Abstract
Glioblastoma (GBM) brain tumor remains among the most lethal and incurable human diseases. Oncogenic microRNA-10b (miR-10b) is strongly and universally upregulated in GBM, and its inhibition by antisense oligonucleotides (ASOs) reduces the growth of heterogeneous glioma cells; therefore, miR-10b represents a unique therapeutic target for GBM. Here we explored the effects of miR-10b gene editing on GBM. Using the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system, we investigated effects of miR-10b gene editing on the growth of cultured human glioma cells, tumor-initiating stem-like cells, and mouse GBM xenografts, as well as the oncogene-induced transformation of normal astrocytes. We show that GBM is strictly “addicted” to miR-10b and that miR-10b gene ablation is lethal for glioma cell cultures and established intracranial tumors. miR-10b loss-of-function mutations lead to the death of glioma, but not other cancer cell lines. We have not detected escaped proliferative clones of GBM cells edited in the miR-10b locus. Finally, neoplastic transformation of normal astrocytes was abolished by the miR-10b-editing vectors. This study demonstrates the feasibility of gene editing for brain tumors in vivo and suggests virus-mediated miR-10b gene ablation as a promising therapeutic approach that permanently eliminates the key regulator essential for tumor growth and survival.
Keywords: microRNA-10b, ncRNA, cancer, glioblastoma, brain tumor, genome editing, gene editing, CRISPR-Cas9, therapy, orthotopic animal models
Graphical Abstract
Oncogenic microRNA-10b (miR-10b) is strongly and universally upregulated in glioblastoma brain tumors. In this issue of Molecular Therapy, El Fatimy and colleagues show that glioblastoma is addicted to miR-10b and that miR-10b gene editing by viral vectors presents a new therapeutic approach for malignant gliomas.
Introduction
Glioblastoma (GBM), the most common malignant brain tumor, remains one of the most lethal human diseases, with a survival period of little more than 1 year that has only marginally changed over the past 25 years. There is an urgent need for new molecular targets, concepts, and approaches to treating this disease. Mounting evidence indicates that GBM growth and invasiveness are closely regulated by microRNAs (miRNAs).1 A decade of work in the field led us to focus on miR-10b, a miRNA embedded within the HOXD genomic locus and implicated in proliferation, invasion, and metastasis of various types of malignancies, including the GBM.2, 3 miR-10b is especially notable in brain tumors due to its unique expression pattern: while virtually undetectable in the normal brain, it becomes extremely abundant in most low- and high-grade gliomas across all subtypes, as well as metastatic brain tumors.3, 4, 5, 6 Breast cancer patients with brain metastases have significantly higher miR-10b levels compared to patients with metastases in other organs.7, 8 Inhibition of miR-10b by chemically modified antisense oligonucleotides (ASOs) reduces growth and invasion of cultured glioma cells4, 9 and metastasis in aggressive cancer models.10, 11 Our work on highly invasive and aggressive intracranial glioma models demonstrated that ASO inhibitors of miR-10b reduce GBM growth in mice.12 However, the effects observed in the orthotopic GBM models were transient, with disease relapse due to both low-efficiency uptake and non-uniform distribution of the ASOs in intracranial GBM.
Despite the association of miR-10b with cancer and the potential as a therapeutic target for GBM, its mechanism of action remains incompletely understood. The predicted miR-10b targets were not enriched among the genes de-repressed by specific miR-10b inhibitors, raising the possibility that miR-10b acts in an unconventional way that is not captured by bioinformatics models.9, 12 Alternatively, the miR-10b inhibitors used may have off-target effects not linked to miR-10b itself. To validate specific loss of miR-10b function as a tumor-inhibiting strategy for GBM, we abrogated miR-10b expression using clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 and investigated the effects of miR-10b gene ablation on glioma growth in vitro and in vivo. This powerful technology facilitates targeted DNA double-strand breaks at specific sites in the mammalian genome and takes advantage of the non-homologous end joining (NHEJ) to introduce insertions or deletions (indels).13, 14, 15 We demonstrate that miR-10b gene editing is deleterious for all glioma cells and GBM-initiating stem-like cells (GSCs) studied, as their viability strictly depends on miR-10b expression. Furthermore, we show that lentivirus-mediated miR-10b editing with CRISPR-Cas9 strongly impairs the growth of orthotopic GBM in mice, supporting targeted miR-10b gene editing as a promising therapeutic approach for GBM.
Results
The Design of sgRNA and Validation of miR-10b Targeting
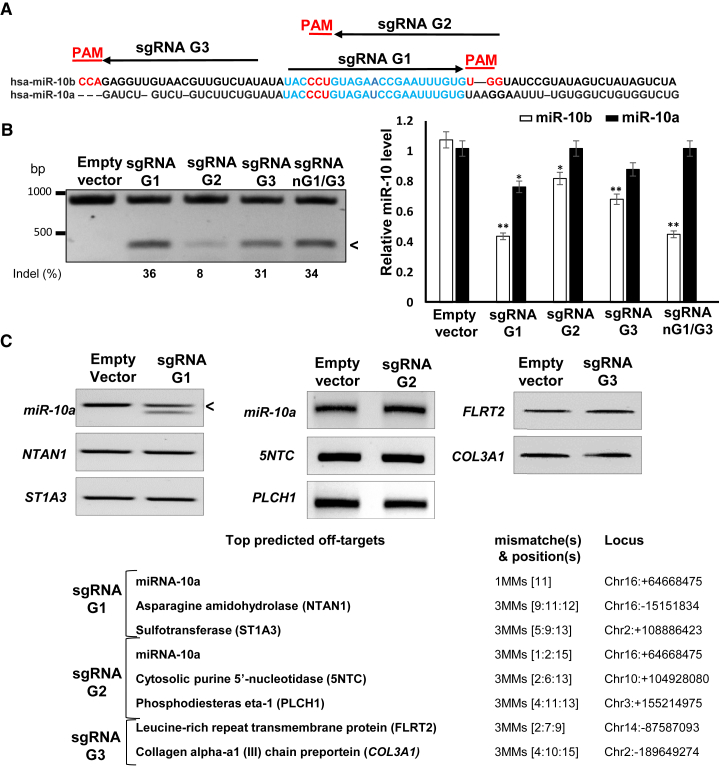
We used the type II CRISPR-Cas9 system derived from Streptococcus pyogenes that induces site-directed double-strand breaks in DNA, leading to disruption or mutation of a targeted site through non-homologous end joining.16 The system requires the protospacer adjacent motif (PAM) sequence of 5′-NGG-3′, located at the immediate 3′ end of the single guide RNA (sgRNA) recognition sequence.17 Alternative sequence-specific sgRNAs (G1–G3) targeting either mature human miR-10b or its precursor pre-miR-10b, and thereby disrupting the pre-miR-10b structure and processing, have been designed using the CRISPRtool (http://crispr.mit.edu) and selected to minimize potential off-target effects (Figure 1A). We have applied the CRISPR-Cas9 system with G1–G3 sgRNAs for mutating miR-10b in tumorigenic glioma LN229 cells. We also used a mutated “nickase” version of the Cas9 enzyme (Cas9n D10A) that, guided by a pair of adjacent, opposite-strand sgRNAs G1 and G3 (nG1/G3), produces double nicks that can be repaired by NHEJ and potentially introduces indels. Double nicking has a potential to reduce unwanted off-target effects greatly.18 Using magnetofection, we achieved plasmid transfection efficiency of 60% in glioma cells. Surveyor cleavage assay indicated that the sgRNAs tested produced 8%–36% editing efficiencies at the miR-10b locus (Figure 1B, left panel), resulting in the measurable downregulation of mature miR-10b expression (Figure 1B, right panel). sgRNA G1 and G3 guided editing, which was more efficient than that of G2, also led to more efficient miR-10b reduction.
