cephalosporins and ciprofloxacin, 15 contained ESBL and PMQR genes. Coexistence of these genes lead to the emergence of MDR and the transmission of them will pose great difficulties in S. Typhimurium treatments. Therefore, surveillance for these MDR isolates should be enhanced.
Keywords: Salmonella, ESBL, ciprofloxacin, azithromycin, PMQR
Introduction
Salmonella infection is a major global public health problem, which has caused food-borne illnesses in many countries. Data from the Foodborne Diseases Active Surveillance Network (Foodnet) report showed that Salmonella has become a leading cause of death as a food-borne bacterial pathogens in the United States (Barton Behravesh et al., 2011). Among the more than 2500 serotypes of Salmonella (Popoff et al., 2001), S. Typhimurium is one of the predominant serotypes in many developed and developing countries, and the global outbreak of food-borne diseases due to infection by S. Typhimurium is impressive. S. Typhimurium infection was very common in the United States; for example, S. Typhimurium enteritis had been diagnosed in 600 persons in 44 states due to peanut butter contamination in 2008 (Maki, 2009). S. Typhimurium infection has also been often reported in China and was found to be the second most prevalent serotype (Ran et al., 2011).
Antimicrobial treatment is needed for infants, the elderly, and immunocompromised individuals with Salmonella infection. Fluoroquinolones and cephalosporins are the primary choice for clinical treatment (Glynn et al., 1998). In addition, azithromycin has gained the approval of the Food and Drug Administration (FDA) as an additional agent to treat Salmonella infections (Sjolund-Karlsson et al., 2011). With the extensive use of such antimicrobials, antimicrobial resistance is increasing at a serious rate in Salmonella isolates. In many countries, more and more Salmonella strains with MDR (defined as resistance to three or more classes of antimicrobials) have been discovered since the 1990’s report of the spread of MDR in S. Typhimurium of definitive phage type 104 (DT104) in the world (Molbak et al., 1999). Relevant monitoring data showed that MDR in the entire Salmonella genus has increased from 20 to 30% in the early 1990s to 70% at the beginning of this century (Su et al., 2004). S. Typhimurium has also been found to show high rates of resistance to the traditional antimicrobials, and resistance to ciprofloxacin or cephalosporins have been found to have emerged in countries such as France and the United States (Weill et al., 2006; Whichard et al., 2007). A mass of S. Typhimurium isolates were also found to be resistant to ciprofloxacin or cephalosporins in many cities of China (Cui et al., 2008; Jiang et al., 2014). In recent years, some cases of azithromycin- resistance in patients with Salmonella infection were reported (Sjolund-Karlsson et al., 2011). Surprisingly, two strains have even been reported to be concurrently resistant to ciprofloxacin, cephalosporins, and azithromycin in China (Wong et al., 2014).
Shanghai is an economic and financial center located in eastern China and is divided into 16 administrative units with more than 24 million people. In order to better monitor public health issues, the Shanghai Center for Disease Control and Prevention (SCDC) became a member of the World Health Organization Global Food-borne Infections Network (WHO-GFN) in 2005 (Zhang et al., 2014). In this study, we aimed to analyze the antimicrobial resistance profiles of S. Typhimurium in Shanghai from 2011 to 2014 and tried to elucidate the molecular mechanisms underlying the emergence of MDR in these isolates. Our findings highlight the importance of increasing surveillance on these isolates with MDR, which will help provide appropriate clinical antimicrobial treatment for patients with S. Typhimurium infection.
Materials and Methods
Specimen Collection and Isolate Identification
Fresh stool samples from clinically suspected patients who had diarrhea were collected from 24 sentinel hospitals and eight regional SCDC diagnostic laboratories in Shanghai as described previously (Zhang et al., 2014). The stool samples were then enriched in Selenite Brilliant Green broth (CHROMagar, China) for 16–22 h at 37°C. The enriched samples were then cultivated on Salmonella-Shigella (SS) agar or xylose lylose deoxycholate agar (XLD; CHROMagar, China) and incubated for 18–24 h at 37°C. Presumptive colonies were screened by testing in triple-sugar-iron agar, motility indole urea agar, L-lysine decarboxylase, and L-galactosidase (o-nitrophenyl-L-D-galactopyranoside; ONPG). One presumptive colony from each sample was stored in semisolid agar and sent to our laboratory for further confirmation. API 20E test strips (bioMerieux Vitek, Marcy-l’Etoile, France) were used to confirm the identity of the isolates. All the isolates were then serotyped by slide agglutination with commercial antiserum (S&A Reagents Laboratory, Thailand) according to the Kauffmann-White scheme (World Health Organization, 2011). This study was approved by Institute of Disease Control and Prevention, Academy of Military Medical Sciences (Beijing, China) and the informed consent was obtained from the subjects involved in this study.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) for 14 antimicrobial agents including cefoxitin (FOX), ceftriaxone (CRO), ceftiofur (XNL), azithromycin (AZI), chloramphenicol (CHL), tetracycline (TET), ciprofloxacin (CIP), gentamicin (GEN), nalidixic acid (NAL), sulfisoxazole (FIS), ampicillin (AMP), streptomycin (STR), amoxicillin/clavulanic acid at a 2:1 ratio (AUG2), and trimethoprim/sulfamethoxazole (SXT) were evaluated by broth microdilution using the Sensititre plate CMV3AGNF (Sensititre, Thermo Fisher Scientific, USA) according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI) (Clinical Laboratory Standards Institute, 2016). An Escherichia coli ATCC 25922 strain was used for quality control.
PCR Amplification and DNA Sequencing
All the cephalosporin-resistant isolates were analyzed using PCR assays for the presence of extended-spectrum β-lactamase (ESBL) genes such as blaCTX-M groups, blaOXA, blaTEM, blaSHV, and blaCMY (Hasman et al., 2005; Weill et al., 2006; Cui et al., 2015). PCR amplification of quinolone resistance-determining regions (QRDRs) of gyrA, gyrB, parC, and parE (Giraud et al., 1999) and plasmid-mediated quinolone resistance (PMQR) determinants [qnrA, qnrB, qnrD, qnrS, and aac(6′)-Ib-cr] (Park et al., 2006; Cui et al., 2015) were performed on the ciprofloxacin-resistant or intermediate resistant isolates. All azithromycin-resistant isolates were subjected to PCR to detect macrolide-resistance genes including mphA, mphB, ermA, ermB, ereA, mefA, and msrA (Phuc Nguyen et al., 2009). The primers used for the above mentioned PCR amplifications are shown in Table 1. All the PCR products were sequenced by the Beijing Genomics Institute (BGI). Sequences data were then analyzed by DNAstar (DNAstar Inc., Madison, WI, USA) and the sequences were compared with reference sequences from NCBI GenBank.
