Abstract
Rapid detection of food-borne pathogens is important in the food industry, to monitor and prevent the spread of these pathogens through contaminated food products. We therefore established a multiplex real-time loop-mediated isothermal amplification (LAMP) assay to simultaneously detect and distinguish Salmonella spp. and Vibrio parahaemolyticus DNA in a single reaction. Two target sequences, one specific for Salmonella and the other specific for Vibrio parahaemolyticus, were amplified by specific LAMP primers in the same reaction tube. After amplification at 65 °C for 60 min, the amplified products were subjected to melting curve analysis and thus could be distinguished based on the different melting temperatures (Tm values) of the two specifically amplified products. The specificity of the multiplex LAMP assay was evaluated using 19 known bacterial strains, including one V. parahaemolyticus and seven Salmonella spp. strains. The multiplex LAMP showed 100% inclusivity and exclusivity, and a detection limit similar to that of multiplex PCR. In addition, we observed and corrected preferential amplification induced by what we call LAMP selection in the multiplex LAMP reaction. In conclusion, our assay was rapid, specific, and quantitative, making it a useful tool for the food industry.
Food-borne diseases, caused mainly by food-borne pathogens, have become a major public health issue in both developed and developing countries. It is estimated that 76 million people fall ill and 5,000 die annually from food-borne illness, and a large proportion of these cases can be attributed to Salmonella spp. and Vibrio parahaemolyticus1. In the active etiological surveillance for foodborne diseases in Guangdong, China, during 2013 and 2014, the detection rate of Salmonella was highest, followed by that of V. parahaemolyticus2. Salmonella spp. has been identified as the most frequent cause of food-borne infection outbreaks in many countries, and V. parahaemolyticus has emerged as a vital food-borne pathogen worldwide due to consumption of raw or undercooked seafood3,4. Thus, rapid and sensitive detection of Salmonella spp. and V. parahaemolyticus is required for prevention and timely treatment.
Various methods have been developed to detect Salmonella spp. and V. parahaemolyticus, including convention culture-based, immunology-based, and molecular methods. Conventional culture-based methods, conducted by selective isolation of bacteria and biochemical identification, are safe but laborious and time-consuming. Immunology-based methods shorten the detection time but are not very effective in terms of detection sensitivity5,6. Molecular methods such as PCR-based approaches and DNA micro-array, which have been used in the detection of numerous food-borne pathogens, including Salmonella and V. parahaemolyticus, are labor-saving, sensitive, and specific7,8, but the need for expensive instruments and trained personnel prevent them from being widely used.
Loop-mediated isothermal amplification (LAMP), invented in 2000 by Notomi et al., is a novel nucleic acid amplification method with high sensitivity, specificity, and rapidity for the low-cost detection of pathogens9. With 4~6 specifically designed primers that target 6~8 distinct regions of the target gene, a large amount of DNA can be synthesized under a constant temperature in less than 60 min, and the amplified products can be detected by the naked eye. As a result, the LAMP assay has been widely applied in the detection of pathogenic bacteria, viruses, and parasites10,11,12,13. However, restriction enzyme analysis of amplified products and probe-based methods, which has been attempted, is laborious and complex14,15,16. Furthermore, the LAMP assay can only detect a gene in a single reaction, and multiplex LAMP assays that can detect and discriminate two or more target genes in a single reaction are limited. In this study, a multiplex LAMP assay was developed to simultaneously detect Salmonella spp. and V. parahaemolyticus based on different melting temperatures determined by melting curve analysis of amplified products.
