Abstract
Cryptococcus neoformans is the most lethal pathogen of the central nervous system. The gold standard treatment of cryptococcosis, a combination of amphotericin B with 5-fluorocytosine, involves broad toxicity, high costs, low efficacy, and limited worldwide availability. Although the need for new antifungals is clear, drug research and development (R&D) is costly and time-consuming. Thus, drug repurposing is an alternative to R&D and to the currently available tools for treating fungal diseases. Here we screened a collection of compounds approved for use in humans seeking for those with anti-cryptococcal activity. We found that benzimidazoles consist of a broad class of chemicals inhibiting C. neoformans growth. Mebendazole and fenbendazole were the most efficient antifungals showing in vitro fungicidal activity. Since previous studies showed that mebendazole reaches the brain in biologically active concentrations, this compound was selected for further studies. Mebendazole showed antifungal activity against phagocytized C. neoformans, affected cryptococcal biofilms profoundly and caused marked morphological alterations in C. neoformans, including reduction of capsular dimensions. Amphotericin B and mebendazole had additive anti-cryptococcal effects. Mebendazole was also active against the C. neoformans sibling species, C. gattii. To further characterize the effects of the drug a random C. gattii mutant library was screened and indicated that the antifungal activity of mebendazole requires previously unknown cryptococcal targets. Our results indicate that mebendazole is as a promising prototype for the future development of anti-cryptococcal drugs.
Keywords: Cryptococcus neoformans, benzimidazoles, mebendazole, antifungal, biofilm, antifungal targets, macrophages
Introduction
Cryptococcus neoformans is a yeast-like pathogen that causes expressive brain damage in immunosuppressed individuals (Colombo and Rodrigues, 2015). The fungus reaches the lungs of humans after inhalation of environmental cells. In the immunosuppressed host, C. neoformans efficiently disseminates to the brain and causes meningitis (Kwon-Chung et al., 2014). Cryptococcal meningitis is a global problem resulting in thousands of deaths annually (Park et al., 2009). Most cases occur among people with HIV/AIDS. Poor and late diagnosis, limited access to antifungals and drug resistance are directly associated to the high fatality rate of cryptococcosis, especially in developing countries (Rodrigues, 2016).
The standard antifungal regimen for cryptococcal meningitis is a combination of amphotericin B with 5-fluorocytosine (Krysan, 2015). Amphotericin B is nephrotoxic and is intravenously administered (Sloan et al., 2009; Micallef et al., 2015), which demands considerable medical infrastructure. A 15-day intravenous treatment with liposomal amphotericin B is estimated to cost from €10.000 to €20.000 in Europe (Ostermann et al., 2014) and 5-fluorocytosine is not widely available outside rich areas (Krysan, 2015). As an alternative, fluconazole is frequently used, although it is associated with poorer outcomes and relapses (Sloan et al., 2009). In South Africa, more than 60% of people with culture-positive relapsed disease had fluconazole resistance (Govender et al., 2011). Hence, the need for new anticryptococcal therapies is clear. In this context, a new class of antifungals targeting the synthesis of fungal sphingolipids has been recently described, but its efficacy in humans is still unknown (Mor et al., 2015).
Drug repurposing has emerged as an alternative to the costly and time-consuming processes of drug discovery and development (Nosengo, 2016). In the field of antifungal development, sertraline, an anti-depressive agent, has been reported to be an in vitro and in vivo fungicidal compound that, in combination with amphotericin B, improves the outcome of cryptococcosis (Zhai et al., 2012; Rhein et al., 2016). Sertraline is now under phase III trial to determine whether adjunctive therapy will lead to improved survival (ClinicalTrials.gov, 2016).
In this manuscript, we aimed at finding anti-cryptococcal activity in a collection of drugs previously approved for use in human diseases. Our results are in agreement with the notion that benzimidazole-like compounds are interesting prototypes for the future development of efficient anti-cryptococcal agents interfering with fungal morphology, biofilm formation, cellular proliferation and intracellular parasitism. This study also supports the hypothesis that the antifungal activity of mebendazole might involve previously unknown cellular targets.
Materials and Methods
Strains and Growth Conditions
Strains H99 of C. neoformans (sorotype A) and R265 of C. gattii (sorotype B) were maintained in Sabouraud’s agar. For capsule size determination and fluorescence microscopy, fungal cells were cultivated in a minimal medium composed of glucose (15 mM), MgSO4 (10 mM), KH2PO4 (29.4 mM), glycine (13 mM), and thiamine-HCl (3 μM), pH5.5 for 48 h at 37°C with shaking. The C. gattii mutant library was maintained in 96-well plates containing yeast peptone-dextrose (YPD) broth with 30% glycerol at -20°C. The cell line J774.16 (murine macrophages) was maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% of Fetal Bovine Serum (FBS), and 1% of penicillin-streptomycin at 37°C in 5% CO2 atmosphere. After four passages in culture medium, the macrophages were plated into 96 well plates for tests of mebendazole intracellular activity.
Screening for Antifungal Activity in a Compound Collection
The National Institutes of Health (NIH) clinical collection (NCC) was screened for antifungal activity against C. neoformans. The NCC consists in a small molecule repository of 727 compounds arrayed in 96-well plates at 10 mM solution in DMSO. These compounds are part of screening library for the NIH Roadmap Molecular Libraries Screening Centers Network (MLSCN) and correspond to a collection of chemically diverse compounds that have been in phase I-III clinical trials. Each compound was first diluted to 1 mM in DMSO and stored at -20°C until use. For initial screening, all compounds were used at 10 μM in 100 μl of RPMI 1640 (two times concentrated) medium buffered with morpholinepropanesulfonic acid (MOPS) at pH 7 containing 2% of glucose, in 96-well plates. Final concentration of DMSO in all samples corresponded to 1%. C. neoformans cells (104) suspended in 100 μl of water were added to each well. The plates were incubated at 37°C with shaking for 48 h. The optical density at 540 nm (OD540) was recorded using the FilterMax 5 microplate reader (Molecular Devices, Sunnyvale, CA, USA). Compounds producing values of OD540 smaller than 0.05 after fungal growth were selected for further studies. As further detailed in the section “Results,” mebendazole was the NCC compound selected for most of the analysis performed in this study.
Analysis of Antifungal Activity of NCC Compounds
Values of minimum inhibitory concentrations (MICs) were determined using the methods proposed by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) with minor modifications. NCC compounds showing antifungal activity were serially diluted (20 to 0.03 μM) in RPMI 1640 (two times concentrated, pH 7; 2% glucose) buffered with MOPS in 96-well plates. The inocula of C. neoformans (strain H99) and C. gattii (strain R265) were prepared following the EUCAST protocol (Subcommittee on Antifungal Susceptibility Testing of the EECfAST, 2008; Arendrup et al., 2012). The plates were incubated at 37°C with shaking for 48 h. MIC values corresponded to the lowest compound concentration producing inhibition of fungal growth. For determination of fungicidal activity, C. neoformans cells were grown overnight in YPD broth at 30°C, washed in PBS and suspended in RPMI 1640 buffered with MOPS, pH 7. The yeast suspension was adjusted to 2 × 104 cells per 10 ml of RPMI 1640 (pH 7; 2% glucose) buffered with MOPS and supplemented with 1.25, 0.3125, and 0.078 μM of antifungal compounds. Final concentration of DMSO corresponded to 0.6% in all samples. The samples were then incubated at 37°C in a rotary shaker at 200 rpm. Aliquots were taken at different time points and plated onto YPD agar plates that were incubated at 37°C for 48 h. The numbers of CFU were then counted and recorded. The minimum fungicidal concentration (MFC) was defined as the lowest drug concentration inhibiting CFU formation in at least 90% in comparison to systems containing no antifungals.