Table 1.
Primers for the PCR detection of antimicrobal-resistance determinants.
| Target | Primer sequence (5′–3′) | Amplicon size (bp) | Reference |
|---|---|---|---|
| β-Lactamases | |||
| blaCTX-M-1 group | F:GGTTAAAAAATCACTGCGTC | 873 | Cui et al., 2015 |
| R:TTACAAACCGTCGGTGACGA | |||
| blaCTX-M-9 group | F:AGAGTGCAACGGATGATG | 868 | Cui et al., 2015 |
| R:CCAGTTACAGCCCTTCGG | |||
| blaCTX-M-2/8/25 group | F:ACCGAGCCSACGCTCAA | 221 | Cui et al., 2015 |
| R:CCGCTGCCGGTTTTATC | |||
| blaOXA-1 group | F:ATGAAAAACACAATACATATC | 830 | Weill et al., 2006 |
| R:AATTTAGTGTGTTTAGAATGG | |||
| blaTEM | F:ATAAAATTCTTGAAGACGAAA | 1080 | Weill et al., 2006 |
| R:GACAGTTACCAATGCTTAATC | |||
| blaSHV | F:TTATCTCCCTGTTAGCCACC | 795 | Weill et al., 2006 |
| R:GATTTGCTGATTTCGCTCGG | |||
| blaCMY | F:GTGGTGGATGCCAGCATCC | 915 | Hasman et al., 2005 |
| R:GGTCGAGCCGGTCTTGTTGAA | |||
| QRDR of Topoisomerase genes | |||
| gyrA | F: TGGGCAATGACTGGAACA | 431 | This study |
| R: GGTTGTGCGGCGGGATA | |||
| gyrB | F: ATGAGCGATATGGCAGAGCG | 309 | Cui et al., 2015 |
| R:GCTGTGATAACGCAGTTTGTCCGGG | |||
| parC | F:ATGAGCGATATGGCAGAGCG | 413 | Giraud et al., 1999 |
| R:TGACCGAGTTCGCTTAACAG | |||
| parE | F:ATGCGTGCGGCTAAAAAAGTG | 290 | Cui et al., 2015 |
| R:TCGTCGCTGTCAGGATCGATAC | |||
| PMQR | |||
| qnrA | F:ATTTCTCACGCCAGGATTTG | 516 | Cui et al., 2015 |
| R:GATCGGCAAAGGTTAGGTCA | |||
| qnrB | F:GATCGTGAAAGCCAGAAAGG | 469 | Cui et al., 2015 |
| R:ACGATGCCTGGTAGTTGTCC | |||
| qnrD | F:CGAGATCAATTTACGGGGAATA | 656 | Cui et al., 2015 |
| R:AACAAGCTGAAGCGCCTG | |||
| qnrS | F:ACGACATTCGTCAACTGCAA | 417 | Cui et al., 2015 |
| R:TAAATTGGCACCCTGTAGGC | |||
| aac(6′)-Ib-cr | F:TTGCGATGCTCTATGAGTGGCTA | 482 | Park et al., 2006 |
| R:CTCGAATGCCTGGCGTGTTT | |||
| Macrolide-resistance Genes | |||
| mphA | F:GTGAGGAGGAGCTTCGCGAG | 403 | Phuc Nguyen et al., 2009 |
| R:TGCCGCAGGACTCGGAGGTC | |||
| mphB | F:GATATTAAACAAGTAATCAGAATAG | 494 | Phuc Nguyen et al., 2009 |
| R:GCTCTTACTGCATCCATACG | |||
| ermA | F:TCTAAAAAGCATGTAAAAGAAA | 533 | Phuc Nguyen et al., 2009 |
| R:CGATACTTTTTGTAGTCCTTC | |||
| ermB | F:GAAAAAGTACTCAACCAAATA | 693 | Phuc Nguyen et al., 2009 |
| R:AATTTAAGTACCGTTACT | |||
| ereA | F:GCCGGTGCTCATGAACTTGAG | 420 | Phuc Nguyen et al., 2009 |
| R:CGACTCTATTCGATCAGAGGC | |||
| mefA | F:AGTATCATTAATCACTAGTGC | 345 | Phuc Nguyen et al., 2009 |
| R:TTCTTCTGGTACTAAAAGTGG | |||
| msrA | F:GCACTTATTGGGGGTAATGG | 384 | Phuc Nguyen et al., 2009 |
| R:GTCTATAAGTGCTCTATCGTG |
Statistical Analysis
Chi-squared test was used for data analysis using SPSS software (SPSS Inc., Chicago, IL, USA; version 17.0); a P-value < 0.05 was considered to indicate statistical significance.
Results
S. Typhimurium Isolates from Human Patients in Shanghai, China, from 2011 to 2014
Between January 2011 and December 2014, 559 S. Typhimurium isolates were cultured from patients with diarrhea in Shanghai, China. The age of the patients ranged from 10 days to 86 years (19 cases unknown) (Figure 1). Children under 5 years of ages, and especially less than 1-year-old children, were highly susceptible to S. Typhimurium infection, which accounted for 49.4% of all the patients (P < 0.05). The ratio of the male to female patients was 1.3:1. Infections occurred mainly in summer and autumn (Figure 1).
FIGURE 1.
