Summary
Background
The control of Clostridium difficile infections is an international clinical challenge. The incidence of C difficile in England declined by roughly 80% after 2006, following the implementation of national control policies; we tested two hypotheses to investigate their role in this decline. First, if C difficile infection declines in England were driven by reductions in use of particular antibiotics, then incidence of C difficile infections caused by resistant isolates should decline faster than that caused by susceptible isolates across multiple genotypes. Second, if C difficile infection declines were driven by improvements in hospital infection control, then transmitted (secondary) cases should decline regardless of susceptibility.
Methods
Regional (Oxfordshire and Leeds, UK) and national data for the incidence of C difficile infections and antimicrobial prescribing data (1998–2014) were combined with whole genome sequences from 4045 national and international C difficile isolates. Genotype (multilocus sequence type) and fluoroquinolone susceptibility were determined from whole genome sequences. The incidence of C difficile infections caused by fluoroquinolone-resistant and fluoroquinolone-susceptible isolates was estimated with negative-binomial regression, overall and per genotype. Selection and transmission were investigated with phylogenetic analyses.
Findings
National fluoroquinolone and cephalosporin prescribing correlated highly with incidence of C difficile infections (cross-correlations >0·88), by contrast with total antibiotic prescribing (cross-correlations <0·59). Regionally, C difficile decline was driven by elimination of fluoroquinolone-resistant isolates (approximately 67% of Oxfordshire infections in September, 2006, falling to approximately 3% in February, 2013; annual incidence rate ratio 0·52, 95% CI 0·48–0·56 vs fluoroquinolone-susceptible isolates: 1·02, 0·97–1·08). C difficile infections caused by fluoroquinolone-resistant isolates declined in four distinct genotypes (p<0·01). The regions of phylogenies containing fluoroquinolone-resistant isolates were short-branched and geographically structured, consistent with selection and rapid transmission. The importance of fluoroquinolone restriction over infection control was shown by significant declines in inferred secondary (transmitted) cases caused by fluoroquinolone-resistant isolates with or without hospital contact (p<0·0001) versus no change in either group of cases caused by fluoroquinolone-susceptible isolates (p>0·2).
Interpretation
Restricting fluoroquinolone prescribing appears to explain the decline in incidence of C difficile infections, above other measures, in Oxfordshire and Leeds, England. Antimicrobial stewardship should be a central component of C difficile infection control programmes.
Funding
UK Clinical Research Collaboration (Medical Research Council, Wellcome Trust, National Institute for Health Research); NIHR Oxford Biomedical Research Centre; NIHR Health Protection Research Unit on Healthcare Associated Infection and Antimicrobial Resistance (Oxford University in partnership with Public Health England [PHE]), and on Modelling Methodology (Imperial College, London in partnership with PHE); and the Health Innovation Challenge Fund.
Introduction
Clostridium difficile infection is a major clinical challenge worldwide.1, 2 At least three antimicrobial classes are deemed to be high-risk C difficile infection triggers,3 including most cephalosporins, to which C difficile is inherently resistant,4 and clindamycin, to which genotypes causing early outbreaks were resistant.5, 6, 7 Global dispersion of hypervirulent NAP1/PCR-ribotype-027 C difficile revealed an association between fluoroquinolone resistance and epidemic spread.8, 9 Accordingly, clindamycin or fluoroquinolone use has been restricted, and combined with other measures aiming to control localised C difficile infection outbreaks.7, 10, 11
Most cases of C difficile infection are temporally associated with health care,2 reflecting a combination of health-care-associated acquisition, and health-care-related triggers including antibiotics. Three UK studies using highly discriminatory whole genome sequences,12, 13, 14 and a US study using alternative high-resolution typing,15 found as few as a third of C difficile infections involved recent acquisition from an active case, leaving the source for two-thirds of infections unexplained.
Research in context.
Evidence before this study
We searched PubMed with the terms “Clostridium difficile” AND “sequencing” for articles published in English on or before Feb 1, 2016. We prioritised articles including whole genome sequences. We also reviewed the references of papers identified by this strategy. In previous studies, whole-genome sequencing of C difficile was done to investigate its transmission and evolution. We identified no studies in which whole genome sequence based phylogenies were used to investigate the specific role of fluoroquinolone susceptibility or selection in the changing molecular epidemiology or incidence of C difficile infection. England is almost unique in experiencing a marked, recent decline in the incidence of health-care-associated C difficile infections. Previous reports showed the decline of one epidemic genotype (PCR-ribotype 027), whereas other genotypes appeared to persist. These changes followed the implementation of a multifaceted national C difficile infection control policy in 2007. However, the relative contributions made by the different interventions that were introduced simultaneously is unknown.
Added value of this study
This study is the first to investigate the contribution of specific public health interventions to the marked national decline in C difficile infection. Our novel approach involved the integrated analysis of multiple, large, concurrent datasets concerning incidence of C difficile infection, antimicrobial prescribing, and, crucially, the whole genome sequences of more than 4000 human C difficile isolates. Our key finding was that the post-interventions decline in C difficile infections reflected the disappearance of fluoroquinolone-resistant isolates (predominantly from four genetically distinct genotypes), whereas the incidence of C difficile infections caused by fluoroquinolone-susceptible isolates (of many different genotypes) remained unchanged. Whole genome sequence-based phylogenetic analyses of the entire C difficile population, with one phylogeny constructed for each genotype, identified shorter, geographically clustered branches, specific to the fluoroquinolone-resistant regions. This finding is consistent with rapid nosocomial transmission preceding the disappearance of fluoroquinolone-resistant isolates. Among the susceptible isolates, the numbers that were closely genetically related (and by inference transmitted, either directly or indirectly) did not change over time. This lack of change was despite the implementation of comprehensive infection prevention and control measures, which would have targeted fluoroquinolone-resistant and susceptible C difficile equally. These data suggest that it was the restriction of fluoroquinolone prescribing, above other interventions (including cephalosporin restriction and infection control precautions), that appears to explain the decline in incidence of C difficile infections.
Implications of all the available evidence
The restriction of fluoroquinolone prescribing should be a cornerstone in the control of epidemic C difficile infections in the UK and worldwide.
By comparison with other countries,1, 2 the incidence of C difficile infection in England decreased markedly over the past decade,16 after the introduction of national C difficile infection prevention and management policies from June, 2007.17, 18 These included recommendations to avoid clindamycin and cephalosporins, minimised use of fluoroquinolone, carbapenem and aminopenicillin, and improved infection prevention and control activities (appendix).17 We investigated the effect of these interventions on C difficile evolution, selection, and transmission, to inform future C difficile infection control policies for this global challenge.
Methods
Study design
This observational study tested two hypotheses. First, if C difficile infection declines in England were driven by reductions in use of particular antibiotics, then incidence of C difficile infection caused by resistant isolates should decline faster than that caused by susceptible isolates across several genotypes (defined by multilocus sequence type). Second, if decreases in C difficile infection were driven by improvements in hospital infection control, then transmitted (secondary) cases should decline regardless of susceptibility.
To confirm that national policies17, 18 affected antibiotic prescribing and C difficile infection incidence, we first compared national antimicrobial prescribing data for hospitals and the community (obtained respectively from IMS Health [Danbury, CT, USA] and the Health & Social Care Information Centre [appendix]) with national incidence of C difficile infection (ie, infections per English population per year, using data from Public Health England).
