Abstract
Background: Previously, we determined that heterogeneous nuclear ribonucleoprotein E1 (hnRNP-E1) functions as an intracellular physiologic sensor of folate deficiency. In this model, l-homocysteine, which accumulates intracellularly in proportion to the extent of folate deficiency, covalently binds to and thereby activates homocysteinylated hnRNP-E1 to interact with folate receptor-α mRNA; this high-affinity interaction triggers the translational upregulation of cell surface folate receptors, which enables cells to optimize folate uptake from the external milieu. However, integral to this model is the need for ongoing generation of hnRNP-E1 to replenish homocysteinylated hnRNP-E1 that is degraded.
Objective: We searched for an interrelated physiologic mechanism that could also maintain the steady-state concentration of hnRNP-E1 during prolonged folate deficiency.
Methods: A novel RNA-protein interaction was functionally characterized by using molecular and biochemical approaches in vitro and in vivo.
Results: l-homocysteine triggered a dose-dependent high-affinity interaction between hnRNP-E1 and a 25-nucleotide cis element within the 5′-untranslated region of hnRNP-E1 mRNA; this led to a proportionate increase in these RNA-protein complexes, and translation of hnRNP-E1 both in vitro and within placental cells. Targeted perturbation of this RNA-protein interaction either by specific 25-nucleotide antisense oligonucleotides or mutation within this cis element or by small interfering RNA to hnRNP-E1 mRNA significantly reduced cellular biosynthesis of hnRNP-E1. Conversely, transfection of hnRNP-E1 mutant proteins that mimicked homocysteinylated hnRNP-E1 stimulated both cellular hnRNP-E1 and folate receptor biosynthesis. In addition, ferrous sulfate heptahydrate [iron(II)], which also binds hnRNP-E1, significantly perturbed this l-homocysteine–triggered RNA-protein interaction in a dose-dependent manner. Finally, folate deficiency induced dual upregulation of hnRNP-E1 and folate receptors in cultured human cells and tumor xenografts, and more selectively in various fetal tissues of folate-deficient dams.
Conclusions: This novel positive feedback loop amplifies hnRNP-E1 during prolonged folate deficiency and thereby maximizes upregulation of folate receptors in order to restore folate homeostasis toward normalcy in placental cells. It will also functionally impact several other mRNAs of the nutrition-sensitive, folate-responsive posttranscriptional RNA operon that is orchestrated by homocysteinylated hnRNP-E1.
Keywords: folate deficiency, l-homocysteine, mRNA-binding protein, posttranscriptional RNA operon, αCP1, poly(C)-binding proteins, glutathione, iron chaperone, folate receptors, nutrition-sensitive
See corresponding commentary on page 471.
Introduction
Folate deficiency triggers the upregulation of folate receptors, which serves to optimize cellular folate uptake and help restore folate homeostasis to normal (1, 2). The molecular basis for upregulation of folate receptors likely involves the progressive homocysteinylation of heterogeneous nuclear ribonucleoprotein E1 (hnRNP-E1)10 in proportion to the degree of folate deficiency (3), with replacement of key cysteine S–S cysteine disulfide bonds in hnRNP-E1 by homocysteine S–S cysteine mixed disulfide bonds, which results in the gradual unmasking of an underlying mRNA-binding site in hnRNP-E1. This allows homocysteinylated hnRNP-E1 to bind with high affinity to an 18-nucleotide cis element in the 5′-untranslated region (5′-UTR) of folate receptor α mRNA, which, in turn, leads to a proportionate translational upregulation of folate receptors (3). Thus, hnRNP-E1, which is able to sense the level of intracellular folate deficiency and proportionately respond by increasing folate receptor α expression, has been incriminated as a physiologic cellular sensor of folate deficiency (2, 3). Because hnRNP-E1 assumes such a critical role to ensure folate homeostasis (2, 3), there is a simultaneous need for ongoing generation of newly synthesized hnRNP-E1 that can replenish homocysteinylated hnRNP-E1, which is degraded with a half-life of 52 h (3), particularly during prolonged periods of folate deficiency. This raises the possibility of an interrelated physiologic mechanism for the coexpression of hnRNP-E1 with folate receptors. Such an occurrence could explain clinical observations in which folate receptors and hnRNP-E1 are concordantly overexpressed in various human and murine tissues (1, 4–13).
The mRNA-binding site within homocysteinylated hnRNP-E1 is promiscuous because there are several diverse mRNAs with common poly(rC)/poly(U)–rich RNA cis elements that can also interact with this protein (14–16). Together, these mRNAs comprise a nutrition-sensitive, folate-responsive, posttranscriptional RNA operon that is orchestrated by homocysteinylated hnRNP-E1 (3). Many of these functionally distinct mRNAs likely contribute to the plethora of progressive pathobiological changes observed as cells experience mild, moderate, and severe folate deficiency. Morphologically, these developing megaloblastic changes, which are most apparent in rapidly proliferating cells, exhibit features of nuclear-cytoplasmic dissociation (involving asynchrony of nuclear and cytoplasmic maturation), with reduced cell proliferation because of varying degrees of cell cycle arrest and apoptosis—the end result of prolonged megaloblastosis (2). These distinct cellular changes during folate deficiency necessitate the coordinated involvement of several functionally diverse genes. In this context, it is therefore plausible that the progressive homocysteinylation of hnRNP-E1 during folate deficiency is also capable of modulating diverse mRNAs that belong to this novel posttranscriptional RNA operon.
Therefore, we focused on determining the mechanism whereby the concentration of hnRNP-E1 was maintained during prolonged folate deficiency. Accordingly, we searched for evidence that favored a specific physiologic interaction of hnRNP-E1 with common poly(rC)/poly(U)–rich RNA cis elements within its own hnRNP-E1 mRNA in vitro, and in response to prolonged folate deficiency in vivo. Then we sought evidence for dual activation of hnRNP-E1 mRNA and folate receptor α mRNA by homocysteinylated hnRNP-E1, both in vitro and in vivo.
Methods
Materials.
All reagents of the highest available purity were purchased from Sigma-Aldrich. All cell culture media and other additives, Dulbecco’s PBS, Escherichia coli DH10B-competent bacteria, and oligonucleotides were from Invitrogen. [α-32P]UTP (specific activity >3000 Curie/mmol) and l-[35S]methionine or l-[35S]cysteine (in vitro translation grade) were from Perkin-Elmer. Restriction endonucleases were from Roche Applied Science. l-homocysteine (98% purity) was from Sigma-Aldrich.
Culture of placental cell lines.
The human placental cell lines (1584, JAR, and CCL-98), obtained from American Type Culture Collection, were propagated long-term in high-folate (HF) media (DMEM-HF), which contained nondialyzed 10% FBS, and 9.1-μmol/L folic acid plus 13.6-nmol/L 5-methyl-tetrahydrofolate (3). These cells were then slowly adapted to growth in physiologically low-folate (LF) media (DMEM-LF), which contained no added folic acid in the media, nondialyzed 10% FBS, and 13.6 nM 5-methyl-tetrahydrofolate, for 14 wk, after which both mRNA and protein for folate receptor and hnRNP-E1 were evaluated (1). Folate receptor α and hnRNP-E1 RNA from HF and LF placental cells was determined by qRT-PCR.
Expression of the glutathione S-transferase fusion protein from recombinant glutathione S-transferase–hnRNP-E1 and its mutants.
Recombinant glutathione S-transferase (GST)–plasmid DNA, pGST–hnRNP-E1, pGST–hnRNP-E2, and related mutated plasmids (3) were transformed into BL21 E. coli from Novagen. After induction by 1 mM isopropyl-1-thio-β-d-galactopyranoside, the GST–hnRNP-E1 and mutated fusion proteins were individually purified with the use of the B-PER GST Fusion Protein Purification Kit (Pierce). The eluted GST fusion proteins were dialyzed against 500 volumes of buffer to remove excess reducing reagents (such as DTT) and assayed by SDS-PAGE and Western blots with the use of anti-peptide hnRNP-E1 antiserum (5). Gel-shift assays with target RNA were carried out as described (3).
Preparation of 25-nucleotide hnRNP-E1 mRNA cis element and related constructs.
To prepare the 25-nucleotide hnRNP-E1 mRNA cis element, pSPT18–25 DNA was obtained by subcloning a pair of oligodeoxynucleotides (5′-g CTC CCG CCC GCT CCC GCT CGC TCC C g-3′ and 5′-aattc GGG AGC GAG CGG GAG CGG GCG GGA Gctgct-3′) into pSPT18 vector linearized with PstI and EcoRI. To prepare the 25-nucleotide hnRNP-E1 mRNA cis element proximal to hnRNP-E1 mRNA, pSPT18–25–hnRNP-E1 DNA was obtained by subcloning the PCR product from Gene Pool cDNA (Human Placenta cDNAs; Invitrogen) into pSPT18. The PCR product from Gene Pool cDNA was digested with PstI and EcoRI, and then ligated to pSPT18 plasmid linearized with PstI and EcoRI. The primers used in PCR were as follows: 5′-tgacctgcag GAC TCC CGC CCG CTC-3′ and 5′-tgacgaattc CTA GCT GCA GGG CAT GC-3′.
To incorporate the 25-nucleotide hnRNP-E1 cis element proximal to a chloramphenicol acetyltransferase (CAT) reporter (pCAT-25) and a control 25-nucleotide scrambled sequence proximal to CAT (pCAT-25-scrambled), 2 pairs of oligodeoxynucleotides were subcloned into pCAT3-control vector that was linearized with NheI and BglII to generate pCAT-25 and pCAT-25-scrambled vectors. Oligodeoxynucleotides for 25-nucleotide hnRNP-E1 cis element inserts were as follows: 5′-ctagc CTC CCG CCC GCT CCC GCT CGC TCC C a-3′ and 5′-gatct GGG AGC GAG CGG GAG CGG GCG GGA G g-3′. Oligodeoxynucleotides for the 25-nucleotide scrambled insert sequence were as follows: 5′-ctagc GCG TCG CTC GCT TCG CAC GTG CGC C a-3′ and 5′-gatct GGC GCA CGT GCG AAG CGA GCG ACG C g-3′.
