Abstract
Francisella tularensis is a highly infectious Gram-negative bacterium and the causative agent of the zoonotic disease tularemia. This bacterial pathogen can infect a broad variety of animal species and can be transmitted to humans in numerous ways with various clinical outcomes. Although, Francisella possesses the capacity to infect numerous mammalian cell types, the macrophage constitutes the main intracellular niche, used for in vivo bacterial dissemination. To survive and multiply within infected macrophages, Francisella must imperatively escape from the phagosomal compartment. In the cytosol, the bacterium needs to control the host innate immune response and adapt its metabolism to this nutrient-restricted niche. Our laboratory has shown that intracellular Francisella mainly relied on host amino acid as major gluconeogenic substrates and provided evidence that the host metabolism was also modified upon Francisella infection. We will review here our current understanding of how Francisella copes with the available nutrient sources provided by the host cell during the course of infection.
Keywords: Francisella tularensis, metabolism, glycolysis, nutrient uptake, virulence
Introduction
Francisella tularensis is a small Gram-negative bacterium, causative agent of the zoonotic disease tularemia (Sjostedt, 2011). This facultative intracellular pathogen can infect humans by different modes, and notably direct contact with sick animals, inhalation, insect bites or ingestion of contaminated water or food (Foley and Nieto, 2010). F. tularensis is able to infect numerous cell types (Jones et al., 2012; Celli and Zahrt, 2013), including dendritic cells, neutrophils, macrophages as well as hepatocytes or endothelial cells but is thought to replicate in vivo mainly in macrophages (Santic et al., 2006). Four major subspecies of F. tularensis are currently listed: tularensis, holarctica, mediasiatica, and novicida (McLendon et al., 2006). These subspecies differ in virulence and geographical origin but all cause a fulminant disease in mice that is similar to tularemia in humans (Kingry and Petersen, 2014). Although, the subspecies novicida (here designated F. novicida) is rarely pathogenic in humans, its genome shares a high degree of nucleotide sequence conservation with the human pathogenic species and is thus widely used as a model to study highly virulent subspecies. The pathogenicity of Francisella is tightly associated to its capacity to multiply in the cytosolic compartment of infected macrophages (Celli and Zahrt, 2013). Different macrophage receptors involved in Francisella uptake have been identified (Moreau and Mann, 2013). After engulfment by phagocytic cells, Francisella transiently resides in a phagosomal compartment (Figure 1) that sequentially displays membrane markers of early (EEA1) and late endosomes/lysosomes (LAMP-1 and -2) but does not acquire the hydrolase cathepsin D or lysosomal tracers (Celli and Zahrt, 2013). Within the phagosome, Francisella must fight against several host antimicrobial defenses, including notably reactive oxygen species (ROS) produced by the NADPH oxidase (Kinkead and Allen, 2016). For this, Francisella is equipped with a series of enzymes that include superoxide dismutase, catalase and acid phosphatases (Jones et al., 2012). Phagosomal escape involves a number of additional factors among which its Type 6 secretion systems (T6SS) (Clemens et al., 2015; Rigard et al., 2016). The precise molecular contribution of the Francisella T6SS apparatus and/or effectors to phagosomal membrane disruption, as well as of additional non FPI-encoded proteins (Eshraghi et al., 2016) is not yet fully understood. The capacity of Francisella adaptation to the host cytosol nutritional environment has been coined “nutritional virulence” (Santic and Abu Kwaik, 2013).
Figure 1.
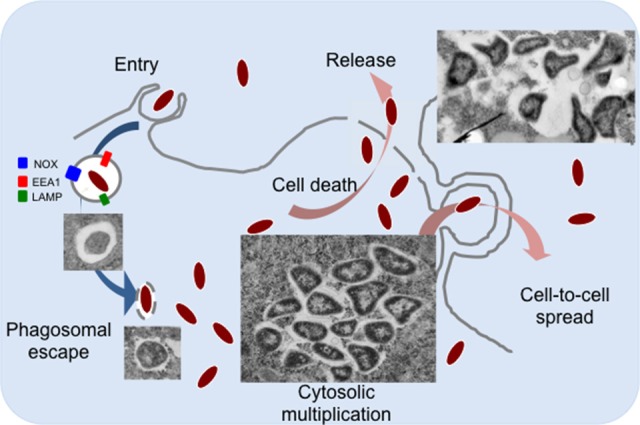
The intracellular life cycle of F. tularensis. Francisella is internalized into macrophages by large pseudopodia. Inside cells, bacteria transiently reside in a phagosomal compartment that partially matures into a late phagocytic compartment, acquiring membrane markers of early (EEA1) and late endosomes/lysosomes (LAMP-1 and -2). Typical transmission electron microscopy images of intracellular Francisella are shown. Once in the host cell cytoplasm, Francisella takes advantage of available nutrients to actively multiply. Bacteria can be released from dead cells (by apoptosis and/or pyroptosis) or can be directly transferred by trogocytosis to neighboring cells.
Francisella belongs to a restricted family of bacteria that exclusively multiply in the cytosolic compartment of infected cells. This family, that notably includes Listeria, Shigella, and Rickettsia, have a direct access to the cytosolic elements necessary for their growth. In contrast, the majority of other (facultative or obligate) intracellular pathogens reside in vacuolar compartments (Creasey and Isberg, 2014) and must therefore first import their host-derived nutrients within this compartment before being able to utilize them. For example, Legionella takes advantage of proteasomal degradation, a natural host degradative pathway (Price et al., 2011), to obtain an abundant source of amino acids to fill-up the vacuolar compartment where it resides. Indeed, Legionella pneumophila has been shown to inject the effector AnkB into the infected host cells (Al-Quadan et al., 2012) which, after lipidation by the host farnesylation machinery, becomes anchored to the vacuolar membrane and serves as a platform for the assembly of Lys48-linked polyubiquitinated proteins. Proteasomal degradation then generates elevated levels of amino acids at the vacuolar membrane, which can be imported into the vacuole.