The primary study dataset comprised whole genome sequences from clinical C difficile isolates cultured from consecutive toxin enzyme immunoassay (EIA)-positive stool samples from symptomatic, unique patients submitted to the Oxford University Hospitals NHS Trust between Sept 12, 2006, and Aug 19, 2013 (n=2021; appendix). A further 261 isolates between Sept 1, 2006, and Feb 26, 2013, where only the sequence type was available were also included. The hospital did all C difficile testing in Oxfordshire, serving general practices, community hospitals, and other providers, so incidence is per Oxfordshire population (approximately 600 000) per year. This culture-positive C difficile infection incidence was compared with Oxfordshire's nationally submitted EIA-positive incidence (incorporating changes in mandatory reporting requirements in 2008) to confirm representativeness of whole genome sequences. The latter was compared with English incidence of C difficile infection to assess generalisability.
Generalisability of Oxfordshire data was also assessed with similar information from Leeds Teaching Hospitals NHS Trust, UK. This comprised whole genome sequences for consecutive clinical, toxin-positive (cytotoxin assay) isolates from symptomatic patients (Aug 2, 2010, to May 1, 2013; n=1020; appendix), Leeds regional incidence of C difficile infection data (nationally submitted) and ribotype prevalence, and antibiotic prescribing data.
Additional genetic context was provided by further regional and international C difficile whole genome sequences (May 9, 2006, to July 12, 2013) of isolates from toxin-EIA-negative clinical samples of symptomatic Oxfordshire patients (n=395), toxin-positive samples representing two clinical trials of fidaxomicin in North America and Europe (n=803),19, 20 and from healthy Oxfordshire infants (non-clinical; n=200; appendix).
Genome sequences and multilocus sequence type identification
Genomes were sequenced using Illumina technology. Velvet de novo assemblies and reference-based assemblies were generated, the latter mapped to C difficile 630 (GenBank AM180355.1; reads submitted to National Center for Biotechnology Information, BioProjectID PRJNA304087; appendix). The sequences of loci defining C difficile sequence types were identified and extracted with BIGSdb;21 sequence types were assigned with the C difficile PubMLST database. The notation ST1(027) indicates, for example, sequence-type-1 (PCR-ribotype-027).
Whole genome sequence-derived fluoroquinolone susceptibility
Isolates were designated fluoroquinolone-susceptible or fluoroquinolone-resistant based on specific non-synonymous substitutions within the quinolone resistance-determining region of gyrA/B genes22, 23 extracted from whole genome sequences.21 gyrA C(245)T[T(82)I] and gyrB G(1276)A [D(426)N] confer high-level fluoroquinolone resistance in C difficile and other species.16, 17 Susceptibility predictions were validated phenotypically for 387 fidaxomicin trial isolates19, 20 (n=191 Canada, n=196 USA), with agar dilution (moxifloxacin minimum inhibitory concentration; appendix).
Statistical analysis
We made univariable comparisons between English antimicrobial prescribing and incidence of C difficile infection with bivariate cross-correlations (appendix). Genotype (sequence type)-specific incidence rates for C difficile infection caused by toxin EIA-positive, culture-positive isolates were calculated with negative binomial regression accounting for missing data by probability weights (appendix). For genotypes with more than 10% fluoroquinolone-resistant isolates, rates were calculated separately for fluoroquinolone-susceptible and fluoroquinolone-resistant isolates. These data were available for isolates from April 2008 to March 2011. Rates were also calculated separately for infections that could plausibly have arisen from secondary spread (transmission) inferred by close genetic relationships to previous infections (two or fewer single nucleotide variants from the original case),12 and also separately for fluoroquinolone-susceptible and fluoroquinolone-resistant isolates. Phylogenetic trees were constructed for each sequence type (or several closely related sequence types), with maximum likelihood, then corrected for recombination using ClonalFrameML (version 1.0–6).24 Trees were time-scaled and made directly comparable post-1990 (appendix). In each tree, the evolutionary distinctiveness (ED) score of each genome was calculated;25 low ED scores indicate closely related genomes, whereas high scores indicate their relative absence (appendix).
Role of the funding source
The study sponsor had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all study data and had final responsibility for the decision to submit for publication.
Results
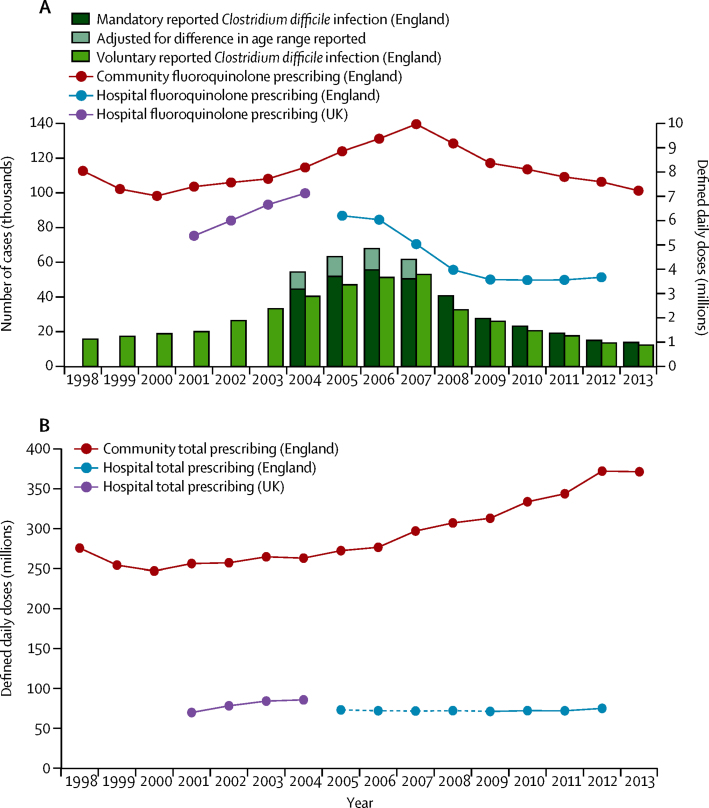
Incidence of C difficile infection in England increased from 1998 to 2006 (p<0·0001) then declined rapidly over the years that followed to 2013 (p<0·0001; figure 1A). C difficile infection declines occurred while total antibiotic prescribing was increasing (by 4·4% per year in the community [p<0·0001, 2006–13], but only 0·5% per year in hospitals [p=0·053, 2006–12]; figure 1B). Between 2005 and 2012 (when data were complete for England), the cross-correlations (CCs) between English incidence of C difficile infection and total English antibiotic prescribing were −0·57 (95% CI −0·67 to −0·41) for hospital and community, −0·59 (−0·68 to −0·44) for community, and 0·29 (−0·19 to 0·60) for hospital prescribing (optimum CC using a 1-year lag; appendix). During the same period, the strongest univariable associations between English incidence of C difficile infection and individual antimicrobials were with cephalosporins (CC=0·97, 95% CI 0·82–0·98 for hospital and community; 0·94, 0·68–0·97 for community; and 0·97, 0·81–0·99 for hopital prescribing; optimum 0-year lag) and fluoroquinolones (CC=1·00, 0·84–1·00 for hospital and community; 0·88, 0·48–0·95 for community; and 0·93, 0·66–0·97 for hospital prescribing; optimum 0-year lag; appendix), although hospital fluoroquinolone prescribing began to decline slightly earlier than community prescribing (p<0·0001 from 2005 to 2009 vs in the community p<0·0001 from 2007 to 2012; figure 1A). Other antibiotics were more weakly associated (appendix).