RNA-protein binding assays.
RNA-protein binding assays involved incubation of one of various radiolabeled target RNAs {1 × 105 counts per minute of [35S]25-nucleotide hnRNP-E1 mRNA cis element, or [35S]18-nucleotide folate receptor α mRNA cis element (3), or human papillomavirus type 16 (HPV16) [35S]L2 RNA cis element (17)} with either purified 0.5-μg dialyzed purified recombinant GST–hnRNP-E1 or its mutants, in standard buffer (3) in the absence or presence of various concentrations of physiologically and nonphysiologically relevant thiols (β-mercaptoethanol, glutathione, methionine, l-cysteine, l-homocysteine, and DTT), as described (1, 3, 17). Parenthetically, there was no significant difference in RNA binding to GST–hnRNP-E1 by use of either 32P-RNA probes (3) or 35S-labeled RNA probes used in this paper (data not shown).
Effects of ferrous sulfate heptahydrate on the homocysteine-induced RNA-protein interaction.
Studies were also conducted to evaluate the effect of ferrous sulfate heptahydrate [iron(II)] or ferric chloride hexahydrate [iron(III)] in modulating RNA-protein interactions during gel-shift assays in either the absence or presence of 10-mmol glutathione/L. The iron preparation, either alone or with 50 μmol deferoxamine/L to define the specificity of iron(II), was incubated first with glutathione and hnRNP-E1 before the addition of other components of the reaction mixture. Briefly, RNA-protein binding and gel-shift assays were carried out with purified recombinant GST–hnRNP-E1 protein and [35S]25-nucleotide hnRNP-E1 mRNA cis element in binding buffer and 5-μmol l-homocysteine/L plus increasing concentrations of iron(II) or iron(III) (0–25 μmol/L). The samples were analyzed by polyacrylamide gel electrophoresis and autoradiography overnight followed by densitometric analysis of the signals representing RNA-protein complexes.
In vitro transcription/translation studies.
In vitro transcription/translation was carried out with the use of the Linked SP6/T7 In Vitro Transcription/Translation Kit (Roche Applied Science) after the addition of one of various targeted DNAs—pSPT18-25–hnRNP-E1 DNA, or PstI-linearized plasmid pSPT18–folate receptor α (5, 18), or HPV16 L2 DNA (17), (0.5 μg/reaction)—to the transcription mix as described (1, 3). In other experiments, the capacity for perturbing the interaction of hnRNP-E1 with the 25-nucleotide hnRNP-E1 cis element during in vitro translation was assessed in the presence of either scrambled oligonucleotides (5′- GCG TCG CTC GCT TCG CAC GTG CGC C-3′) or specific antisense oligonucleotides (5′-GGG AGC GAG CGG GCG GGA G-3′) generated against the 25-nucleotide hnRNP-E1 mRNA cis element (GenScript USA). Glutathione was not added when the effect of iron(II) was assessed during in vitro translation because the commercial reticulocyte lysate contained 4.1-mmol β-mercaptoethanol/L.
Dissociation constant of various RNA-protein interactions.
The influence of either l-homocysteine or l-cysteine on the dissociation constant (KD) of the RNA-protein interaction was assessed in the absence and presence of physiologic concentrations of glutathione (10-mmol/L) with the use of previously described methods (3). Specific binding was determined by subtracting values of nonspecific binding with GST from those of total binding with purified recombinant GST–hnRNP-E1; however, nonspecific binding with the use of BSA was subtracted from total binding to GST–hnRNP-E1 to determine specific binding in Tables 1 and 2. The KD was calculated from a Scatchard plot with the use of GraphPad Prism 6, as described (3).
TABLE 1.
Comparison of the KD of the interaction of recombinant purified GST–hnRNP-E1 or its mutant protein for the [35S]25-nucleotide hnRNP-E1 mRNA cis-element in the absence or presence of l-homocysteine1
| Protein | l-homocysteine, μM | KD, nM |
| GST–hnRNP-E1 | 0 | 1.93 ± 0.18a |
| GST–hnRNP-E1 | 10 | 1.14 ± 0.17b |
| GST–hnRNP-E1 | 50 | 0.62 ± 0.10c |
| GST–hnRNP-E1(G292A) | 0 | 2.02 ± 0.19a |
| GST–hnRNP-E1(C293S) | 0 | 0.39 ± 0.04d |
Values are means ± SDs, n = 3 (means of triplicates). Labeled means without a common superscript letter differ, P < 0.05. GST, glutathione S-transferase; hnRNP-E1, wild-type heterogeneous nuclear ribonucleoprotein E1; hnRNP-E1(C293S), highest affinity–heterogeneous nuclear ribonucleoprotein E1 (C293S)–mutant protein; hnRNP-E1(G292A), wild-type–like heterogeneous nuclear ribonucleoprotein E1 (G292A)–mutant protein; KD, dissociation constant.
TABLE 2.
Dose-dependent influence of iron(II) on the KD involving the l-homocysteine–triggered RNA-protein interaction between the [35S]25-nucleotide hnRNP-E1 mRNA cis element and purified recombinant GST–hnRNP-E1 protein1
| Protein | Iron(II), μM | l-homocysteine, μM | KD, nM |
| GST–hnRNP-E1 | 0 | 25 | 2.74 ± 0.52a |
| GST–hnRNP-E1 | 5 | 25 | 3.72 ± 0.34b |
| GST–hnRNP-E1 | 25 | 25 | 6.50 ± 0.83c |
Values are means ± SDs, n = 3 (means of triplicates). Labeled means without a common superscript letter differ, P < 0.05. GST, glutathione S-transferase; hnRNP-E1, wild-type heterogeneous nuclear ribonucleoprotein E1; iron(II), ferrous sulfate heptahydrate; KD, dissociation constant.
qRT-PCR to determine RNA expression of hnRNP-E1 and folate receptor α in cells.
Total RNA was separately extracted from placental 1584-HF and 1584-LF cells. Gene expression was determined on triplicate RNA samples (50 ng/reaction) with the use of an Invitrogen SuperScript III Platinum One-Step qRT-PCR kit and an ABI 7900HT Sequence Detection System (PE Biosystems). The cycle conditions were as follows: after a first step of 15 min at 50°C and 10 min at 95°C, the samples were cycled 40 times at 95°C for 15 s and at 60°C for 60 s. For all quantitative analyses, we used the comparative cycle threshold method by following the PE Biosystems protocol. The specific primers were obtained from Invitrogen with fluorogenic labels either 5′6-carboxyfluorescein (FAM) or 6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein (JOE). Primers for hnRNP-E1 were 5′-GAC GCC GGA GAC TGG GAG AGC G [FAM] C-3′and 5′-GGA TAT GCT GCC CAA CTC CA-3′. Primers for folate receptor α were 5′-GAA CCT ATG AGG AGG TGG CGA GG [FAM] TC-3′ and 5′-TAG GGC CAG GCT AAG CAG GA-3′. Primers for GAPDH were 5′-CAA CAG GAG GAG TGG GTG TCG CTG (JOE) TG -3′ and 5′-GGC ATC CTG GGC TAC ACT GA-3′. Primers to the hnRNP-E1 gene or folate receptor α gene (labeled with FAM) and primers to a housekeeping gene GAPDH (labeled with JOE) were run in parallel, respectively, to standardize the input amount. Controls consisting of ribonuclease-free water were negative in all runs.
Effects of l-homocysteine on various CAT reporter constructs transfected into placental 1584-HF cells.
To prepare pCAT-25 or pCAT-25-mutant, 2 pairs of oligonucleotides, 1 for pCAT-25 (5′-tagcaggtac AAG CTT CTC CCG CCC GCT CCC GCT CGC TCC CCA TGG tgtaactagct-3′ 5′-agctagttaca CCA TGG GGA GCG AGC GGG AGC GGG CGG GAG AAG CTT gtacctgcta-3′), and 1 for pCAT-25 mutant no. 6 (5′-tagcaggtac AAG CTT CTT CCG CCC GCT CCC GCT CGC TTC CCA TGG tgtaactagct-3′ 5′-agctagttaca CCA TGG GGA GCG AGC GGG AGC GGG CGG GAG AAG CTT gtacctgcta-3′), were first linearized with HindIII and NcoI and subcloned into pCAT3 control DNA digested with HindIII and NcoI. After transient transfection of either pCAT-25 or pCAT-25 mutant no. 6 plasmid DNA together with pSV-β-gal DNA into placental 1584-HF cells, these cells were incubated with physiologically relevant concentrations of l-homocysteine, and net CAT expression was assessed as described (3).
Capture of RNA-protein complexes within cells.
Slot-blot hybridization was carried out under high stringency conditions to detect intracellular RNA-protein complexes (composed of endogenous hnRNP-E1 RNA cis element–bound to cellular hnRNP-E1 proteins) in placental 1584 cells under experimental conditions that modulated the intracellular concentration of homocysteine. We used principles similar to those used to capture folate receptor α mRNA cis element–bound hnRNP-E1 complexes in HeLa-IU1 cells (3). Briefly, after l-homocysteine treatment, the extant intracellular RNA-protein complexes were UV-crosslinked and isolated on anti–hnRNP-E1 antiserum–linked agarose. After ribonuclease treatment and proteolysis, equal aliquots of the released small mRNA cis element fragments were probed for evidence of enrichment of hnRNP-E1 mRNA cis element in experimental cells with the use of a specific [35S]-labeled antisense hnRNP-E1 mRNA cis element probe (3).