The host cytosol was initially considered as a nutrient-replete cellular compartment (Ray et al., 2009). However, numerous studies have now clearly established that it contains a number of nutrients in limiting amounts (Fonseca and Swanson, 2014; Abu Kwaik and Bumann, 2015; Eisenreich and Heuner, 2016). Invading intracellular pathogens have therefore evolved various strategies to take advantage of the available nutrient-limiting resources (Abu Kwaik and Bumann, 2013; Zhang and Rubin, 2013; Gouzy et al., 2014b,c; Miller and Celli, 2016).
After several rounds of active multiplication in the host cytosol, Francisella dissemination to adjacent cells occurs mainly after their release from lysed cells (Jones et al., 2012). Host guanylate-binding proteins (GBPs) have been shown to be involved in pyroptotic cell death by lysing cytosolic Francisella thereby, leading to the activation of the Absent in Melanoma 2 (AIM2) inflammasome (Meunier et al., 2015). Interestingly, a novel cell-to-cell dissemination mechanism has been also described very recently, by which Francisella can infect adjacent cells by trogocytosis (Steele et al., 2016). In this review, we will address the role of nutrient acquisition in Francisella intracellular adaptation and multiplication, focusing on amino acids and carbohydrates as nutrient supplies.
Amino acids constitute a major carbon source for intracellular Francisella
Genome-scale genetic screens, performed in different cellular and/or animal models, have repeatedly identified genes encoding either metabolic pathways or predicted membrane proteins, highlighting the importance of metabolic adaptation and nutrient acquisition in intracellular survival of Francisella (Pechous et al., 2009; Meibom and Charbit, 2010). In 2009, we showed that Francisella used glutathione (a cysteine-containing tripeptide) as a source of intracellular cysteine to compensate its natural auxotrophy for cysteine (Alkhuder et al., 2009), thus providing the first demonstration that this pathogenic bacterium relied on host-derived compounds for intracellular survival. Because of its multiple auxotrophies (arginine, histidine, lysine, tyrosine, methionine, and cysteine), resulting from genetic alterations impairing biosynthetic pathways (Larsson et al., 2005), Francisella must acquire many other amino acids, including some available only in limiting concentrations within infected host cells. For this, the bacterium possesses high affinity dedicated uptake systems. Our previous genome analyses revealed that F. tularensis is mainly equipped with secondary carriers (Meibom and Charbit, 2010). This family of transporters encompasses several major families, including amino acid transporters, such as the amino acid-polyamine-organocation transporters (APC), the proton-dependent oligopeptide transporters (POT); the hydroxy/aromatic amino acid permeases (HAAAP); as well as the major facilitator superfamily (MFS) proteins, which is involved in a variety of transport functions, including amino acid uptake.
Since 2014, our laboratory has been able to characterize four amino acid transporters (two MFS members, AnsP and IleP; and two APC members, GadC, and ArgP) and to elucidate their contribution to the major steps of Francisella intracellular life cycle. We will first briefly recall below their respective properties and then discuss how amino acid availability could be used by the bacterium to sense its intracellular environment.
GadC, AnsP, IleP, and ArgP: all for one…We first functionally characterized the glutamate permease GadC (Ramond et al., 2014) and showed that intracellular multiplication of a ΔgadC mutant was essentially abolished because the mutant bacteria remained trapped within the phagosomal compartment. Specifically, our experimental data revealed that GadC contributed, within this compartment, to resistance to reactive oxygen species (ROS). Direct quantification, of tricarboxylic acid (TCA) cycle intermediates present in the cytoplasm of the wild-type and ΔgadC strains, showed that inactivation of the gadC gene significantly affected succinate, fumarate, and oxoglutarate contents. These data supported the notion that imported glutamate is used by Francisella to feed the TCA cycle within the phagosomal compartment (Ramond et al., 2014). Imported glutamate can be converted into various compounds like glutamine, glutathione, GABA or oxoglutarate (OG), which is known to be a potent anti-oxidant molecule (Mailloux et al., 2009). Of note, we had previously shown that the AAA+ chaperone MoxR of Francisella (Dieppedale et al., 2013) interacted physically with the enzymes pyruvate dehydrogenase and oxoglutarate dehydrogenase and that this interaction was required for optimal activity of these two enzymes. Since moxR gene inactivation also impaired bacterial intracellular viability and stress resistance, one may assume that the activity of the TCA cycle contributes to stress defense and bacterial virulence. The role of the TCA cycle in stress defenses is currently investigated in other pathogenic bacterial species and a direct role of the oxoglutarate dehydrogenase in resistance to nitrosative stress has been recently demonstrated in M. tuberculosis (Maksymiuk et al., 2015).
In a parallel study, we identified a second permease, designated AnsP, specifically involved in asparagine uptake. In sharp contrast to GadC, AnsP appeared to be exclusively required for bacterial cytosolic multiplication. Impaired intracellular growth of the F. novicida ΔansP mutant could be fully suppressed upon supplementation with an excess of asparagine, in vitro as well as in vivo (Gesbert et al., 2014). Of note, Neyrolles and co-workers have shown that M. tuberculosis, which is prototrophic for all 20 amino acids in broth, also relied on two paralogous amino acid transporters, one for aspartate and one for asparagine (designated AnsP1 and AnsP2, respectively) for intracellular survival and multiplication (Gouzy et al., 2013, 2014a,b,c). Interestingly, genetic inactivation of the gene encoding AnsP1 led to a severe decrease of mycobacterial fitness in vivo. In contrast, inactivation of the asparagine transporter AnsP2, although specifically involved in asparagine uptake at acidic pH, did not affect bacterial virulence in vivo.