Figure 1.
National incidence of Clostridium difficile infections and fluoroquinolone prescribing (A) and national antibiotic prescribing overall (B)
(A) Mandatory incidence of C difficile infections corresponds to all infections reported for individuals older than 2 years (from 2004 to 2007, infections were only reported for individuals older than 65 years, and are upweighted to provide similar estimates in individuals older than 2 years; appendix). Since mandatory reporting was only introduced in 2004, we have also included voluntary-reported C difficile infections to give an indication of trends before that date. (B) Dotted lines are estimates (appendix).
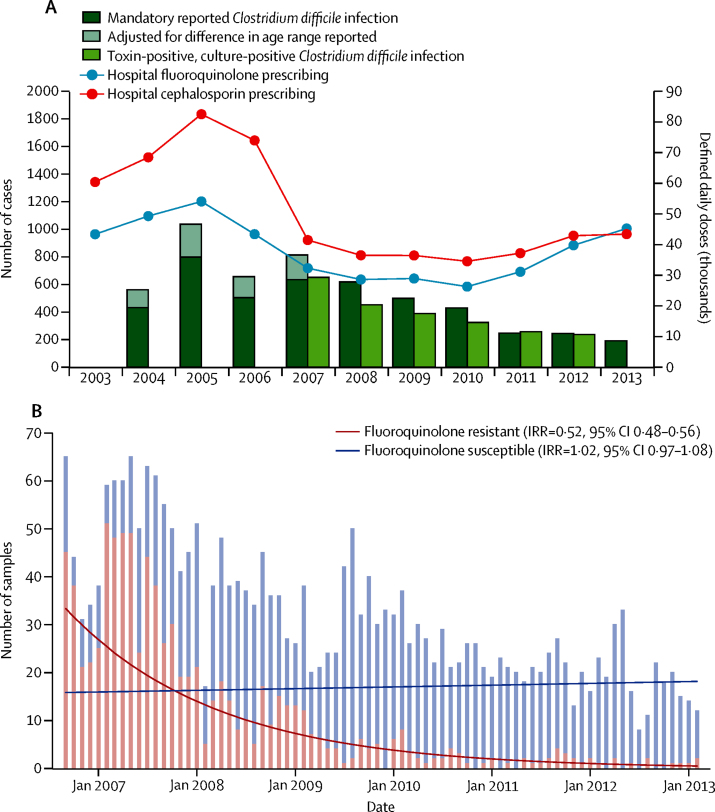
Similar to English incidence of C difficile infection, Oxfordshire rates also decreased from 2007 (when isolate-level fluoroquinolone-susceptibility could be determined; p<0·0001; figure 2A). Fluoroquinolone prescribing in Oxfordshire hospitals declined from a peak in 2005 until 2010 (p<0·0001), when use began to increase again (p<0·0001 from 2010 to 2013). Hospital cephalosporin and fluoroquinolone prescribing were also positively associated with incidence of C difficile infection (CC=0·73, 0·15 to 0·86, and 0·62, −0·09 to 0·81; appendix), but associations were estimated much less precisely given the much smaller population (approximately 1% of England). Positive associations were also observed between C difficile infection decline and decline in extended spectrum penicillins (0·84, 0·24 to 0·90) and beta-lactamase-resistant penicillins (0·67, −0·04 to 0·81; appendix). Community prescribing data were not available.
Figure 2.
Incidence of Clostridium difficile infections together with fluoroquinolone and cephalosporin prescribing for Oxfordshire (A) and incidence of C difficile infections by fluoroquinolone susceptibility for Oxfordshire (B)
(A) Mandatory incidence of C difficile infections corresponds to all cases reported for individuals older than 2 years (from 2004 to 2007, cases were only reported for individuals older than 65 years, and are upweighted to provide similar estimates in individuals older than 2 years; appendix). Only toxin-positive culture-positive samples were used in the genotype-specific and phylogenetic analyses. (B) C difficile is inherently resistant to most cephalosporins.4 IRR=annual incidence rate ratio.
Paired fluoroquinolone susceptibility phenotype and gyrA/B DNA sequences were assessed for 387 isolates from the two clinical trials of fidaxomicin in North America and Europe,19, 20 representing 53 sequence types. Phenotype and whole genome sequences were 98·7% concordant (appendix; sensitivity 97·8%, specificity 99·5%); only one of 185 isolates predicted as resistant by whole genome sequences22, 23 lacked an elevated minimum inhibitory concentration (MIC). Conversely, only four of 202 isolates lacking resistance-associated substitutions22, 23 had raised MICs (16 mg/L). gyrA/B sequence therefore reliably predicts the fluoroquinolone resistance phenotype.
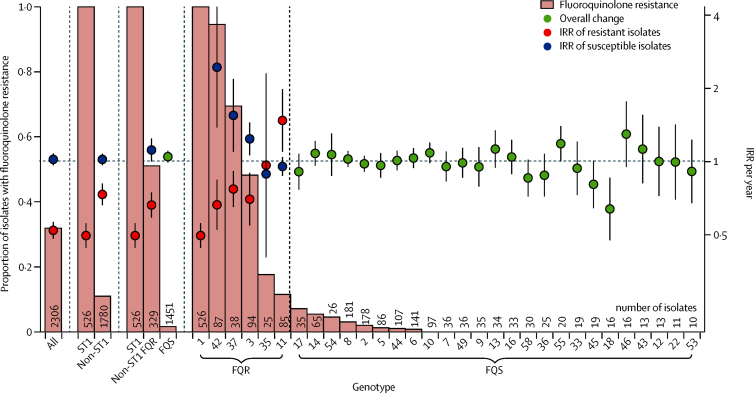
The decrease in Oxfordshire C difficile infections was solely due to a decline in C difficile infection caused by fluoroquinolone-resistant isolates, estimated at approximately 67% of all Oxfordshire C difficile infections in September, 2006, falling to approximately 3% by February, 2013 (annual incidence rate ratio [aIRR] 0·52, 95% CI 0·48–0·56, p<0·0001; figure 2B). Most (62%) fluoroquinolone-resistant isolates were from genotype ST1(027), but the decline persisted even when excluding ST1(027) and pooling the remaining fluoroquinolone-resistant isolates (aIRR 0·73, 0·66–0·81, p<0·0001 for all non-ST1; 0·66, 0·59–0·75, p<0·0001 for all non-ST1 with >10% resistant isolates; figure 3, appendix). Considering genotypes containing more than 10% resistant isolates separately, C difficile infection caused by fluoroquinolone-resistant isolates declined significantly for four genotypes from three distinct chromosomal backgrounds:26 clade 1 ST42(106) (p=0·00076), ST3(001) (p=0·0054); clade 2 ST1(027) (p<0·0001) and clade 4 ST37(017) (p=0·0027; figure 3, figure 4A, B, appendix).
Figure 3.