Transfection of antisense oligonucleotides to the 25-nucleotide hnRNP-E1 cis element on the biosynthetic rate of hnRNP-E1 proteins in cells.
On the day before transfection, placental 1584-HF cells were trypsinized, counted, and plated in 6-well plates at 2 × 105 cells per well in 2.5 mL MEM-HF medium, so that cells would be 70–80% confluent after overnight culture. Cells were then transfected with either wild-type or scrambled DNA or antisense DNA to 25-nucleotide hnRNP-E1 cis element with the use of Lipofectamine 2000 DNA transfection reagent and the manufacturer’s protocol. One well of cells was trypsinized and harvested on day 3 [2 d after transfection] as a control. Cells in the remaining 5 wells were starved of cysteine with the use of cysteine-free MEM-HF for 4 h, after which the rate of biosynthesis of hnRNP-E1 protein was determined in transfected cells by specifically immunoprecipitating and quantifying newly synthesized [35S]hnRNP-E1 (3).
RNA interference of hnRNP-E1 mRNA on the biosynthetic rate of hnRNP-E1 and folate receptor proteins in cells.
Before the day of small interfering RNA (siRNA) transfection, placental 1584-HF cells were trypsinized, counted, and plated in 6-well plates at 1.4 × 105 cells per well (∼30–40% confluence) in 2.5 mL MEM-HF medium, so that cells were 60–80% confluent after overnight culture. Cells were then transfected over 2 d with either 10 nmol predesigned Stealth RNA interference/L [siRNA–hnRNP-E1/poly(rC)-binding protein 1 (PCBP1)] or 10 nmol scrambled negative stealth RNA interference control/L with the use of Lipofectamine RNAiMAX transfection reagent and a minor modification of the manufacturer’s protocol (3). One of these wells was trypsinized and harvested 2 d after transfection, after which the RNA was purified for qRT-PCR analysis of hnRNP-E1 RNA expression. Cells in the remaining 5 wells were starved of cysteine for 4 h and the rate of biosynthesis of newly synthesized [35S]hnRNP-E1 and [35S]folate receptor proteins was determined, as described (3).
Determination of the biosynthetic rate of folate receptors or hnRNP-E1 proteins in cells transfected with either wild-type or various mutant hnRNP-E1 proteins.
The transfection of purified recombinant wild-type or various mutant hnRNP-E1 proteins into cells was achieved with the use of the Xfect Protein Transfection Kit (Clontech). Briefly, placental 1584-HF cells in 10-cm dishes at 70–80% confluence were transfected with 2.4 mL serum-free medium and Xfect Protein Transfection Reagent (1.2 mL) containing 20 μg either wild-type hnRNP-E1-, or highest-affinity (HA) hnRNP-E1(C293S) mutant, or wild-type–like hnRNP-E1(G292A) mutant proteins (described in Supplemental Methods, Supplemental Results, Supplemental Discussion, and Supplemental References) and 2 μg β-galactosidase at 37°C for 60 min. After cysteine starvation for 4 h, an aliquot of cells was assessed for β-galactosidase activity (to measure the efficiency of cotransfection of β-galactosidase), and the remaining cells were pulsed with l-[35S]cysteine (250 μCurie) and assessed for specific [35S]cysteine incorporated into folate receptor to determine the biosynthetic rate of this protein (3). A similar protocol was used for the analysis of newly synthesized [35S]hnRNP-E1 with the use of anti–hnRNP-E1 antiserum–coupled agarose and control agarose in order to derive values for specific [35S]cysteine incorporated into hnRNP-E1 and thereby determine the biosynthetic rate of this protein (3).
Mouse protocols and care.
All mouse care procedures conformed to the Guide for the Care and Use of Laboratory Animals (13). The protocols for the use of athymic mice in experiments involving tumor xenografts were approved by the Institutional Animal Care and Use Committee at Indiana University–Purdue University at Indianapolis. To evaluate for upregulation of folate receptors and hnRNP-E1 within cervical cancer xenograft tumors that were generated in athymic mice, 6 athymic mice were fed a standard folate-replete diet (1200 nmol folate/kg diet) and 5 athymic mice were fed a severely folate-restricted diet (120 nmol folate/kg diet) for 4 wk (13) before injection of 1 million HeLa-IU1 cells into their flanks (19). The tumors were subsequently examined for hnRNP-E1 and folate receptor expression by immunohistochemistry, northern blots, and western blots. For western blot analyses of folate receptor and hnRNP-E1 expression in tumors, 50 μg protein from HeLa-IU1–derived tumors was subjected to 10% SDS-PAGE and western transfer followed by probing of nitrocellulose-bound proteins with either anti-folate receptor antiserum or anti–hnRNP-E1 antiserum or anti-GAPDH antibodies (3). Total RNA was obtained from HeLa-IU1–derived tumors in mice fed a normal or LF diet. The hnRNP-E1 probe was excised from plasmid pGEX-4T-1–hnRNP-E1 with the use of BamH1 and Not1 digestion. The folate receptor α plasmid was digested with HindIII and EcoRI to liberate its cloned insert. DNA fragments were gel-purified and labeled with the use of the Random Primed Labeling Kit (Roche Applied Science) and [α-32P]deoxy-ATP (1). Northern blots were carried out by electrophoresis of 20 μg RNA in standard formaldehyde-agarose gels followed by transfer to Hybond-N+ nylon membranes (Amersham/GE Healthcare Biosciences) and UV crosslinking. Membranes were hybridized with the 32P-labeled probes and detected according to the manufacturer's instructions (Roche Applied Science). Ethidium bromide staining of 28S was monitored for RNA quality, and β-actin was used as a loading control.
Mouse experiments with the use of CD-1 mice were conducted at the National Center for Toxicological Research; these mouse studies were approved by each of the local institutional animal care and use committees in Arkansas and Indiana. The protocols for the procurement and feeding of CD-1 mice (Charles River) with either a folate-replete diet (1200 nmol folate/kg diet) or folate-deficient diet (400 nmol folate/kg diet), breeding and dams killed on gestation day 17, removal and killing of fetuses and fixation in formalin, histochemical staining, and analysis of paraffin-embedded fetal tissues (13) were as described (3). The placentas from dams that were fed a folate-deficient compared with a folate-replete diet (3) were also examined for morphologic differences after sectioning and staining. Images were captured with a Leitz DMLB light microscope equipped with a Diagnostic Instruments Spot digital camera (Diagnostic Instruments) and processed with the use of Adobe Photoshop software. The localization of tissues in gestation day 17 fetuses was based on Kaufman’s atlas (20).
Supplemental Methods.
The following experiments can be found in the Supplemental Methods: Site-directed mutagenesis of hnRNP-E1; HPV16 L2 RNA-related gel-shift and in vitro translation; Determination of the concentration of homocysteine, cystathionine, cysteine, and methionine in placental cells; and Transfection of various CAT reporters linked to hnRNP-E1 DNA or its mutants into Hela-IU1-HF cells.
Statistical analyses.
All of the statistical analyses were conducted with the use of GraphPad Prism 6. Unless otherwise specified, results are expressed as means ± SDs, n = 3 (means of triplicates). Comparisons between 2 groups were analyzed by Student’s t test and comparisons between multiple groups was analyzed with the use of 1-factor ANOVA followed by a Tukey’s test. The statistical significance was set at P < 0.05.
Results
Characterization of a 25-nucleotide cis element in the 5′-UTR of hnRNP-E1 mRNA that interacts with hnRNP-E1.
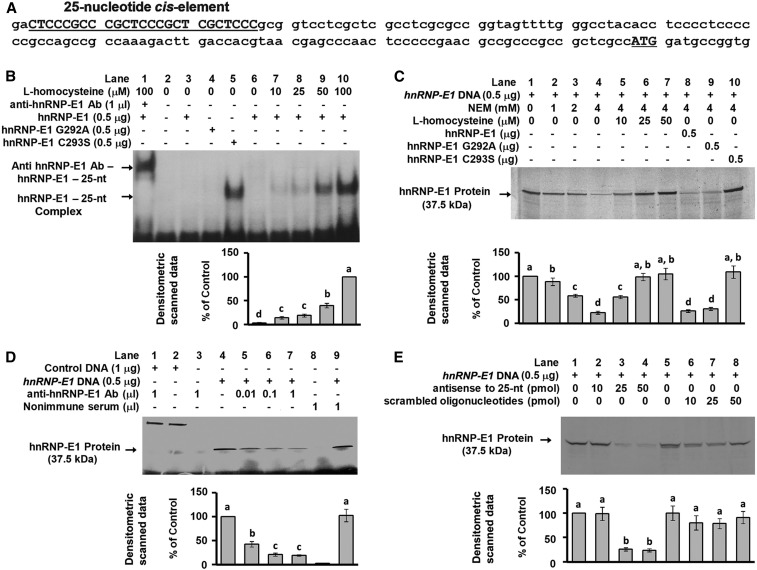
The gene sequence of hnRNP-E1 revealed a candidate 25-nucleotide poly(rC)-rich region in the 5′-UTR of hnRNP-E1, from −145 to −120 upstream of the start site of the coding sequence of hnRNP-E1 (Figure 1A). This candidate hnRNP-E1 cis element had a structural organization similar to that noted within the 18-nucleotide folate receptor α mRNA cis element (18), which consisted of a tandem CUCC sequence separated by 6–8 bases as a putative protein binding motif; likewise, the hnRNP-E1 mRNA cis element contained 3 such CUCC sequence motifs. Therefore, we focused on the potential for binding between homocysteinylated hnRNP-E1 and this 25-nucleotide cis element in the 5′-UTR of hnRNP-E1 mRNA to determine if this RNA-protein interaction led to enhanced translation of hnRNP-E1 in vitro and in cultured placental cells.
FIGURE 1.