Results
Optimization of primer concentration in the multiplex LAMP
The simultaneous amplification of Salmonella and V. parahaemolyticus DNA targets was evaluated. Figure 1A shows the results when equal concentrations of Salmonella and V. parahaemolyticus primers were used. For S. typhimurium DNA, the mean Cq and Tm values were 13.96 min and 86.25 ± 0.06 °C, respectively (Fig. 1A, well 1), whereas for V. parahaemolyticus DNA, the mean Cq and Tm values were 12.68 min and 84.05 ± 0.06 °C, respectively (Fig. 1A, well 2). For a mixture of S. typhimurium and V. parahaemolyticus DNAs, the mean Cq value was 12.14 min and the mean Tm values for S. typhimurium and V. parahaemolyticus were 85.63 ± 0.15 °C and 82.88 ± 0.21 °C, respectively (Fig. 1A, well 3). No amplification occurred in the negative controls. These results indicate that the multiplex LAMP reaction with two sets of primers for S. typhimurium and V. parahaemolyticus successfully amplified the two target genes, and the amplified products showed different Tm values (86.25 ± 0.06 °C and 84.05 ± 0.06 °C). In addition, the simultaneous amplification of the target genes from a mixture of S. typhimurium and V. parahaemolyticus DNAs resulted in two detection peaks, which could be differentiated by their different Tm values (85.63 ± 0.15 °C and 82.88 ± 0.21 °C). However, the fluorescence produced by V. parahaemolyticus DNA amplification was clearly weaker than that produced by S. typhimurium DNA amplification, suggesting that there was preferential amplification of the S. typhimurium target sequence over the V. parahaemolyticus target sequence in the multiplex LAMP reaction. Thus, an adjustment of the relative concentration of the primers was required. The concentrations of S. typhimurium primers were reduced to half (1.3 μM) and concentrations of V. parahaemolyticus primers were kept unchanged, enabling similar amplification efficiency and more intuitive identification of the two target sequences (Fig. 1B). Therefore, these reaction concentrations (1.3 μM for S. typhimurium primers and 2.6 μM for V. parahaemolyticus primers) were selected for subsequent analysis.
Figure 1. Effect of primer concentration on the discrimination of Salmonella typhimurium and Vibrio parahaemolyticus target amplification by different Tm values generated by melting curve analysis of the multiplex LAMP assay.

(A) Equal concentrations of Salmonella and V. parahaemolyticus primers. (B) Halved concentrations of Salmonella primers. The melting curve shows temperature on the X-axis and fluorescence on the Y-axis. Well 1 (purple), S. typhimurium; well 2 (red), V. parahaemolyticus; well 3 (yellow), S. typhimurium and V. parahaemolyticus; well 4 (green), negative control.
Specificity of the multiplex LAMP assay
To evaluate the specificity of the multiplex LAMP assay, all 19 bacterial strains were subjected to the multiplex LAMP reaction. The assay successfully detected Salmonella spp. and V. parahaemolyticus. The Tm value for V. parahaemolyticus was 83.8 °C and the Tm values for seven Salmonella spp. consistently fell between 86.5 °C and 86.8 °C, with an average of 86.61 ± 0.11 °C. For non-Salmonella spp. and non-V. parahaemolyticus strains, no Tm value was obtained, suggesting that no amplification occurred. We conclude that the two sets of primers are highly specific for the detection of Salmonella spp. and V. parahaemolyticus (Table 1).
Table 1. Bacterial strains used in this study along with the specificity of the multiplex LAMP assay for the detection of Salmonella spp. and Vibrio parahaemolyticus.
| No. | Bacterial species | Strain serial | Multiplex LAMP |
|---|---|---|---|
| 1 | Vibrio parahaemolyticus | ATCC17802 | + |
| 2 | Salmonella typhimurium | CICC21483 | + |
| 3 | Salmonella typhimurium | ATCC14208 | + |
| 4 | Salmonella enteritidis | 1655 | + |
| 5 | Salmonella enteritidis | CMCC(B)50335 | + |
| 6 | Salmonella choleraesuis | SH1055 | + |
| 7 | Salmonella aberdeen | SF080 | + |
| 8 | Salmonella senftenberg | 2638 | + |
| 9 | Listeria monocytogenes | ATCC19115 | − |
| 10 | Shigella flexneri | 4536 | − |
| 11 | Shigella sonnei | 2531 | − |
| 12 | Shigella dysenteriae | CMCC(B)51105 | − |
| 13 | Staphylococcus aureus | CMCC(B)26003 | − |
| 14 | Escherichia coli | ATCC25992 | − |
| 15 | Escherichia coli O157 | NCTC12900 | − |
| 16 | Stenotrophomonas maltophilia | K279a | − |
| 17 | Proteus vulgaris | CMCC49027 | − |
| 18 | Vibrio cholera | 3802 | − |
| 19 | Yersinia enterocolitica | 027 | − |
Sensitivity of the multiplex LAMP assay
To estimate the detection limit of the multiplex LAMP assay, 10-fold serial dilutions of S. typhimurium CICC 21483 and V. parahaemolyticus ATCC 17802 DNA templates with concentrations ranging from 100 ng/μL to 0.1 pg/μL were used in the amplification reaction. The assay sensitivity for both the simultaneous detection of S. typhimurium and V. parahaemolyticus and the multiplex PCR was found to be 10 pg/μL (Fig. 2).