Figure 1.
miR-10b Gene Is Specifically Edited by CRISPR-Cas9
(A) Design of alternative sgRNA guides for CRISPR-Cas9 miR-10b editing. The closely related hsa-pre-miR-10b and hsa-pre-miR-10a are aligned. The respective mature sequences are indicated in blue. sgRNA G1–G3 are marked by arrows, and the corresponding PAMs are shown in red. sgRNA G1 and G2 were designed to target the mature miR-10b, and G3 was designed to target its precursor pre-miR-10b. (B) CRISPR-Cas9-mediated editing of the miR-10b locus in LN229 glioma cells, 48 hr post-transfection. The efficiency of miR-10b gene editing with alternative sgRNAs was estimated by Surveyor cleavage assay and bands densitometry (left panel). Cleavage products, indicative of the edited gene, are marked with an arrowhead. miR-10b editing results in a significant downregulation of mature miR-10b expression (right panel). miR-10b/a levels were analyzed by TaqMan qRT-PCR and normalized to the geometrical mean of unaffected miR-99a, miR-125a, and miR-148a. Error bars depict SEM, n = 6, *p < 0.01, **p < 0.005, Student’s t test. (C) Assessment of putative off-target effects. Bioinformatically predicted off-targets with a maximum of three mismatches for sgRNA G1, G2, and G3 (right panel). miR-10a represents the major off-target, because it differs from miR-10b by a single nucleotide. Surveyor cleavage assay depicts miR-10a editing by sgRNA G1, but not G2 or G3, and the lack of editing of other top predicted genes.
Because the off-target effects of the Cas9 activity represent the major concern for the use of the CRISPR-Cas9 system, we assessed potential off-targets for G1–G3 sgRNAs by employing several computational algorithms. miR-10a, the most closely related miR-10 family member that differs from miR-10b by a single nucleotide, represents the top off-target for both G1- and G2-directed targeting (Figures 1A and 1C). As expected, despite the strong similarity between mature miR-10a and miR-10b, the miR-10a locus was not targeted by CRISPR-Cas9 with G3 sgRNA that was designed for the less similar pre-miR-10b precursor (Figure 1B). Additional predicted top protein-coding off-targets were not edited (Figure 1C). In addition, expression of the adjacent HOXD3 and HOXD4 genes was unaffected by CRISPR-Cas9 with G1–G3 sgRNAs (Figure S1).
miR-10b Expression Is Essential for Viability of Glioma Cells
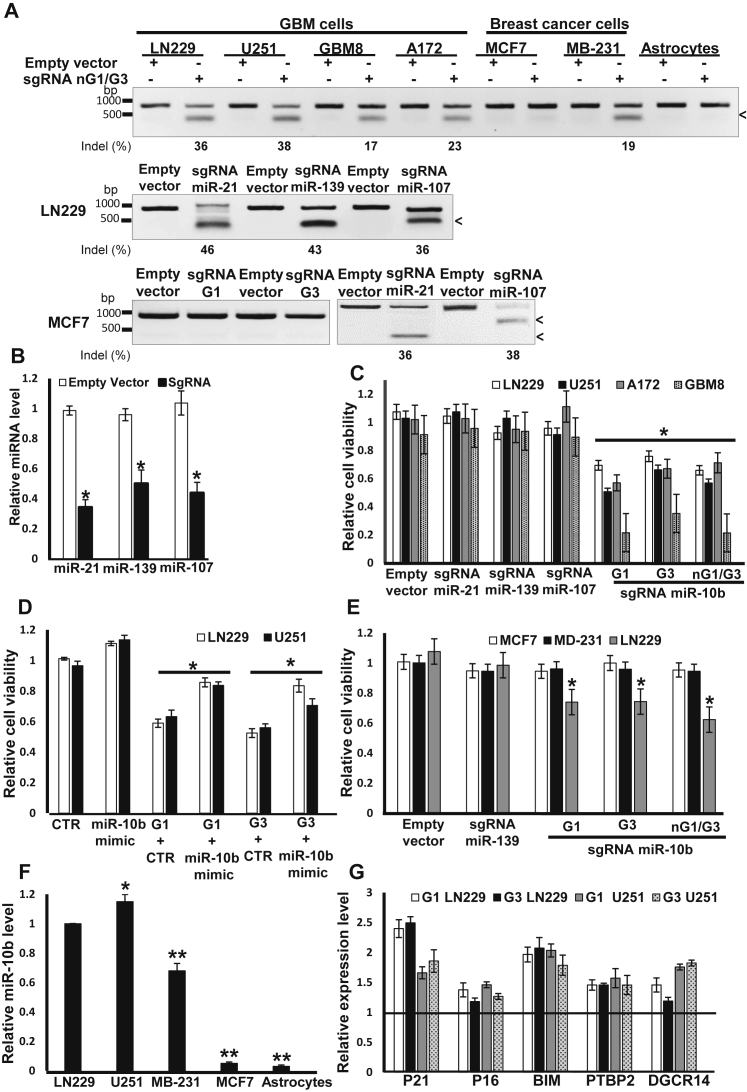
Although the CRISPR-Cas9 editing of miR-10b and other highly expressed glioma miRNA genes such as miR-21, miR-139, and miR-107 proved efficient and reduced the levels of the respective miRNAs, only miR-10b editing impaired the viability of all tested glioma cell lines and GSC cultures (Figures 2A–2C). Overall, we observed correlation between the efficacy of miR-10b gene editing and the viability of monolayer GBM cell lines, with the exception of low-passage GSCs (GBM8) cultured in neurospheres, which were extremely sensitive to even less-efficient miR-10b editing (Figure 2C). Reduced viability was rescued by sequential transfections with the miR-10b synthetic mimic, indicating that the phenotype observed in miR-10b-targeted cultures was caused by its loss (Figure 2D). This rescue was partial, possibly due to “imperfect” intracellular trafficking and incorporation of the synthetic mimic to the functional RNA-induced silencing complex (RISC) complex, not fully mimicking the endogenous miR-10b activity, as well as additional unknown off-target effects. Efficient miR-10b gene editing in metastatic, triple-negative (ER−/PR−/HER2low) breast carcinoma line MDA-MB-231 reduced cell migration but not viability (Figure 2E; Figure S2), consistent with the established role of miR-10b in breast cancer metastasis but not survival.10, 11 A similar CRISPR-Cas9 strategy failed to edit the miR-10b gene in the cell types not expressing miR-10b, such as non-metastatic breast carcinoma MCF7 and primary astrocytes (Figures 2A and 2F). Although miR-10b editing affected only a part of cells in targeted glioma cultures, it led to elevated expression of the previously validated miR-10b targets,4, 12 including the mediator of apoptosis BIM, cell-cycle inhibitors P21 and P16, and splicing regulator PTBP2 (Figure 2G).