Characterization of the interaction of a 25-nucleotide cis element in the 5′-UTR of hnRNP-E1 with hnRNP-E1 in vitro. Location of the candidate 25-nucleotide cis element (bold, capitalized, and underlined) in the 5′-UTR of hnRNP-E1 from −145 to −120 from the ATG start site (A). Gel-shift analysis of the interaction of radiolabeled 25-nucleotide hnRNP-E1 mRNA cis element and purified recombinant GST–hnRNP-E1 (lanes 1, 3, and 6–10) or hnRNP-E1 mutants (lanes 4 and 5) in the absence or presence of l-homocysteine; supershift of the RNA-protein signal with anti–hnRNP-E1 antiserum (lane 1) (B). In vitro translation of hnRNP-E1 after quenching excess thiols in the reaction mixture by NEM (lane 2–4) to assess the effect of increasing l-homocysteine (lanes 5–7), and comparison of the independent effect of wild-type or mutant hnRNP-E1 (lanes 8–10) (C). Demonstration of the key role of hnRNP-E1 in mediating in vitro translation of hnRNP-E1 protein by the addition of anti–hnRNP-E1 antiserum (lanes 4–7) (D). Lanes 1 and 2 were internal protein controls. Comparison of the effectiveness of antisense oligonucleotides to the 25-nucleotide hnRNP-E1 cis element and scrambled oligonucleotides in quenching the translation of hnRNP-E1 in vitro (E). The autoradiograms shown in B, C, D, and E are representative of 3 separate experiments, and pooled densitometric scanned data of signals are compared with the 100% value. Values are means ± SDs, n = 3 (means of triplicates). Labeled means without a common letter differ, P < 0.05. Ab, antiserum; GST, glutathione S-transferase; hnRNP-E1, wild-type heterogeneous nuclear ribonucleoprotein E1; hnRNP-E1(G292A), wild-type–like heterogeneous nuclear ribonucleoprotein E1 (G292A) mutant; hnRNP-E1(C293S), highest affinity–heterogeneous nuclear ribonucleoprotein E1 (C2923S) mutant; NEM, N-ethylmaleimide; nt, nucleotide; 5′-UTR, 5′-untranslated region.
Gel-shift assays showed a dose-dependent increase of RNA-protein signal with increasing physiologically relevant concentrations of l-homocysteine (Figure 1B, lanes 6–10). The involvement of hnRNP-E1 in RNA-protein complexes was shown by a supershift of the RNA-protein signal with anti–hnRNP-E1 antiserum (Figure 1B, lane 1); no such supershift was noted when nonimmune serum was used (not shown). Supplemental Figure 1 and Supplemental Results contain data that confirm the specificity of the anti–hnRNP-E1 antiserum used in these studies. The reticulocyte lysate mixture used for in vitro translation contains small quantities of hnRNP-E1 (8, 9, 11, 12, 21). Accordingly, we evaluated the functional consequences of interaction of the 25-nucleotide hnRNP-E1 mRNA cis element and (endogenous) hnRNP-E1 during in vitro translation. Quenching excess β-mercaptoethanol in the translation mixture with N-ethylmaleimide (Figure 1C, lane 2–4) led to a progressive reduction in the amount of hnRNP-E1 translated compared with the baseline hnRNP-E1 signal (Figure 1C, lane 1). Maintaining N-ethylmaleimide in the reaction mixture improved the sensitivity of the system to the subsequent addition of physiologic concentrations of l-homocysteine, which led to a progressively increased translation of hnRNP-E1 (Figure 1C, lanes 5–7). The specific involvement of endogenous hnRNP-E1 in mediating the translation of hnRNP-E1 was confirmed by the dose-dependent quenching of the signal with the addition of increasing concentrations of anti–hnRNP-E1 antiserum (Figure 1D, lanes 5–7); by contrast, there was no effect of anti–hnRNP-E1 antiserum on the translation of internal control DNA (lane 1 compared with 2), and no reduction in translation of hnRNP-E1 by nonimmune serum (lane 9).
The experiments that led to the selection of the most effective molecular mimic of homocysteinylated hnRNP-E1 and its control that was used to further characterize the interaction of hnRNP-E1 with its own cis element are described in the Supplemental Methods, Supplemental Results, and Supplemental Table 1. Thus, Supplemental Figure 2 demonstrated that the mutation of cysteine 293 to serine in the third K-homology domain of hnRNP-E1 resulted in the highest affinity for the folate receptor α mRNA cis element and for HPV16 L2 cis element, even in the absence of l-homocysteine, which also led to expected functional outcomes (3, 17); this mutant is hereafter referred to as the (HA)-hnRNP-E1(C293S) mutant. By contrast, mutation of the adjacent glycine-292 to alanine failed to confer such properties, and this mutant protein behaved similarly to wild-type hnRNP-E1 in that interaction with these target cis elements was increased only upon the addition of l-homocysteine; hence, this control was referred to as a wild-type–like hnRNP-E1(G292A) mutant. Thus, as also shown in Figure 1B, incubation of 25-nucleotide hnRNP-E1 RNA cis element with wild-type hnRNP-E1 and wild-type–like hnRNP-E1(G292A) mutant in the absence of l-homocysteine (lanes 3 and 4) revealed no RNA-protein gel-shift signals, but there was a strong signal when 25-nucleotide hnRNP-E1 mRNA cis element reacted with (HA)-hnRNP-E1(C293S) mutant (lane 5), even in the absence of l-homocysteine. Moreover, after β-mercaptoethanol was neutralized by N-ethylmaleimide, only the (HA)-hnRNP-E1(C293S) mutant markedly stimulated the translation of hnRNP-E1 in vitro (Figure 1C, lane 10) compared with wild-type hnRNP-E1 and wild-type–like hnRNP-E1(G292A) mutant (lanes 8, 9).
Specific antisense oligonucleotides to the 25-nucleotide hnRNP-E1 mRNA cis element led to a dose-dependent inhibition of translation of hnRNP-E1, as shown in Figure 1E (lanes 2–4). By contrast, scrambled oligonucleotides failed to quench the translation of hnRNP-E1 (Figure 1E, lanes 6–8). Thus, several lines of evidence suggested that interaction of homocysteinylated hnRNP-E1 and the 25-nucleotide cis element in the 5′-UTR of hnRNP-E1 mRNA led to an increase in biosynthesis of hnRNP-E1 in vitro, and that the (HA)-hnRNP-E1(C293S) mutant was also capable of mediating similar effects even in the absence of homocysteine. Because (HA)-hnRNP-E1(C293S) mutant similarly reacted with the 18-nucleotide folate receptor α mRNA cis element in the absence of homocysteine (Supplemental Figure 2), we quantified and compared the RNA-protein interactions involving these distinct cis elements by formal dissociation constant studies (Tables 1 and 3). When purified wild-type hnRNP-E1 was reacted with the 25-nucleotide hnRNP-E1 mRNA cis element in the presence of increasing (physiologically relevant) concentrations of l-homocysteine, there was a dose-dependent increase in binding affinity (Table 1); thus, the KD progressively decreased from a basal value of 1.93 nmol/L (in the absence of l-homocysteine) to a KD of 1.14 nmol/L and 0.62 nmol/L in the presence of 10 μmol l-homocysteine/L and 50 μmol l-homocysteine/L, respectively (P < 0.05). Moreover, although the difference in basal KD values between wild-type hnRNP-E1 and wild-type–like hnRNP-E1(G292A) mutant (in the absence of l-homocysteine) was not significant (1.93 nmol/L compared with 2.02 nmol/L, respectively), the (HA)-hnRNP-E1(C293S) mutant exhibited a significantly higher affinity for the 25-nucleotide hnRNP-E1 mRNA cis element even in the absence of l-homocysteine (KD = 0.39 nM). This reflected a significantly higher affinity than that observed when the wild-type hnRNP-E1 protein was reacted with the 25-nucleotide hnRNP-E1 mRNA cis element in the presence of 50 μmol l-homocysteine/L (KD = 0.62 nM).
TABLE 3.
Comparison of the KD of the interaction of recombinant purified GST–hnRNP-E1 or its mutant protein for the [35S]18-nucleotide folate receptor α mRNA cis element in the absence or presence of l-homocysteine1
| Protein | l-homocysteine, μM | KD, nM |
| GST–hnRNP-E1 | 0 | 1.47 ± 0.16a |
| GST–hnRNP-E1 | 10 | 0.89 ± 0.16b |
| GST–hnRNP-E1 | 50 | 0.47 ± 0.09c |
| GST–hnRNP-E1(G292A) | 0 | 1.57 ± 0.16a |
| GST–hnRNP-E1(C293S) | 0 | 0.27 ± 0.04d |
Values are means ± SDs, n = 3 (means of triplicates). Labeled means without a common superscript letter differ, P < 0.05. GST, glutathione S-transferase; hnRNP-E1, wild-type heterogeneous nuclear ribonucleoprotein E1; hnRNP-E1(C293S), highest affinity–heterogeneous nuclear ribonucleoprotein E1 (C293S)–mutant protein; hnRNP-E1(G292A), wild-type–like heterogeneous nuclear ribonucleoprotein E1 (G292A)–mutant protein; KD, dissociation constant.
As expected (3), there was a progressive increase in binding affinity between wild-type hnRNP-E1 and folate receptor α mRNA cis element in the presence of increasing concentrations of l-homocysteine (Table 3). Here, too, in the absence of l-homocysteine, the KD of both wild-type hnRNP-E1 and wild-type–like hnRNP-E1(G292A) mutant for folate receptor α mRNA cis element was comparable, i.e., KD = 1.47-nmol/L compared with 1.57-nmol/L, respectively. However, (HA)-hnRNP-E1(C293S) mutant exhibited a significantly higher affinity for folate receptor α mRNA cis element even in the absence of l-homocysteine. Again, this value (KD = 0.27 nmol/L) reflected a significantly higher affinity than the value of the wild-type hnRNP-E1 for this cis element in the presence of 50 μmol l-homocysteine/L (KD = 0.47 nmol/L). Taken together, these data confirmed that 1) as with its interaction with folate receptor α mRNA cis element (3), homocysteinylated hnRNP-E1 also bound the 25-nucleotide hnRNP-E1 mRNA cis element with high affinity at physiologic concentrations of l-homocysteine, as would be found in mild-to-moderate folate deficiency in vivo (2, 22, 23); and 2) the (HA)-hnRNP-E1(C293S) mutant was capable of binding both the hnRNP-E1 mRNA cis element, as well as the folate receptor α mRNA cis element, with similarly high (but not identical) affinity, even in the absence of homocysteine.