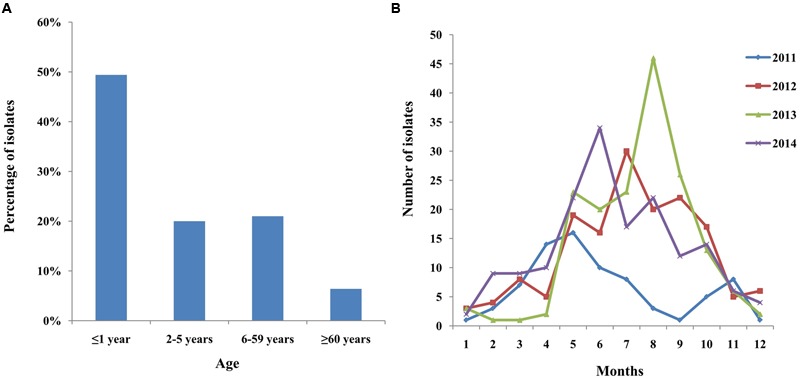
Distribution of human S. Typhimurium isolates in patients by (A) age and (B) months (2011–2014) in Shanghai, China.
Antimicrobial Susceptibility Testing
Among the 559 isolates, only 26 (4.7%) were susceptible to all 14 antimicrobials. Resistance to tetracycline was the most common (83.7%), followed by ampicillin (80.1%), nalidixic acid (79.2%), sulfisoxazole (77.1%), chloramphenicol (54.9%), streptomycin (53.7%), trimethoprim/sulfamethoxazole (47.4%), gentamicin (37%), and amoxicillin/clavulanic acid (7.9%) (Table 2). In addition, resistance to ceftiofur, ceftriaxone, and cefoxitin was found in 20, 20, and 5% of isolates, respectively. Notably, 3.6% (20/599) of the isolates displayed resistance (MIC ≥ 4 μg/mL) to ciprofloxacin, and 9.1% (51/599) exhibited intermediate resistance to ciprofloxacin (MICs = 2 μg/mL). More importantly, 22 of the isolates showed co-resistance to cephalosporins. Moreover, 31 (5.5%) isolates were showed resistance to azithromycin, and 6 of them were co-resistant to cephalosporins and ciprofloxacin.
Table 2.
Antimicrobial resistance of S. Typhimurium isolates in Shanghai from 2011 to 2014.
|
N (%) of resistant isolates |
|||||
|---|---|---|---|---|---|
| 2011 (N = 77) | 2012 (N = 155) | 2013 (N = 167) | 2014 (N = 160) | Total (N = 559) | |
| Pan-susceptible | 4 (5.2) | 8 (5.2) | 4 (2.4) | 10 (6.3) | 26 (4.7) |
| Nalidixic Acid | 67 (87) | 107 (60) | 145 (86.8) | 124 (77.5) | 443 (79.2) |
| Ciprofloxacin | 7 (9.1) | 4 (2.6) | 6 (3.6) | 3 (1.9) | 20 (3.6) |
| Ampicillin | 54 (70.1) | 124 (80) | 150 (89.8) | 120 (75) | 448 (80.1) |
| Ceftiofur | 23 (29.9) | 37 (23.9) | 30 (18) | 22 (13.8) | 112 (20) |
| Cefoxitin | 8 (10.4) | 17 (10.3) | 1 (0.6) | 2 (3.8) | 28 (5) |
| Ceftriaxone | 23 (29.9) | 37 (23.9) | 30 (18) | 22 (13.8) | 112 (20) |
| Tetracycline | 58 (75.3) | 124 (80) | 156 (93.4) | 130 (81.3) | 468 (83.7) |
| Streptomycin | 33 (42.9) | 74 (47.7) | 97 (58.1) | 96 (60) | 300 (53.7) |
| Gentamicin | 29 (37.7) | 62 (40) | 81 (48.5) | 35 (21.9) | 207 (37) |
| Chloramphenicol | 31 (40.3) | 86 (55.5) | 124 (74.3) | 66 (41.3) | 307 (54.9) |
| Azithromycin | 1 (1.3) | 13 (8.4) | 4 (2.4) | 13 (8.1) | 31 (5.5) |
| Sulfisoxazole | 27 (35.1) | 126 (81.3) | 156 (93.4) | 122 (76.3) | 431 (77.1) |
| trimethoprim/sulfamethoxazole | 31 (40.3) | 76 (49) | 97 (58.1) | 61 (38.1) | 265 (47.4) |
| amox-icillin/clavulanic acid | 9 (11.7) | 20 (12.9) | 5 (3) | 10 (6.3) | 44 (7.9) |
| MDR pattern | |||||
| ≥3 antimicrobials | 56 (72.7) | 130 (83.9) | 159 (95.2) | 128 (80) | 473 (84.6) |
| ≥4 antimicrobials | 51 (66.2) | 121 (78.1) | 147 (88) | 116 (72.5) | 435 (77.8) |
| ≥5 antimicrobials | 40 (51.9) | 91 (58.7) | 125 (74.9) | 85 (53.1) | 341 (61) |
| ≥6 antimicrobials | 25 (32.5) | 52 (33.5) | 91 (54.5) | 47(29.4) | 215 (38.5) |
| ≥7 antimicrobials | 1 (1.3) | 0 (0) | 1 (0.6) | 4 (2.5) | 6 (1.1) |
| ACSSuT | 15 (19.5) | 42 (27.1) | 66 (39.5) | 38 (23.8) | 161 (28.8) |
| ACSSuT+CIP | 5 (6.5) | 6 (3.9) | 18 (10.8) | 11 (6.9) | 40 (7.2) |
| ACSSuT+CEP | 7 (9.1) | 15 (9.7) | 8 (4.8) | 6 (3.8) | 36 (6.4) |
| ACSSuT+AZI | 1 (1.3) | 2 (1.3) | 1 (0.6) | 6 (3.8) | 10 (1.8) |
| ACSSuT+CIP+CEP | 2 (2.6) | 0 (0) | 2 (1.2) | 2 (1.3) | 6 (1.1) |
| ACSSuT+CIP+CEP+AZI | 1 (0.2) | 0 (0) | 0 (0) | 0 (0) | 1 (0.2) |
CIP, Ciprofloxacin; CEP, cephalosporins; AZI, Azithromycin.