Oxfordshire Clostridium difficile IRR by fluoroquinolone resistance and genotype
For genotypes with more than 10% resistant isolates (denoted FQR), rates were calculated separately for C difficile infections caused by fluoroquinolone-susceptible and resistant isolates. To show that the difference in IRR for resistant and susceptible isolates is not driven solely by the decline in ST1(027), rates were also calculated for all non-ST1(027) genotypes together, as well as for all genotypes with more than 10% resistant isolates (excluding ST1(027)) and for all genotypes with 10% or less resistant isolates (FQS). Heterogeneity between IRRs in C difficile infections caused by fluoroquinolone-resistant versus fluoroquinolone-susceptible isolates: all p<0·0001, non-ST1 p<0·0001, non-ST1 FQR p<0·0001, ST42 p<0·0001, ST37 p=0·00015, ST3 p=0·00070, ST35 p=0·92, ST11 p=0·0053. The dotted vertical lines separate out the different ways the isolates were divided for the different analyses. The horizontal dotted line represents an IRR of 1. IRR=annual incidence rate ratio.
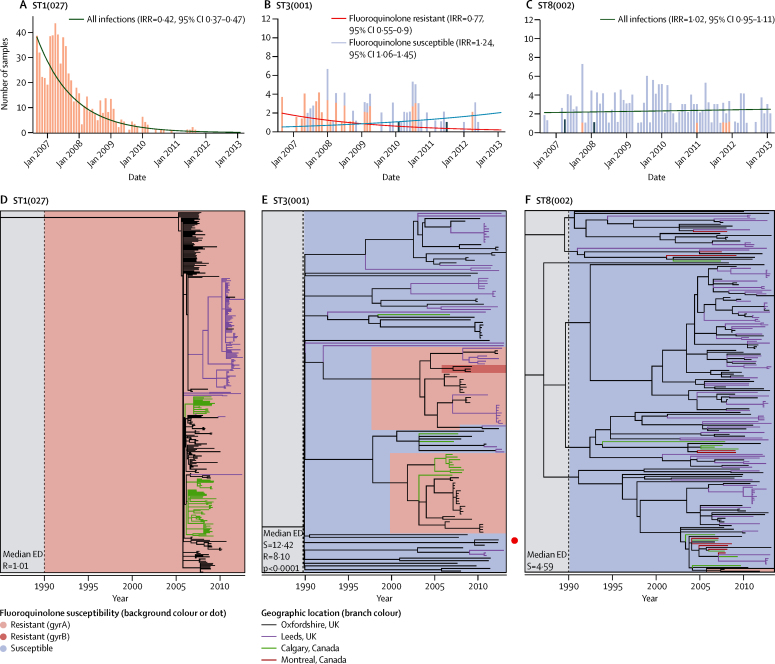
Figure 4.
Contrasting incidence of Clostridium difficile infections (Oxfordshire) and whole-genome sequence phylogenies representing the fluoroquinolone-resistant genotype ST1(027), the mixed resistant and susceptible genotype ST3(001), and the almost entirely fluoroquinolone-susceptible genotype ST8(002)
(A) Incidence of C difficile infections by fluoroquinolone susceptibility for genotype ST1(027) in Oxfordshire. Red bars indicate fluoroquinolone-resistant isolates, blue bars indicate fluoroquinolone-susceptible isolates, grey bars indicate resistance not determined. (B) Incidence of C difficile infections by fluoroquinolone susceptibility for genotype ST3(001) in Oxfordshire. (C) Incidence of C difficile infections by fluoroquinolone susceptibility for genotype ST8(002) in Oxfordshire. (D) Time-scaled phylogeny for ST1(027) generated with ClonalFrameML.24 Every third Oxfordshire isolate (by date) is shown. Phylogenies were scaled to be directly similar post-1990; the grey shaded regions before 1990 represent the regions of the phylogenies that should not be compared because they are not scaled identically. Background colour indicates fluoroquinolone susceptibility; branch colour indicates geographic location. (E) Time-scaled phylogeny for the mixed fluoroquinolone resistant or susceptible genotype, ST3(001), generated using ClonalFrameML.24 Two fluoroquinolone-resistant areas of the phylogeny are indicated by red shading within the blue susceptible region. Rapid clonal expansion after resistance emergence is supported by significantly lower ED scores for resistant versus susceptible areas. (F) Time-scaled phylogeny for ST8(002) generated using ClonalFrameML.24 Every second Oxfordshire isolate (by date) is shown. Two fluoroquinolone-resistant isolates are indicated at the bottom of the panel. IRR=annual incidence rate ratio. ED=evolutionary distinctiveness.25 R=fluoroquinolone resistant. S=fluoroquinolone susceptible.
The incidence of C difficile infection caused by fluoroquinolone-susceptible isolates remained unchanged (aIRR 1·02, 95% CI 0·97–1·08, p=0·45; figure 2B; heterogeneity p<0·0001 vs fluoroquinolone-resistant), and actually increased in three of the five genotypes with more than 10% but less than 99% resistant isolates (figure 3, figure 4B, appendix). More limited data for Leeds, representing a geographically independent region, were broadly similar (aIRR0·55, 0·49–0·61, p<0·0001 pooling predominantly fluoroquinolone-resistant ribotypes versus 1·03, 1·01–1·05, p=0·0031 pooling fluoroquinolone-susceptible ribotypes; appendix), as were national ribotyping data,27 supporting generalisability.
19 phylogenies were constructed representing the 22 most common C difficile genotypes in Oxfordshire and Leeds (figure 4D–F, appendix). The phylogeny of each genotype containing more than 10% fluoroquinolone-resistant isolates (figure 4D, E, appendix) showed rapid, geographically structured clonal expansions associated with resistance. This observation was reproduced internationally in parts of the phylogenies representing Calgary, Canada (figure 4D, E) and in isolates from three cities in northern Italy: Modena, Turin, and Arsizio (appendix). We recorded significantly lower ED scores for resistant versus susceptible areas of phylogenies containing both fluoroquinolone-resistant and fluoroquinolone-susceptible isolates (eg, ST3 p<0·0001, figure 4E; ST37 p<0·0001, appendix). By contrast, the phylogenies of genotypes consisting primarily of susceptible isolates (figure 4F, appendix) were geographically unstructured and had longer branches. This was also seen internationally in susceptible isolates from Calgary and Montreal, Canada (figure 4E, appendix). In fluoroquinolone-susceptible genotypes, the ED scores (and, by inference, transmission) did not differ significantly between Oxfordshire and Leeds clinical isolates (p>0·1; appendix).
Additional phylogenies for three prevalent fluoroquinolone-susceptible genotypes revealed similar branch lengths irrespective of sampling region size (appendix). Oxfordshire phylogenies (appendix), containing genomes from toxin EIA-positive and EIA-negative samples, plus genomes from healthy, asymptomatic, community infants, showed a lack of structure by source, even within a single region. ED scores were generally lower for clinical toxin EIA-positive genomes than for infant and EIA-negative genomes, especially in ST8(002) (p=0·0033) and ST2(014/020) (p=0·0014; appendix), consistent with greater transmission in the former.