Antifungal Activity of Mebendazole against Intracellular Cryptococci
To assess antifungal activity against intracellular C. neoformans, the fungus was first opsonized by incubation (20 min, 37°C, with shaking) in DMEM containing 10% FBS and 10 μg/ml of the 18B7 IgG1, an opsonic monoclonal antibody to GXM (Casadevall et al., 1998) kindly donated by Dr. Arturo Casadevall (Johns Hopkins University). The fungus was washed with fresh DMEM and incubated with J774.16 macrophages (1:1 ratio, 5 × 105 cells/well of 96 well-plates) for 2 h in DMEM containing 10% FBS, 0.3 μg/ml LPS and 0.005 μg/ml IFNγ (37°C, 5% CO2). After interaction of C. neoformans with macrophages, the systems were washed with DMEM to remove extracellular fungal cells and fresh DMEM containing 10% FBS and variable concentrations of mebendazole (0.25, 0.5, and 1 μM, 200 μl/well) was added to each well. The plates were incubated at 37°C with 5% CO2. After 8 or 24 h, supernatants were collected for inoculation of YPD agar plates and subsequent CFU counting. Alternatively, infected macrophages were lysed with cold, distillated water and the resulting lysates were plated onto YPD agar plates for CFU counting. To evaluate the toxicity of mebendazole for J774.16 macrophages, 5 × 105 macrophages suspended in DMEM containing 10% FBS were plated in each well of 96 wells plates and incubated overnight at 37°C with 5% CO2. The medium was supplemented with fresh DMEM containing 10% FBS and mebendazole (0.25, 0.5, and 1 μM) or 0.5% DMSO. The plate was incubated at 37°C in a 5% CO2 atmosphere. After 48 h, the systems were washed with DMEM and 50 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) at 5 mg/ml was added to each well, for further incubation for 4 h (37°C, 5% CO2) under light protection (Mosmann, 1983). Supernatants were removed and 200 μl DMSO was added for dissolution of formazan crystals. Absorbances were recorded using the FilterMax 5 microplate reader (Molecular Devices, Sunnyvale, CA, USA) at 570 nm.
Analysis of Synergistic Effects
Synergistic activity between mebendazole and standard antifungal drugs was determined on the basis of the calculation of the fractional inhibitory index (FIC) (Mor et al., 2015). Briefly, mebendazole (denominated drug A) was serially diluted (0.38–0.006 μg/ml, 8 dilutions) in 96-well plates. Standard antifungals (denominated drugs B) were serially diluted (11 dilutions) from 16 to 0.015 μg/ml (amphotericin B) or 64 to 0.06 μg/ml (fluconazole). The FIC was defined as:
Synergism was categorized as follows: strongly synergistic effect, FIC < 0.5; synergistic effect, FIC < 1; additive effect, FIC = 1; no effect, 1 < FIC < 2; antagonistic effect, FIC > 2 (Mor et al., 2015).
Effects of Mebendazole on Glucuronoxylomannan (GXM) Release
Cryptococcus neoformans cells (105/well of 96-well plates, final volume of 200 μl, duplicates) were cultivated in RPMI 1640 buffered with MOPS, pH 7. The medium was supplemented with mebendazole (0.3125–0.001 μM). After 48 h of incubation at 37°C with shaking, the optical density of 540 nm (OD540) was recorded using the FilterMax 5 microplate reader (Molecular Devices, Sunnyvale, CA, USA). The plate was centrifuged for 10 min and supernatants were used for GXM quantification by ELISA using the protocol described by Casadevall et al. (1992). C. neoformans viability was monitored by propidium iodide (PI) staining of fungal cells. For this analysis, fungal cells obtained after exposure to 0.3125, 0.15625, and 0.078 μM mebendazole as described above were stained with 1 mg/ml PI for 5 min on ice and analyzed by flow cytometry in a FACS Cabibur (BD Biosciences, CA, USA). The percentage of stained (non-viable) cells was obtained with the FlowJo 7 software.
Analysis of Capsular Size and Morphology
Cryptococcus neoformans was grown overnight in YPD broth and washed twice with PBS. The fungus was then suspended in minimal medium containing sub-inhibitory concentrations of mebendazole and incubated for 48 h at 37°C with shaking. C. neoformans cells were collected by centrifugation, washed in PBS and analyzed by microscopic approaches. For capsule size determination, the suspension was counterstained with India ink and placed onto glass slides. The suspensions were covered with glass coverslips and analyzed with an Axioplan 2 (Zeiss, Germany) microscope. Capsule size, calculated with the ImageJ Software, was defined as the distance between the cell wall and the outer border of the capsule. Cell diameters were determined using the same software. For additional analysis of capsular morphology, cellular suspensions were processed for fluorescence microscopy. Staining reagents used in this analysis-included calcofluor white (cell wall chitin, blue fluorescence) and the monoclonal antibody 18B7 (Casadevall et al., 1998). C. neoformans cells were prepared for fluorescence microscopy following the protocols established by our laboratory for routinely analysis of surface architecture (Rodrigues et al., 2008).
Effects of Mebendazole on C. neoformans Biofilms
Cryptococcus neoformans was grown in Sabouraud’s dextrose broth for 24 h at 30°C with shaking. The cells were centrifuged at 3,000 g for 5 min, washed twice with PBS, suspended in minimal medium and adjusted to a density of 107 cells/ml. Cell suspensions (100 μl) were added into quadruplicate wells of polystyrene 96-well plates (Greiner Bio-One, Australia), following incubation at 37°C for 48 h. The wells containing biofilms were washed three times with PBS to remove non-adhered cryptococcal cells. Fungal cells that remained attached to the wells were considered mature biofilms. To evaluate the susceptibility of C. neoformans biofilms to mebendazole, 100 μl solutions (31.25, 15.63, 3.13, 1.56, 0.31, and 0.16 μM) were added to each well. Amphotericin B and fluconazole (2 and 8 μg/ml, respectively) were used as control systems of antifungal activity. Negative controls corresponded to wells containing only water and untreated biofilms. Mature biofilms and antifungal drugs were incubated at 37°C for 24 h, washed three times with PBS and the biofilm metabolic activity quantified by the 2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide (XTT) reduction assay (Meshulam et al., 1995). Prior studies demonstrated that the XTT reduction assay measurements correlate with biofilm and fungal cell number (Martinez and Casadevall, 2007). In addition to testing the effects of mebendazole on established biofilms, we evaluated whether this compound would inhibit biofilm formation. Cryptococcal cells were suspended in minimal medium and adjusted to a density of 107 cells/ml in the presence or absence of mebendazole (31.25, 15.63, 3.13, 1.56, 0.31, and 0.16 μM). These suspensions were added in quadruplicates to the wells of polystyrene 96-well plates, following incubation at 37°C for 48 h. Amphotericin B and fluconazole (2 and 8 μg/ml, respectively) were used as antifungal controls. In negative controls, wells contained only ultrapure water. The wells were washed three times with PBS and biofilm formation was quantified by the XTT assay.
Analysis of the Antifungal Activity of Mebendazole against a Collection of C. gattii Mutants
A collection of randomly generated C. gattii mutants was screened for identification of possible cellular targets for antifungal activity. Mutants (n = 7,569) were generated by insertional mutagenesis after incubation of C. gattii with Agrobacterium tumefaciens as previously described (Idnurm et al., 2004). All colonies that grew on YPD hygromycin plates were selected and maintained at -20°C in 96 wells plates containing 200 μl/well of YPD broth. Before exposure to mebendazole, mutant cells were first grown for 72 h (30°C) in 200 μl of YPD distributed into the wells of 96 wells plates. The antifungal activity of mebendazole against C. neoformans was reproduced in the C. gattii R265 strain. The mutants were tested for their ability to grow in the presence of RPMI 1640 supplemented with MOPS, 2% of glucose, 1% DMSO and 10 μM mebendazole. Resistance phenotypes (A540 > 0.3) were selected for dose-response tests of antifungal activity as described above and potential target identification was performed as detailed below.