Multidrug resistance was observed in 84.6% of the isolates, in which ≥3, ≥4, ≥5, ≥6, and ≥7 classes of antimicrobials were found in 84.6, 77.8, 61, 38.5, and 1.1% of the MDR isolates, respectively. Among the MDR isolates, 161 (28.8%) showed the ACSSuT resistance pattern (defined as resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline); among them, 40 (7.2%) isolates exhibited reduced susceptibility to ciprofloxacin, 36 (6.4%) were resistant to cephalosporins, and 10 (1.8%) were resistant to azithromycin. It is noteworthy that 6 (1.1%) isolates with ACSSuT resistance pattern were concurrently resistant to ciprofloxacin and cephalosporins, and one of them was also co-resistant to azithromycin (Table 2).
PCR Detection of Antimicrobial Drug Resistance Genes and DNA Sequencing
Resistance genes including blaOXA, blaTEM, blaSHV, blaCMY, and blaCTX-M in the 115 cephalosporin-resistance isolates were detected by PCR. PCR screenings revealed that 75 (65.2%), 52 (45.2%), 43 (37.4%), and 3 (2.6%) isolates contained the blaCTX, blaTEM, blaOXA, and blaSHV genes, respectively. All the strains were negative for blaCMY. Fifty-two isolates contained two of the above described genes (blaCTX-M+blaTEM/blaCTX-M+blaOXA/blaTEM+blaSHV), and nine isolates concurrently contained blaCTX-M, blaTEM, and blaOXA. Sequencing of the PCR products revealed that blaTEM-1, blaOXA-1, and blaSHV -12 were the main genotypes. blaCTX-M included eight subtypes: blaCTX-M-14, blaCTX-M-15, blaCTX-M-27, blaCTX-M-55, blaCTX-M-65, blaCTX-M-104, blaCTX-M-123, and blaCTX-M-125, among which blaCTX-M-14 and blaCTX-M-55 were the most common subtypes (Figure 2).
FIGURE 2.
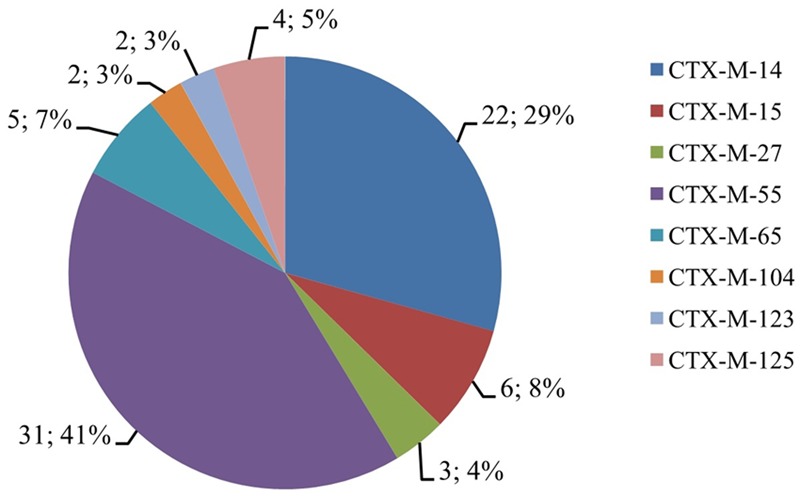
The CTX-M proportions of cephalosporin-resistant S. Typhimurium isolates. The digit before “;” indicates the number of isolates that contained blaCTX-M genes, and the digit after “;” indicates the percentage of isolates that contained blaCTX-M genes.
We identified 71 isolates with decreased susceptibility to ciprofloxacin, and 65 of them contained at least one gyrA mutation which mainly occurred at encode 87 (D87Y, D87N, or D87G). Among them, six had double mutations (S83F/D87N or D87G) in the gyrA gene and a single mutation (S80R) in the parC gene; 1 isolate with four mutations, including two in gyrA gene (S83F/D87N) and two in parC gene (T57S/S80R); 58 isolates with only one gyrA mutation (D87Y or D87N). No point mutations in gyrB and parE were found. Here, three species of PMQR determinants including aac(6′)-Ib-cr (n = 50), qnrB (qnrB2, n = 2; qnrB4, n = 4), and qnrS1 (n = 1) were detected. Among the 49 isolates with no mutation or only one mutation in gyrA gene, one or two PMQR genes were detected (Table 3).
Table 3.
Distribution of PMQR genes and mutations in gyrA, gyrB, parC, and parE genes in S. Typhimurium isolates with decreased susceptibility to ciprofloxacin in Shanghai, China.
| QRDR mutation |
PMQR |
N |
Total | ||||
|---|---|---|---|---|---|---|---|
| gyrA | gyrB | parC | parE | I∗ | R∗∗ | ||
| WT | WT | WT | WT | qnrS | 1 | 0 | 1 |
| WT | WT | WT | WT | aac(6′)-Ib-cr | 1 | 0 | 1 |
| WT | WT | WT | WT | aac(6′)-Ib-cr,qnrB | 1 | 3 | 4 |
| D87Y | WT | WT | WT | WT | 15 | 0 | 15 |
| D87Y | WT | WT | WT | aac(6′)-Ib-cr | 20 | 4 | 24 |
| D87N | WT | WT | WT | aac(6′)-Ib-cr | 13 | 4 | 17 |
| D87Y | WT | WT | WT | aac(6′)-Ib-cr,qnrB | 0 | 2 | 2 |
| S83F/D87N | WT | S80R | WT | WT | 0 | 4 | 4 |
| S83F/D87N | WT | S80R | WT | aac(6′)-Ib-cr | 0 | 1 | 1 |
| S83F/D87G | WT | S80R | WT | WT | 0 | 1 | 1 |
| S83F/D87N | WT | T57S/S80R | WT | aac(6′)-Ib-cr | 0 | 1 | 1 |
∗S. Typhimurium isolates with intermediate resistance to CIP (MIC = 2 μg/mL); ∗∗S. Typhimurium isolates with resistance to CIP (MIC ≥ 4 μg/mL); WT, wild type.
In addition, PCR results showed that 15 out of the 31 azithromycin-resistant isolates (MIC ≥ 128 μg/mL) harbored mphA genes, while the other 16 (MIC: 16–64 μg/mL) did not. All the azithromycin-resistant isolates were negative for mphB, ermA, ermB, ereA, mefA, and msrA.