The fact that Francisella is prototrophic for both glutamate and asparagine during growth in synthetic medium (and possesses intact biosynthetic pathways) suggests that the bacterium becomes “phenotypically” auxotrophic in infected cells, in agreement with the notion that uptake is generally preferred to synthesis. Whereas AnsP-dependent asparagine uptake was more specifically dedicated to protein synthesis, GadC-dependent glutamate transport was required for oxidative stress defense, indicating that amino acid acquisition contributes to multiple aspects of intracellular bacterial adaptation. The mechanisms down-regulating (or not up-regulating) the corresponding biogenesis pathways remain to be elucidated in intracellular bacteria.
More recently, we showed that two other amino acid permeases, IleP, and ArgP, were required in both phagosomal and cytosolic compartments. The orthologues of IleP, in the highly virulent strain F. tularensis Schu S4 and in F. tularensis LVS, had been shown to be required for normal bacterial replication in the hepatocytic human cell line HepG2 (Qin and Mann, 2006; Marohn et al., 2012). We found that the MFS permease IleP of Francisella mediated isoleucine uptake and was vital for bacterial intracellular multiplication and virulence (Gesbert et al., 2015). Remarkably, inactivation of the ileP gene in both F. novicida and F. tularensis LVS led to a delayed bacterial phagosomal escape and to a reduced cytosolic multiplication.
Genome comparisons showed that pathogenic Francisella subspecies possessed defective branched-chain amino acid (BCAA) pathways involved in leucine, isoleucine, and valine biosynthesis, relying thus exclusively on transporter-mediated acquisition of these amino acids from the host. At the opposite, F. novicida was equipped with an intact BCAA pathway and could use threonine as a precursor for their synthesis.
Of note, AnsP, and IleP proteins both belong to the Phagosomal transporter (Pht) subclass of MFS that is exclusively found among intracellular pathogenic bacteria (Chen et al., 2008). In L. pneumophila, the Pht protein PhtA has been shown to be required for threonine uptake in the Legionella-containing vacuole (Sauer et al., 2005). The L. pneumophila genome encodes 10 additional PhtA paralogues, some of which are also required during intracellular replication (Fonseca and Swanson, 2014). Interestingly, whereas PhtJ is required for acquisition of valine (Sauer et al., 2005), PhtC and PhtD were more recently shown to contribute to protecting L. pneumophila from dTMP starvation (Fonseca et al., 2014), indicating that Pht transporters are not strictly devoted to amino acid uptake.
We also showed that the permease ArgP of Francisella was a high affinity arginine transporter (Ramond et al., 2015). ArgP-mediated arginine uptake appeared to be crucial for efficient phagosomal escape. Arginine constitutes an essential amino acid for Francisella since the metabolic pathways, leading to- or coming to arginine, are predicted to be inactive, thus, highlighting the importance of essential amino acids during early stage infection. By using high-resolution mass spectrometry, we found that arginine limitation affected biogenesis of the majority of the ribosomal proteins. Indeed, in bacteria grown under arginine limiting conditions, the majority of the ribosomal proteins identified (app. 80%) were present in lower amount in the ΔargP mutant compared to wild-type, suggesting possible links between ribosomal proteins amounts and phagosomal escape (Ramond et al., 2015). Of note, ArgP is the closest paralogue of GadC within the APC family of transporter (Meibom and Charbit, 2010).
The contribution of the glycine cleavage system (GCS) to the pathogenesis of Francisella was addressed by Gerard Nau and co-workers (Brown et al., 2014). Genes encoding the GCS have been identified in genome-wide genetic screens developed to identify novel Francisella virulence genes (Meibom and Charbit, 2010). This pathway facilitates the degradation of glycine to acquire 5,10-methylene-tetrahydrofolate, a one carbon donor utilized in the production of serine, thymidine, and purines. Hence, the GCS is expected to contribute to pathogen fitness in conditions where these metabolites are limiting. In F. tularensis Schu S4, inactivation of the glycine cleavage system aminomethyltransferase T (GcvT) leads to serine auxotrophy, thus implying that the intact serABC pathway still present in the ΔgcvT mutant strain is either insufficiently active in vitro or not involved in serine biosynthesis in Francisella. The authors demonstrated that the F. tularensis GCS was essential for intracellular multiplication in conditions of serine limitation and contributed to in vivo pathogenesis. Of note, the GCS has been previously associated to persistence during chronic bacterial infection with Brucella abortus (Hong et al., 2000).
Could amino acids serve as sensors of the intracellular milieu?
The metabolism of branched chain amino acid lies at the crossroads of several other bacterial metabolic pathways in living cells. BCAAs are essential amino acids for humans and therefore must be supplied in the diet. Yet, BCAAs are among the most abundant amino acids in proteins; maintaining their pools is, thus, a prerequisite for high level synthesis of proteins. As mentioned above, in the pathogenic subsps holarctica and tularensis, BCAA degradation pathways were predicted to be nonfunctional, suggesting that exogenously acquired BCAAs could be used mainly for protein synthesis in these species. Interestingly, we found that the F. tularensis LVS triggered the uptake of important amounts of BCAAs upon entry into THP-1 macrophages (Gesbert et al., 2015). Indeed, the intracellular BCAA concentration sharply increased after 1 h of infection and strongly decreased after 24 h, suggesting that these amino acids had been consumed during the course of intracellular bacterial multiplication. We found that infection with L. monocytogenes EGD-e strain also triggered a significant (10-fold) rise in the concentration of each of the BCAAs in these cells but their concentration varied only very moderately over the course of infection. Of note, it has been previously reported that L. monocytogenes infection induced the BCAA pathway in macrophages (Lobel et al., 2012), suggesting that L. monocytogenes encountered limited amounts of BCAAs in the host cytosol. Furthermore, the pleiotropic isoleucine-responsive regulator CodY was found to be responsible for the upregulation of L. monocytogenes virulence genes under limiting concentrations of BCAAs in chemically defined medium. These observations led Herskovits and co-workers to propose that the limiting intracellular concentrations of BCAAs could represent a signal for the bacteria to sense their subcellular localization. More recently, these authors demonstrated that CodY directly bound the proximal portion of the coding sequence of the master virulence activator gene, prfA, and that this binding resulted in up-regulation of prfA transcription specifically under low concentrations of BCAA (Lobel et al., 2015), linking directly metabolism and virulence in this pathogen. Francisella genomes do not encode any CodY orthologue. Hence, one may speculate that intracellular isoleucine concentration influences Francisella genes expression via another, yet unidentified, regulatory mechanism.