Identification of Potential Cellular Targets Required for the Antifungal Activity of Mebendazole
Based on clear resistant phenotypes, two C. gattii mutant strains were selected for target identification by polymerase chain reaction (PCR). The fungus was cultivated overnight in 10 ml YPD at 30°C with constant rotation. Each culture (2 ml) was centrifuged for 2 min at 4,000 × g and cell pellets were washed twice with PBS and collected for DNA extraction (Bolano et al., 2001). The cells were suspended in 400 μl of lysis buffer (50 mM Tris-HCl 1 mM EDTA, 200 mM NaCl, 2% Triton X100, 0.5% SDS, pH 7.5), for further addition of 1 volume of a phenol-chloroform mixture (pH 8) and 100 μl of 2-μm, acid washed glass beads. Mechanical disruption was performed by alternate 1-min cycles of vortexing and ice incubation. Lysates were centrifuged 4,000 × g for 20 min at 4°C. Supernatants were collected and DNA was ethanol-precipitated overnight at -20°C for subsequent treatment with RNAse (Bolano et al., 2001). Identification of missing genes in the mutants was performed using inverse PCR (Pavlopoulos, 2011). DNA was quantified using the Qubit reagent (Invitrogen) and 1 μg of each sample was cleaved separately with BglII, SalI, or StuI (Promega) restriction enzymes. The cleavage product was submitted to T4 DNA ligase reaction (New England) followed by PCR using primers for inverse PCR. Amplicons were gel-purified with the QIAquick Gel Extraction Kit (Qiagen). For the DNA sequencing reaction, 50 ng of each sample and 5 pmol of each primer were used. Sequences were obtained in an ABI-Prism 3500 Genetic Analyzer (Applied Biosystems) and their qualities were determined by a electropherogram analysis based on phred1. The identification of genes interrupted by the T-DNA was performed by comparison of each sequenced DNA fragment, which correspond to the T-DNA flanks, with the genomic sequence of C. neoformans H99 strain, available in the Cryptococcus genome databases (Broad Institute2) using BLASTn. Orthologs distribution was evaluated using the OrthoMCL database (Chen et al., 2006).
Mutants showing resistance to mebendazole were analyzed for their ability to produce melanin and extracellular GXM as previously described by our group (Rodrigues et al., 2015).
Statistical Analyses
Statistics were obtained with the GraphPad Prism 6.0 software. Unpaired t student test was used for mebendazole toxicity analysis. The variance two-way ANOVA was carried out using Tukey’s and Bonferroni’s comparisons test for fungicidal activity and intracellular activity of mebendazole in macrophages. For biofilm analyses, one-way ANOVA was performed using Dunnett’s multiple comparisons.
Results
Selection of Mebendazole as a Potential Anti-cryptococcal Agent
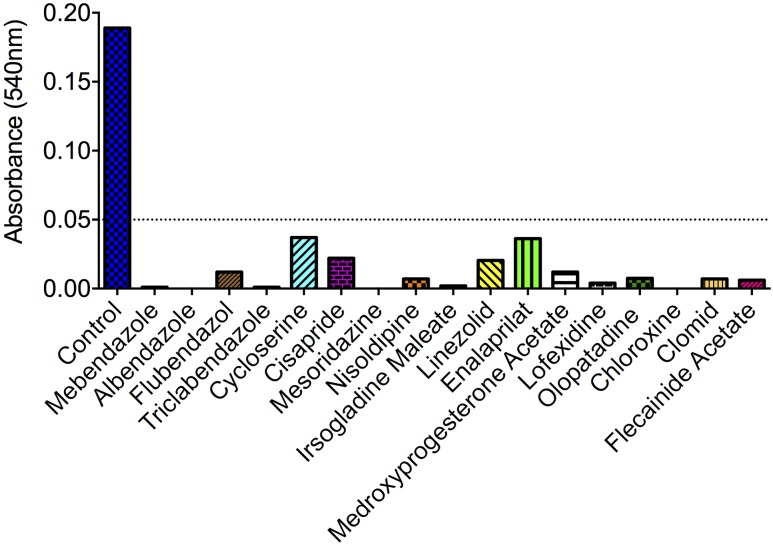
Of the 727 drugs tested at 10 μM, 17 compounds were active against C. neoformans, including antibacterials (chloroxine, cycloserine, and linezolid), a neuroleptic drug (mezoridazine), calcium channel blockers (nisoldipine and enalaprilat), antiarrhythmic agents (flecainide acetate), drugs for gastrointestinal malignances (irsogladine maleate and cisapride), gynecologic regulators (medroxyprogesterone acetate and clomid), an anti-histaminic (olopatadine) and the anti-helminthic benzimidazoles (mebendazole, albendazole, flubendazole, and triclabendazole) (Figure 1). Noteworthy, none of the molecules showing antifungal activity were structurally related, except the benzimidazoles. Considering the currently observed efficacy in inhibiting the growth of C. neoformans, their structural similarity and previously described anti-cryptococcal activity (Cruz et al., 1994), we selected benzimidazoles for further investigation in our model.
FIGURE 1.
Identification of small-molecule inhibitors of Cryptococcus neoformans via drug-repurposing screening. Out of 727 compounds tested, 17 molecules induced A540 values smaller than 0.05 (dotted line). Compounds with no antifungal activity are not shown. For description of the primary activity of each molecule, see section “Results.” Data illustrate a representative experiment of two independent screenings.
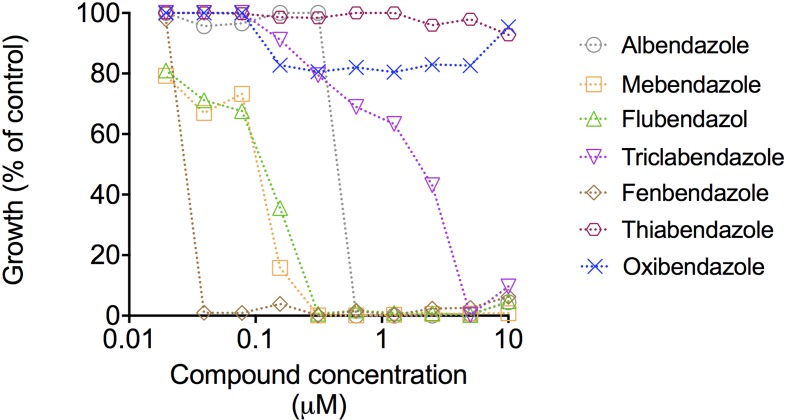
We extended the results obtained with the compound collection to dose-response tests using the four benzimidazoles showing antifungal activity and other related molecules, including thiabendazole, oxibendazole, and fenbendazole. The most active compounds were mebendazole, fenbendazole, and flubendazole (Figure 2). Mebendazole and flubendazole produced the same MIC values (0.3125 μM). Fenbendazole was the most potent compound, with a MIC corresponding to 0.039 μM. Mebendazole, however, efficiently penetrates the brain in animal models (Bai et al., 2015) and is in clinical trial for the treatment of pediatric gliomas in humans (ClinicalTrials.gov, 2013). Considering that the worse clinical outcome of cryptococcosis includes colonization of the brain, we selected mebendazole as the molecular prototype for our subsequent analyses of fungicidal effects, ability to kill intracellular cryptococci, effects on fungal morphology, interference on fungal biofilms and identification of cellular targets.
FIGURE 2.
Dose-response curves of the antifungal effects of benzimidazoles against C. neoformans. Curves represent C. neoformans growth in comparison with vehicle-treated cells (DMSO). Data illustrate a representative experiment of three independent replicates.
Mebendazole Is Fungicidal against C. neoformans
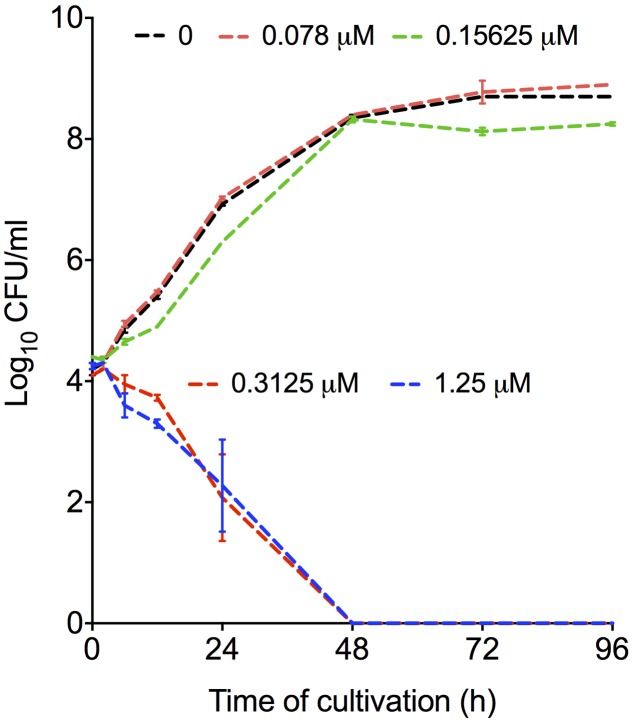
To test the fungicidal activity of mebendazole, C. neoformans was exposed to different drug concentrations for different periods of time. Mebendazole concentrations lower than 0.3125 μM had no effects on C. neoformans (Figure 3). At 0.3125 μM or higher, however, significant antifungal activity was observed after 6 to 12 h of exposure of C. neoformans to the drug. After 48 h, mebendazole killed 100% of C. neoformans cells.