Figure 2.
CRISPR-Cas9 Targeting Reveals that miR-10b Expression Is Essential for Glioma Viability
(A) miR-10b is efficiently edited in heterogeneous human glioma cell lines and GSCs, but not in the non-expressing normal astrocytes and MCF7 cells, as determined by Surveyor assay. Efficient editing of other miRNAs in MCF7 cells is shown as a control. (B) Editing of miR-21, miR-139, and miR-107 results in significant downregulation of the corresponding mature miRNAs, as analyzed by qRT-PCR. The data were normalized to the geometrical mean of three unaffected miRNAs (miR-99a, miR-125a, and miR-148a). Error bars depict SEM, n = 6, *p < 0.005, Student’s t test. (C) miR-10b gene editing reduces viability of glioma cells, as determined by WST1 assays 48 hr post-transfection for glioma lines and 5 days post-transfection for GSCs. n = 6, *p < 0.001, Student’s t test. (D) Viability of miR-10b-edited glioma LN229 and U251 cells (edited by lentiviral CRISPR-Cas9, guided by either G1 or G3 sgRNAs) is rescued by the miR-10b mimic transfected at 25 nM, as monitored by WST1 assays 48 hr post-transfection. n = 6, *p < 0.05. (E) miR-10b does not affect the viability of breast cancer cell lines MDA-MB-231 and MCF7, as determined by WST1 assays. n = 6, *p < 0.001. (F) qRT-PCR analysis demonstrates negligible miR-10b expression in primary astrocytes and MCF7 cells. The data were normalized to the geometrical mean of unaffected miR-99a, miR-125a, and miR-148a. Error bars depict SEM, n = 6, *p < 0.05, **p < 0.001, Student’s t test. (G) qRT-PCR analysis of established miR-10b targets BIM, CDKN1A/p21 and CDKN2A/p16, PTBP2, and DGCR14 demonstrates their de-repression in edited LN229 cells. mRNA expression levels were normalized to the geometrical mean of three unaffected genes (GAPDH, 18S rRNA, and SERAC1). Error bars depict SEM, n = 6, *p < 0.05, Student’s t test.
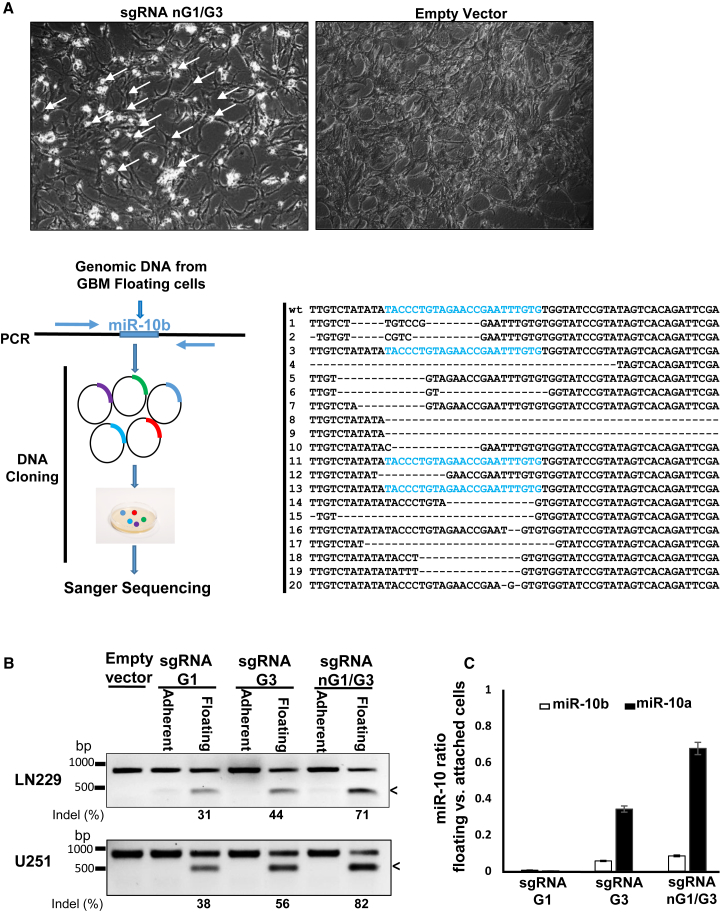
Due to the imperfect efficacy of CRISPR-Cas9 editing, the G1- and G3-targeted glioma cultures were expected to contain a variety of miR-10b mutants and indels, as well as the cells with the wild-type (WT) miR-10b gene. Correspondingly, miR-10b editing of glioma cell lines that normally grew in a monolayer resulted in the production of a mixed cell population containing distinctly apoptotic round floating cells, as well as unaffected attached cells with normal morphology (Figure 3A). To investigate whether miR-10b editing leads to glioma cell death, we analyzed the cells of these mixed cultures. Glioma cells from the miR-10b-targeted cultures were plated as single cells in 96 individual wells, which led to the growth of 53 single-cell-derived clones. The DNA was extracted from these clones, and the miR-10b gene was sequenced. Strikingly, there was no mutation found among the viable clones examined (0/53). In contrast, clonal analysis of the DNA collected from the floating apoptotic cells in the targeted parental cultures revealed an 85% mutation rate in the miR-10b locus (Figure 3A). Consistent with these findings, in the parental cultures, the miR-10b gene was efficiently edited in the floating pro-apoptotic cells but unedited in the attached viable cells (Figure 3B). Correspondingly, miR-10b levels were 20- to 30-fold lower in floating pro-apoptotic cells than in attached viable cells (Figure 3C). Collectively, these results indicate that glioma cells are addicted to miR-10b and expression of this molecule is essential for glioma viability and survival.