Macrophages are able to synthesize their own arginine and to import it via constitutive and inducible cationic amino acid transporters CAT-1 (or SLC7A1) and CAT-2 (or SLC7A2), respectively (Ramond et al., 2015). Arginine serves either to produce nitrogen reactive species (NO, via NO synthases) or convert it to ornithine and urea (via type 1 arginase). The phagosome into which Francisella transiently resides is a dynamic compartment whose size and internal composition may considerably vary during the limited period of time spent by the bacterium. The arginine concentration available to Francisella in the phagosome might also progressively decrease during its maturation. One may suggest that intracellular Francisella respond to variations of arginine availability in this compartment by regulating their own ribosomal protein biogenesis. Indeed, repression of ribosomal protein synthesis in response to stresses, such as nutritional limitation, has been observed in all kingdoms of life (Conrad et al., 2014).
Linking carbohydrate metabolism and amino acid uptake
In mammalian cells, glycolysis and the oxidative branch of the pentose-phosphate pathway occur in the cytosol as the anabolic reactions (gluconeogenesis; and amino acid, nucleotide, fatty acids biosynthesis). In contrast, the TCA cycle and the electron-transfer chain, leading to oxidative phosphorylation, take place exclusively in the mitochondria and metabolites, such as pyruvate, are transferred from the cytosol to the mitochondria to feed the TCA. While several intracellular bacterial pathogens (such as enteroinvasive Escherichia coli and Brucella) mainly rely on glucose as a preferred carbon source for their intracellular metabolism (Abu Kwaik and Bumann, 2015), others simultaneously use multiple carbon sources (Abu Kwaik and Bumann, 2015). For example, L. monocytogenes has been shown to rely on two major carbon substrates, glycerol, and glucose-6P (Eisenreich et al., 2010; Grubmuller et al., 2014), but preferentially uses glycerol during its intracellular replication. Like L. monocytogenes, Francisella can use glucose as a carbon and energy sources. However, L. monocytogenes possesses a transporter (UhpT) specifically mediating the uptake of host glucose-6P that constitutes a key element of its cytosolic multiplication (Chico-Calero et al., 2002). Francisella genomes do not encode any orthologue of the UhpT sugar phosphate transporter family and F. novicida and F. tularensis LVS are even unable to ferment glucose-6P (Gesbert et al., 2014). This suggests that host-derived glucose-6P might not be utilizable as a source of carbohydrate by Francisella during intracellular multiplication. In contrast, the non-pathogenic related species Francisella philomiragia, which has an environmental habitat, possesses a uhpT orthologous gene (Fphi_0883, unpublished observation), suggesting that this bacterium is able to utilize this sugar during its planktonic life.
Of note, Francisella genomes encode a glycerol uptake transporter (GlpF) and a glycerol-3 phosphate transporter (GlpT). Hence, in the absence of glucose, Francisella probably mainly uses the available carbon sources (such as pyruvate, glycerol and glycerol-3P), in addition to amino acids, as intracellular carbon and nitrogen sources. The mechanisms by which glucose or glycerol are taken up by Francisella are still unknown. Indeed, genome analyses indicate that Francisella is devoid of any carbohydrate PEP-dependent phosphotransferase (PTS) system (Meibom and Charbit, 2010) or other non-PTS putative glucose permease.
Gluconeogenesis, which allows glucose synthesis from non-sugar compounds such as amino acids or TCA cycle intermediates, is involved in virulence of several intracellular bacterial pathogens, including M. tuberculosis (Marrero et al., 2010; Puckett et al., 2014) but is dispensable for other bacteria such as Brucella abortus (Zuniga-Ripa et al., 2014). In Francisella, we recently showed that gluconeogenesis constituted a major pathway required for pathogenesis (Brissac et al., 2015). Indeed, inactivation of the gene glpX, encoding the unique class II fructose biphosphatase (FBPase) of Francisella, severely impaired intra-macrophagic bacterial multiplication, in the presence of gluconeogenic substrates and considerably attenuated virulence in the mouse mode. The strictly gluconeogenic enzyme is responsible for the conversion of fructose 1,6-bisphosphate into fructose 6-phosphate (Figure 2). A severe intracellular multiplication defect of a ΔglpX mutant was also observed when cells were supplemented with a gluconeogenic substrate (e.g., pyruvate or glycerol). In contrast, wild-type multiplication was restored when the medium was supplemented with glucose. In chemically defined medium (CDM), inactivation of glpX also led to a severe growth defect in all the media containing gluconeogenic substrates. Growth of the wild-type strain in CDM lacking glucose (CDMΔGlc) was significantly reduced but still detectable, due to the presence of the amino acids present in the 3 mM range whereas growth of the ΔglpX mutant was essentially abolished. Supplementation of CDMΔGlc with individually added excess amino acid (25 mM) improved to variable extents growth of the wild-type strain in the absence of glucose. Remarkably, alanine was the only amino acid to restore wild-type growth (Brissac et al., 2015) although no gene encoding a putative alanine dehydrogenase (catalyzing the conversion of L-alanine to pyruvate) could be predicted in the F. novicida genome (unpublished observation). As expected, supplementation of the CDMΔGlc medium with any of the (individually added) twenty amino acids failed to restore growth of the ΔglpX mutant, strongly suggesting that amino acids served as gluconeogenic substrates by Francisella.