FIGURE 3.
Fungicidal activity of mebendazole against C. neoformans. Fungal cells were exposed to mebendazole (0.078–1.25 μM) for periods varying from 0 to 96 h. Fungicidal activity was evident after 48 h and required a minimum concentration of 0.3125 μM mebendazole. In comparison to control systems (no drug), mebendazole concentrations of 0.3125 and 1.25 μM significantly affected fungal growth in all incubation periods. All the other drug concentrations resulted in fungal growth that was similar to that observed in the absence of mebendazole. Data illustrate a representative experiment of three independent replicates.
Activity of Mebendazole against Intracellular C. neoformans
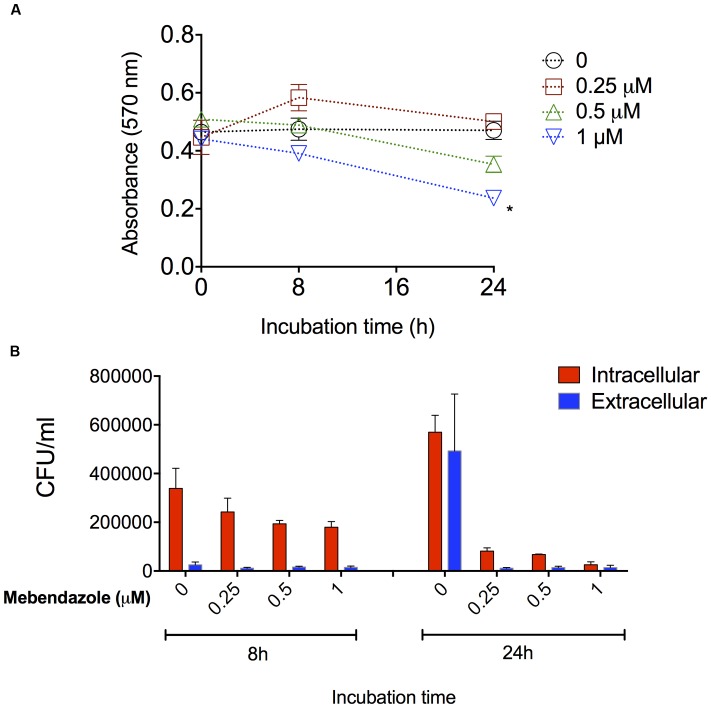
Cryptococcus neoformans is a facultative intracellular pathogen and this characteristic likely has a negative impact on the anti-cryptococcal treatment (Feldmesser et al., 2000). We then evaluated the ability of mebendazole to kill intracellular fungi. J774.16 macrophages were first infected with C. neoformans and then the cultures were treated with mebendazole for different periods (8 and 24 h) at variable drug concentrations (1, 0.5, and 0.25 μM). Macrophage viability was monitored by treating non-infected J774.16 cells with mebendazole alone (Figure 4A). After 8 h, no differences were observed between the viability of untreated macrophages and mebendazole-treated cells (P > 0.1). After 24 h, macrophage viability was affected by higher mebendazole concentrations (P = 0.0071 for 1 μM mebendazole), but no differences were observed between untreated phagocytes and cells that were exposed to 0.25 μM mebendazole (P = 0.3225). In infected macrophages, all mebendazole concentrations showed antifungal activity against both extracellular and intracellular fungi (Figure 4B). The concentration showing the lowest impact on macrophage viability (0.25 μM) was efficient in killing both intracellular and extracellular C. neoformans.
FIGURE 4.
Activity of mebendazole against intracellular C. neoformans. (A) Viability of drug-treated, non-infected macrophages. After 8 h, all systems had similar viability levels (P > 0.1). After 24 h, cell viability was only affected by 1 μM mebendazole concentration (P = 0.0071). (B) Activity of mebendazole against intracellular (red bars) or extracellular (blue bars) C. neoformans. In intracellular assays, statistical differences (P < 0.05) between no drug (0) and drug-treated systems were always observed, except when the 0.25 μM concentration of mebendazole was used in the 8 h incubation period. In extracellular assays, statistical differences (P < 0.05) between no drug (0) and drug-treated systems were observed for all mebendazole concentrations, but only after the 24 h incubation. Comparative analysis of fungal loads obtained from intracellular and extracellular assays revealed statistical differences (P < 0.05) only in the 8 h period of incubation, suggesting that mebendazole is initially more effective against extracellular fungi, but similarly active against both intracellular and extracellular cryptococci after prolonged (24 h) periods of exposure to infected macrophages. Data illustrate a representative experiment of three independent replicates.
Analysis of Potential Synergism between Mebendazole and Standard Antifungals
To evaluate whether the association of mebendazole with amphotericin B or fluconazole results in improved anti-cryptococcal activity, checkerboard assays were performed for calculation of the FIC index (Table 1). We found that mebendazole had additive activity against C. neoformans when combined with amphotericin B. Fluconazole did not show any improved effect in combination with mebendazole.
Table 1.
Impact of the association of mebendazole with amphotericin B (AmB) or fluconazole (FLC) on antifungal activity.
| Drug A | Drug B | FIC∗ | ||||
|---|---|---|---|---|---|---|
| MIC Alone (μg/ml) | MIC Combined (μg/ml) | MIC Alone (μg/ml) | MIC Combined (μg/ml) | |||
| Mebendazole | 0.095 | 0.0475 | AmB | 0.25 | 0.125 | 1 |
| Mebendazole | 0.095 | 0.0475 | Fluconazole | 2 | 2 | 1.5 |
∗Fractional inhibitory concentration index (FIC); Strongly synergistic: FIC < 0.5; Synergistic: FIC < 1; Additive: FIC = 1; No effect: 1 < FIC < 2; Antagonistic: FIC > 2.
Mebendazole Affects Capsule Size and Fungal Morphology
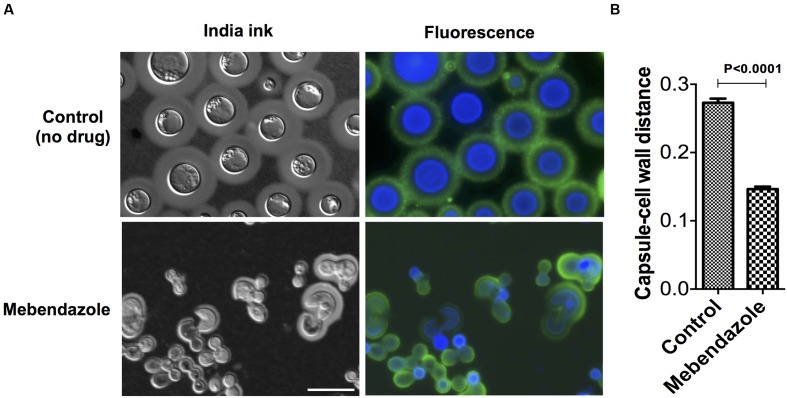
To evaluate whether mebendazole affected key cellular structures of C. neoformans during regular growth, we cultivated the fungus for 48 h in the presence of a sub-inhibitory concentration (IC50; 0.22 μM) of the drug for further analysis of morphology and capsule size by a combination of fluorescence microscopy and India ink counterstaining. Fungal cells cultivated in the presence of mebendazole presented marked morphological alterations, including loss of spherical shape, intracellular furrows (Figure 5A) and reduced capsular dimensions (Figure 5B, P < 0.05).
FIGURE 5.
Morphological alterations of C. neoformans cells treated with mebendazole. (A) India ink counterstaining (left panels) and fluorescence microscopy (right panels) showing GXM (green fluorescence) and chitin (blue fluorescence) detection. Scale bar represents 10 μm. (B) Capsule measurement of India ink counterstained cells showing reduction in capsular dimensions by mebendazole. Data illustrate a representative experiment of three independent replicates.