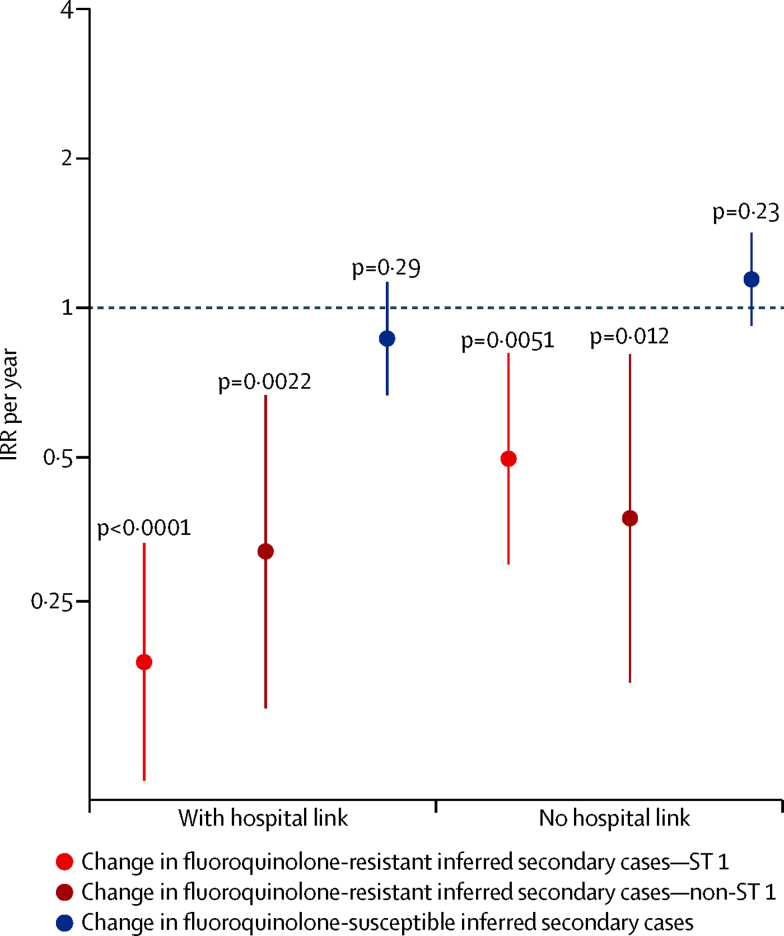
Fluoroquinolone restriction and multiple enhanced infection control measures were introduced simultaneously in England in 2007.17 Therefore, we investigated the hypothesis that infection control, not antimicrobial stewardship, reduced incidence of C difficile infection by reducing transmission (eg, that fluoroquinolone-resistant isolates were simply more prevalent in hospitals where infection control efforts were concentrated). Secondary spread (transmission) was inferred when subsequent infections had closely genetically related isolates. We estimated the Oxfordshire incidence of inferred secondary cases separately for fluoroquinolone-resistance versus fluoroquinolone-susceptibility, and also for infections where hospital-based contact occurred between primary and secondary cases.12 There was strong evidence for declines in secondary C difficile infections caused by fluoroquinolone-resistant isolates, both with hospital contact with a previous case (aIRR 0·21, 95% CI 0·13–0·34, p<0·0001) and without (0·45, 0·29–0·71, p<0·0001; figure 5). Declines occurred in secondary cases caused by fluoroquinolone-resistant isolates of ST1(027) and non-ST1(027) genotypes (p≤0·012, appendix). By contrast, there was no evidence of declines in secondary cases caused by fluoroquinolone-susceptible isolates, either with hospital contact with a previous infection (0·87, 0·67–1·13, p=0·29) or without (1·14, 0·92–1·42, p=0·23), supporting the importance of fluoroquinolone restriction over infection control interventions.
Figure 5.
Incidence trends of inferred secondary Clostridium difficile cases in Oxfordshire from April 2008 to March 2011
Inferred secondary cases are those caused by C difficile isolates that are genetically closely related (≤two single nucleotide variants) to isolates recovered from a previous case, and therefore potentially transmitted. Incidence trends were calculated separately for inferred secondary cases caused by fluoroquinolone-resistant ST1(027), fluoroquinolone-resistant non-ST1(027), and fluoroquinolone-susceptible isolates, stratified by with versus without hospital-based contact. Horizontal dotted line shows an IRR per year of 1 (ie, no change over time) against which the 95% CI bars are compared to determine statistical significance of any change. The p values are a test of the IRR against the null hypothesis of no change over time (IRR=1). IRR=annual incidence rate ratio.
Discussion
Our analysis of multiple whole genome sequence datasets shows that reductions in the incidence of C difficile infections caused by fluoroquinolone-resistant isolates (of multiple genotypes) plausibly has driven the decline in C difficile infections in Oxfordshire and Leeds, England, from 2007. Declines occurred alongside significant reductions in fluoroquinolone use in hospitals and the community. Extensive whole genome sequence phylogenies show that acquisition of fluoroquinolone resistance preceded the emergence of multiple, prevalent genotypes (figure 4, appendix); after fluoroquinolone prescribing was controlled, incidence declines were specific to C difficile infections caused by fluoroquinolone-resistant isolates of these same genotypes (figure 3, figure 4, appendix). By contrast, the incidence of C difficile infections from multiple fluoroquinolone-susceptible genotypes remained constant (figure 3, figure 4C, appendix), unaffected by changes in fluoroquinolone use or other national policy measures, such as restricted cephalosporin prescribing and enhanced infection control interventions, irrespective of genotype (figure 5, appendix).17 Crucially, there was no evidence of a decline in plausibly nosocomially transmitted secondary cases caused by fluoroquinolone-susceptible C difficile, which would be expected if improved infection control had made a major contribution to C difficile infection declines, whereas secondary cases caused by fluoroquinolone-resistant C difficile decreased markedly (figure 5, appendix).
The phylogenetically estimated date of fluoroquinolone resistance emergence preceded the clinical emergence of several problematic C difficile genotypes of different phylogenetic clades:26 ST1(027),9 ST42(106), ST3(001), and ST37(017) (figure 4, appendix).28, 29 The recent emergence of fluoroquinolone-resistant ST17(018) in Italy (appendix) also followed high fluoroquinolone use.30 Our greater sampling density9 revealed short-branched, geographically structured phylogenies of fluoroquinolone-resistant C difficile consistent with rapid spread within hospitals, and occasional transmission between them (figure 4D–F, appendix). Inclusion of international isolates allowed us to show generalisability of our findings outside of the UK. Although fluoroquinolone-susceptible, limited ST8(002) and ST2(014/020) transmission plausibly occurred, as indicated by small, short-branched clusters, and lower ED scores for clinical-toxin EIA-positive isolates than for infant/EIA-negative isolates (appendix). However, the absence of large-scale geographic structure in the long-branched phylogenies of all fluoroquinolone-susceptible genotypes (appendix) suggests that most were introduced independently into the clinical environment from alternative potential reservoirs.31, 32 Fluoroquinolone-susceptible C difficile might therefore represent a population lacking large-scale adaptation to antimicrobial selection pressures of clinical environments.