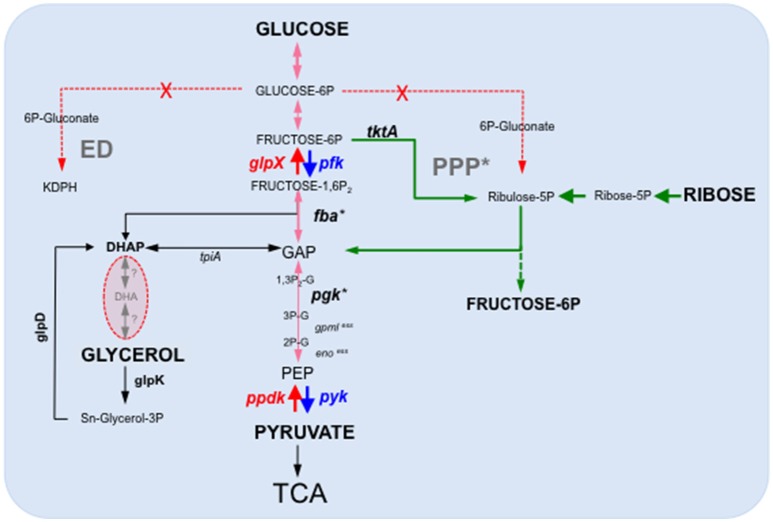
Figure 2.
Schematic depiction of the Francisella glycolytic and gluconeogenic pathways. F. tularensis possesses a complete glycolysis and gluconeogenesis pathways as well as an intact TCA cycle. In contrast, it lacks a functional Entner-Doudoroff (ED) pathway and the oxidative branch of the Pentose phosphate pathway (PPP), in red-dotted arrows. PPP*, the non-oxidative branch of the PPP is still functional. Ppdk and FBPase (in red) represent only two enzymatic steps specifically acting in the gluconeogenic direction. The dotted green arrows indicate the non-oxidative part of the Pentose Phosphate Pathway. Gene numbers in F. novicida: pgm (FTN_0514); pgi (FTN_0663); gpml (FTN_0648); eno (FTN_0621); glpX (FTN_0298); pfk (FTN_1210); (FTN_1631); glpD (FTN_1584); glpK (FTN_1585). xess, essential genes. *Expression of fba and pgK have shown to be up-regulated in BMM (Wehrly et al., 2009). F. tularensis Schu S4 and F. novicida U112 are able to ferment glycerol but the enzymes involved are unknown. F. tularensis LVS is unable to ferment glycerol. DHAP, Dihydroxyacetone phosphate (or Glycerone-Phosphate); GAP, glyceraldehyde 3-phosphate; PEP, phosphoenolpyruvate.
Isotopic profiling, using either 13C-labeled glucose or 13C-labeled pyruvate, revealed that Francisella possessed both active glycolysis and gluconeogenesis pathways in CDM (Brissac et al., 2015). The existence of a complete and functional glycolytic pathway in Francisella is in agreement with a genome-wide study which notably revealed that gene FTN_1210 in F. novicida most likely encoded a phosphofructokinase, responsible for the conversion fructose-6P to fructose-1,6P (Enstrom et al., 2012). Indeed, this study, aimed at attributing novel non-predictable metabolic functions to non-essential genes, showed that inactivation of gene FTN_1210 abolished growth on sugars but not on short-chain carbon sources, suggesting a block in glycolysis. Furthermore, the gene FTN_1210 was shown to functionally complement the growth defect of an E. coli pfk mutant on sorbitol as the carbon source. Of note, the gene FTN_1210, now designated pfk, is still erroneously annotated as encoding a putative ribokinase in the KEGG database.
Our metabolomics analyses further indicated that the enzyme of the non-oxidative PPP (transketolase, transaldolase) were functional (Figure 2). However, 6-phosphogluconate (6-PG) and pentose phosphates were not detected, suggesting an absence of the oxidative part of PPP, from 6-PG to pentose-5-phospate, suggesting a major role of the Embden–Meyerhof (glycolysis) pathway in glucose catabolism in Francisella. Remarkably, a reduction in the intracellular glucose concentration was recorded in both J774.1 and THP-1 macrophages infected for 24 h with F. tularensis LVS (Brissac et al., 2015). The need for cytosolic Francisella to possess an active gluconeogenic pathway is thus consistent with the significant reduction in the available intracellular glucose pool observed upon infection. The control of the intracellular glucose homeostasis is likely to be a key issue for proper bacterial multiplication. Infected cells might modify their metabolism to reduce the available glucose intracellular pool and, hence, limit bacterial proliferation. Of particular interest, a very recent study (Sanman et al., 2016) demonstrated that Salmonella typhimium activated the NLRP3 inflammasome by disrupting the glycolytic flux upon infection of bone marrow-derived macrophages. The authors showed that this trigger occurred because intracellular bacteria were using the macrophage supply of glycolysis precursor molecules. This study suggests that glycolytic disruption may constitute a more general mechanism of inflammasome activation triggered in response to metabolic parasitism by microbes.
In conclusion, our recent studies have shown that amino acid uptake systems and carbohydrate metabolic pathways played both an important and complementary role in Francisella pathogenesis (Figure 3). Novel approaches are developed to translate multi-omics data into functional metabolic programs. For example, constraints-based systems analysis has been used on Francisella to integrate existing high-throughput data, in silico and experimental information (Raghunathan et al., 2010). This approach allowed the reconstruction of a genome-scale metabolic model, which identified significant changes of metabolism during Francisella intracellular growth in the infected macrophage. Altogether, a switch from oxidative metabolism, in the initial stages of infection, to glycolysis, fatty acid oxidation, and gluconeogenesis, during the later stages, could be deduced from their analyses.