Notably, among the 22 isolates that concurrently exhibited reduced susceptibility to cephalosporins and ciprofloxacin, 15 contained ESBL and PMQR genes. Two isolates contained four types of antimicrobial-resistance genes: qnrB/aac(6′)-Ib-cr/blaCTX-M/blaTEM (n = 1) and aac(6′)-Ib-cr/blaOXA/blaCTX-M/blaTEM (n = 1). Nine isolates harbored three types of antimicrobial resistant genes, including qnrB /aac(6′)-Ib-cr/blaOXA (n = 2), aac(6′)-Ib-cr/blaOXA/blaCTX-M (n = 4), and aac(6′)-Ib-cr/blaCTX-M/blaTEM (n = 3). Four isolates contained two types of antimicrobial-resistant genes: aac(6′)-Ib-cr/blaTEM (n = 2) and aac(6′)-Ib-cr/blaCTX-M (n = 2). In addition, 2 out of the 15 isolates that were co-resistant to azithromycin concurrently harbored ESBL, PMQR, and mphA genes: aac(6′)-Ib-cr/blaCTX-M/blaTEM/mphA (Table 4).
Table 4.
Summary of phenotypes of S. Typhimurium isolates showing concurrently decreased susceptibility to ciprofloxacin and cephalosporins and their corresponding resistance genes.
| Strain | MIC (μg/mL) |
QRDR mutation |
PMQR | ESBL | mphA | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| FOX | CRO | XNL | CIP | AZI | gyrA | parC | ||||
| SHI 1G663 | >32 | 4 | 8 | 4 | 4 | WT | WT | qnrB/aac(6′)-Ib-cr | ||
| SHI 1G664 | >32 | 8 | >8 | 4 | 2 | WT | WT | qnrB/aac(6′)-Ib-cr | blaOXA | |
| SHI 1G665 | >32 | 32 | 8 | 4 | 4 | WT | WT | qnrB/aac(6′)-Ib-cr | blaOXA | |
| SH11G680 | 4 | >64 | >8 | >4 | 2 | S83F/D87N | S80R | blaOXA/blaCTX-M-55 | ||
| SHI 1G993 | >32 | 16 | >8 | >4 | 256 | S83F/D87N | S80R | blaOXA | mphA | |
| SH11G1304 | 2 | >64 | >8 | 2 | 4 | D87Y | WT | aac(6′)-Ib-cr | blaOXA/blaCTX-M-27 | |
| SH11G1306 | 2 | >64 | >8 | 2 | 4 | D87Y | WT | |||
| SHI 2G734 | 32 | <0.25 | 2 | 2 | 4 | D87Y | WT | aac(6′)-Ib-cr | blaTEM | |
| SHI 2G889 | 32 | <0.25 | 2 | 4 | 8 | D87Y | WT | aac(6′)-Ib-cr | blaOXA/blaCTX-M-14/blaTEM | |
| SH12G978 | 32 | >64 | >8 | 2 | 64 | WT | WT | qnrB/aac(6′)-Ib-cr | blaCTX-M-15/blaTEM | |
| SHI 2G1005 | 8 | >64 | >8 | >4 | 64 | S83F/D87N | T57S/S80R | aac(6′)-Ib-cr | blaCTX-M-65/blaTEM | |
| SHI 3G355 | 2 | >64 | >8 | 2 | 4 | D87N | WT | aac(6′)-Ib-cr | blaTEM | |
| SHI 3G769 | 2 | >64 | >8 | 2 | 4 | D87N | WT | aac(6′)-Ib-cr | blaCTX-M-14 | |
| SHI 3G938 | 16 | >64 | >8 | 2 | 4 | D87N | WT | blaOXA/blaCTX-M-55 | ||
| SHI 3G939 | 16 | >64 | >8 | 2 | 4 | D87N | WT | blaOXA/blaCTX-M-55 | ||
| SH13G1219 | 8 | >64 | >8 | >4 | 128 | D87Y | WT | aac(6′)-Ib-cr | blaCTX-M-15/blaTEM | mphA |
| SHI 3G1825 | 2 | >64 | >8 | >4 | 128 | D87Y | WT | aac(6′)-Ib-cr | blaCTX-M-15/blaTEM | mphA |
| SHI 4G287 | 4 | >64 | >8 | 2 | 8 | WT | WT | aac(6′)-Ib-cr | blaOXA/blaCTX-M-125 | |
| SHI 4G294 | 8 | 64 | >8 | 2 | 8 | D87N | WT | aac(6′)-Ib-cr | blaOXA/blaCTX-M-125 | |
| SHI 4G300 | 4 | 64 | >8 | 2 | 64 | D87N | WT | aac(6′)-Ib-cr | blaOXA/blaCTX-M-125 | |
| SHI 4G1345 | 4 | >64 | >8 | 2 | 4 | D87Y | WT | blaOXA/blaCTX-M-55/blaTEM | ||
| SH14G1601 | 2 | >64 | >8 | 2 | 4 | D87N | WT | aac(6′)-Ib-cr | blaCTX-M-55 | |
FOX, cefoxitin; CRO, ceftriaxone; XNL, ceftiofur; CIP, ciprofloxacin; AZI, azithromycin; WT, wild type.
Discussion
In present study, we found that S. Typhimurium isolates in Shanghai, China, exhibited high rates of resistance to traditional antimicrobials, such as tetracycline, ampicillin, nalidixic acid, and sulfisoxazole. Moreover, resistance to cephalosporins, ciprofloxacin and azithromycin were found in 20.6, 3.6, and 5.5% of the isolates, respectively. Contradictory to this result from the current study, the report of the NARMS indicated that the number of cephalosporin-resistant, ciprofloxacin-resistant and azithromycin-resistant isolates was lower than 6, 1, and 1% (2011–2014), respectively (Centers for Disease Control and Prevention, 2016). More importantly, 84.6% of the isolates were found to exhibit MDR in our study, which is significantly higher than the percentage observed from the surveillance data during 2005 to 2010 in Shanghai and that indicated in the NARMS report for the same period (P < 0.05) (Zhang et al., 2014; Centers for Disease Control and Prevention, 2016). Notably, in the current study, we found that 22 MDR isolates with decreased susceptibility to ciprofloxacin were co-resistant to cephalosporins. Six of the 22 isolates also showed resistance to azithromycin. This phenomenon has only been reported previously in two clinical S. Typhimurium strains in China (Wong et al., 2014). If these MDR strains are as prevalent as S. Typhimurium DT104 all over the world, it will pose a great threat to global public health. Our findings indicate that it is therefore necessary to continue monitoring the antimicrobial resistance of S. Typhimurium isolates to help determine the appropriate antimicrobial therapy for patients with S. Typhimurium infection. More importantly, it is necessary to study the mechanisms underlying the antimicrobial resistance in these isolates.