The decline in incidence of C difficile infection after national restriction of high-risk antimicrobials is consistent with previously successful small-scale interventions restricting high-risk antimicrobials as part of control packages.7, 10, 11 However, our study showed conclusively that Oxfordshire declines of C difficile infection were due to the parallel disappearance of fluoroquinolone-resistant isolates of multiple genotypes (figure 2, figure 3), suggesting that any selective advantage specific to resistant isolates might be lost when the antimicrobial is withdrawn. In England, additional antimicrobials were also targeted for restriction.17 However, only cephalosporin use also fell (figure 2A, appendix). Because all C difficile is inherently resistant to most cephalosporins,4 their restriction cannot explain the fluoroquinolone-susceptibility-specific declines in incidence observed. Similarly, if an ST1(027)-specific factor had led to its decline, there would be no reason for C difficile infection caused by fluoroquinolone-resistant isolates of several other genotypes—ST42(106), ST3(001), and ST37(017)—in two other C difficile clades (1 and 3)26 to decline concurrently (figure 3, figure 5). Although univariate cross-correlations between decline of C difficile infection and hospital-prescribed extended-spectrum penicillins (mostly amoxicillin alone) and beta-lactamase resistant penicillins (mostly flucloxacillin alone) were stronger than for fluoroquinolones in Oxfordshire, the use of many antibiotics in these groups actually rose because they were instead used in combinations, such as co-amoxiclav. Penicillins generally have a lesser risk of provoking C difficile infections than other classes of antibiotics,8, 33 and when taking community prescribing into account, (which forms a larger proportion of overall antimicrobial use than hospital prescribing) the correlation between these penicillin groups and incidence of C difficile infection in England disappears. Unfortunately, community prescribing data were not available for Oxfordshire for comparison. Finally, the much smaller population size meant that these univariate cross-correlations were estimated imprecisely compared with those for England as a whole. Our study therefore clarifies the issue of whether fluoroquinolone or cephalosporin restriction alone or in combination is key to C difficile infection control.34, 35, 36 However, changes in dominant genotypes over time have been reported in a single centre in the absence of antimicrobial restriction policies.37 ST1(027) outbreak control has also been achieved when total antimicrobial (not only fluoroquinolone) use was reduced,38 although this change could still predominantly reflect the effect of fluoroquinolones.
Similar to cephalosporin restriction, enhanced infection control measures17 such as isolation, contact precautions, and enhanced environmental cleaning do not target specific C difficile genotypes and should therefore reduce numbers of symptomatic patients infected with transmitted strains, irrespective of fluoroquinolone susceptibility. Analysis of closely related C difficile genomes from different patients (ie, representing possible transmissions12 potentially preventable by infection control measures) clearly showed that incidence only fell for secondary cases caused by fluoroquinolone-resistant C difficile, irrespective of hospital contact with a previous closely genetically related case, with no change in secondary cases caused by fluoroquinolone-susceptible isolates (figure 5, appendix). This finding is consistent with previous work38 finding no change in incidence of C difficile infection after infection control procedures were strengthened. This finding supports the greater importance of fluoroquinolone restriction in both hospitals and the community over enhanced infection control in recent reductions in English incidence of C difficile infection.
Antimicrobial stewardship targeted all patients in hospitals and the community,17 so clinically adapted resistant C difficile might conceivably have been eliminated from asymptomatic carriers and cases. If fluoroquinolone-resistant C difficile persisted in carriers, outbreak conditions should have returned rapidly once fluoroquinolone prescribing increased. This did not occur even after post-2010 increases in hospital fluoroquinolone prescribing in Oxford and Leeds (figure 2A, appendix). However, whereas before 2007 fluoroquinolones were prescribed widely, including in elderly people, increases after 2010 do not necessarily equate to increased exposure of patients with high risk of C difficile infection. Instead, these increases might reflect new, specific indications such as neutropenic prophylaxis (see appendix for Leeds; equivalent data not available in Oxford), consistent with observations that fluoroquinolone use is not a risk factor under non-outbreak conditions.39 The lack of national rise in fluoroquinolone-resistant C difficile infections also supports their almost complete eradication from both symptomatic patients and asymptomatic carriers in England, consistent with regional (Oxfordshire) findings that by late 2011, fluoroquinolone-resistant isolates of the commonest incidence genotype (ST1(027)) had disappeared from asymptomatic colonisation as well as infection.31
The genotypes ST1(027), ST42(106), ST3(001), and ST37(017), accounting for most fluoroquinolone-resistant isolates, represent three divergent C difficile clades,26 each with a genetically distinct, toxin-encoding pathogenicity locus.26 These genotypes could therefore differ in virulence or transmissibility due to varying gene content. ST1(027), for example, is almost four times likelier than other genotypes to cause symptomatic infection40 (although this could reflect its fluoroquinolone-resistant phenotype in settings with high fluoroquinolone prescribing). It seems unlikely that other gene content should be completely confounded with fluoroquinolone resistance, particularly within the large clade 126 (containing ST42(106), ST3(001), and Italian ST17(018)). However, even if additional virulence factors are associated with ST1(027), the overall diversity of outbreak-associated genetic backgrounds in which fluoroquinolone resistance is found suggests that this phenotype alone might be sufficient to confer outbreak potential.
A few sporadic fluoroquinolone-resistant isolates were identified in otherwise susceptible genotypes (appendix), suggesting that chance, combined with regional antibiotic prescribing policies, could trigger localised spread. ST11(078) was unusual, in that fluoroquinolone resistance occurred in 24 (13%) of 182 isolates, distributed throughout the phylogeny (appendix). ST11(078) can be transmitted zoonotically,32 and the unstructured pattern of fluoroquinolone resistance within this phylogeny could reflect the sporadic emergence of resistance either during agricultural fluoroquinolone use, or after human colonisation and antibiotic exposure.
The main study limitation was being primarily based in one, albeit large (population of approximately 600 000 people) region, where 7 years of individual-isolate whole genome sequences enabled us to predict fluoroquinolone susceptibility. Whole genome sequence data from Leeds were available for less than 3 years, precluding a similar analysis to figure 2 in another region. Different datasets from different sources were used for incidence of C difficile infections and antibiotic use because no one dataset was collected consistently across the entire period from a single source. Comparisons of incidence of C difficile infections and antibiotic use are ecological, and therefore prone to unmeasured confounding. English hospital-level antibiotic data are not available before 2013 (only subsequently),41 so we were unable to investigate associations between fluoroquinolone use and C difficile infections across Trusts in a broader ecological analysis. However, our key characteristics, fluoroquinolone susceptibility and genotype, were unknown when the C difficile infections occurred and were not part of the inclusion or exclusion criteria. Therefore, the phylogenetic analyses are representative of the genotypes circulating in the locations studied when sampled.
In summary, fluoroquinolone resistance occurs in several genetically divergent C difficile genotypes.26 The contrasting phylogenies of fluoroquinolone-resistant and fluoroquinolone-susceptible C difficile probably reflect increased potential for health-care-associated selection and epidemic spread of fluoroquinolone-resistant bacteria. Thus, the C difficile genotypes causing infections at any given time and location, and the relative importance of different transmission routes (nosocomial person-to-person versus multiple introductions) might be a direct consequence of antimicrobial prescribing policies. The multifaceted approach to C difficile infection control adopted by England successfully curtailed transmission. Whole genome sequence data suggest that fluoroquinolone restriction plausibly played the most important part in this success. Appropriate antimicrobial stewardship therefore is, and will likely remain, central to the control of C difficile infections.
Acknowledgments
Acknowledgments
This study was supported by the UK Clinical Research Collaboration (Wellcome Trust [grant 087646/Z/08/Z], Medical Research Council, National Institute for Health Research [NIHR grant G0800778]), NIHR Biomedical Research Centre, NIHR Oxford Health Protection Research Units on Healthcare Associated Infection and Antimicrobial Resistance at University of Oxford in partnership with Public Health England (grant HPRU-2012-10041), NIHR Health Protection Unit in Modelling at Imperial College London in partnership with Public Health England (grant HPRU-2012-10080), and the Health Innovation Challenge Fund (a parallel funding partnership between the Wellcome Trust [grant WT098615/Z/12/Z] and the Department of Health [grants WT098615 and HICF-T5-358]). The authors acknowledge the research collaboration of IMS Health, within the HPRU grant HPRU-2012-10041. The funders had no role in the writing of the manuscript or the decision to submit it for publication. DWC and TEAP are NIHR senior investigators. DJW is a Sir Henry Dale Fellow, jointly funded by the Wellcome Trust and the Royal Society (grant 101237/Z/13/Z). The authors acknowledge the contribution of the Modernising Medical Microbiology Informatics Group comprising Carlos Del Ojo Elias, Charles Crichton, Vasiliki Kostiou, and Adam Giess (Nuffield Department of Clinical Medicine, University of Oxford, UK), and Jim Davies, (Department of Computer Science, University of Oxford, UK). This report presents independent research funded by the NlHR, Wellcome Trust, and the Department of Health. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the NIHR, Wellcome Trust, the Department of Health, or Public Health England.