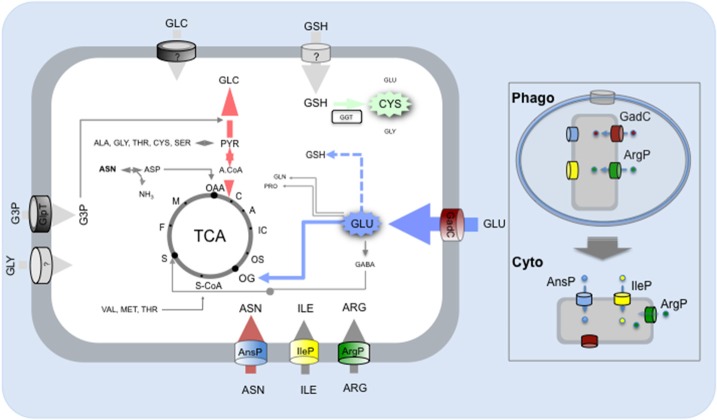
Figure 3.
Amino acid and carbohydrate uptake systems involved in Francisella intracellular survival. Schematic depiction of the carbohydrate and amino acid transporters (predicted or characterized) of Francisella and contribution of the imported amino acid and carbohydrates to metabolic pathways. Amino acids (ASN, asparagine; ILE, Isoleucine; ARG, Arginine; GLU, Glutamate) are taken up by Francisella via dedicated amino acid transporters and used to feed the TCA cycle (GLU) and/or as gluconeogenic substrates and/or as building blocks for protein synthesis (ASN, ARG, ILE). The imported carbohydrates (GLC, Glucose; GLY, Glycerol; G3P, Glyceraldehyde-3-Phosphate) are used to feed the glycolysis/gluconeogenesis pathways. Protein numbers of the represented transporters in F. novicida: GadC (FTN_ 0571), AnsP (FTN_1586), IleP (FTN_1654), ArgP (FTN-0848); G3P transporter GlpT (FTN_0636). Boxed to the right of the Figure, the respective contributions of the amino acid transporters to: (i) phagosomal survival and escape (upper part); (ii) cytosolic multiplication (lower part).
Understanding the subtle interplay between bacterial and host metabolism will be a major challenge for future studies. Many important questions remain to be answered on the bacterial side to assess the contribution of other nutrient sources (such as lipids, ions) during intracellular multiplication. The molecular dissection of the links between metabolic alterations triggered by the pathogen and the modulation of the innate immune response constitutes a novel and very promising field of investigation.
Author contributions
AC and JZ wrote the paper, MB edited the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
These studies were supported by INSERM, CNRS and Université Paris Descartes Paris Cité Sorbonne. JZ was funded by a fellowship from the “Délégation Générale à l'Armement.” The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Abu Kwaik Y., Bumann D. (2013). Microbial quest for food in vivo: 'Nutritional virulence' as an emerging paradigm. Cell. Microbiol. 15, 882–890. 10.1111/cmi.12138 [DOI] [PubMed] [Google Scholar]
- Abu Kwaik Y., Bumann D. (2015). Host delivery of favorite meals for intracellular pathogens. PLoS Pathog. 11:e1004866. 10.1371/journal.ppat.1004866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhuder K., Meibom K. L., Dubail I., Dupuis M., Charbit A. (2009). Glutathione provides a source of cysteine essential for intracellular multiplication of Francisella tularensis. PLoS Pathog. 5:e1000284. 10.1371/journal.ppat.1000284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Quadan T., Price C. T., Abu Kwaik Y. (2012). Exploitation of evolutionarily conserved amoeba and mammalian processes by Legionella. Trends Microbiol. 20, 299–306. 10.1016/j.tim.2012.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissac T., Ziveri J., Ramond E., Tros F., Kock S., Dupuis M., et al. (2015). Gluconeogenesis, an essential metabolic pathway for pathogenic Francisella. Mol. Microbiol. 98, 518–534. 10.1111/mmi.13139 [DOI] [PubMed] [Google Scholar]
- Brown M. J., Russo B. C., O'Dee D. M., Schmitt D. M., Nau G. J. (2014). The contribution of the glycine cleavage system to the pathogenesis of Francisella tularensis. Microbes Infect. 16, 300–309. 10.1016/j.micinf.2013.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J., Zahrt T. C. (2013). Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb. Perspect. Med. 3:a010314. 10.1101/cshperspect.a010314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. E., Podell S., Sauer J. D., Swanson M. S., Saier M. H., Jr. (2008). The phagosomal nutrient transporter (Pht) family. Microbiology 154(Pt 1), 42–53. 10.1099/mic.0.2007/010611-0 [DOI] [PubMed] [Google Scholar]
- Chico-Calero I., Suarez M., Gonzalez-Zorn B., Scortti M., Slaghuis J., Goebel W., et al. (2002). Hpt, a bacterial homolog of the microsomal glucose- 6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc. Natl. Acad. Sci. U.S.A. 99, 431–436. 10.1073/pnas.012363899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens D. L., Ge P., Lee B. Y., Horwitz M. A., Zhou Z. H. (2015). Atomic structure of T6SS reveals interlaced array essential to function. Cell 160, 940–951. 10.1016/j.cell.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M., Schothorst J., Kankipati H. N., Van Zeebroeck G., Rubio-Texeira M., Thevelein J. M. (2014). Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 38, 254–299. 10.1111/1574-6976.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey E. A., Isberg R. R. (2014). Maintenance of vacuole integrity by bacterial pathogens. Curr. Opin. Microbiol. 17, 46–52. 10.1016/j.mib.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppedale J., Gesbert G., Ramond E., Chhuon C., Dubail I., Dupuis M., et al. (2013). Possible links between stress defense and the tricarboxylic acid (TCA) cycle in Francisella pathogenesis. Mol. Cell. Proteomics 12, 2278–2292. 10.1074/mcp.M112.024794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenreich W., Dandekar T., Heesemann J., Goebel W. (2010). Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8, 401–412. 10.1038/nrmicro2351 [DOI] [PubMed] [Google Scholar]
- Eisenreich W., Heuner K. (2016). The life stage-specific pathometabolism of Legionella pneumophila. FEBS Lett. 590, 3868–3886. 10.1002/1873-3468.12326 [DOI] [PubMed] [Google Scholar]