Posttranscriptional upregulation of hnRNP-E1 in placental cells involves interaction of the 25-nucleotide hnRNP-E1 cis-element and homocysteinylated hnRNP-E1.
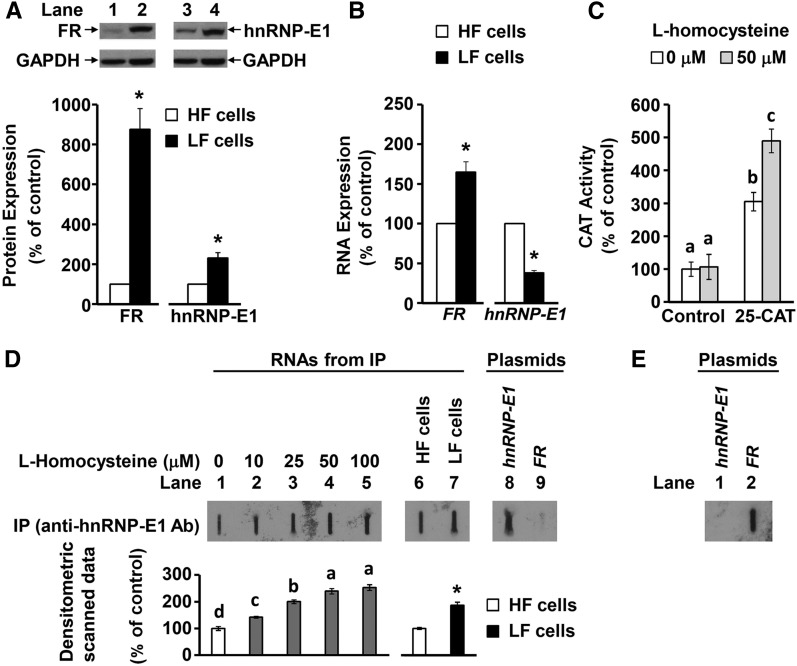
Both folate receptors and hnRNP-E1 were comparably upregulated at the posttranscriptional level under folate-depleted conditions in placental JAR and 1584 cell lines (Supplemental Figure 3). Accordingly, the latter cell line was selected to further characterize the role of a specific RNA-protein interaction in the posttranscriptional upregulation of hnRNP-E1 within cells. As shown in Figure 2A and B folate-depleted placental 1584-LF cells exhibited a 9-fold overexpression of folate receptor proteins, with only a 1.6-fold increase in folate receptor mRNA over basal values found in folate-replete placental 1584-HF cells. Likewise, there was a >2-fold increase in hnRNP-E1 protein in folate-depleted cells, which was associated with only one-third the basal expression of hnRNP-E1 mRNA found in folate-replete cells. Thus, the disproportionately higher folate receptor and hnRNP-E1 protein overexpression compared with folate receptor and hnRNP-E1 mRNA transcripts suggested that both of these proteins were upregulated at the posttranscriptional level.
FIGURE 2.
Evidence of posttranscriptional upregulation of hnRNP-E1 (A and B) and an RNA-protein interaction involving a 25-nucleotide hnRNP-E1 mRNA cis element and endogenous hnRNP-E1 proteins in response to homocysteine in placental 1584 cells (C–E). Western blots of placental 1584-HF cells (lanes 1 and 3) and 1584-LF cells (lanes 2 and 4) probed for FR and hnRNP-E1 (A). One representative blot of 3 independent experiments is shown; results of densitometric scanning analysis are presented below the gels as means ± SDs, n = 3. Comparison of the FR RNA and hnRNP-E1 RNA from 1584-HF and 1584-LF cells (B). CAT reporter activity after transfection of either a pCAT construct (denoted “control”) or a 25-nucleotide hnRNP-E1 cis element–driven CAT reporter construct (denoted “25-CAT”) into 1584-HF cells before (open bars) and after (shaded bars) exposure to 50 μmol l-homocysteine/L (C). Results are presented as means ± SDs, n = 3 (means of triplicates). Slot-blot hybridization analysis with the use of a [35S]-labeled antisense probe to the 25-nucleotide hnRNP-E1 RNA cis element (to detect intracellular RNA-protein complexes composed of the 25-nucleotide hnRNP-E1 RNA cis element bound to hnRNP-E1) after a 2-h exposure of placental 1584-HF cells to increasing concentrations of l-homocysteine (lanes 1–5) (3), or under basal conditions in 1584-HF and 1584-LF cells (lanes 6 and 7) (D). The 25-nucleotide antisense probe was reacted with hnRNP-E1 plasmid and an FR plasmid (lanes 8 and 9). Reaction of a [35S]-18-nucleotide FR mRNA cis element probe with an FR plasmid (lane 2) or hnRNP-E1 plasmid (lane 1) (E). Results of densitometric scans of the signals are shown below the slot-blot and are presented as means ± SDs, n = 3. Labeled means without a common letter differ, P < 0.05. *Different from control, P < 0.05. Ab, antiserum; CAT, chloramphenicol acetyltransferase; FR, folate receptor α HF, high folate; hnRNP-E1, heterogeneous nuclear ribonucleoprotein E1; IP, immunoprecipitation with anti–heterogeneous nuclear ribonucleoprotein E1 antiserum; LF, low folate.
Transfection of a control pCAT construct into folate-replete placental 1584-HF cells did not show a rise in CAT activity under basal conditions or after incubation with 50-μmol l-homocysteine/L for 3 h at 37°C, as shown in Figure 2C. By contrast, 25-nucleotide hnRNP-E1 cis element–driven CAT reporter constructs led to a 3-fold greater CAT activity over control CAT constructs (even without added homocysteine). This suggested that the basal concentration of thiols (including cysteine and glutathione and l-homocysteine) extant in folate-replete cells (1), could be responsible for facilitating constitutive interaction of endogenous hnRNP-E1 with the 25-nucleotide hnRNP-E1 cis element–linked CAT reporter (Figure 2C). Furthermore, when cells transfected with the 25-nucleotide hnRNP-E1 cis element–driven CAT reporter construct were exposed to 50 μmoll-homocysteine/L for 3 h at 37°C, there was a further significant increase in CAT activity over baseline (Figure 2C). This confirmed that the interaction of endogenous hnRNP-E1 with the 25-nucleotide hnRNP-E1 cis element would be further increased by accumulated intracellular l-homocysteine found in clinical folate deficiency (2). Taken together, these data on placental 1584-HF cells, which were comparable with earlier studies on the interaction of homocysteinylated hnRNP-E1 with 18-nucleotide folate receptor α mRNA cis element in HeLa-IU1 cells (3), predicted that folate deficiency would similarly trigger the translational upregulation of hnRNP-E1 through this RNA-protein interaction.
Next, we sought to capture endogenous RNA-protein complexes composed of hnRNP-E1 mRNA cis element–bound hnRNP-E1 from either folate-replete placental 1584-HF or folate-depleted 1584-LF cells, as well as from 1584-HF cells acutely exposed to l-homocysteine. The specificity of our [35S]labeled antisense hnRNP-E1 mRNA cis element probe, which reacted only with hnRNP-E1 plasmids (lane 8), but not with a plasmid-containing folate receptor α DNA (lane 9), is demonstrated in Figure 2D. In addition, the 18-nucleotide folate receptor α mRNA cis element reacted with the folate receptor α plasmid, but failed to react with the hnRNP-E1 plasmid (Figure 2E); this confirmed that there were no common sequences that could result in false positive signals with the probe used in Figure 2D. There was evidence of hnRNP-E1–bound hnRNP-E1 mRNA cis element signals signifying the constitutive existence of RNA-protein complexes within 1584-HF cells, as shown in Figure 2D, lane 1. Furthermore, experimental induction of the accumulation of l-homocysteine within placental 1584-HF cells led to a progressive dose-dependent increase in signal (Figure 2D, lanes 2–5) compared with baseline values. This reflected the dynamic responsiveness of this system to l-homocysteine (by formation of additional RNA-protein complexes). Because 1584-LF cells accumulated more l-homocysteine than did 1584-HF cells (Figure 3D), this predicted significantly more intracellular RNA-protein complexes captured from 1584-LF cells, as confirmed in Figure 2D (lane 7 compared with lane 6). Taken together, these results demonstrated increased specific hybridization signals with a radiolabeled antisense 25-nucleotide hnRNP-E1 mRNA cis element probe, which reflected the capture of 25-nucleotide hnRNP-E1 mRNA cis element–bound hnRNP-E1 protein complexes within placental 1584-HF cells, both constitutively and in response to l-homocysteine.
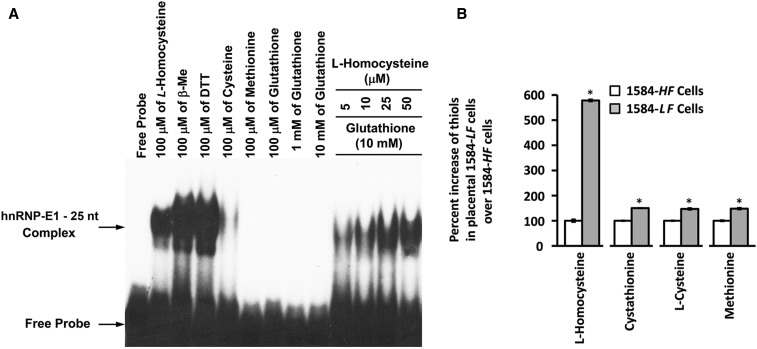
FIGURE 3.