Contributors
MHW, TEAP, ASW, and DWC contributed equally to the work. KED, XD, and TPQ contributed equally to the work. KED, XD, TPQ, MHW, TEAP, ASW, and DWC designed the study with input from DWE, NS, TG, RMH, and DJW. KED, DWE, NS, DG, AV, SJO, WNF, JF, KM, and JM collected specimens from Oxfordshire and Leeds, cultured C difficile, and extracted chromosomal C difficile DNA for whole genome sequences. SG, EJCG, and DMC contributed the fidaxomicin clinical trial isolate collection. EJCG and DMC did fluoroquinolone susceptibility testing. The Modernising Microbiology Informatics Group, JMF, TG, and DHW optimised or did the assembly of short DNA sequence reads. KED derived genotype data from whole genome sequences and identified genomes for the construction of ClonalFrameML dual-scaled phylogenies. All phylogenies were done by XD. KED combined phylogenetic and fluoroquinolone resistance genotype data. PH, SH, MHW, and DWC obtained antimicrobial prescribing data. RH and APJ obtained national incidence data for C difficile infection. TPQ, ASW, and TEAP did the biostatistical analysis of Oxfordshire and national incidence and antimicrobial prescribing data. WNF, TPQ, ASW, and MHW did the biostatistical analysis of Leeds incidence and antimicrobial prescribing data. DWE, TPQ, ASW, and TEAP did the inferred secondary cases analysis. KED, XD, TPQ, MHW, TEAP, ASW, and DWC wrote the first draft of the article and all authors contributed to and had final approval of the Article.
Declaration of interests
Relevant to the submitted work, MHW has received both grants and personal fees from Actelion, Cubist, Astellas, Merck, Sanofi-Pasteur, Summit, Biomerieux, and Qiagen; personal frees only from Optimer and Synthetic Biologics; and grants, personal fees, and other funding were received from Alere (the latter including consulting fees, research funding, and a grant to department). Outside the submitted work, MHW received grants and personal fees from Cerexa, Abbott, Da Volterra, and European Tissue Symposium; and personal fees only from AstraZeneca, Durata, Nabriva, Pfizer, Roche, The Medicines Company, VH Squared, Basilea, Bayer, MotifBio, and Paratek. EJCG reports the following relationships: 2016 advisory boards for Merck & Co, Bayer Pharmaceuticals, BioK+, Sanofi-Adventis, Summit Corp PLC, Kindred Healthcare Corp, Novartis, Sankyo-Daichi, Rempex; speakers' bureau for Bayer Inc and Merck & Co; and research grants from Merck & Co, Theravance Inc. Pfizer Inc, Astellas Inc, Cerexa, Forrest Pharmaceuticals, Impex Pharmaceuticals, Novartis, Clinical Microbiology Institute, Genzyme, Nanopacific Holdings Inc, Romark Laboratories LC, Viroxis Corp, Warner Chilcott, Avidbiotics Corp, GLSynthesis Inc, Immunome Inc, Toltec Pharma LLC, Salix, Summit Corp PLC, GlaxoSmithKline, Rempex Pharmaceuticals, Symbiomix Therapeutics, Toltec Pharmaceuticals LLC, Amicrobe Inc, Durata, Gynuity Health Projects, and Medicines Company. SH is affiliated with the National Institute for Health Research Health Protection Research Units (NIHR HPRU) in Healthcare Associated Infection and Antimicrobial Resistance at Imperial College London in partnership with Public Health England and the NIHR HPRU in Healthcare Associated Infection and Antimicrobial Resistance at University of Oxford in partnership with Public Health England. The views expressed are those of the author and not necessarily those of the NHS, the NIHR, the Department of Health, or Public Health England. JF reports grants from Astellas Pharma Europe, Melinta Therapeutics, and Morphochem AG, outside the submitted work. PH has received speaker's fees from Astellas, advisory board fee from Merck Sharp & Dohme, conference and travel fees from Eumedica, and speaker's fees from Gilead, outside the submitted work. SG reports previous employment with Optimer Pharmaceticals and Cubist, as well as several patents with Optimer Pharmaceticals (mostly expired, no income). ASW reports grants from Wellcome Trust, grants from National Institutes of Health UK, grants from Medical Research Council UK, grants from Department of Health UK, during the conduct of the study. The other authors declare no competing interests.
Contributor Information
Kate E Dingle, Email: kate.dingle@ndcls.ox.ac.uk.
Modernising Medical Microbiology Informatics Group:
Carlos Del Ojo Elias, Charles Crichton, Vasiliki Kostiou, Adam Giess, and Jim Davies
Supplementary Material
References
- 1.Davies KA, Longshaw CM, Davis GL. Underdiagnosis of Clostridium difficile across Europe: the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID) Lancet Infect Dis. 2014;14:1208–1219. doi: 10.1016/S1473-3099(14)70991-0. [DOI] [PubMed] [Google Scholar]
- 2.Lessa FC, Mu Y, Bamberg WM. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Owens RC, Jr, Donskey CJ, Gaynes RP, LooVG, Muto CA. Antimicrobial-associated risk factors for Clostridium difficile infection. Clin Infect Dis. 2008;46:S19–S31. doi: 10.1086/521859. [DOI] [PubMed] [Google Scholar]
- 4.Shuttleworth R, Taylor M, Jones DM. Antimicrobial susceptibilities of Clostridium difficile. J Clin Pathol. 1980;33:1002–1005. doi: 10.1136/jcp.33.10.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spigaglia P. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther Adv Infect Dis. 2016;3:23–42. doi: 10.1177/2049936115622891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson S, Samore MH, Farrow KA. Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals. N Engl J Med. 1999;341:1645–1651. doi: 10.1056/NEJM199911253412203. [DOI] [PubMed] [Google Scholar]
- 7.Pear SM, Williamson TH, Bettin KM, Gerding DN, Galgiani JN. Decrease in nosocomial Clostridium difficile-associated diarrhea by restricting clindamycin use. Ann Intern Med. 1994;120:272–277. doi: 10.7326/0003-4819-120-4-199402150-00003. [DOI] [PubMed] [Google Scholar]
- 8.Loo VG, Poirier L, Miller MA. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med. 2005;353:2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 9.He M, Miyajima F, Roberts P. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kallen AJ, Thompson A, Ristaino P. Complete restriction of fluoroquinolone use to control an outbreak of Clostridium difficile infection at a community hospital. Infect Control Hosp Epidemiol. 2009;30:264–272. doi: 10.1086/595694. [DOI] [PubMed] [Google Scholar]
- 11.Muto CA, Blank MK, Marsh JW. Control of an outbreak of infection with the hypervirulent Clostridium difficile BI strain in a university hospital using a comprehensive “bundle” approach. Clin Infect Dis. 2007;45:1266–1273. doi: 10.1086/522654. [DOI] [PubMed] [Google Scholar]
- 12.Eyre DW, Cule ML, Wilson DJ. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med. 2013;369:1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin J, Eyre DW, Fawley WN, Walker AS, Crook DW, Wilcox MH. 2016 C. difficile (CD) ribotypes exhibit variable patient-to-patient transmission rates, as determined by whole-genome sequencing (WGS), suggesting differing reservoirs and modes of acquisition. ECCMID; Amsterdam; April 9–12, 2016. Abstract O557.