Figure 2. Sensitivity of the multiplex LAMP compared with multiplex PCR for the simultaneous detection of Salmonella typhimurium and Vibrio parahaemolyticus.
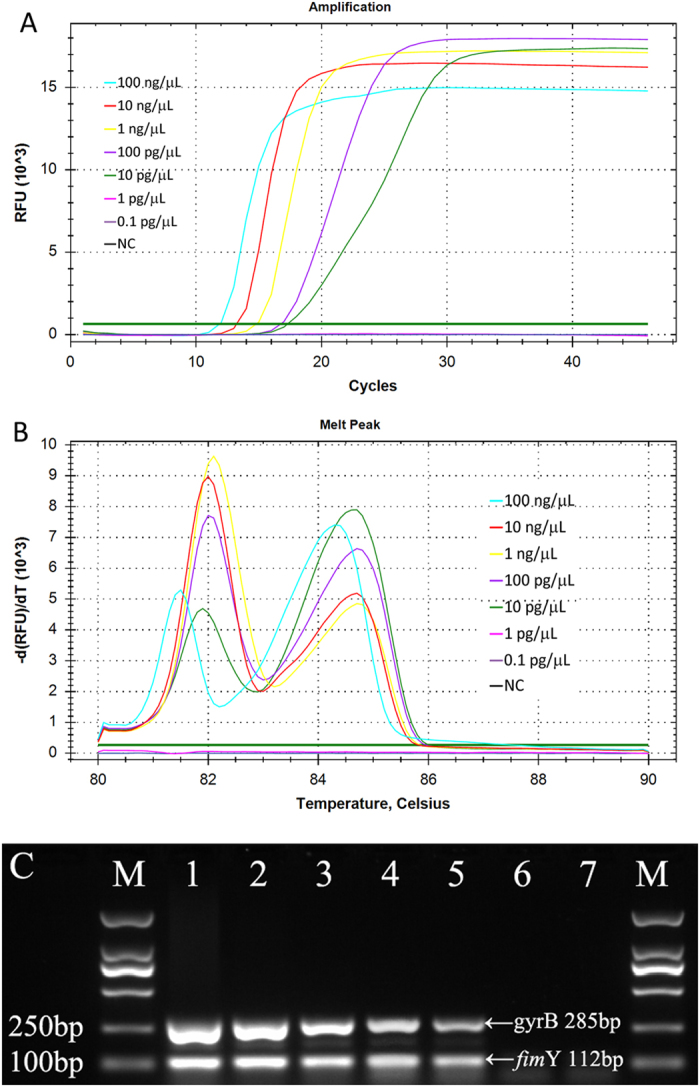
10-fold serial dilutions of S. typhimurium (CICC 21483) and V. parahaemolyticus (ATCC 17802) DNA templates from 100 ng/μL to 0.1 pg/μL were tested. (A) The amplification curve generated from 100 ng/μL to 10 pg/μL shows time on the X-axis and fluorescence on the Y-axis. (B) Two melting peaks (84.62 ± 0.18 °C for S. typhimurium and 81.90 ± 0.23 °C for V. parahaemolyticus) distinguishing S. typhimurium and V. parahaemolyticus amplification products were generated by melting curve analysis. (C) Sensitivity of the multiplex PCR for simultaneous detection of S. typhimurium and V. Parahaemolyticus. Lanes 1–7: S. typhimurium and V. parahaemolyticus DNA concentrations of 100 ng/μL, 10 ng/μL, 1 ng/μL, 100 pg/μL, 10 pg/μL, 1 pg/μL and negative control.
Quantitative capability of the multiplex LAMP assay
The quantitative capability of the multiplex LAMP assay was also evaluated. Figure 3 shows the standard curve generated when simultaneously detecting S. typhimurium and V. parahaemolyticus in three independent replicates. In the multiplex LAMP assay, the mean Cq values decreased linearly with increasing target concentrations, and the quantification equation for the assay was validated to be Y = −3.688x + 32.734, with a correlation coefficient (R2) of 0.9314. Moreover, comparison of Cq values for each concentration in all three independent experiments showed minimal variation, indicating that the multiplex LAMP amplification is highly reproducible.