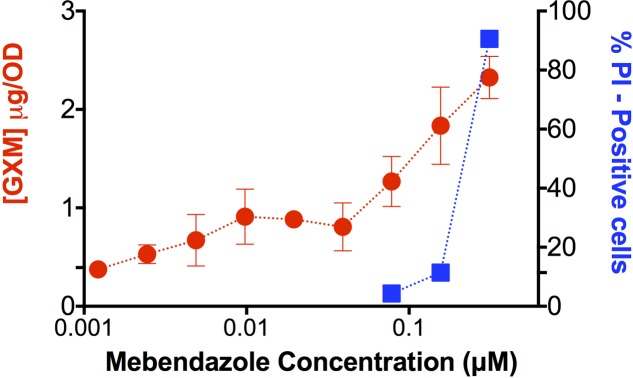
Effect of Mebendazole on GXM Release
Since mebendazole interfered with capsule size (Figure 5B), we asked whether GXM release was affected by exposure of C. neoformans to the drug. Supernatants of fungal cells cultivated in the presence of mebendazole were used for GXM quantification by ELISA (Figure 6). Unexpectedly, supernatants of mebendazole-treated cells had increased GXM concentration, mainly in the dose range required for fungal killing. Since the polysaccharide is synthesized intracellularly (Yoneda and Doering, 2006), we hypothesized that the increased GXM detection would result from leakage induced by membrane damage. To address this possibility, mebendazole-treated cells were stained with PI. Stained cells varied from 70 to 90% in the dose range generating cell death, which was compatible with membrane damage and polysaccharide leakage. Noteworthy, these results and those described in Figure 5 are also compatible with the hypothesis of GXM release from the cell surface, so we cannot rule out the possibility that mebendazole promotes detachment of capsular structures in C. neoformans.
FIGURE 6.
Extracellular release of GXM by C. neoformans cells treated with different concentrations of mebendazole. GXM extracellular detection correlated with increasing mebendazole concentrations. Staining of fungal cells with PI after exposure to fungicidal doses of mebendazole revealed a similar profile, suggesting polysaccharide leakage. Data illustrate a representative experiment of three independent replicates.
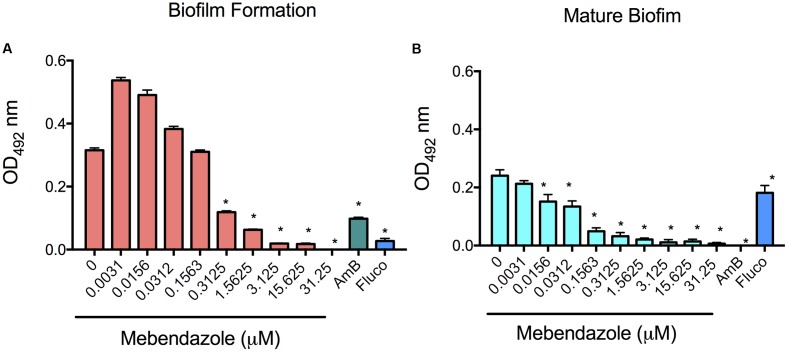
Activity of Mebendazole against C. neoformans Biofilms
Biofilm formation causes well-known difficulties in the treatment of a number of infectious diseases, including cryptococcosis (Martinez and Casadevall, 2006; Borghi et al., 2016). Based on this observation, we evaluated whether co-incubation of mebendazole with yeast cells prevented C. neoformans biofilm formation or caused damage to mature biofilms. The metabolic activity was measured by XTT reduction assay (Figure 7). Fluconazole is known to have no effects on mature biofilms, in contrast to amphotericin B (Martinez and Casadevall, 2006). Therefore, these drugs were used as negative and positive controls, respectively. Mebendazole at MIC (0.3125 μM) affected biofilm formation (Figure 7A; P < 0.0001) and damaged mature biofilms (Figure 7B; P < 0.0001). Lower concentrations of mebendazole similarly affected C. neoformans mature biofilms (Figure 7B; P < 0.0001). As expected, higher concentrations of mebendazole had even clearer impacts on C. neoformans biofilms.
FIGURE 7.
Effects of mebendazole on cryptococcal biofilms. Biofilm formation and mature biofilm activity were measured indirectly by XTT reduction assay. (A) Incubation of C. neformans with mebendazole before biofilm formation. (B) Treatment of mature biofilms with mebendazole. AmB, amphotericin B (2 μg/ml); Fluco, fluconazole (8 μg/ml). Statistical significances after comparison with control systems are indicated by asterisks (P < 0.0001). Data illustrate a representative experiment of three independent replicates.
Identification of Potential Cellular Targets for Mebendazole
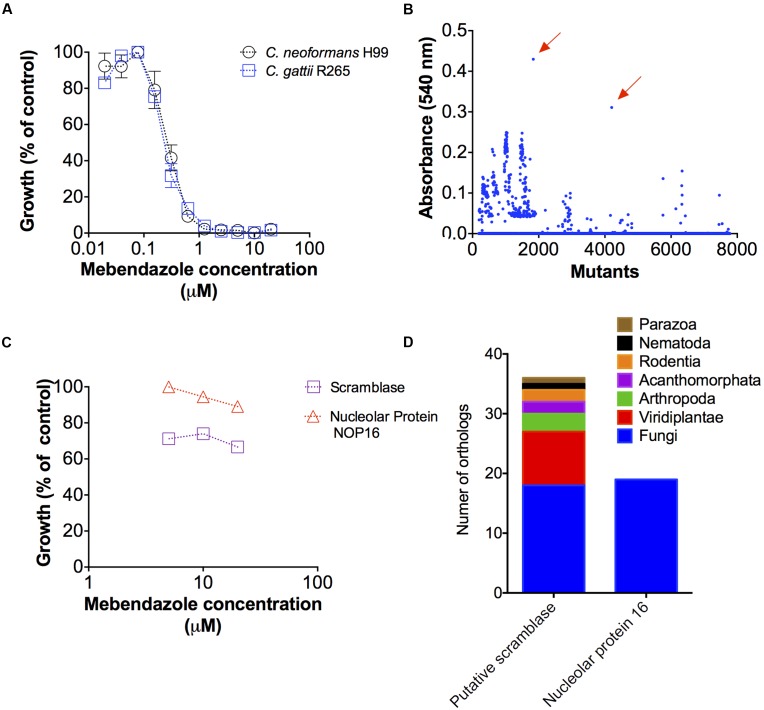
Antifungal activities of benzimidazoles were described before (Cruz et al., 1994), but the mechanisms by which these drugs affected cryptococcal growth remained unknown. In order to identify potential targets for mebendazole in Cryptococcus spp., we first evaluated whether the drug affected the growth of C. gattii and C. neoformans in a similar fashion. In fact, growth inhibition curves were identical for both pathogens (Figure 8A).
FIGURE 8.
Potential cellular targets required for mebendazole activity. (A) Comparative analysis showing that C. neoformans and C. gattii are similarly susceptible to mebendazole. (B) Screening of a C. gattii mutant library in the presence of 10 μM of mebendazole, revealing two highly resistant strains (arrows). (C) Growth of the two mutants selected in panel B in the presence of variable concentrations of mebendazole. (D) Sequence identification by PCR and distribution of cellular proteins required for mebendazole activity.
A mutant collection generated by co-incubation of C. gattii and A. tumefaciens (Idnurm et al., 2004) was produced by our group for general purposes involving development of antifungals and pathogenic studies. In the present study, these mutants were screened for resistance phenotypes in the presence of 10 μM mebendazole, based on the assumption that, in the absence of a cellular target required for antifungal activity, the drug would lack anti-cryptococcal properties. Most of the mutants were sensitive to mebendazole and a number of strains were partially resistant to the drug (Figure 8B). However, two of the mutant strains clearly stood out from the entire collection, showing highest levels of resistance even in higher mebendazole concentrations (Figure 8C). Interrupted regions in these two mutants were identified by inverse PCR (Figure 8D). In the most mebendazole-resistant mutant, the protein codified by the interrupted gene contained a scramblase domain (PF03803 – CNBG_3981). In the second most resistant mutant, the gene coding for ribosome biogenesis protein Nop16 (PF09420 – CNBG_3695) was interrupted. These domains were found in diverse phylogenetic groups, according to the PFAM database (Finn et al., 2016). However, orthologs for these genes were found in a narrow group of organisms and absent in human cells (Figure 8D), according to OrthoMCL database (Chen et al., 2006). These results indicate that at least two novel cellular targets are involved in the antifungal activity of mebendazole.