- 14.Kumar N, Miyajima F, He M. Genome-based infection tracking reveals dynamics of Clostridium difficile transmission and disease recurrence. Clin Infect Dis. 2016;62:746–752. doi: 10.1093/cid/civ1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curry SR, Muto CA, Schlackman JL. Use of multilocus variable number of tandem repeats analysis genotyping to determine the role of asymptomatic carriers in Clostridium difficile transmission. Clin Infect Dis. 2013;57:1094–1102. doi: 10.1093/cid/cit475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilcox MH, Shetty N, Fawley WN. Changing epidemiology of Clostridium difficile infection following the introduction of a national ribotyping-based surveillance scheme in England. Clin Infect Dis. 2013;55:1056–1063. doi: 10.1093/cid/cis614. [DOI] [PubMed] [Google Scholar]
- 17.Public Health England and the Department of Health Clostridium difficile infection: How to deal with the problem. http://www.hpa.org.uk/webc/HPAwebFile/HPAweb_C/1232006607827 (accessed Feb 16, 2016).
- 18.Department of Health. NHS Saving Lives: reducing infection, delivering clean and safe care, antimicrobial prescribing, a summary of best practice. 2007. http://webarchive.nationalarchives.gov.uk/20130107105354/http://www.dh.gov.uk/prod_consum_dh/groups/dh_digitalassets/@dh/@en/documents/digitalasset/dh_078117.pdf (accessed Feb 16, 2016).
- 19.Cornely OA, Crook DW, Esposito R. Fidaxomicin versus vancomycin for infection with Clostridium difficile in Europe, Canada, and the USA: a double-blind, non-inferiority, randomised controlled trial. Lancet Infect Dis. 2012;12:281–289. doi: 10.1016/S1473-3099(11)70374-7. [DOI] [PubMed] [Google Scholar]
- 20.Louie TJ, Miller MA, Mullane KM. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med. 2011;364:422–431. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 21.Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drudy D, Quinn T, O'Mahony R, Kyne L, O'Gaora P, Fanning S. High-level resistance to moxifloxacin and gatifloxacin associated with a novel mutation in gyrB in toxin-A-negative, toxin-B-positive Clostridium difficile. J Antimicrob Chemother. 2006;58:1264–1267. doi: 10.1093/jac/dkl398. [DOI] [PubMed] [Google Scholar]
- 23.Spigaglia P, Barbanti F, Mastrantonio P. Fluoroquinolone resistance in Clostridium difficile isolates from a prospective study of C. difficile infections in Europe. J Med Microbiol. 2008;57:784–789. doi: 10.1099/jmm.0.47738-0. [DOI] [PubMed] [Google Scholar]
- 24.Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol. 2015;11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isaac NJB, Turvey ST, Collen B, Waterman C, Baillie JEM. Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS One. 2007;2:e296. doi: 10.1371/journal.pone.0000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dingle KE, Elliott B, Robinson E. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evol. 2014;6:36–52. doi: 10.1093/gbe/evt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Public Health England Clostridium difficile Ribotyping Network (CDRN) for England and Northern Ireland, 2011–2013 Report. https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/329156/C_difficile_ribotyping_network_CDRN_report.pdf (accessed Feb 1, 2016).
- 28.Drudy D, Fanning S, Kyne L. Toxin A-negative, toxin B-positive Clostridium difficile. Int J Infect Dis. 2007;11:5–10. doi: 10.1016/j.ijid.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Borgmann S, Kist M, Jakobiak T. Increased number of Clostridium difficile infections and prevalence of Clostridium difficile PCR ribotype 001 in southern Germany. Euro Surveill. 2008;13:19057. [PubMed] [Google Scholar]
- 30.Spigaglia P, Barbanti F, Dionisi AM, Mastrantonio P. Clostridium difficile isolates resistant to fluoroquinolones in Italy: emergence of PCR ribotype 018. J Clin Microbiol. 2010;48:2892–2896. doi: 10.1128/JCM.02482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eyre DW, Griffiths D, Vaughan A. Asymptomatic Clostridium difficile colonisation and onward transmission. PLoS One. 2013;12:e78445. doi: 10.1371/journal.pone.0078445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bauer MP, Kuijper EJ. Potential sources of Clostridium difficile in human infection. Infect Dis Clin North Am. 2015;29:29–35. doi: 10.1016/j.idc.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Brown KA, Khanafer N, Daneman N, Fisman DM. Meta-analysis of antibiotics and the risk of community-associated Clostridium difficile infection. Antimicrob Agents Chemother. 2013;57:2326–2332. doi: 10.1128/AAC.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talpaert MJ, Rao GG, Cooper BS, Wade P. Impact of guidelines and enhanced antibiotic stewardship on reducing broad-spectrum antibiotic usage and its effect on incidence of Clostridium difficile infection. J Antimicrob Chemother. 2011;66:2168–2174. doi: 10.1093/jac/dkr253. [DOI] [PubMed] [Google Scholar]
- 35.Sarma JB, Marshall B, Cleeve V, Tate D, Oswald T, Woolfrey S. Effects of fluoroquinolone restriction (from 2007 to 2012) on Clostridium difficile infections: interrupted time-series analysis. J Hosp Infect. 2015;91:74–80. doi: 10.1016/j.jhin.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 36.Feazel LM, Malhotra A, Perencevich EN, Kaboli P, Diekema DJ, Schweizer ML. Effect of antibiotic stewardship programmes on Clostridium difficile incidence: a systematic review and meta-analysis. J Antimicrob Chemother. 2014;69:1748–1754. doi: 10.1093/jac/dku046. [DOI] [PubMed] [Google Scholar]
- 37.Belmares J, Johnson S, Parada JP. Molecular epidemiology of Clostridium difficile over the course of 10 years in a tertiary care hospital. Clin Infect Dis. 2009;49:1141–1147. doi: 10.1086/605638. [DOI] [PubMed] [Google Scholar]
- 38.Valiquette L, Cossette B, Garant MP, Diab H, Pépin J. Impact of a reduction in the use of high-risk antibiotics on the course of an epidemic of Clostridium difficile-associated disease caused by the hypervirulent NAP1/027 strain. Clin Infect Dis. 2007;45(suppl 2):S112–S121. doi: 10.1086/519258. [DOI] [PubMed] [Google Scholar]
- 39.Hensgens MP, Goorhuis A, van Kinschot CM, Crobach MJ, Harmanus C, Kuijper EJ. Clostridium difficile infection in an endemic setting in the Netherlands. Eur J Clin Microbiol Infect Dis. 2011;30:587–593. doi: 10.1007/s10096-010-1127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loo VG, Bourgault AM, Poirier L. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med. 2011;365:1693–1703. doi: 10.1056/NEJMoa1012413. [DOI] [PubMed] [Google Scholar]
- 41.English surveillance programme for antimicrobial utilisation and resistance (ESPAUR) 2010 to 2014, Report 2015. https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/477962/ESPAUR_Report_2015.pdf (accessed Feb 16, 2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.