- Enstrom M., Held K., Ramage B., Brittnacher M., Gallagher L., Manoil C. (2012). Genotype-phenotype associations in a nonmodel prokaryote. mBio 3:e00001–12. 10.1128/mbio.00001-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshraghi A., Kim J., Walls A. C., Ledvina H. E., Miller C. N., Ramsey K. M., et al. (2016). Secreted effectors encoded within and outside of the Francisella Pathogenicity Island promote intramacrophage growth. Cell Host Microbe 20, 573–583. 10.1016/j.chom.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J. E., Nieto N. C. (2010). Tularemia. Vet. Microbiol. 140, 332–338. 10.1016/j.vetmic.2009.07.017 [DOI] [PubMed] [Google Scholar]
- Fonseca M. V., Sauer J. D., Crepin S., Byrne B., Swanson M. S. (2014). The phtC-phtD locus equips Legionella pneumophila for thymidine salvage and replication in macrophages. Infect. Immun. 82, 720–730. 10.1128/IAI.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca M. V., Swanson M. S. (2014). Nutrient salvaging and metabolism by the intracellular pathogen Legionella pneumophila. Front. Cell. Infect. Microbiol. 4:12. 10.3389/fcimb.2014.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesbert G., Ramond E., Rigard M., Frapy E., Dupuis M., Dubail I., et al. (2014). Asparagine assimilation is critical for intracellular replication and dissemination of Francisella. Cell. Microbiol. 16, 434–449. 10.1111/cmi.12227 [DOI] [PubMed] [Google Scholar]
- Gesbert G., Ramond E., Tros F., Dairou J., Frapy E., Barel M., et al. (2015). Importance of branched-chain amino acid utilization in Francisella intracellular adaptation. Infect. Immun. 83, 173–183. 10.1128/IAI.02579-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzy A., Larrouy-Maumus G., Bottai D., Levillain F., Dumas A., Wallach J. B., et al. (2014a). Mycobacterium tuberculosis exploits asparagine to assimilate nitrogen and resist acid stress during infection. PLoS Pathog. 10:e1003928. 10.1371/journal.ppat.1003928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzy A., Larrouy-Maumus G., Wu T. D., Peixoto A., Levillain F., Lugo-Villarino G., et al. (2013). Mycobacterium tuberculosis nitrogen assimilation and host colonization require aspartate. Nat. Chem. Biol. 9, 674–676. 10.1038/nchembio.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzy A., Poquet Y., Neyrolles O. (2014b). Amino acid capture and utilization within the Mycobacterium tuberculosis phagosome. Future Microbiol. 9, 631–637. 10.2217/fmb.14.28 [DOI] [PubMed] [Google Scholar]
- Gouzy A., Poquet Y., Neyrolles O. (2014c). Nitrogen metabolism in Mycobacterium tuberculosis physiology and virulence. Nat. Rev. Microbiol. 12, 729–737. 10.1038/nrmicro3349 [DOI] [PubMed] [Google Scholar]
- Grubmuller S., Schauer K., Goebel W., Fuchs T. M., Eisenreich W. (2014). Analysis of carbon substrates used by Listeria monocytogenes during growth in J774A.1 macrophages suggests a bipartite intracellular metabolism. Front. Cell. Infect. Microbiol. 4:156. 10.3389/fcimb.2014.00156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P. C., Tsolis R. M., Ficht T. A. (2000). Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun. 68, 4102–4107. 10.1128/IAI.68.7.4102-4107.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. L., Napier B. A., Sampson T. R., Llewellyn A. C., Schroeder M. R., Weiss D. S. (2012). Subversion of host recognition and defense systems by Francisella spp. Microbiol. Mol. Biol. Rev. 76, 383–404. 10.1128/MMBR.05027-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingry L. C., Petersen J. M. (2014). Comparative review of Francisella tularensis and Francisella novicida. Front. Cell. Infect. Microbiol. 4:35. 10.3389/fcimb.2014.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkead L. C., Allen L. A. (2016). Multifaceted effects of Francisella tularensis on human neutrophil function and lifespan. Immunol. Rev. 273, 266–281. 10.1111/imr.12445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P., Oyston P. C., Chain P., Chu M. C., Duffield M., Fuxelius H. H., et al. (2005). The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37, 153–159. 10.1038/ng1499 [DOI] [PubMed] [Google Scholar]
- Lobel L., Sigal N., Borovok I., Belitsky B. R., Sonenshein A. L., Herskovits A. A. (2015). The metabolic regulator CodY links Listeria monocytogenes metabolism to virulence by directly activating the virulence regulatory gene prfA. Mol. Microbiol. 95, 624–644. 10.1111/mmi.12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L., Sigal N., Borovok I., Ruppin E., Herskovits A. A. (2012). Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence. PLoS Genet. 8:e1002887. 10.1371/journal.pgen.1002887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailloux R. J., Singh R., Brewer G., Auger C., Lemire J., Appanna V. D. (2009). α-ketoglutarate dehydrogenase and glutamate dehydrogenase work in tandem to modulate the antioxidant α-ketoglutarate during oxidative stress in Pseudomonas fluorescens. J. Bacteriol. 191, 3804–3810. 10.1128/JB.00046-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksymiuk C., Balakrishnan A., Bryk R., Rhee K. Y., Nathan C. F. (2015). E1 of α-ketoglutarate dehydrogenase defends Mycobacterium tuberculosis against glutamate anaplerosis and nitroxidative stress. Proc. Natl. Acad. Sci. U.S.A. 112, E5834–E5843. 10.1073/pnas.1510932112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marohn M. E., Santiago A. E., Shirey K. A., Lipsky M., Vogel S. N., Barry E. M. (2012). Members of the Francisella tularensis phagosomal transporter subfamily of major facilitator superfamily transporters are critical for pathogenesis. Infect. Immun. 80, 2390–2401. 10.1128/IAI.00144-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero J., Rhee K. Y., Schnappinger D., Pethe K., Ehrt S. (2010). Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. U.S.A. 107, 9819–9824. 