Figure 3.
CRISPR-Cas9 Editing Reveals that miR-10b Expression Is Essential for Glioma Viability
(A) Light microscopy images of glioma cells transfected with either the control empty vector or the miR-10b-targeting vectors demonstrate the appearance of floating apoptotic cells in the edited cultures (upper panels). Schematic view of the analysis of miR-10b DNA locus in the floating cells. The DNA was isolated from the sgRNA nG1/G3-targeted cultures, and the miR-10b genomic locus was amplified and sequenced. The sequencing results reveal a range of miR-10b mutants, with 17 of 20 clones mutated in the miR-10b locus. (B) Surveyor cleavage assay of the attached and floating populations of LN229 and U251 glioma cells demonstrates that miR-10b is edited preferentially in floating cells, whereas the unedited cells remain attached. (C) miR-10b levels are reduced in the floating apoptotic, but not in the attached viable, LN229 cells.
miR-10b Editing Impairs Tumor Growth in Intracranial GBM Models
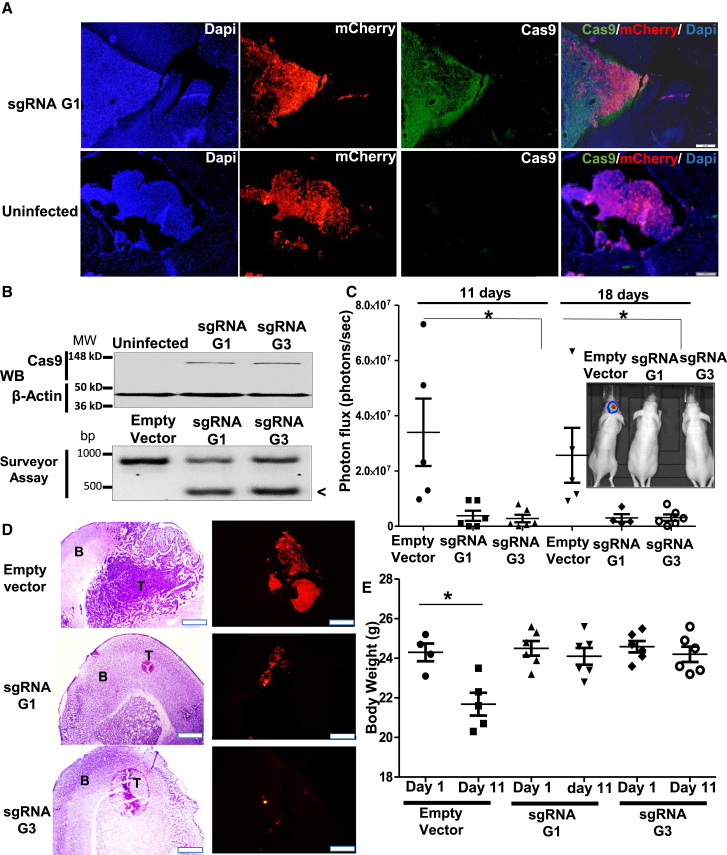
To investigate the effects of miR-10b gene editing in orthotopic GBM models in vivo, we produced miR-10b-targeting lentiCRISPR v2 plasmid based on Shalem et al.19 and Sanjana et al.,20 a single vector expressing Cas9, either G1 or G3 sgRNA, and a puromycin selection marker and then packaged it into a vesicular stomatitis virus G protein (VSV-G)-pseudotyped lentivirus. High-titer (108 transducing units [TU]/mL) viral miR-10b targeting resulted in efficient editing and reduced cell viability of various genetically distinct glioma cell lines and GSC cultures (Figure S3). Intratumoral injections of miR-10b-targeting virus to the established exponentially growing orthotopic LN229-formed GBM xenografts resulted in tumor-specific Cas9 expression and efficient miR-10b editing in the tumor tissue, with little Cas9 immunostaining in surrounding brain parenchyma (Figures 4A and 4B). Tumor growth, monitored by in vivo imaging, was strongly reduced in miR-10b-targeted G1 and G3 groups relative to the control group injected with the corresponding empty virus that expresses Cas9 but lacks miR-10b-targeting sgRNA (Figure 4C). Histological analysis of the brains harvested on day 18 after a viral injection revealed barely visible tumors in both G1 and G3 treatment groups, while large tumors were found in controls (Figure 4D). Both treatment groups also had better maintenance of body weight compared with controls (Figure 4E). Similar results were obtained on a highly invasive GBM8 xenograft model treated with the mutated nickase version of the virus-encoded Cas9n D10A enzyme and guided by the pair of G1 and G3 sgRNAs (nG1/G3) (Figures S4 and S5). Single injection of the miR-10b-editing vector effectively blocked the growth of orthotopic GBM8 and rescued the body weight of the animals.
Figure 4.