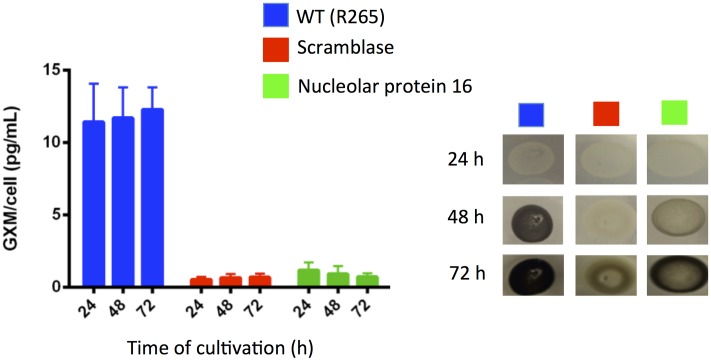
Neutralizing cryptococcal virulence factors is also likely to be beneficial for the control of cryptococcosis. In this context, we evaluated whether the mutants lacking potential targets for activity of mebendazole had normal production of the most-well characterized cryptococcal virulence factors. The two mutants had normal urease activity (not shown). Analysis of extracellular GXM and pigmentation, however, showed important differences between WT and mutant cells (Figure 9). Mutants disrupted for expression of the putative scramblase and of nucleolar protein 16 had decreased contents of extracellular GXM, in comparison with WT cells (P < 0.001). The kinetics of melanin production was also negatively affected in the mutants.
FIGURE 9.
Cryptococcus gattii mutants lacking potential targets for mebendazole activity show defective capacity to produce well-known cryptococcal virulence factors. GXM determination by ELISA (left panel) indicated that the putative scramblase and nucleolar protein 16 were required for polysaccharide export. Both proteins were apparently involved in the kinetics of pigmentation in C. gattii (right panel).
Discussion
Processes of drug research and development are costly, time consuming and have questionable success (Kaitin, 2010; Kaitin and DiMasi, 2011). In this scenario, drug repurposing has provided a potential boost to the drug pipeline combating health emergencies and assisting neglected populations. Recent studies have provided evidence of successful drug repurposing to combat Cryptococcus (ClinicalTrials.gov, 2016; Rhein et al., 2016) and Zika virus (Veljkovic and Paessler, 2016; Xu et al., 2016; Sacramento et al., 2017) infections with promising results. The need for additional anti-infectious agents, however, is clear.
In this study, we identified small-molecule inhibitors of C. neoformans via a drug-repurposing screen. Our findings demonstrated antifungal activity in a group of anti-helminthic benzimidazoles and suggested potential targets for development of novel antifungals. Benzimidazoles are heterocyclic aromatic bis-nitrogen azoles that are considered promising anchors for development of new therapeutic agents (Bansal and Silakari, 2012). Benzimidazole derivatives have been associated to the control of infectious diseases through antiviral, antifungal, antimicrobial, and antiprotozoal properties, but they also manifest antiinflammatory, anticancer, antioxidant, anticoagulant, antidiabetic and antihypertensive activities (Ates-Alagoz, 2016). The anti-cryptococcal activity of benzimidazoles was demonstrated two decades ago (Cruz et al., 1994), but the effects of these compounds on fungal morphology and biofilm formation were not explored. Similarly, their cellular targets and ability to kill intracellular fungi remained unknown. Mebendazole, one of the benzimidazoles showing antifungal activity, is in clinical trial for the treatment of human pediatric glioma (ClinicalTrials.gov, 2013). Since effective anti-cryptococcal agents obligatorily need to reach the central nervous system at biologically active concentrations, we selected mebendazole for our experiments of antifungal activity. This compound affected cryptococcal growth, morphology, biofilms and macrophage infection.
The pathogenic mechanisms used by C. neoformans during infection bring significant complexities in the management of cryptococcosis. Conditions favoring biofilm formation are thought to contribute to cryptococcal virulence (Benaducci et al., 2016). Like with other pathogens, C. neoformans biofilms are resistant to antimicrobial agents and host defense mechanisms, causing significant morbidity and mortality (Martinez and Casadevall, 2015). These characteristics are especially relevant in a scenario of increasing use of ventriculoperitoneal shunts to manage intracranial hypertension associated with cryptococcal meningoencephalitis (Martinez and Casadevall, 2015). In our model, mebendazole was an efficient antifungal agent against C. neoformans biofilms. The minimum mebendazole concentration required for antifungal activity against planktonic cells (0.325 μM) was much higher than the doses required for activity against mature biofilms (0.0156–0.0312 μM). The reason for this discrepancy is unclear. Since capsular polysaccharides have well described roles on the assembly of cryptococcal biofilms (Martinez and Casadevall, 2005), we hypothesize that the herein described impact of mebendazole on capsular architecture and GXM release may affect biofilm stability.
An additional complexity in the treatment of cryptococcosis is the ability of the fungus to reside inside phagocytes. In fact, persistent pulmonary infection is associated with the intracellular parasitism of C. neoformans (Goldman et al., 2000). In this context, targeting intracellular survival and growth and/or cryptococcal virulence factors expressed during intracellular parasitism might offer new strategies to improve anticryptococcal treatment, as reviewed by Voelz and May (2010). Mebendazole was effective against phagocytized fungi in our model. The fact that the anti-helminthic compound had an additive effect to AmB also suggests that benzimidazole-like compounds could be used in therapeutic protocols against cryptococcosis.
The primary mechanism of anthelmintic activity of mebendazole relies on binding the β-subunit of tubulin before dimerization with α-tubulin, with subsequent blocking of microtubule formation (Gardner and Hill, 2001). Tubulin may also be the target of mebendazole in C. neoformans, but our studies suggest the possibility that the antifungal effects of mebendazole may involve additional targets. Mebendazole induced membrane permeabilization, as concluded from increased levels of PI staining after exposure of C. neoformans to the drug. In addition, mutants lacking genes coding for a putative scramblase and the nucleolar protein Nop16 were highly resistant to mebendazole. Both mutants had defective formation of important virulence factors. Remarkably, sequences showing similarity to these two proteins were absent in human cells, suggesting a great potential for these two proteins as novel antifungal specific targets. Scramblases are ATP-independent enzymes that act to randomize lipid distribution by bidirectionally translocating lipids between leaflets (Hankins et al., 2015). Lipid-translocating enzymes, in fact, are fundamental for cryptococcal pathogenesis and GXM export (Hu and Kronstad, 2010; Rizzo et al., 2014). The functions of Nop16 in C. neoformans are unknown, but in S. cerevisiae this protein is a component of 66S pre-ribosomal particles required for 60S ribosomal subunit biogenesis (Harnpicharnchai et al., 2001; Horsey et al., 2004). Although there is no evidence in the literature linking tubulin polymerization, membrane permeability and cellular functions of these two potential targets, our studies suggest that a connection may exist and these proteins may be functionally integrated in fungi.
Cryptococcosis affects regions where health infrastructure resources are extremely limited. Considering the high mortality rates associated with this disease and the socio-economic scenario behind cryptococcosis, low-cost and efficient antifungal alternatives are urgent. Clinical use of novel drugs, however, depends on a number of properties of the molecular candidates. In this regard, the potential use of mebendazole against human cryptococcosis raises several concerns. For instance, the use of mebendazole at large doses may cause bone marrow suppression (Fernandez-Banares et al., 1986) and it is unclear if the compound is safe in pregnancy (Torp-Pedersen et al., 2012). Benzimidazoles have only limited water solubility, which impacts the rate and extent of their absorption and, consequently, systemic bioavailability, maximal plasma concentration, and tissue distribution (McKellar and Scott, 1990). Animal studies with mebendazole demonstrated drug distribution through all the organs, including the central nervous system (Lanusse and Prichard, 1993). However, the drug and its metabolites were concentrated mainly in the liver, where they remain for at least 15 days post treatment (Lanusse and Prichard, 1993). Mebendazole is thought to be the active form of the drug rather than its metabolites (Gottschall et al., 1990). However, in the liver, benzimidazoles are mostly modified by the enzymatic system of hepatic microsomal oxidases, which are involved in sulfoxidation, demethylation, and hydroxylation (Short et al., 1988; Lanusse and Prichard, 1993). In fact, benzimidazoles are usually short lived and metabolic products predominate in plasma and all tissues and excreta of the host (Fetterer and Rew, 1984).