Figure 3. Standard curve generated when simultaneously detecting Salmonella typhimurium and Vibrio parahaemolyticus.
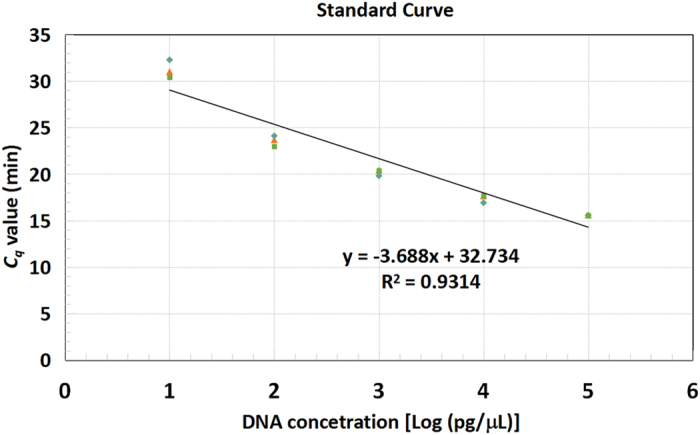
DNA templates with concentrations ranging from 100 ng/μL to 10 pg/μL were tested in triplicate. The standard curve shows DNA concentrations in log scale on the X -axis and Cq values (min) on the Y -axis.
Discussion
In this report, a simple and rapid protocol for simultaneous detection of Salmonella spp. and V. parahaemolyticus based on the multiplex real-time LAMP technique was developed. This protocol incorporates the melting curve analysis, which allows the detection and discrimination of amplified products from a mixture by their different Tm values within 60 min. In addition, the phenomenon of amplification bias, defined as uneven or preferential amplification of one target gene over another in a multiplex reaction, which was induced by what we call LAMP selection and can be balanced by primer concentration adjustments, was also observed in the present study.
Considerable time and effort can be saved by simultaneously amplifying and detecting two or more target sequences in a single reaction. However, multiplex LAMP in which multiple sequences can be amplified is limited owing to the difficulty of distinguishing the origin or specificity of the amplified products from the mixture. Several methods have been tried to resolve this difficulty, such as species-specific identification after restriction enzyme digestion14,15, visual detection by addition of fluorescence-labeled probes or primers coupled with cationic polymers16,17,18, colorimetric distinction using an immunochromatographic strip19, and melting curve analysis of amplified products20. The last approach was chosen in this study because of its simplicity, rapidity, and specificity. The detection assay takes advantage of the different Tm values of target sequences, which are relatively stable and determined by melting curve analysis. Unlike PCR, the Tm values of LAMP products are closely related to GC content rather than the length of target sequences. As shown in Fig. 1, two target sequences were simultaneously amplified with mean Tm values for S. typhimurium and V. parahaemolyticus in mixed products of 85.63 ± 0.15 °C and 82.88 ± 0.21 °C, respectively, and were thus clearly distinguished.
The presence of two or more primer pairs in the multiplex LAMP increases the chance of obtaining false positive amplification, primarily because of 4~6 primers contained in a single LAMP reaction, which is another factor limiting the development of multiplex LAMP. Although there is no theoretical limit to the number of primers to be used in a multiplex LAMP reaction, spurious-prone amplification among primers limits the establishment of specific reactions. A multiplex LAMP assay combining 21 primers targeting four Candida species has been reported21. The high specificity of primers used in this study has been reported and validated previously10,12, and there was no spurious amplification products in our multiplex LAMP reaction, which showed 100% inclusivity and 100% exclusivity for the detection of 19 bacterial strains including Salmonella spp. and V. parahaemolyticus. The detection limit for simultaneous detection of S. typhimurium and V. parahaemolyticus was found to be 10 pg/μL, which was identical to that of LAMP amplification only for S. typhimurium or V. parahaemolyticus. Additionally, the sensitivity of multiplex LAMP is similar to that of multiplex PCR. The feasibility of the multiplex LAMP was evaluated and validated by the artificially inoculating Salmonella and V. parahaemolyticus into pasteurizad milk sample. The result showed that the assay could detect and distinguish Salmonella and V. parahaemolyticus contamination of food samples. Further work to optimize the detection of these two pathogens from clinical samples will also be important.