Intratumoral Injections of 105 TU of Lentiviral miR-10b-Editing Vectors Strongly Impair Tumor Growth of Established Orthotopic LN229 GBM
(A) Immunohistochemistry of brain sections exhibits specific Cas9 staining in the tumor areas, marked by mCherry fluorescence. (B) Western blot analysis (top panel) and Surveyor cleavage assay (bottom panel) demonstrate, correspondingly, Cas9 expression and efficient miR-10b editing in infected tumor xenografts, but not control tumors, 3 days after infections with G1 and G3 sgRNAs. Cleavage products, indicative of the edited miR-10b gene, are marked with an arrowhead. (C) Tumor growth was monitored by luciferase imaging in vivo. There were six or seven mice per group at the treatment initiation, and each dot represents an animal or tumor. The insert illustrates tumor imaging in representative animals. *p < 0.005 by unpaired ANOVA test. (D) H&E histology and mCherry fluorescence of the LN229 intracranial GBM demonstrate markedly reduced tumors in G1 and G3 sgRNA-targeted groups. Scale bars, 500 μm for H&E and 200 μm for IF. T, tumor; B, brain tissue. (E) miR-10b gene editing helps maintain the body weight in mice bearing intracranial tumors. n = 6 animals per group, *p < 0.005, Student’s t test.
miR-10b Editing Abolishes Transformation of Normal Astrocytes
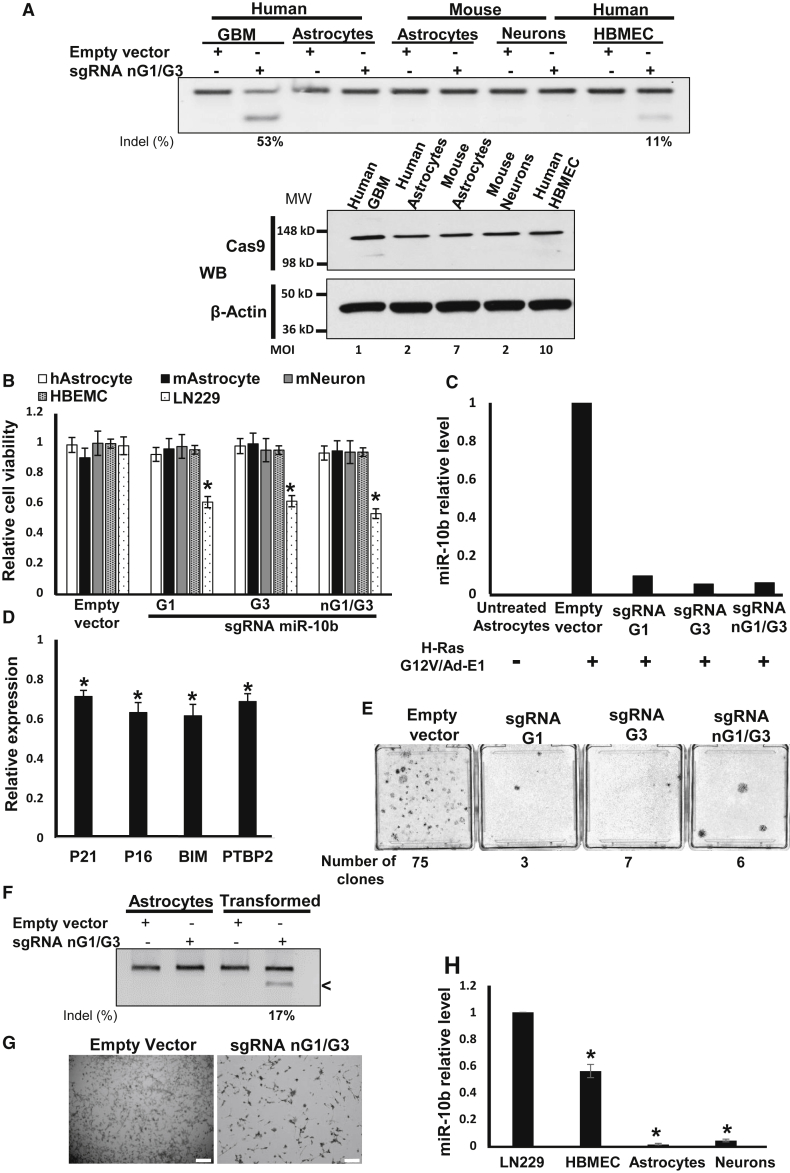
Primary mouse and human astrocytes do not express miR-10b.4 Transductions of human and mouse primary astrocytes, as well as mouse primary neurons with miR-10b-editing lentivirus at the MOI range that led to similar levels of Cas9 expression in those cells, resulted in neither miR-10b editing nor phenotypic effects on these cells (Figures 5A and 5B). Similarly, CRISPR-Cas9 vectors at the fixed titer of 3×105 TU were highly efficient in glioma, but not in normal human or murine neuroglial cells (Figure S6). However, when primary mouse astrocytes underwent oncogenic transformation by H-RasG12V/Ad-E1 or SV40 large T antigen oncogenes, they strongly upregulated miR-10b (Figure 5C) and downregulated the levels of validated miR-10b targets P21, P16, BIM, and PTBP2 (Figure 5D). miR-10b upregulation was abolished by transduction with miR-10b-editing lentiCRISPR vectors, indicating that transformed astrocytes become editable in this locus (Figure 5C). Furthermore, miR-10b editing in oncogene-induced astrocytes markedly reduced the number of transformed colonies, suggesting that miR-10b is required for transformation or essential for the survival of transformed astrocytes (Figure 5E). When miR-10b editing was performed post-transformation, it caused death of the transformed cells, similar to the effect on glioma cells (Figures 5F and 5G). The only principal type of normal brain cells expressing miR-10b are human brain microvascular endothelial cells (HBMECs) (Figure 5H). The efficiency of miR-10b gene editing in these cells, however, was much lower than in glioma cells, and even the high-titer virus has not affected their viability and morphology (Figures 5A and 5B).
Figure 5.
Lentivirus-Mediated miR-10b Gene Editing Abolishes Neoplastic Transformation of Oncogene-Induced Astrocytes
(A) Transductions of human and mouse primary astrocytes and neurons with miR-10b-editing lentivirus at the MOI levels that led to similar levels of Cas9 expression, as assessed by western blot with Cas9 antibody (low panel), does not cause miR-10b gene editing. In these conditions, 100% of glioma LN229 cells were Cas9 positive. Human brain microvascular endothelial cells (HBMECs) were edited in the miR-10b gene by high-titer virus with low efficiency (11% versus 53% in glioma cells, at a 10-fold higher viral titer). The relative MOI required for similar Cas9 expression in these cells is indicated. (B) miR-10b gene editing reduces the viability of glioma cells, but not human and mouse primary astrocytes, neurons, and HBMECs, as determined by WST1 assays 48 hr post-transduction. Transduction conditions and MOI match those used in (A). n = 6, *p < 0.001, Student’s t test. (C) miR-10b levels in mouse primary astrocytes induced for transformation by H-RasG12V/Ad-E1 and subsequently transduced with miR-10b-editing vectors for 2 weeks, as determined by qRT-PCR and normalized to the geometrical mean of unaffected miR-99a, miR-125a, and miR-148a. (D) Transformed primary astrocytes exhibit reduced levels of miR-10b targets p21, p16, BIM, and PTBP2, relative to the corresponding naive cultures. qRT-PCR data were normalized to the geometrical mean of three unaffected genes (GAPDH, 18S rRNA, and SERAC1). Error bars depict SEM, n = 3, *p < 0.05, Student’s t test. (E) miR-10b editing reduces the number of transformed colonies. Crystal violet staining and quantification of the colonies are shown 2 weeks after infections with miR-10b-editing vectors. (F) Transformed, miR-10b-expressing mouse astrocytes become editable in the miR-10b locus. (G) miR-10b editing of transformed astrocytes induces cell death, similar to the effect on glioma cell lines. Scale bar, 20 μm. (H) Relative miR-10b levels in glioma and various brain-derived cell types were assessed by qRT-PCR analysis, and the data were normalized to the geometrical mean of unaffected miR-99a, miR-125a, and miR-148a. n = 6, *p < 0.001, Student’s t test.