Although the difficulties associated with the clinical use of mebendazole are clear, our results combine a multiple antifungal activity with molecular targets that are absent in human cells, which encourages further development of benzimidazole-like molecules against C. neoformans. In fact, a number of reports have suggested that the benzimidazole core represents a promising scaffold for development of new therapeutic agents (Yadav and Ganguly, 2015). Montresor et al. (2010) estimated that the cost to procure one million doses of standard benzimidazoles (500 mg each) would be approximately US$ 20,000, including international transport. In this context, the ability of mebendazole to penetrate the brain (Bai et al., 2015) and to cause expressive damage in C. neoformans cells suggest great potential as a prototype for development of novel anti-cryptococcal agents.
Author Contributions
LJ, CS, LK, AS, MDP, MV, and MR prepared the experimental design. LJ, RS, WL, RA, and CS performed the experiments. LJ, CS, LK, AS, MDP, MV, and MR discussed the results, wrote and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Part of the data presented here is also the subject of a pending patent application (Ref.: BR1020170047300, National Institute of Industrial Property - INPI, Brazil). The reviewer JNDAJ and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
This work was supported by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), by grants numbers 443586/2014-4, 300699/2013-1, E-26/102.835/2012 and 210.918/2015, and by NIH grant AI-116420 to MDP, MR is the recipient of a Pathfinder Award from the Wellcome Trust (United Kingdom; grant no. WT103212MF) and the author also acknowledges support from the Instituto Nacional de Ciência e Tecnologia de Inovação em Doenças Negligenciadas (INCT-IDN).
Footnotes
References
- Arendrup M. C., Cuenca-Estrella M., Lass-Florl C., Hope W., Eucast A. (2012). EUCAST technical note on the EUCAST definitive document EDef 7.2: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts EDef 7.2 (EUCAST-AFST). Clin. Microbiol. Infect. 18 E246–E247. 10.1111/j.1469-0691.2012.03880.x [DOI] [PubMed] [Google Scholar]
- Ates-Alagoz Z. (2016). Antimicrobial activities of 1-H-benzimidazole-based molecules. Curr. Top. Med. Chem. 16 2953–2962. 10.2174/1568026616666160506130226 [DOI] [PubMed] [Google Scholar]
- Bai R. Y., Staedtke V., Wanjiku T., Rudek M. A., Joshi A., Gallia G. L., et al. (2015). Brain penetration and efficacy of different mebendazole polymorphs in a mouse brain tumor model. Clin. Cancer Res. 21 3462–3470. 10.1158/1078-0432.CCR-14-2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal Y., Silakari O. (2012). The therapeutic journey of benzimidazoles: a review. Bioorg. Med. Chem. 20 6208–6236. 10.1016/j.bmc.2012.09.013 [DOI] [PubMed] [Google Scholar]
- Benaducci T., Sardi Jde C., Lourencetti N. M., Scorzoni L., Gullo F. P., Rossi S. A., et al. (2016). Virulence of Cryptococcus sp. biofilms in vitro and in vivo using Galleria mellonella as an alternative model. Front. Microbiol. 7:290 10.3389/fmicb.2016.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolano A., Stinchi S., Preziosi R., Bistoni F., Allegrucci M., Baldelli F., et al. (2001). Rapid methods to extract DNA and RNA from Cryptococcus neoformans. FEMS Yeast Res. 1 221–224. [DOI] [PubMed] [Google Scholar]
- Borghi E., Borgo F., Morace G. (2016). Fungal biofilms: update on resistance. Adv. Exp. Med. Biol. 931 37–47. 10.1007/5584_2016_7 [DOI] [PubMed] [Google Scholar]
- Casadevall A., Cleare W., Feldmesser M., Glatman-Freedman A., Goldman D. L., Kozel T. R., et al. (1998). Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob. Agents Chemother. 42 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Mukherjee J., Scharff M. D. (1992). Monoclonal antibody based ELISAs for cryptococcal polysaccharide. J. Immunol. Methods 154 27–35. 10.1016/0022-1759(92)90209-C [DOI] [PubMed] [Google Scholar]
- Chen F., Mackey A. J., Stoeckert C. J., Jr., Roos D. S. (2006). OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34 D363–D368. 10.1093/nar/gkj123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ClinicalTrials.gov (2013). Adjunctive Sertraline for the Treatment of HIV-Associated Cryptococcal Meningitis (ASTRO-CM). Available at: https://clinicaltrials.gov/ct2/show/NCT01802385 [Google Scholar]
- ClinicalTrials.gov (2016). A Phase I Study of Mebendazole for the Treatment of Pediatric Gliomas. Available at: https://clinicaltrials.gov/ct2/show/NCT01837862 [Google Scholar]
- Colombo A. C., Rodrigues M. L. (2015). Fungal colonization of the brain: anatomopathological aspects of neurological cryptococcosis. An. Acad. Bras. Cienc. 87 1293–1309. 10.1590/0001-3765201520140704 [DOI] [PubMed] [Google Scholar]
- Cruz M. C., Bartlett M. S., Edlind T. D. (1994). In vitro susceptibility of the opportunistic fungus Cryptococcus neoformans to anthelmintic benzimidazoles. Antimicrob. Agents Chemother. 38 378–380. 10.1128/AAC.38.2.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmesser M., Kress Y., Novikoff P., Casadevall A. (2000). Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 68 4225–4237. 10.1128/IAI.68.7.4225-4237.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Banares F., Gonzalez-Huix F., Xiol X., Catala I., Miro J., Lopez N., et al. (1986). Marrow aplasia during high dose mebendazole treatment. Am. J. Trop. Med. Hyg. 35 350–351. [DOI] [PubMed] [Google Scholar]
- Fetterer R. H., Rew R. S. (1984). Interaction of Fasciola hepatica with albendazole and its metabolites. J. Vet. Pharmacol. Ther. 7 113–118. 10.1111/j.1365-2885.1984.tb00886.x [DOI] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44 D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner T. B., Hill D. R. (2001). Treatment of giardiasis. Clin. Microbiol. Rev. 14 114–128. 10.1128/CMR.14.1.114-128.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D. L., Lee S. C., Mednick A. J., Montella L., Casadevall A. (2000). Persistent Cryptococcus neoformans pulmonary infection in the rat is associated with intracellular parasitism, decreased inducible nitric oxide synthase expression, and altered antibody responsiveness to cryptococcal polysaccharide. Infect. Immun. 68 832–838. 10.1128/IAI.68.2.832-838.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschall D. W., Theodorides V. J., Wang R. (1990). The metabolism of benzimidazole anthelmintics. Parasitol. Today 6 115–124. 10.1016/0169-4758(90)90228-V [DOI] [PubMed] [Google Scholar]
- Govender N. P., Patel J., van Wyk M., Chiller T. M., Lockhart S. R. Group for Enteric Respiratory and Meningeal Disease Surveillance in South Africa (GERMS-SA) (2011). Trends in antifungal drug susceptibility of Cryptococcus neoformans isolates obtained through population-based surveillance in South Africa in 2002-2003 and 2007-2008. Antimicrob. Agents Chemother. 55 2606–2611. 10.1128/AAC.00048-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankins H. M., Baldridge R. D., Xu P., Graham T. R. (2015). Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic 16 35–47. 10.1111/tra.12233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnpicharnchai P., Jakovljevic J., Horsey E., Miles T., Roman J., Rout M., et al. (2001). Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8 505–515. 10.1016/S1097-2765(01)00344-6 [DOI] [PubMed] [Google Scholar]
- Horsey E. W., Jakovljevic J., Miles T. D., Harnpicharnchai P., Woolford J. L., Jr. (2004). Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA 10 813–827. 10.1261/rna.5255804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Kronstad J. W. (2010). A putative P-type ATPase, Apt1, is involved in stress tolerance and virulence in Cryptococcus neoformans. Eukaryot. Cell 9 74–83. 10.1128/EC.00289-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A., Reedy J. L., Nussbaum J. C., Heitman J. (2004). Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot. Cell 3 420–429. 10.1128/EC.3.2.420-429.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitin K. I. (2010). Deconstructing the drug development process: the new face of innovation. Clin. Pharmacol. Ther. 87 356–361. 10.1038/clpt.2009.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitin K. I., DiMasi J. A. (2011). Pharmaceutical innovation in the 21st century: new drug approvals in the first decade, 2000-2009. Clin. Pharmacol. Ther. 89 183–188. 10.1038/clpt.2010.286 [DOI] [PubMed] [Google Scholar]