Characterization of the influence of various thiols on the interaction of hnRNP-E1 with the 25-nucleotide hnRNP-E1 mRNA cis element and the accumulation of various thiol amino acids in placental 1584-LF compared with 1584-HF cells. Gel-shift assays on the interaction of 0.5 μg dialyzed purified recombinant GST–hnRNP-E1 with 1 × 105 counts/min of [35S]25-nucleotide hnRNP-E1 mRNA cis element in the presence of various indicated thiols (A). The result is representative of 2 studies that gave comparable results. Comparison of the percentage increase in various thiols in placental 1584-LF over 1584-HF cells (B); the data were taken from Table 5 to highlight the percentage increase in homocysteine over other thiols in folate-depleted cells. *Different from control, P < 0.05. GST, glutathione S-transferase; HF, high folate; hnRNP-E1, wild-type heterogeneous nuclear ribonucleoprotein E1; LF, low folate; nt, nucleotide; β-Me, β-mercaptoethanol.
Comparison of the effect of various thiols on RNA-protein interactions.
There were much stronger RNA-protein signals with 100 μmol nonphysiologic thiols/L (DTT and β-mercaptoethanol) than with physiologic l-homocysteine, as shown in Figure 3A; however, by contrast, signals with equimolar concentrations of other physiologic thiols, such as l-cysteine, were far weaker, and there were no detectable signals with either glutathione or methionine. Despite the inclusion of physiologically relevant concentrations of glutathione (10-mmol/L) in the reaction mixture, there was a dose-dependent increase in RNA-protein complex formation with even 5 μmol/L l-homocysteine. [During the course of these studies, we noted that recent batches of commercially available l-homocysteine led to RNA-protein interactions between hnRNP-E1 and 25-nucleotide hnRNP-E1 mRNA cis element at lower concentrations than previously noted (3)].
When the KD of the RNA-protein interaction was tested in the presence of 10 mmol glutathione/L and equimolar concentrations of l-homocysteine and l-cysteine, there was a significantly higher affinity obtained with l-homocysteine (quantified by a lower KD) (Table 4). These studies were generally comparable with RNA-protein interactions involving folate receptor α mRNA binding to hnRNP-E1 (3).
TABLE 4.
KD of the RNA-protein interaction of the [35S]25-nucleotide hnRNP-E1 mRNA cis element and purified recombinant GST–hnRNP-E1 protein in the presence of 10 mmol glutathione/L and either 15 μmol l-homocysteine/L or 15 μmol l-cysteine/L1
| Thiol, 15 μM | KD, nM |
| l-Homocysteine | 1.18 ± 0.27a |
| l-Cysteine | 2.38 ± 0.56b |
Values are means ± SDs, n = 3 (means of triplicates). Labeled means without a common superscript letter differ, P < 0.05. GST, glutathione S-transferase; hnRNP-E1, wild-type heterogeneous nuclear ribonucleoprotein E1; KD, dissociation constant.
There was a <2-fold increase in other thiols (cystathionine, cysteine, and methionine) in 1584-LF cells compared with in1584-HF cells. However, there was a 5.75-fold increase in homocysteine in 1584-LF cells compared with in 1584-HF cells (184-μmol/L compared with 32-μmol/L, respectively) (Figure 3B; Table 5). Thus, although the basal concentration of cysteine in 1584-HF cells was high (327 μmol/L), and rose only 1.5-fold more in 1584-LF cells, based on data in Figure 3A, it was not likely that these concentrations would have had as much influence in increasing the binding affinity of the RNA-protein interaction as l-homocysteine, which rose nearly 6-fold in folate-deficient cells.
TABLE 5.
Concentration of various thiol amino acids in placental 1584-HF cells and 1584-LF cells1
| Thiol amino acids, μM | 1584-HF cells | 1584-LF cells |
| Homocysteine | 31.9 ± 1.8 | 184.4 ± 1.5* |
| Cystathionine | 19.4 ± 0.3 | 29.1 ± 0.3* |
| Cysteine | 327.3 ± 5.2 | 481.2 ± 12.6* |
| Methionine | 46.7 ± 1.3 | 69.2 ± 1.9* |
Values are means ± SDs, n = 3 (means of triplicates). *Different from control (1584-HF cells), P < 0.05. HF, high folate; LF, low folate.
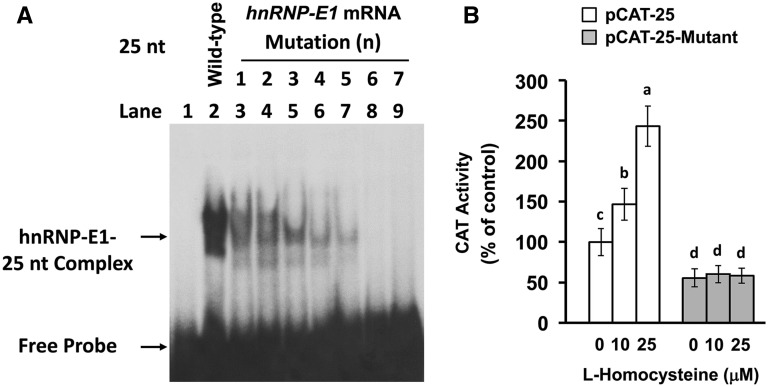
Effect of mutation of the 25-nucleotide hnRNP-E1 cis element on the RNA-protein Interaction.
Studies with 18-nucleotide folate receptor α mRNA cis element suggested that the sequence motif of CUCC in tandem (with intervening nucleotides) could be important for interaction with hnRNP-E1 (5, 18). Because the 25-nucleotide hnRNP-E1 mRNA cis element contained 3 such distinct CUCC sequences, we evaluated the effect of mutating a single nucleotide within each of these CUCC sequences on the specificity of binding of hnRNP-E1 to the hnRNP-E1 mRNA cis element with the use of gel-shift assays. We noted that various mutations (numbered 1–5) (Figure 4A and Table 6) led to progressively quenched RNA-protein interaction signals. Of significance, mutation no. 6 (involving the first and third CUCC sequences) led to complete quenching of the signal. Thus, the integrity of the first and third CUCC sequence motif in the 25-nucleotide hnRNP-E1 mRNA cis element was critical for interaction with homocysteinylated hnRNP-E1 in vitro. Moreover, although wild-type 25-nucleotide hnRNP-E1 cis element placed proximal to the CAT reporter led to progressively increased and significant CAT reporter signals in response to exogenously added l-homocysteine (Figure 4B), mutation no. 6 of the hnRNP-E1 mRNA cis element placed proximal to CAT reporters yielded no such response. Thus, these mutations in the 25-nucleotide hnRNP-E1 cis element were also functionally relevant within placental cells.
FIGURE 4.
Effect of specific mutations within the 25-nucleotide hnRNP-E1 mRNA cis element on the RNA-protein interaction, and CAT reporter activity after transfection into placental 1584-HF cells. Gel-shift assays to assess the capacity of individual indicated 25-nucleotide hnRNP-E1 RNA mutants generated in Table 6 (numbered from 1 to 7) to bind to purified recombinant GST–hnRNP-E1 in the presence of 15 μmol l-homocysteine/L (A). Comparison of CAT reporter activity after transfection of either a wild-type 25-nucleotide hnRNP-E1 cis element–driven CAT reporter construct or a mutated 25-nucleotide hnRNP-E1 cis element–bearing mutant no. 6–driven CAT reporter construct into placental 1584-HF cells (B). Values are means ± SDs, n = 3 (means of triplicates). Labeled means without a common letter differ, P < 0.05. CAT, chloramphenicol acetyltransferase; GST, glutathione S-transferase; HF, high folate; hnRNP-E1, wild-type heterogeneous nuclear ribonucleoprotein E1; nt, nucleotide.
TABLE 6.
Primer sequences for various point mutations within the 25-nucleotide hnRNP-E1 mRNA cis element1
| Primer | Sequence |
| Wild-type | |
| Primer 0F | 5′-cacggaattCTCCCGCCCGCTCCCGCTCGCTCCCaagcttgggt-3′ |
| Primer 0R | 5′-acccaagcttGGGAGCGAGCGGGAGCGGGCGGGAgaattccgtg-3′ |
| Mutation 1 | |
| Primer 1F | 5′-cacggaattCTTCCGCCCGCTCCCGCTCGCTCCCaagcttgggt-3′ |
| Primer 1R | 5′-acccaagcttGGGAGCGAGCGGGAGCGGGCGGAAgaattccgtg-3′ |
| Mutation 2 | |
| Primer 2F | 5′-cacggaattCTCCCGCCCGCTTCCGCTCGCTCCCaagcttgggt-3′ |
| Primer 2R | 5′-acccaagcttGGGAGCGAGCGGAAGCGGGCGGGAgaattccgtg-3′ |
| Mutation 3 | |
| Primer 3F | 5′-cacggaattCTCCCGCCCGCTCCCGCTCGCTTCCaagcttgggt-3′ |
| Primer 3R | 5′-acccaagcttGGAAGCGAGCGGGAGCGGGCGGGAgaattccgtg-3′ |
| Mutation 4 | |
| Primer 4F | 5′-cacggaattCTTCCGCCCGCTTCCGCTCGCTCCCaagcttgggt-3′ |
| Primer 4R | 5′-acccaagcttGGGAGCGAGCGGAAGCGGGCGGAAgaattccgtg-3′ |
| Mutation 5 | |
| Primer 5F | 5′-cacggaattCTCCCGCCCGCTTCCGCTCGCTTCCaagcttgggt-3′ |
| Primer 5R | 5′-acccaagcttGGAAGCGAGCGGAAGCGGGCGGGAgaattccgtg-3′ |
| Mutation 6 | |
| Primer 6F | 5′-cacggaattCTTCCGCCCGCTCCCGCTCGCTTCCaagcttgggt-3′ |
| Primer 6R | 5′-acccaagcttGGAAGCGAGCGGGAGCGGGCGGAAgaattccgtg-3′ |
| Mutation 7 | |
| Primer 7F | 5′-cacggaattCTTCCGCCCGCTTCCGCTCGCTTCCaagcttgggt-3′ |
| Primer 7R | 5′-acccaagcttGGAAGCGAGCGGAAGCGGGCGGAAgaattccgtg-3′ |
The point mutation sites (numbered Mutations 1–7) are shown in bold capital letters, and are also underlined. aagctt, restriction enzyme HindIII site; gaattc, restriction enzyme EcoRI site; hnRNP-E1, heterogeneous nuclear ribonucleoprotein E1.