The main mechanisms of quinolone-resistance in Salmonella have been attributed to several point mutations in the QRDRs of the gyrA and parC genes and the PMQR genes (Hopkins et al., 2007; Kim et al., 2013). Mutations at different sites contribute to different levels of resistance to ciprofloxacin, and simultaneous mutations in both gyrA and parC genes produce high levels of ciprofloxacin resistance. Besides, most isolates with decreased susceptibility to ciprofloxacin contain a mutation in gyrA or PMQR genes (Ling et al., 2003; Lin et al., 2015). In the present study, we found that six isolates had double mutations (S83F/D87N or D87G) in the gyrA gene and a single mutation (S80R) in the parC gene. Additionally, one isolate with four mutations, including two in gyrA gene (S83F/D87N) and two in parC gene (T57S/S80R). These mutations have been previously reported to be most common in ciprofloxacin-resistant S. Typhimurium strains (Wasyl et al., 2014). PMQR was initially identified in Klebsiella pneumonia in 1998 (Martinez-Martinez et al., 1998), since then, various types of PMQR genes have been detected in Salmonella all over the world (Cattoir et al., 2007; Cavaco et al., 2009; Kim et al., 2013). Previous studies have shown that the presence of qnr genes increase the MICs for ciprofloxacin by 8–16-fold (Kim et al., 2013). In addition, the presence of aac(6′)-Ib-cr genes can increase the selection of chromosomal mutants that cause ciprofloxacin resistance (Robicsek et al., 2006). In the current study, we also showed that the existence of PMQR genes remarkably enhances the MICs (≥2 μg/mL) of ciprofloxacin in the presence of a single or no point mutation in the gyrA gene in S. Typhimurium isolates. The emergence of PMQR genes described in this report has also been frequently reported previously in Salmonella isolates in other parts of China (Lu et al., 2011; Jiang et al., 2014; Wong et al., 2014), which suggests that the PMQR genes have been prevalent in China.
Since the discovery of blaCTX-M genes in Japan, Europe, and South America in the 1980s (Matsumoto et al., 1988; Bauernfeind et al., 1990, 1992), they have quickly replaced blaTEM and blaSHV as the major ESBL (D’Andrea et al., 2013). In the present study, of the 115 cephalosporin-resistant isolates, 62.5% harbored blaCTX-M genes, in which blaCTX-M-55 was the most common gene, followed by blaCTX-M-14. This phenomenon proved that the blaCTX-M-1 and blaCTX-M-9 groups are popular types in S. Typhimurium isolates in Shanghai. Besides, the subtypes of blaCTX-M-123 and blaCTX-M-125 have also been identified in E. coli (Rao et al., 2014), but have not been found in Salmonella. To the best of our knowledge, this study is the first to report blaCTX-M-123 and blaCTX-M-125 in S. Typhimurium. In addition, 52 (45.2%), 43 (37.4%), and 3 (2.6%) isolates were found to contain the blaTEM-1, blaOXA-1, and blaSHV -12 genes, respectively. Because the ESBL genes are usually located on the antimicrobial-resistance plasmid of the bacteria, they can be easily disseminated between different species of bacteria with the transfer of the drug-resistance plasmid (Arlet et al., 2006).
As the number of cephalosporin-resistant and ciprofloxacin-resistant Salmonella strains gradually increased, an alternative antimicrobial class was needed to manage Salmonella infections. Azithromycin showed excellent ability to accumulating at high intracellular concentrations, and it achieved intracellular concentrations of 50 to 100 times greater than the serum levels (Panteix et al., 1993). Therefore, azithromycin was recommended for the treatment of invasive Salmonella infections by the FAD (Sjolund-Karlsson et al., 2011). However, in the present study, 31(5.5%) isolates were found to be resistant to azithromycin. Previously, Salmonella strains resistant to azithromycin have also been found in other countries (Sjolund-Karlsson et al., 2011; Kalonji et al., 2015). The plasmid-borne mphA gene was reported as one of the reasons of azithromycin-resistance (Glynn et al., 1998). In the present study as well, 15 high-level azithromycin-resistant isolates (MIC ≥ 128 μg/mL) harbored the mphA gene, while other 16 (MIC: 16–64 μg/mL) did not. All azithromycin-resistant isolates were negative for mphB, ermA, ermB, ereA, mefA, and msrA. These results indicate that the mphA gene may mediate a high level of resistance to azithromycin in Salmonella, as described previously in Shigella in China (Zhang et al., 2016). In addition, azithromycin resistance may arise from other possible mechanisms, such as mutations in the rlpD and rlpV genes (Roberts, 2008). At present, few studies have been focused on investigating azithromycin resistance mechanisms in Salmonella. The findings of the present study and those of other previous studies emphasize the need for investigating the further mechanisms underlying azithromycin resistance in Salmonella isolates.
Antimicrobial-resistance genes are usually located on plasmids and therefore can be easily disseminated; for example the ESBL genes often coexist with the genes encoding resistance to other antimicrobial agents in plasmids, which can easily lead to a MDR phenotype in ESBL-producing bacteria (Jiang et al., 2014). In this study, we found that 15 isolates showed both ESBL and PMQR genes, in which concurrently displayed decreased susceptibility to cephalosporins and ciprofloxacin. Besides, 2 of the 15 isolates co-contained mphA genes, which were resistant to azithromycin.