The quantitative capability of LAMP has been examined previously12,22. In the present study, a standard curve was generated when detecting S. typhimurium and V. parahaemolyticus DNAs. In our multiplex LAMP assay, the Cq values decreased linearly with increasing initial concentrations of template DNAs from 10 pg/μL to 100 ng/μL. The standard curve had a linear relationship with R2 value of 0.9314, indicating the quantitative capability of the multiplex LAMP assay.
Ideally, all target sequences in a multiplex detection assay should have similar amplification efficiencies so that greater accuracy of species identification can be obtained. To our knowledge, very few reports have focused on the amplification efficiencies in the multiple amplification system. Amplification bias of one target gene over another was observed in this paper when equimolar primer concentrations were used. The results showed the preferential amplification of the Salmonella target sequence over that of V. parahaemolyticus, which was induced by what we call LAMP selection23. Therefore, adjustment of primer concentration in the multiplex reaction was required, and we found that halving the concentrations of the Salmonella primers led to equal amplification efficiency of both target sequences. Generally, optimal concentration of the primers in a multiplex detection reaction may vary greatly between targets and is established empirically24.
For the past few years, diseases caused by food-borne pathogens have become a significant public health issue globally1. As a leading cause of food-borne disease, Salmonella spp. and V. parahaemolyticus contamination of food products has become a vital concern for food safety. The multiplex real-time LAMP assay described here can simultaneously detect and distinguish Salmonella spp. and V. parahaemolyticus based on their different melting temperatures. We also observed preferential amplification in the multiplex LAMP reaction, which was balanced by primer optimization. The assay showed sensitivity similar to that of PCR and was rapid, specific, and quantitative, making it a useful tool for the food industry.
Materials and Methods
Bacterial strains, culture conditions, and DNA preparation
In total, 19 known bacterial strains, including a V. parahaemolyticus reference strain (ATCC 17802) and seven Salmonella sp. strains, were analyzed in this study (Table 1). All strains were stored in 10% (w/v) glycerol broth at −70 °C. Salmonella typhimurium CICC 21483 and V. parahaemolyticus ATCC 17802 strains were used for primer optimization and sensitivity testing. Vibrio strains were cultured in trypticase soy broth supplemented with 2% NaCl at 35 °C overnight. Non Vibrio strains were grown in Luria-Bertani broth at 37 °C.
Genomic DNA extraction was conducted using the boiling method. Briefly, 1 ml of overnight bacterial culture was centrifuged at 5,000 × g for 10 min and the pellet was suspended in 200 μl of TE buffer [10 mM Tris, 0.1 mM EDTA (pH 8.0)]. The bacterial suspension was boiled for 10 min and centrifuged at 12,000 × g for 5 min at 4 °C. The supernatant was used as the DNA template for multiplex LAMP and multiplex PCR assays. The genomic DNA of S. typhimurium CICC 21483 and V. parahaemolyticus ATCC 17802 was extracted with the Genomic DNA Isolation Kit (Sangon Biotechnology, Shanghai, China) following the manufacturer’s instructions.
LAMP primers and the multiplex LAMP reaction
We used previously reported Salmonella- and V. parahaemolyticus-specific primers10,12 (Table 2). The primers were synthesized by Sangon Biotech (Shanghai). Aliquots (50 μM) of each primer were prepared and used for the multiplex LAMP reaction.
Table 2. Primer sequences for the multiplex LAMP assay.