10.1073/pnas.1000715107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon M. K., Apicella M. A., Allen L. A. (2006). Francisella tularensis: taxonomy, genetics, and Immunopathogenesis of a potential agent of biowarfare. Annu. Rev. Microbiol. 60, 167–185. 10.1146/annurev.micro.60.080805.142126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom K. L., Charbit A. (2010). Francisella tularensis metabolism and its relation to virulence. Front. Microbiol. 1:140. 10.3389/fmicb.2010.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E., Wallet P., Dreier R. F., Costanzo S., Anton L., Ruhl S., et al. (2015). Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 16, 476–484. 10.1038/ni.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C., Celli J. (2016). Avoidance and subversion of eukaryotic homeostatic autophagy mechanisms by bacterial pathogens. J. Mol. Biol. 428, 3387–3398. 10.1016/j.jmb.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau G. B., Mann B. J. (2013). Adherence and uptake of Francisella into host cells. Virulence 4, 826–832. 10.4161/viru.25629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechous R. D., McCarthy T. R., Zahrt T. C. (2009). Working toward the future: insights into Francisella tularensis pathogenesis and vaccine development. Microbiol. Mol. Biol. Rev. 73, 684–711. 10.1128/MMBR.00028-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. T., Al-Quadan T., Santic M., Rosenshine I., Abu Kwaik Y. (2011). Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science 334, 1553–1557. 10.1126/science.1212868 [DOI] [PubMed] [Google Scholar]
- Puckett S., Trujillo C., Eoh H., Marrero J., Spencer J., Jackson M., et al. (2014). Inactivation of fructose-1,6-bisphosphate aldolase prevents optimal co-catabolism of glycolytic and gluconeogenic carbon substrates in Mycobacterium tuberculosis. PLoS Pathog. 10:e1004144. 10.1371/journal.ppat.1004144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A., Mann B. J. (2006). Identification of transposon insertion mutants of Francisella tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 31:69 10.1186/1471-2180-6-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan A., Shin S., Daefler S. (2010). Systems approach to investigating host-pathogen interactions in infections with the biothreat agent Francisella. Constraints-based model of Francisella tularensis. BMC Syst. Biol. 4:118. 10.1186/1752-0509-4-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramond E., Gesbert G., Guerrera I. C., Chhuon C., Dupuis M., Rigard M., et al. (2015). Importance of host cell arginine uptake in Francisella phagosomal escape and ribosomal protein amounts. Mol. Cell. Proteomics 14, 870–881. 10.1074/mcp.M114.044552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramond E., Gesbert G., Rigard M., Dairou J., Dupuis M., Dubail I., et al. (2014). Glutamate utilization couples oxidative stress defense and the tricarboxylic acid cycle in Francisella phagosomal escape. PLoS Pathog. 10:e1003893. 10.1371/journal.ppat.1003893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K., Marteyn B., Sansonetti P. J., Tang C. M. (2009). Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat. Rev. Microbiol. 7, 333–340. 10.1038/nrmicro2112 [DOI] [PubMed] [Google Scholar]
- Rigard M., Broms J. E., Mosnier A., Hologne M., Martin A., Lindgren L., et al. (2016). Francisella tularensis IglG belongs to a novel family of PAAR-Like T6SS proteins and harbors a unique N-terminal extension required for virulence. PLoS Pathog. 12:e1005821. 10.1371/journal.ppat.1005821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanman L. E., Qian Y., Eisele N. A., Ng T. M., van der Linden W. A., Monack D. M., et al. (2016). Disruption of glycolytic flux is a signal for inflammasome signaling and pyroptotic cell death. Elife 5:e13663. 10.7554/elife.13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M., Abu Kwaik Y. (2013). Nutritional virulence of Francisella tularensis. Front. Cell. Infect. Microbiol. 3:112. 10.3389/fcimb.2013.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M., Molmeret M., Klose K. E., Abu Kwaik Y. (2006). Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 14, 37–44. 10.1016/j.tim.2005.11.008 [DOI] [PubMed] [Google Scholar]
- Sauer J. D., Bachman M. A., Swanson M. S. (2005). The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc. Natl. Acad. Sci. U.S.A. 102, 9924–9929. 10.1073/pnas.0502767102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostedt A. (2011). Special topic on Francisella tularensis and tularemia. Front. Microbio. 2:86. 10.3389/fmicb.2011.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele S., Radlinski L., Taft-Benz S., Brunton J., Kawula T. H. (2016). Trogocytosis-associated cell to cell spread of intracellular bacterial pathogens. Elife 5:e10625. 10.7554/elife.10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrly T. D., Chong A., Virtaneva K., Sturdevant D. E., Child R., Edwards J. A., et al. (2009). Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell. Microbiol. 11, 1128–1150. 10.1111/j.1462-5822.2009.01316.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. J., Rubin E. J. (2013). Feast or famine: the host–pathogen battle over amino acids. Cell. Microbiol. 15, 1079–1087. 10.1111/cmi.12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga-Ripa A., Barbier T., Conde-Alvarez R., Martinez-Gomez E., Palacios-Chaves L., Gil-Ramirez Y., et al. (2014). Brucella abortus depends on pyruvate phosphate dikinase and malic enzyme but not on Fbp and GlpX fructose-1,6-bisphosphatases for full virulence in laboratory models. J. Bacteriol. 196, 3045–3057. 10.1128/JB.01663-14 [DOI] [PMC free article] [PubMed] [Google Scholar]