Conclusion
In this study, we reported the common occurrence of MDR in S. Typhimurium isolates in Shanghai, China. We found that resistance to ciprofloxacin, cephalosporins, and azithromycin was most prevalent among the isolates. Among the MDR isolates, we found various transferrable antimicrobial-resistance genes, which included ESBL, PMQR and mphA genes, and some isolates even contained at least two types of these genes. The dissemination of these genes poses a huge threat to the control of S. Typhimurium infection in the world. Our findings indicate that it is imperative to continue monitoring the prevalence of the above-mentioned three types of antimicrobial resistance and their resistant genes in S. Typhimurium isolates. Future studies should be focused on identifying ways to prevent the dissemination of these antimicrobial-resistance genes.
Author Contributions
SQ, HS, and JW designed the study, XX participated in the collection of the samples. BL, FW, XY, CS, XH, and JX completed identification and preservation of samples, YL, XD, HaL, and JW were responsible for the experiments. JW, QM, HoL, RH, and CY analyzed the data. JW wrote the manuscript, and SQ and HS provided academic revision for the manuscript. All the authors have read and approved the final draft of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by the National Key Program for Infectious Diseases of China (nos. 2012ZX10004215 and AWS15J006), and the National Nature Science Foundation of China (nos. 81673237, 81373053, and 81371854).
References
- Arlet G., Barrett T. J., Butaye P., Cloeckaert A., Mulvey M. R., White D. G. (2006). Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8 1945–1954. 10.1016/j.micinf.2005.12.029 [DOI] [PubMed] [Google Scholar]
- Barton Behravesh C., Jones T. F., Vugia D. J., Long C., Marcus R., Smith K., et al. (2011). Deaths associated with bacterial pathogens transmitted commonly through food: foodborne diseases active surveillance network (FoodNet), 1996-2005. J. Infect. Dis. 204 263–267. 10.1093/infdis/jir263 [DOI] [PubMed] [Google Scholar]
- Bauernfeind A., Casellas J. M., Goldberg M., Holley M., Jungwirth R., Mangold P., et al. (1992). A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection 20 158–163. 10.1007/BF01704610 [DOI] [PubMed] [Google Scholar]
- Bauernfeind A., Grimm H., Schweighart S. (1990). A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18 294–298. 10.1007/BF01647010 [DOI] [PubMed] [Google Scholar]
- Cattoir V., Weill F. X., Poirel L., Fabre L., Soussy C. J., Nordmann P. (2007). Prevalence of qnr genes in Salmonella in France. J. Antimicrob. Chemother. 59 751–754. 10.1093/jac/dkl547 [DOI] [PubMed] [Google Scholar]
- Cavaco L. M., Hasman H., Xia S., Aarestrup F. M. (2009). qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 53 603–608. 10.1128/AAC.00997-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2016). NARMS Human Isolates Surveillance Report for 2014 (Final Report). Available from http://www.cdc.gov/narms/pdf/2014-annual-report-narms-508c.pdf [Google Scholar]
- Clinical Laboratory Standards Institute (2016). Performance Standards for Antimicrobial Susceptibility Testing: 26th Informational Supplement. Document M 100-S26. Wayne, PA: CLSI. [Google Scholar]
- Cui S., Li J., Sun Z., Hu C., Jin S., Guo Y., et al. (2008). Ciprofloxacin-resistant Salmonella enterica serotype Typhimurium. China. Emerg. Infect. Dis. 14 493–495. 10.3201/eid1403.070857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Wang J., Yang C., Liang B., Ma Q., Yi S., et al. (2015). Prevalence and antimicrobial resistance of Shigella flexneri serotype 2 variant in China. Front. Microbiol. 6:435 10.3389/fmicb.2015.00435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea M. M., Arena F., Pallecchi L., Rossolini G. M. (2013). CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int. J. Med. Microbiol. 303 305–317. 10.1016/j.ijmm.2013.02.008 [DOI] [PubMed] [Google Scholar]
- Giraud E., Brisabois A., Martel J. L., Chaslus-Dancla E. (1999). Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob. Agents Chemother. 43 2131–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn M. K., Bopp C., Dewitt W., Dabney P., Mokhtar M., Angulo F. J. (1998). Emergence of multidrug-resistant Salmonella enterica serotype typhimurium DT104 infections in the United States. N. Engl. J. Med. 338 1333–1338. 10.1056/NEJM199805073381901 [DOI] [PubMed] [Google Scholar]
- Hasman H., Mevius D., Veldman K., Olesen I., Aarestrup F. M. (2005). β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56 115–121. 10.1093/jac/dki190 [DOI] [PubMed] [Google Scholar]
- Hopkins K. L., Arnold C., Threlfall E. J. (2007). Rapid detection of gyrA and parC mutations in quinolone-resistant Salmonella enterica using Pyrosequencing technology. J. Microbiol. Methods 68 163–171. 10.1016/j.mimet.2006.07.006 [DOI] [PubMed] [Google Scholar]
- Jiang H. X., Song L., Liu J., Zhang X. H., Ren Y. N., Zhang W. H., et al. (2014). Multiple transmissible genes encoding fluoroquinolone and third-generation cephalosporin resistance co-located in non-typhoidal Salmonella isolated from food-producing animals in China. Int. J. Antimicrob. Agents 43 242–247. 10.1016/j.ijantimicag.2013.12.005 [DOI] [PubMed] [Google Scholar]
- Kalonji L. M., Post A., Phoba M. F., Falay D., Ngbonda D., Muyembe J. J., et al. (2015). Invasive Salmonella infections at multiple surveillance sites in the democratic republic of the congo, 2011-2014. Clin. Infect. Dis. 61(Suppl. 4) S346–S353. 10.1093/cid/civ713 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Cho J. K., Kim K. S. (2013). Prevalence and characterization of plasmid-mediated quinolone resistance genes in Salmonella isolated from poultry in Korea. Avian. Pathol. 42 221–229. 10.1080/03079457.2013.779636 [DOI] [PubMed] [Google Scholar]