Effect of transfection of specific antisense oligonucleotides on the biosynthesis of hnRNP-E1.
Transfection of wild-type and scrambled oligonucleotides did not significantly alter the biosynthetic rate of hnRNP-E1 (similar rates of 1.0 fmol [35S]cysteine · mg protein−1 · h−1 incorporated into hnRNP-E1); by contrast, transfection of antisense oligonucleotides to the 25-nucleotide mRNA cis element of hnRNP-E1 led to a significant reduction in the biosynthetic rate of newly synthesized hnRNP-E1 of 0.62 fmol [35S]cysteine · mg protein−1 · h−1 incorporated into hnRNP-E1 (Table 7).
TABLE 7.
Effect of transfection of wild-type, scrambled, and antisense oligonucleotides to the 25-nucleotide hnRNP-E1 mRNA cis element on the rate of biosynthesis of [35S]cysteine–hnRNP-E1 protein in placental 1584-HF cells1
| Oligonucleotide transfected into placental 1584-HF cells | [35S]cysteine–hnRNP-E1 protein, fmol · mg protein−1 · h−1 |
| Wild-type | 1.04 ± 0.02a |
| Scrambled | 1.01 ± 0.03a |
| Antisense | 0.62 ± 0.02b |
Values are means ± SDs, n = 3 (means of triplicates). Means in a column without a common superscript letter differ, P < 0.05. HF, high folate; hnRNP-E1, wild-type heterogeneous nuclear ribonucleoprotein E1.
Effect of RNA interference of hnRNP-E1 mRNA on the biosynthesis of hnRNP-E1.
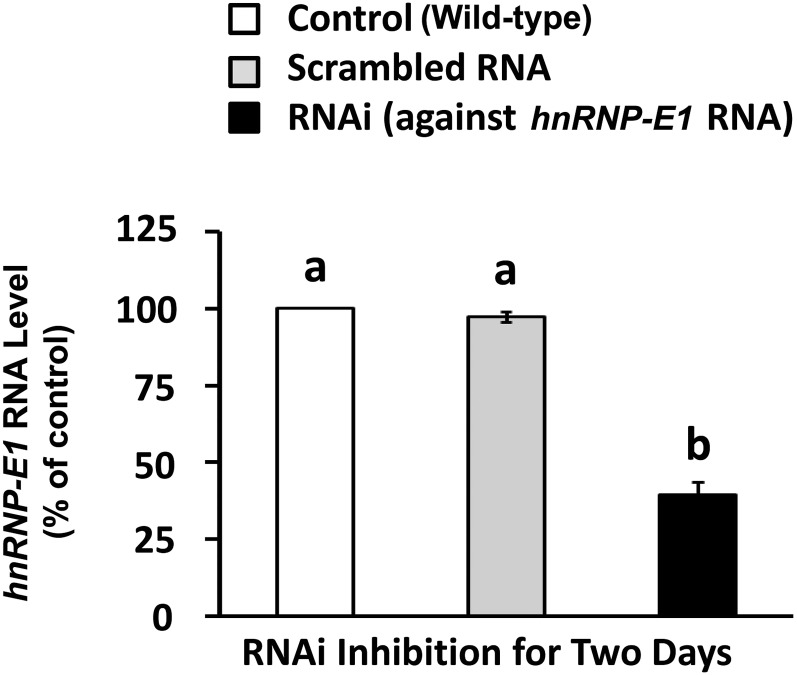
The transfection of siRNA directed specifically to hnRNP-E1 mRNA, which did not have an impact on hnRNP-E2 (3), led to less cell death after 2 d, and achieved an ∼60% reduction of hnRNP-E1 mRNA (Figure 5). Compared with basal control values that used scrambled RNA that had comparable values as wild-type cells, there was a significant reduction in rate of biosynthesis of hnRNP-E1 and folate receptor proteins (Table 8). Specifically, there was a reduction in the rate of newly synthesized hnRNP-E1 from 0.95 to 0.32 fmol l-[35S]cysteine incorporated into hnRNP-E1 · mg protein−1 · h−1 in scrambled compared with siRNA-treated cells, respectively (Table 8). Likewise, there was a reduction in the rate of newly synthesized folate receptor from 1.57 to 0.35 fmol l-[35S]cysteine incorporated into folate receptor · mg protein−1 · h−1 in scrambled compared with siRNA-treated cells, respectively (Table 8). This pointed to the specificity of effects of RNA interference of hnRNP-E1 mRNA on the biosynthesis of hnRNP-E1 within cells. Moreover, because folate receptor biosynthesis is mediated by hnRNP-E1 (5), the coreduction of biosynthesis of folate receptor also verified the effects of RNA interference of hnRNP-E1 mRNA on the reduction of hnRNP-E1 (3). These studies unambiguously confirmed that hnRNP-E1 was directly involved in the cellular biosynthesis of both hnRNP-E1 and folate receptor proteins. Moreover, because there was a reduction in the basal rate of hnRNP-E1 (and folate receptor) biosynthesis even in placental 1584-HF cells, these data suggest that the constitutive expression of hnRNP-E1 (and folate receptors) in these cells is also mediated by hnRNP-E1 interaction with both 25-nucleotide hnRNP-E1 mRNA cis elements and 18-nucleotide folate receptor α mRNA cis elements. Together, the data in Figures 4 and 5 and Tables 7 and 8 further confirm the specificity of the RNA-protein interaction (involving hnRNP-E1 binding to its own mRNA cis element) for the biosynthesis of hnRNP-E1 at the posttranscriptional level in placental 1584-HF cells.
FIGURE 5.
Evidence of the specificity of RNAi against hnRNP-E1 mRNA in placental 1584-HF cells. Values are means ± SDs, n = 3 (means of triplicates). Labeled means without a common letter differ, P < 0.05. HF, high folate; hnRNP-E1, heterogeneous nuclear ribonucleoprotein E1; RNAi, RNA interference.
TABLE 8.
Evidence of the specificity of RNAi against hnRNP-E1 mRNA in perturbing the biosynthetic rate of newly synthesized hnRNP-E1 and folate receptor proteins in placental 1584-HF cells1
| RNAi transfected in placental 1584-HF cells | [35S]cysteine–hnRNP-E1, fmol · mg protein−1 · h−1 | [35S]cysteine–folate receptor, fmol · mg protein−1 · h−1 |
| Wild-type 25-nucleotide hnRNP-E1 RNA | 1.02 ± 0.07a | 1.58 ± 0.02a |
| Scrambled 25-nucleotide RNA | 0.95 ± 0.04a | 1.57 ± 0.05a |
| RNAi against 25-nucleotide hnRNP-E1 mRNA | 0.32 ± 0.02b | 0.35 ± 0.02b |
Values are means ± SDs, n = 3 (means of triplicates). Means in a column without a common superscript letter differ, P < 0.05. HF, high folate; hnRNP-E1, heterogeneous nuclear ribonucleoprotein E1; RNAi, RNA interference.
Triggering the biosynthesis of folate receptor and hnRNP-E1 proteins by the introduction of (HA)-hnRNP-E1(C293S) mutant proteins into cells.
(HA)-hnRNP-E1(C293S) mutant proteins interact with both the 25-nucleotide hnRNP-E1 RNA cis element and 18-nucleotide folate receptor α mRNA cis element in the absence of l-homocysteine in vitro (Figure 1, Supplemental Figure 2, and Tables 1 and 3). Therefore, we tested the potential of liposome-transfected (HA)-hnRNP-E1(C293S) mutant proteins to bind to both of these endogenous hnRNP-E1 and folate receptor mRNA cis elements and trigger the biosynthesis of hnRNP-E1 and folate receptors in 1584-HF cells (where homocysteine concentrations are at basal levels). Wild-type hnRNP-E1 proteins induced a significant increase in the biosynthesis of [35S]hnRNP-E1 compared with basal rates (1.12 compared with 0.53 fmol [35S]cysteine incorporated into hnRNP-E1 · mg protein−1 · h−1, respectively) (Table 9). Similarly, the wild-type–like hnRNP-E1(G292A) mutant also resulted in a comparable rise in rates of [35S]cysteine–hnRNP-E1 biosynthesis (1.03 fmol · mg protein−1 · h−1) as wild-type hnRNP-E1. By contrast, the introduction of (HA)-hnRNP-E1(C293S) mutant proteins led to a significant (>6-fold) increase in the rate of biosynthesis of [35S]cysteine–hnRNP-E1 (to 6.70-fmol · mg protein−1 · h−1). There were also comparable results in the stimulation of the biosynthesis of newly synthesized [35S]cysteine–folate receptors after transfection of these proteins into placental 1584-HF cells (Table 9). Whereas control wild-type hnRNP-E1 and wild-type–like hnRNP-E1(G292A) mutant proteins comparably stimulated the biosynthesis of [35S]cysteine–folate receptors (1.62 compared with 1.79 fmol · mg protein−1 · h−1, respectively) over basal values, the (HA)-hnRNP-E1(C293S) mutant significantly induced a nearly 4-fold increase of [35S]cysteine–folate receptors (up to 6.62-fmol · mg protein−1 · h−1) over controls (Table 9). Thus, the transfection of liposomes bearing purified wild-type, wild-type–like hnRNP-E1(G292A) mutant, and (HA)-hnRNP-E1(C293S) mutant proteins into cells led to functional interactions with both endogenous 25-nucleotide hnRNP-E1 mRNA cis elements and 18-nucleotide folate receptor mRNA cis elements, leading to significantly increased biosynthesis of hnRNP-E1 and folate receptor proteins, respectively.