- Krysan D. J. (2015). Toward improved anti-cryptococcal drugs: novel molecules and repurposed drugs. Fungal Genet. Biol. 78 93–98. 10.1016/j.fgb.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K. J., Fraser J. A., Doering T. L., Wang Z., Janbon G., Idnurm A., et al. (2014). Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb. Perspect. Med. 4:a019760 10.1101/cshperspect.a019760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanusse C. E., Prichard R. K. (1993). Clinical pharmacokinetics and metabolism of benzimidazole anthelmintics in ruminants. Drug Metab. Rev. 25 235–279. 10.3109/03602539308993977 [DOI] [PubMed] [Google Scholar]
- Martinez L. R., Casadevall A. (2005). Specific antibody can prevent fungal biofilm formation and this effect correlates with protective efficacy. Infect. Immun. 73 6350–6362. 10.1128/IAI.73.10.6350-6362.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez L. R., Casadevall A. (2006). Susceptibility of Cryptococcus neoformans biofilms to antifungal agents in vitro. Antimicrob. Agents Chemother. 50 1021–1033. 10.1128/AAC.50.3.1021-1033.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez L. R., Casadevall A. (2007). Cryptococcus neoformans biofilm formation depends on surface support and carbon source and reduces fungal cell susceptibility to heat, cold, and UV light. Appl. Environ. Microbiol. 73 4592–4601. 10.1128/AEM.02506-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez L. R., Casadevall A. (2015). Biofilm formation by Cryptococcus neoformans. Microbiol. Spectr. 3 10.1128/microbiolspec.MB-0006-2014 [DOI] [PubMed] [Google Scholar]
- McKellar Q. A., Scott E. W. (1990). The benzimidazole anthelmintic agents–a review. J. Vet. Pharmacol. Ther. 13 223–247. 10.1111/j.1365-2885.1990.tb00773.x [DOI] [PubMed] [Google Scholar]
- Meshulam T., Levitz S. M., Christin L., Diamond R. D. (1995). A simplified new assay for assessment of fungal cell damage with the tetrazolium dye, (2,3)-bis-(2-methoxy-4-nitro-5-sulphenyl)-(2H)-tetrazolium-5-carboxanil ide (XTT). J. Infect. Dis. 172 1153–1156. 10.1093/infdis/172.4.1153 [DOI] [PubMed] [Google Scholar]
- Micallef C., Aliyu S. H., Santos R., Brown N. M., Rosembert D., Enoch D. A. (2015). Introduction of an antifungal stewardship programme targeting high-cost antifungals at a tertiary hospital in Cambridge, England. J. Antimicrob. Chemother. 70 1908–1911. 10.1093/jac/dkv040 [DOI] [PubMed] [Google Scholar]
- Montresor A., Gabrielli A. F., Diarra A., Engels D. (2010). Estimation of the cost of large-scale school deworming programmes with benzimidazoles. Trans. R. Soc. Trop. Med. Hyg. 104 129–132. 10.1016/j.trstmh.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor V., Rella A., Farnoud A. M., Singh A., Munshi M., Bryan A., et al. (2015). Identification of a new class of antifungals targeting the synthesis of fungal sphingolipids. mBio 6:e00647 10.1128/mBio.00647-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. (1983). Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65 55–63. 10.1016/0022-1759(83)90303-4 [DOI] [PubMed] [Google Scholar]
- Nosengo N. (2016). Can you teach old drugs new tricks? Nature 534 314–316. 10.1038/534314a [DOI] [PubMed] [Google Scholar]
- Ostermann H., Solano C., Jarque I., Garcia-Vidal C., Gao X., Barrueta J. A., et al. (2014). Cost analysis of voriconazole versus liposomal amphotericin B for primary therapy of invasive aspergillosis among patients with haematological disorders in Germany and Spain. BMC Pharmacol. Toxicol. 15:52 10.1186/2050-6511-15-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B. J., Wannemuehler K. A., Marston B. J., Govender N., Pappas P. G., Chiller T. M. (2009). Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23 525–530. 10.1097/QAD.0b013e328322ffac [DOI] [PubMed] [Google Scholar]
- Pavlopoulos A. (2011). Identification of DNA sequences that flank a known region by inverse PCR. Methods Mol. Biol. 772 267–275. 10.1007/978-1-61779-228-1_16 [DOI] [PubMed] [Google Scholar]
- Rhein J., Morawski B. M., Hullsiek K. H., Nabeta H. W., Kiggundu R., Tugume L., et al. (2016). Efficacy of adjunctive sertraline for the treatment of HIV-associated cryptococcal meningitis: an open-label dose-ranging study. Lancet Infect. Dis. 16 809–818. 10.1016/S1473-3099(16)00074-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo J., Oliveira D. L., Joffe L. S., Hu G., Gazos-Lopes F., Fonseca F. L., et al. (2014). Role of the Apt1 protein in polysaccharide secretion by Cryptococcus neoformans. Eukaryot. Cell 13 715–726. 10.1128/EC.00273-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues J., Fonseca F. L., Schneider R. O., Godinho R. M., Firacative C., Maszewska K., et al. (2015). Pathogenic diversity amongst serotype C VGIII and VGIV Cryptococcus gattii isolates. Sci. Rep. 5:11717 10.1038/srep11717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M. L. (2016). Funding and innovation in diseases of neglected populations: the paradox of cryptococcal meningitis. PLoS Negl. Trop. Dis. 10:e0004429 10.1371/journal.pntd.0004429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M. L., Alvarez M., Fonseca F. L., Casadevall A. (2008). Binding of the wheat germ lectin to Cryptococcus neoformans suggests an association of chitinlike structures with yeast budding and capsular glucuronoxylomannan. Eukaryot. Cell 7 602–609. 10.1128/EC.00307-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacramento C. Q., de Melo G. R., de Freitas C. S., Rocha N., Hoelz L. V., Miranda M., et al. (2017). The clinically approved antiviral drug sofosbuvir inhibits Zika virus replication. Sci Rep. 7:40920 10.1038/srep40920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short C. R., Flory W., Hsieh L. C., Barker S. A. (1988). The oxidative metabolism of fenbendazole: a comparative study. J. Vet. Pharmacol. Ther. 11 50–55. 10.1111/j.1365-2885.1988.tb00120.x [DOI] [PubMed] [Google Scholar]
- Sloan D. J., Dedicoat M. J., Lalloo D. G. (2009). Treatment of cryptococcal meningitis in resource limited settings. Curr. Opin. Infect. Dis. 22 455–463. 10.1097/QCO.0b013e32832fa214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subcommittee on Antifungal Susceptibility Testing of the EECfAST (2008). EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin. Microbiol. Infect. 14 398–405. 10.1111/j.1469-0691.2007.01935.x [DOI] [PubMed] [Google Scholar]
- Torp-Pedersen A., Jimenez-Solem E., Andersen J. T., Broedbaek K., Torp-Pedersen C., Poulsen H. E. (2012). Exposure to mebendazole and pyrvinium during pregnancy: a Danish nationwide cohort study. Infect. Dis. Obstet. Gynecol. 2012:769851 10.1155/2012/769851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veljkovic V., Paessler S. (2016). Possible repurposing of seasonal influenza vaccine for prevention of Zika virus infection. F1000Research 5:190 10.12688/f1000research.8102.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelz K., May R. C. (2010). Cryptococcal interactions with the host immune system. Eukaryot. Cell 9 835–846. 10.1128/EC.00039-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Lee E. M., Wen Z., Cheng Y., Huang W. K., Qian X., et al. (2016). Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 22 1101–1107. 10.1038/nm.4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav G., Ganguly S. (2015). Structure activity relationship (SAR) study of benzimidazole scaffold for different biological activities: a mini-review. Eur. J. Med. Chem. 97 419–443. 10.1016/j.ejmech.2014.11.053 [DOI] [PubMed] [Google Scholar]
- Yoneda A., Doering T. L. (2006). A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol. Biol. Cell 17 5131–5140. 10.1091/mbc.E06-08-0701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B., Wu C., Wang L., Sachs M. S., Lin X. (2012). The antidepressant sertraline provides a promising therapeutic option for neurotropic cryptococcal infections. Antimicrob. Agents Chemother. 56 3758–3766. 10.1128/AAC.00212-12 [DOI] [PMC free article] [PubMed] [Google Scholar]