| Primer name | Primer Sequence (5′–3′) | Reference |
|---|---|---|
| bcfD-F3 | CCGGACAAACGATTCTGGTA | 10 |
| bcfD-B3 | CCGACATCGGCATTATCCG | |
| bcfD-FIP | TGCACTTTACCGGTACGCTGAA-TACAGCGGCAATTTCAACCA | |
| bcfD-BIP | CGGTCTGGATTCGCAGGTCAAA-GCGATAGCCTGGGGAAC | |
| bcfD-LF | TACCCCCTCCGGCTTTTG | |
| bcfD-LB | ACAATGCGTCTTATCGCTACG | |
| toxR-F3 | CGAAGTTGTACGATTAGGAAG | 12 |
| toxR-B3 | AAACTCTGGAGATTTGGTTG | |
| toxR-FIP | GCTCGTTACGGGTTAAAACTTCG-CAACGAAAGCCGTATACTCC | |
| toxR-BIP | GGCGTGAGCAAGGTTTTGAG-CCTTCAACATCTTACGCAG | |
| toxR-LF | GGTCTCTCCGCCAACATCA | |
| toxR-LB | GTGGATGACTCAAGCCTGACT |
The multiplex LAMP reaction was carried out in a final volume of 25 μL containing 12.5 μL 2 × LAMP reaction buffer [20 mM Tris-HCl, 10 mM KCl, 10 mM (NH4)2SO4, 8 mM MgSO4, 0.1% Tween 20, 0.8 M betaine (Sigma-Aldrich), 1.4 mM of each dNTP], 8 U of Bst DNA polymerase (New England Biolabs, Ipswich, MA), the bcfD and toxR primer mix (0.8 μM of FIP and BIP, 0.4 μM of LB and LF, 0.1 μM of F3 and B3), and 2 μL of DNA template. Additionally, 1 μL EvaGreen (20× in water) (Yeasen Biotech, Shanghai, China) was added. The reaction was performed at 65 °C for 40 min, followed by melting curve analysis from 80 °C to 90 °C with 0.1 °C increment per second. Each experiment was performed in triplicate.
Primer optimization
Initially, equimolar primer concentrations of 2.6 μM each were tested in the multiplex LAMP reaction. In addition, equimolar template concentrations at 100 ng/μL of DNA isolated from S. typhimurium CICC 21483 and V. parahaemolyticus ATCC 17802 were provided to the reaction to keep the same primer-to-template ratio. The amplification was performed in quadruplicate and the results are shown as Cq values, which represent the cycle in which fluorescence is first detected, and Tm (melting temperature) values determined by the melting curve analysis. A no-template control, in which double-distilled water was substituted for template DNA, was used in each amplification.
Multiplex PCR amplification
Multiplex PCR was performed with the specific primers for simultaneous detection of Salmonella spp. and V. parahaemolyticus. PCR primers for Salmonella were fimY-F (CCGTATGGCTGGGCGTTT) and fimY-R (AGTACGGCTAAAGCTTTCCGATAAG), and PCR primers for V. Parahaemolyticus were gyrB-F (CGGCGTGGGTGTTTCGGTAGT) and gyrB-R (TCCGCTTCGCGCTCATCAATA). The reaction was carried out in a 25 μL volume with 12.5 μL Taq Mix (TaKaRa, Dalian, China), 1 μL each of forward and reverse primers (10 μM), and 2 μL of DNA templates at 100 ng/μL to 1 pg/μL. The multiplex PCR amplification was conducted as follows: initial denaturation at 95 °C for 5 min, 35 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 5 min. The amplified products were analyzed by electrophoresis on 1.5% agarose gels.
Additional Information
How to cite this article: Liu, N. et al. Development of a multiplex loop-mediated isothermal amplification method for the simultaneous detection of Salmonella spp. and Vibrio parahaemolyticus. Sci. Rep. 7, 45601; doi: 10.1038/srep45601 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
This work is supported by the National Natural Science Foundation of China (81501836).
Footnotes
The authors declare no competing financial interests.
Author Contributions N.L. prepared materials & bacterial strains and performed the experiments; D.Z. and D.D. analyzed the data and performed the discussion; Z.Y. and D.A. prepared Figures 1–3; W.L. and L.H. designed the study and wrote the manuscript. All authors reviewed the manuscript.