Discussion
Despite the firmly established growth-promoting functions for many specific miRNAs, there are only a few examples of true oncogenic miRNA (onco-miR) dependencies known for cancer cells. Our data indicate that high expression of the WT miR-10b gene is essential for glioma, whereas loss-of-function mutations lead to the lethality of heterogeneous glioma cells and tumor-initiating GSCs. Alternative sgRNA guides targeting miR-10b either alone or with its closely related paralog miR-10a produced a diverse range of mutants, none of which were viable. The loss-of-function mutations in miR-10b alone were sufficient to cause the lethality, validating the key role of miR-10b in the sustained growth and survival of glioma. Specifically, mutated nCas9 guided by the G1/G3 sgRNAs has detrimental effects on glioma cells by reducing the levels of miR-10b without affecting the levels of miR-10a gene, suggesting the efficacy of the miR-10b single-gene targeting approach for GBM. Because miR-10a and miR-10b differ in one nucleotide and are largely functionally redundant, the relative efficacy of miR-10a targeting remains to be evaluated. miR-10b is expressed in normal extracranial tissues; nevertheless, its activity in these tissues seems to be dispensable, because the initial analysis of miR-10b knockout mice has no apparent pathological phenotype (https://www.jax.org/strain/016950). Therefore, glioma addiction to miR-10b appears to be the tumor-specific trait, probably associated with de-repression of the gene in the brain microenvironment, where it is normally silenced. Unique onco-miR dependence of GBM also suggests that the tumor could be potentially eradicated by targeting a single miRNA gene.
Administration of synthetic miR-10b inhibitors caused potent but transient effects on orthotopic GBM in the aggressive GSC-based models.12 This was probably due to both poor uptake and distribution of the ASOs in intracranial GBM and dilution of the drug in the actively growing tumor. Gene editing, based on permanent miR-10b inactivation, may provide an alternative strategy, eliminating the need for continuous delivery of miR-10b inhibitors to intracranial brain tumors and improving the efficacy of tumor cell destruction. Even moderately efficient miR-10b gene editing of GBM8 glioma stem cells led to disaggregation and massive death of glioma spheres, suggesting that disruption of this core cell population may have detrimental effects on the tumor growth. Using lentiviral CRISPR-Cas9 targeting, we have examined the effects of miR-10b ablation on highly aggressive human GBM xenografts. Efficient Cas9 expression and miR-10b editing throughout the tumor resulted in the permanent ablation of miR-10b and near-eradication of orthotopic GBM tumors. The data suggest that less than 100% efficient editing and miR-10b ablation are sufficient for potent inhibition of GBM growth. The possibility of bystander effects remains to be investigated. Most of our in vitro experiments used transfected plasmids, whereas the in vivo experiments relied on lentivirus vectors. The same lentiviral editing vector caused strong effects on glioma growth both in vitro and in vivo. The effects may appear stronger in vivo due to the longer duration of the experiment; however, miR-10b editing in cultures also resulted in death of the entire population when analyzed over a longer time. Overall, these data provide proof of principle for the therapeutic strategy for malignant gliomas based on single-target gene editing and may apply to other miR-10b-dependent metastatic cancers.11, 21
A major limitation of the CRISPR-Cas9 technology, and particularly its clinical application, is associated with its restricted specificity.22 Our bioinformatics analysis suggested only a few potential high-ranked protein-coding off-targets for the designed miR-10b sgRNAs (Figure 1C), and none of them was edited in our experiments. To start evaluating the therapeutic potential of miR-10b editing in the brain and assess its safety, we tested the effects of miR-10b ablation in the normal cells of the brain tumor microenvironment in vitro. Major cell types of the brain, including neurons, astrocytes, microglia, and neuroprogenitors, express very low or undetectable levels of miR-10b while exhibiting low levels of miR-10a.4 Although the CRISPR-Cas9 system can target genes in any cell type, including postmitotic neurons,23, 24 the efficacy of editing genes that are not actively transcribed in a specific cellular context, and might be less accessible by Cas9-sgRNA due to their epigenetic state and chromatin structure, is presently unknown and expected to be low.25 Our data indicate that CRISPR-Cas9 plasmid- and virus-mediated miR-10b targeting does not cause the locus editing in normal brain cells and does not affect the viability of mouse primary astrocytes or neurons (Figures 2A and 5A); neither do the miR-10b ASO inhibitors.4 Additional experiments on human MCF7 cells that express only negligible miR-10b levels also demonstrate the lack of miR-10b editing and no visible phenotypic effects, despite the efficient editing of other highly expressed miRNAs in these cells (Figure 2A). These data suggest that miR-10b is not edited in normal neuroglial and other non-expressing cells due to the compact chromatin structure of the locus, and not merely lower efficiency of transfection or transduction. Human brain-derived microvascular endothelial cells do express substantial levels of miR-10b. While the functional role of miR-10b in these cells requires further investigation, our results demonstrate that miR-10b gene editing (albeit less efficient than in glioma cells) does not affect their phenotype. The lack of toxicity for normal brain cells suggests the therapeutic window for miR-10b editing in glioma in vivo, further validates the high targeting specificity, and paves the way for its clinical development.