- Lin D., Chen K., Wai-Chi Chan E., Chen S. (2015). Increasing prevalence of ciprofloxacin-resistant food-borne Salmonella strains harboring multiple PMQR elements but not target gene mutations. Sci. Rep. 5:14754 10.1038/srep14754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J. M., Chan E. W., Lam A. W., Cheng A. F. (2003). Mutations in topoisomerase genes of fluoroquinolone-resistant salmonellae in Hong Kong. Antimicrob. Agents Chemother. 47 3567–3573. 10.1128/AAC.47.11.3567-3573.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Wu C. M., Wu G. J., Zhao H. Y., He T., Cao X. Y., et al. (2011). Prevalence of antimicrobial resistance among Salmonella isolates from chicken in China. Foodborne Pathog. Dis. 8 45–53. 10.1089/fpd.2010.0605 [DOI] [PubMed] [Google Scholar]
- Maki D. G. (2009). Coming to grips with foodborne infection–peanut butter, peppers, and nationwide Salmonella outbreaks. N. Engl. J. Med. 360 949–953. 10.1056/NEJMp0806575 [DOI] [PubMed] [Google Scholar]
- Martinez-Martinez L., Pascual A., Jacoby G. A. (1998). Quinolone resistance from a transferable plasmid. Lancet 351 797–799. 10.1016/S0140-6736(97)07322-4 [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Ikeda F., Kamimura T., Yokota Y., Mine Y. (1988). Novel plasmid-mediated β-lactamase from Escherichia coli that inactivates oxyimino-cephalosporins. Antimicrob. Agents Chemother. 32 1243–1246. 10.1128/AAC.32.8.1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molbak K., Baggesen D. L., Aarestrup F. M., Ebbesen J. M., Engberg J., Frydendahl K., et al. (1999). An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype typhimurium DT104. N. Engl. J. Med. 341 1420–1425. 10.1056/NEJM199911043411902 [DOI] [PubMed] [Google Scholar]
- Panteix G., Guillaumond B., Harf R., Desbos A., Sapin V., Leclercq M., et al. (1993). In-vitro concentration of azithromycin in human phagocytic cells. J. Antimicrob. Chemother. 31(Suppl. E) 1–4. 10.1093/jac/31.suppl_E.1 [DOI] [PubMed] [Google Scholar]
- Park C. H., Robicsek A., Jacoby G. A., Sahm D., Hooper D. C. (2006). Prevalence in the United States of aac(6’)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50 3953–3955. 10.1128/AAC.00915-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuc Nguyen M. C., Woerther P. L., Bouvet M., Andremont A., Leclercq R., Canu A. (2009). Escherichia coli as reservoir for macrolide resistance genes. Emerg. Infect. Dis. 15 1648–1650. 10.3201/eid1510.090696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoff M. Y., Bockemuhl J., Brenner F. W., Gheesling L. L. (2001). Supplement 2000 (no. 44) to the Kauffmann-White scheme. Res. Microbiol. 152 907–909. 10.1016/S0923-2508(01)01274-8 [DOI] [PubMed] [Google Scholar]
- Ran L., Wu S., Gao Y., Zhang X., Feng Z., Wang Z., et al. (2011). Laboratory-based surveillance of nontyphoidal Salmonella infections in China. Foodborne Pathog. Dis. 8 921–927. 10.1089/fpd.2010.0827 [DOI] [PubMed] [Google Scholar]
- Rao L., Lv L., Zeng Z., Chen S., He D., Chen X., et al. (2014). Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003-2012. Vet. Microbiol. 172 534–541. 10.1016/j.vetmic.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Roberts M. C. (2008). Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 282 147–159. 10.1111/j.1574-6968.2008.01145.x [DOI] [PubMed] [Google Scholar]
- Robicsek A., Strahilevitz J., Jacoby G. A., Macielag M., Abbanat D., Park C. H., et al. (2006). Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12 83–88. 10.1038/nm1347 [DOI] [PubMed] [Google Scholar]
- Sjolund-Karlsson M., Joyce K., Blickenstaff K., Ball T., Haro J., Medalla F. M., et al. (2011). Antimicrobial susceptibility to azithromycin among Salmonella enterica isolates from the United States. Antimicrob. Agents Chemother. 55 3985–3989. 10.1128/AAC.00590-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L. H., Chiu C. H., Chu C., Ou J. T. (2004). Antimicrobial resistance in nontyphoid Salmonella serotypes: a global challenge. Clin. Infect. Dis. 39 546–551. 10.1086/422726 [DOI] [PubMed] [Google Scholar]
- Wasyl D., Hoszowski A., Zajac M. (2014). Prevalence and characterisation of quinolone resistance mechanisms in Salmonella spp. Vet. Microbiol. 171 307–314. 10.1016/j.vetmic.2014.01.040 [DOI] [PubMed] [Google Scholar]
- Weill F. X., Guesnier F., Guibert V., Timinouni M., Demartin M., Polomack L., et al. (2006). Multidrug resistance in Salmonella enterica serotype Typhimurium from humans in France (1993 to 2003). J. Clin. Microbiol. 44 700–708. 10.1128/JCM.44.3.700-708.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whichard J. M., Gay K., Stevenson J. E., Joyce K. J., Cooper K. L., Omondi M., et al. (2007). Human Salmonella and concurrent decreased susceptibility to quinolones and extended-spectrum cephalosporins. Emerg. Infect. Dis. 13 1681–1688. 10.3201/eid1311.061438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. H., Yan M., Chan E. W., Biao K., Chen S. (2014). Emergence of clinical Salmonella enterica serovar Typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrob. Agents Chemother. 58 3752–3756. 10.1128/AAC.02770-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2011). Collaborating Centre for Reference and Research on Salmonella Antigenic Formulae of Salmonella serovars. Paris: Pasteur Institute. [Google Scholar]
- Zhang C., Zhang R., Yu Q., Chu X., Sun J., Liu Q. (2016). Decreased susceptibility to azithromycin among clinical Shigella isolates from China. Microb. Drug Resist. 10.1089/mdr.2016.0134 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Zhang J., Jin H., Hu J., Yuan Z., Shi W., Ran L., et al. (2014). Serovars and antimicrobial resistance of non-typhoidal Salmonella from human patients in Shanghai, China, 2006-2010. Epidemiol. Infect. 142 826–832. 10.1017/S0950268813001659 [DOI] [PMC free article] [PubMed] [Google Scholar]