References
- Oliver S. P., Jayarao B. M. & Almeida R. A. Foodborne Pathogens in Milk and the Dairy FarmEnvironment: Food Safety and Public Health Implications. Foodborne Pathog Dis. 2, 115–129 (2005). [DOI] [PubMed] [Google Scholar]
- Ke B. X. et al. Active etiological surveillance for foodborne diseases in Guangdong province. Zhonghua Liu Xing Bing Xue Za Zhi. 37, 1373–1358 (2016). [DOI] [PubMed] [Google Scholar]
- Birgitta D. J. & Ekdahl K. The comparative burden of salmonellosis in the European Union member states, associated and candidate countries. BMC Public Health. 6, 1–4 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung P. S. & Boor K. J. Epidemiology, pathogenesis, and prevention of foodborne Vibrio parahaemolyticus infections. Foodborne Pathog Dis. 1, 74–88 (2004). [DOI] [PubMed] [Google Scholar]
- Kumar B. K. et al. Development of monoclonal antibody based sandwich ELISA for the rapid detection of pathogenic Vibrio parahaemolyticus in seafood. Int J Food Microbiol. 145, 244–249 (2011). [DOI] [PubMed] [Google Scholar]
- Bolton F. J. et al. Rapid enzyme-linked immunoassay for the detection of Salmonella in food and feed products: performance testing program. J AOAC Int. 83, 299–303 (2000). [PubMed] [Google Scholar]
- Chen W. et al. Molecular beacons: a real time polymerase chain reaction assay for detecting Salmonella. Anal Biochem. 280, 166–172 (2000). [DOI] [PubMed] [Google Scholar]
- Chiang Y. C. et al. Multiplex PCR and a chromogenic DNA macroarray for the detection of Listeria monocytogenes, Staphylococcus aureus, Streptococcus agalactiae, Enterobacter sakazakii, Escherichia coli O157:H7, Vibrio parahaemolyticus, Salmonella spp. and Pseudomonas fluorescens in milk and meat samples. J Microbiol Methods. 88, 110–116 (2011). [DOI] [PubMed] [Google Scholar]
- Notomi T. et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28, E63 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang L. et al. Detection of Salmonella spp. by a loop-mediated isothermal amplification (LAMP) method targeting bcfD gene. Lett Appl Microbiol. 59, 658–664 (2014). [DOI] [PubMed] [Google Scholar]
- Ranjan R. et al. Development and evaluation of a one step reverse transcription-loop mediated isothermal amplification assay (RT-LAMP) for rapid detection of foot and mouth disease virus in India. Virus Disease. 25, 358–364 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. & Ge B. Development of a toxR-based loop-mediated isothermal amplification assay for detecting Vibrio parahaemolyticus. BMC Microbiol. 10, 32–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q. M. et al. Loop-mediated isothermal amplification (LAMP): early detection of Toxoplasma gondii infection in mice. Parasit Vectors. 5, 1–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y. et al. Development of multiplex loop-mediated isothermal amplification-RFLP (mLAMP-RFLP) to detect Salmonella spp. and Shigella spp. in milk. Int J Food Microbiol. 148, 75–79 (2011). [DOI] [PubMed] [Google Scholar]
- Iseki H. et al. Development of a multiplex loop-mediated isothermal amplification (mLAMP) method for the simultaneous detection of bovine Babesia parasites. J Microbiol Methods. 71, 281–287 (2007). [DOI] [PubMed] [Google Scholar]
- Mori Y., Hirano T. & Notomi T. Sequence specific visual detection of LAMP reactions by addition of cationic polymers. BMC Biotechnol. 6, 1–3 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamlor T. et al. Bovine embryo sex determination by multiplex loop-mediated isothermal amplification. Theriogenology. 83, 891–896 (2015). [DOI] [PubMed] [Google Scholar]
- Aonuma H. et al. A single fluorescence-based LAMP reaction for identifying multiple parasites in mosquitoes. Exp Parasitol. 125, 179–183 (2010). [DOI] [PubMed] [Google Scholar]
- Jung J. H. et al. Combination of multiplex reverse-transcription loop-mediated isothermal amplification with an immunochromatographic strip for subtyping influenza A virus. Anal Chim Acta. 853, 541–547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T. H., Gwo J. C. & Lin K. H. Rapid sex identification of papaya (Carica papaya) using multiplex loop-mediated isothermal amplification (mLAMP). Planta. 236, 1239–1246 (2012). [DOI] [PubMed] [Google Scholar]
- Kasahara K. et al. Development of multiplex loop-mediated isothermal amplification assays to detect medically important yeasts in dairy products. FEMS Microbiol Lett. 357, 208–216 (2014). [DOI] [PubMed] [Google Scholar]
- Wang F., Jiang L. & Ge B. Loop-mediated isothermal amplification assays for detecting shiga toxin-producing Escherichia coli in ground beef and human stools. J Clin Microbiol. 50, 91–97 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. & Pendleton J. Surveys of gene families using polymerase chain reaction: PCR selection and PCR drift. Systematic Biology. 43, 250–261 (1994). [Google Scholar]
- Markoulatos P. et al. Development of a quadriplex polymerase chain reaction for human cytomegalovirus. J Clin Lab Anal. 13, 99–105 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]