We envision the clinical application of miR-10b-editing viral therapy for GBM patients as a one-time treatment with local administration of the viral vector to the surgical bed immediately after tumor resection. The lentiviral vectors used in our experiments in vivo transduce dividing and quiescent cells. This can be viewed as a major advantage for cancer gene therapy in general, because within a short treatment window, most tumor cells (especially GSCs) do not divide. Because miR-10b editing prevents neoplastic transformation of astrocytes and selectively eradicates the transforming cells (Figure 5), in addition to malignant tumor cells, this approach may target the brain cells undergoing early stages of gliomagenesis. Therapeutic gene editing using high viral titers applied locally to the surgical cavity may also prove to be effective for targeting infiltrating tumor cells.26, 27 An additional advantage of the locally applied lentivirus pseudotyped with the VSV-G glycoprotein is that its inactivation by human serum28 would reduce systemic effects. Although the application of human lentiviral gene therapy is hampered by the risk of carcinogenesis by random proviral integration into the genome of normal somatic cells, future studies should determine whether this risk is acceptable for local GBM treatment, given the lack of efficacious drugs and poor life expectancy of patients with the disease. The identification of Staphylococcus aureus (SaCas9) and other smaller Cas9 enzymes that can be packaged into adeno-associated viral vectors, which are highly stable and effective in vivo,29, 30, 31 are easily produced, have been approved by the U.S. Food and Drug Administration (FDA) for other applications, and have been tested in multiple clinical trials, paves new avenues for therapeutic gene editing. Further optimization of the targeting vectors with increased tropism for glioma cells, as well as in-depth investigation of potential neurotoxic effects, must be performed before clinical applications of this promising new strategy.
Materials and Methods
CRISPR-Cas9 Plasmid Construction and Lentivirus Production
The sgRNA guide sequences were designed and cloned into plasmids PX330, PX335, and lentiCRISPR v2 (a gift from Feng Zhang, Addgene plasmids #42230, #42335, and #52961), based on Sanjana et al.20 and Cong et al.32 LentiCRISPR v2 plasmid was used as a template for site-directed mutagenesis by inverse PCR to generate lentiCRISPR v2 nCas9 (D10A mutant Cas9, A changed to C at position 146 from the ATG). The sequences used for miRNA targeting are listed in Table S1. For lentivirus production, the lentiCRISPR v2 plasmids were co-transfected with packaging psPAX2 plasmids and VSV-G envelope-expressing plasmid (Addgene plasmids #12259 and #12260) as described,20 and viruses were concentrated by additional ultracentrifugation at 25,000 rpm. Lentivirus functional titer was determined by serial dilution in LN229 cells using immunofluorescence for Cas9 with Novusbio 7A9-3A3 antibody. Positive cells were counted, and the titer was estimated using the following formula: titer (TU/mL) = number of transduced cells in day 1 × percentage of fluorescent-positive cells × 1,000/volume of lentivirus used (μL).
Surveyor Assay for Genome Editing
Genomic regions surrounding the CRISPR-Cas9 target sites were amplified using Q5 polymerase (NEB), and Surveyor nuclease assay was performed as described.32 Efficiency of editing was estimated based on relative band intensities as a percentage of gene modification: indels % = 100 × (1 − (1 − fraction cleaved)1/2). PCR primers used for genomic amplification are listed in Table S1.
Cell Cultures and Transfections
Human glioma LN229, U251, and A172 and breast cancer MCF7 and MDA-MB-231 cell lines were maintained as described.4 Patient-derived low-passage GBM8 cells growing as neurospheres were maintained in Neurobasal medium as previously described.33 HBMECs were cultured in human endothelial culture medium with the Complete Growth Medium supplement kit (Cell Biologics). Primary mouse and human astrocytes were maintained in DMEM-F12 supplemented with 10% fetal bovine serum (FBS). Primary mouse neurons were maintained in Neurobasal supplemented with B27 (Invitrogen). The cells were seeded in 24-well plates at 60% confluence and transfected the next day with 800 ng of plasmids, with or without 20 nM of oligonucleotide (oligo) mimics (Ambion) using the NeuroMag transfection protocol, according to the manufacturer’s instructions (OZ Bioscience). RNA isolation, qRT-PCR, and protein analysis by western blotting were performed as previously described.34 Cell viability was assessed using WST1 (Roche), according to the manufacturer’s instructions, 2 days post-transfection for the monolayer cultures and 5 days post-transfection for neurospheres. Wound healing assay was performed as described.35
Transformation Assay
Primary P1 mouse astrocytes plated at 10% density in 25 cm2 flasks were transfected with 10 μg of RasG12V/Ad-E1 plasmids or infected by SV40 large T antigen lentivirus; 24 hr later, they were infected by lentivirus expressing CRISPR-Cas9. Total RNA was extracted, and supplemental culture plates were fixed with 4% formaldehyde and stained with crystal violet 2 weeks post-transformation.
Stereotaxic Injections of Tumor Cells, Whole-Body Imaging, and Lentivirus Injection
LN229 and GBM8 cells (105) expressing firefly luciferase and mCherry were stereotactically implanted into the striatal area (coordinates: P–A, 0.5 mm; C–L, 1.7 mm; and D–V, 2.3 mm) of 8-week-old athymic nu/nu mice (Jackson Laboratory), and the growth of intracranial tumors was monitored by Fluc bioluminescence imaging.34 When bioluminescence reached the exponential phase with signal of 106 photons/s (10 days after LN229 and 19 days after GBM8 implantation), the lentiviral CRISPR-Cas9 constructs (3 × 105 TU) were injected intratumorally to the same coordinates. The animals were randomized to the treatment and control groups based on the whole-body imaging (WBI), with similar average bioluminescence signal and tumor growth rates per group. All animal studies have been approved and performed in accordance with Harvard Medical Area Standing Committee guidelines.
Immunohistochemistry and H&E Staining
Intracranial tumors were fixed with 4% formaldehyde, embedded, and cryo-sectioned. Staining of 20-μm-thick sections was performed using Cas9 antibody (7A9-3A3, Novus Biologicals), DAPI, and H&E as described.34
Statistical Analysis
The unpaired, two-tailed Student’s t test was used for comparison between two groups, and the unpaired ANOVA test was used for comparison of three groups. All values were presented as mean ± SEM. The adequate sample sizes were calculated based on the resource equation method.36
Author Contributions
R.E.F. and A.M.K. conceived the project, R.E.F. performed the experiments, S.S. and E.J.U. assisted with the experiments, and R.E.F. and A.M.K. wrote the manuscript. All authors analyzed the data and critically reviewed the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
We thank Drs. Feng Zhang and Didier Trono for providing plasmids and members of A.M.K. laboratory for valuable discussions. This work was supported by grants from the NIH/NCI (RO1CA138734), Sontag Foundation Distinguished Scientist Award, and Brain Science Foundation (to A.M.K. and E.J.U.).
Footnotes
Supplemental Information includes six figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2016.11.004.
Supplemental Information
References
- 1.Floyd D., Purow B. Micro-masters of glioblastoma biology and therapy: increasingly recognized roles for microRNAs. Neuro-oncol. 2014;16:622–627. doi: 10.1093/neuonc/nou049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biagioni F., Bossel Ben-Moshe N., Fontemaggi G., Yarden Y., Domany E., Blandino G. The locus of microRNA-10b: a critical target for breast cancer insurgence and dissemination. Cell Cycle. 2013;12:2371–2375. doi: 10.4161/cc.25380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tehler D., Høyland-Kroghsbo N.M., Lund A.H. The miR-10 microRNA precursor family. RNA Biol. 2011;8:728–734. doi: 10.4161/rna.8.5.16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabriely G., Yi M., Narayan R.S., Niers J.M., Wurdinger T., Imitola J., Ligon K.L., Kesari S., Esau C., Stephens R.M. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71:3563–3572. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teplyuk N.M., Mollenhauer B., Gabriely G., Giese A., Kim E., Smolsky M., Kim R.Y., Saria M.G., Pastorino S., Kesari S., Krichevsky A.M. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro-oncol. 2012;14:689–700. doi: 10.1093/neuonc/nos074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun L., Yan W., Wang Y., Sun G., Luo H., Zhang J., Wang X., You Y., Yang Z., Liu N. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res. 2011;1389:9–18. doi: 10.1016/j.brainres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Ahmad A., Sethi S., Chen W., Ali-Fehmi R., Mittal S., Sarkar F.H. Up-regulation of microRNA-10b is associated with the development of breast cancer brain metastasis. Am. J. Transl. Res. 2014;6:384–390. [PMC free article] [PubMed] [Google Scholar]
- 8.Parrella P., Barbano R., Pasculli B., Fontana A., Copetti M., Valori V.M., Poeta M.L., Perrone G., Righi D., Castelvetere M. Evaluation of microRNA-10b prognostic significance in a prospective cohort of breast cancer patients. Mol. Cancer. 2014;13:142. doi: 10.1186/1476-4598-13-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guessous F., Alvarado-Velez M., Marcinkiewicz L., Zhang Y., Kim J., Heister S., Kefas B., Godlewski J., Schiff D., Purow B., Abounader R. Oncogenic effects of miR-10b in glioblastoma stem cells. J. Neurooncol. 2013;112:153–163. doi: 10.1007/s11060-013-1047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma L., Reinhardt F., Pan E., Soutschek J., Bhat B., Marcusson E.G., Teruya-Feldstein J., Bell G.W., Weinberg R.A. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma L., Teruya-Feldstein J., Weinberg R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 12.Teplyuk N.M., Uhlmann E.J., Gabriely G., Volfovsky N., Wang Y., Teng J., Karmali P., Marcusson E., Peter M., Mohan A. Therapeutic potential of targeting microRNA-10b in established intracranial glioblastoma: first steps toward the clinic. EMBO Mol. Med. 2016;8:268–287. doi: 10.15252/emmm.201505495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 14.Swiech L., Heidenreich M., Banerjee A., Habib N., Li Y., Trombetta J., Sur M., Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis L., Maizels N. Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc. Natl. Acad. Sci. USA. 2014;111:E924–E932. doi: 10.1073/pnas.1400236111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian Y., Luo A., Cai Y., Su Q., Ding F., Chen H., Liu Z. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J. Biol. Chem. 2010;285:7986–7994. doi: 10.1074/jbc.M109.062877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X.H., Tee L.Y., Wang X.G., Huang Q.S., Yang S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Incontro S., Asensio C.S., Edwards R.H., Nicoll R.A. Efficient, complete deletion of synaptic proteins using CRISPR. Neuron. 2014;83:1051–1057. doi: 10.1016/j.neuron.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Straub C., Granger A.J., Saulnier J.L., Sabatini B.L. CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PLoS ONE. 2014;9:e105584. doi: 10.1371/journal.pone.0105584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldberg G.W., Jiang W., Bikard D., Marraffini L.A. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huszthy P.C., Giroglou T., Tsinkalovsky O., Euskirchen P., Skaftnesmo K.O., Bjerkvig R., von Laer D., Miletic H. Remission of invasive, cancer stem-like glioblastoma xenografts using lentiviral vector-mediated suicide gene therapy. PLoS ONE. 2009;4:e6314. doi: 10.1371/journal.pone.0006314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayin N.S., Modrek A.S., Dietrich A., Lebowitz J., Abel T., Song H.R., Schober M., Zagzag D., Buchholz C.J., Chao M.V., Placantonakis D.G. Selective lentiviral gene delivery to CD133-expressing human glioblastoma stem cells. PLoS ONE. 2014;9:e116114. doi: 10.1371/journal.pone.0116114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DePolo N.J., Reed J.D., Sheridan P.L., Townsend K., Sauter S.L., Jolly D.J., Dubensky T.W., Jr. VSV-G pseudotyped lentiviral vector particles produced in human cells are inactivated by human serum. Mol. Ther. 2000;2:218–222. doi: 10.1006/mthe.2000.0116. [DOI] [PubMed] [Google Scholar]
- 29.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiani S., Chavez A., Tuttle M., Hall R.N., Chari R., Ter-Ovanesyan D., Qian J., Pruitt B.W., Beal J., Vora S. Cas9 gRNA engineering for genome editing, activation and repression. Nat. Methods. 2015;12:1051–1054. doi: 10.1038/nmeth.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakimoto H., Mohapatra G., Kanai R., Curry W.T., Jr., Yip S., Nitta M., Patel A.P., Barnard Z.R., Stemmer-Rachamimov A.O., Louis D.N. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro-oncol. 2012;14:132–144. doi: 10.1093/neuonc/nor195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong H.K., Fatimy R.E., Onodera C., Wei Z., Yi M., Mohan A., Gowrisankaran S., Karmali P., Marcusson E., Wakimoto H. The Cancer Genome Atlas analysis predicts microRNA for targeting cancer growth and vascularization in glioblastoma. Mol. Ther. 2015;23:1234–1247. doi: 10.1038/mt.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez L.G., Wu X., Guan J.L. Wound-healing assay. Methods Mol. Biol. 2005;294:23–29. doi: 10.1385/1-59259-860-9:023. [DOI] [PubMed] [Google Scholar]
- 36.Festing M.F., Altman D.G. Guidelines for the design and statistical analysis of experiments using laboratory animals. ILAR J. 2002;43:244–258. doi: 10.1093/ilar.43.4.244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.