TABLE 9.
Effect of the introduction of equivalent amounts of various purified recombinant wild-type or mutant GST–hnRNP-E1 proteins into placental 1584-HF cells on the biosynthetic rate of newly synthesized [35S]cysteine–hnRNP-E1 and [35S]cysteine–folate receptor proteins1
| Protein transfected in placental 1584-HF cells | [35S]cysteine–hnRNP-E1, fmol · mg protein−1 · h−1 | [35S]cysteine–folate receptor, fmol · mg protein−1 · h−1 |
| No transfection | 0.53 ± 0.17a | 0.95 ± 0.12a |
| GST–hnRNP-E1 | 1.12 ± 0.23b | 1.62 ± 0.11b |
| GST–hnRNP-E1(G292A) | 1.03 ± 0.21b | 1.79 ± 0.13b |
| GST–hnRNP-E1(C293S) | 6.70 ± 1.74c | 6.62 ± 0.74c |
Values are means ± SDs, n = 3 (means of triplicates). Means without a common superscript letter differ, P < 0.05. GST, glutathione S-transferase; HF, high folate; hnRNP-E1, wild-type heterogeneous nuclear ribonucleoprotein E1; hnRNP-E1(C293S), highest affinity–heterogeneous nuclear ribonucleoprotein E1 (C293S)–mutant protein; hnRNP-E1(G292A), wild-type–like heterogeneous nuclear ribonucleoprotein E1 (G292A)–mutant protein.
Effect of iron(II) on the interaction between hnRNP-E1 and the 25-nucleotide hnRNP-E1 mRNA cis element.
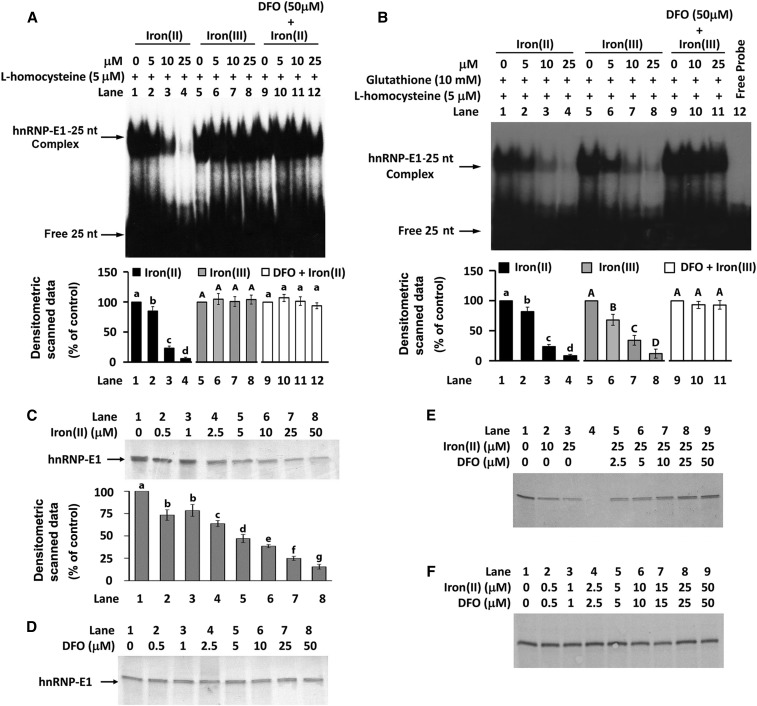
hnRNP-E1 is an iron(II)-binding protein that directly binds ferrous iron with micromolar affinity (3:1 ratio of iron to hnRNP-E1) (24, 25). So we evaluated whether iron(II) had a specific modulating effect in the homocysteine-induced RNA-protein interaction. The sensitivity of the system was markedly improved by the use of fresh radiolabeled [35S]25-nucleotide hnRNP-E1 cis element, freshly opened vials of commercially purchased l-homocysteine and iron(II), and incubating iron(II) with dialyzed, purified GST–hnRNP-E1 before the addition of radiolabeled RNA and l-homocysteine. In the presence of a fixed concentration of l-homocysteine, iron(II) exerted a dose-dependent inhibitory effect on the formation of these RNA-protein complexes (Figure 6A, lanes 2–4). By contrast, iron(III) had no such influence (Figure 6A, lanes 6–8). The quenching effect of iron(II) was nearly completely blocked by the addition of 50-μmol deferoxamine/L to the reaction mixture (Figure 6A, lanes 10–12); this confirmed the specificity of the inhibitory effect of iron(II) on the formation of these RNA-protein complexes in the presence of l-homocysteine. To eliminate the potential of iron(II) to catalyze oxidation changes in proteins (hnRNP-E1) and potentially nucleic acids (hnRNP-E1 mRNA cis element) that could have also led to reduced RNA-protein interaction, these experiments were repeated in the presence of physiologically relevant concentrations of glutathione; under these conditions, iron(III) is converted to iron(II) at pH 7.0 (26, 27). Accordingly, when 10 mmol glutathione/L was included in all reaction mixtures, both iron(II) and iron(III) exerted a comparable dose-dependent reduction in RNA-complex formation on gel-shift assays (Figure 6B, lanes 2–4 compared with lanes 6–8). The ability of deferoxamine to eliminate the quenching effect of iron(III) in the presence of glutathione confirmed the specificity of effect of iron(III) that was converted to iron(II) (Figure 6B, lanes 10–11). Thus, the trivial possibility of direct oxidative effects of iron(II) on hnRNP-E1 or RNA was unlikely. To confirm that this was a general effect of iron(II) on the capacity of hnRNP-E1 to interact with other target mRNAs in the presence of l-homocysteine, we were able to reproduce this effect of iron(II) when 18-nucleotide folate receptor α mRNA cis element was substituted for the 25-nucleotide hnRNP-E1 mRNA cis element (data not shown).
FIGURE 6.
Characterization of the specific effect of iron(II) or iron(III) on the l-homocysteine–triggered RNA-protein interaction involving 25-nucleotide hnRNP-E1 mRNA cis element and purified recombinant GST–hnRNP-E1 by gel-shift assay (A and B) and in vitro translation assay (C–F). Effect of iron(II) or iron(III) on the RNA-protein interaction in the absence (A) or presence (B) of 10 mM glutathione. Effect of the addition of either iron(II) (C), deferoxamine (D), or varying combinations of iron(II) and deferoxamine (E and F) on the translation of hnRNP-E1 in vitro. Each of these representative gels is from 3 independent sets of experiments that gave comparable data with <10% variation. The pooled densitometric scanned data from 3 independent experiments (A–C) are shown as a bar graph below one representative gel; values are means ± SDs, n = 3. Labeled means without a common letter differ, P < 0.05. In each panel, uppercase letters are compared only with other uppercase letters, and lowercase letters are compared only with other lowercase letters. DFO, deferoxamine; GST, glutathione S-transferase; hnRNP-E1, wild-type heterogeneous nuclear ribonucleoprotein E1; iron(II), ferrous sulfate heptahydrate; iron(III), ferric chloride hexahydrate; nt, nucleotide.
Iron(II) also had a dose-dependent impact in quenching the in vitro translation of hnRNP-E1 in the presence of 4.1-mmol β-mercaptoethanol/L, as shown in Figure 6C; of significance, even 0.5–1 μmol iron(II)/L consistently led to a negative effect compared with basal levels, suggesting that there could be a physiologic effect within cells. Conversely, deferoxamine alone did not independently influence in vitro translation of hnRNP-E1 (Figure 6D); this suggested that there was little free iron(II) available for chelation within the reticulocyte-rich lysates. However, deferoxamine, in a dose-dependent manner, was capable of progressively reversing the (quenching) effect of 25 μmol iron(II)/L on the translation of hnRNP-E1 (Figure 6E, lanes 5–9). An equimolar concentration of deferoxamine added to iron(II) was also effective in preventing an inhibitory effect of iron(II) on the translation of hnRNP-E1 (Figure 6F). Finally, iron(II) induced a dose-dependent reduction in RNA-protein–binding affinity, as indicated by the progressive increase in KD (Table 2). Taken together, these data suggested that whereas hnRNP-E1 readily reacted with target mRNA cis elements in the presence l-homocysteine, the prior binding of iron(II) to hnRNP-E1 (24, 25) significantly quenched this l-homocysteine–triggered RNA-protein interaction.
Dual upregulation of hnRNP-E1 and folate receptors in tumor xenografts propagated in folate-deficient mice.
Because hnRNP-E1 could bind 2 distinct cis elements in cultured placental cells, we pursued investigations into whether there was dual upregulation of folate receptors and hnRNP-E1 in vivo in placentas of pregnant mice fed a folate-deficient diet compared with folate-replete dams (13). However, there was significant apoptosis of megaloblastic placental trophoblastic cells in situ, which led to significant architectural changes only in placentas of folate-deficient dams (Supplemental Figure 4). Because the trophoblastic cell mass is also the major locus of expression of folate receptors and hnRNP-E1, it was not meaningful to directly compare the expression of folate receptors or the hnRNP-E1 of placentas between folate-replete and folate-deficient dams.
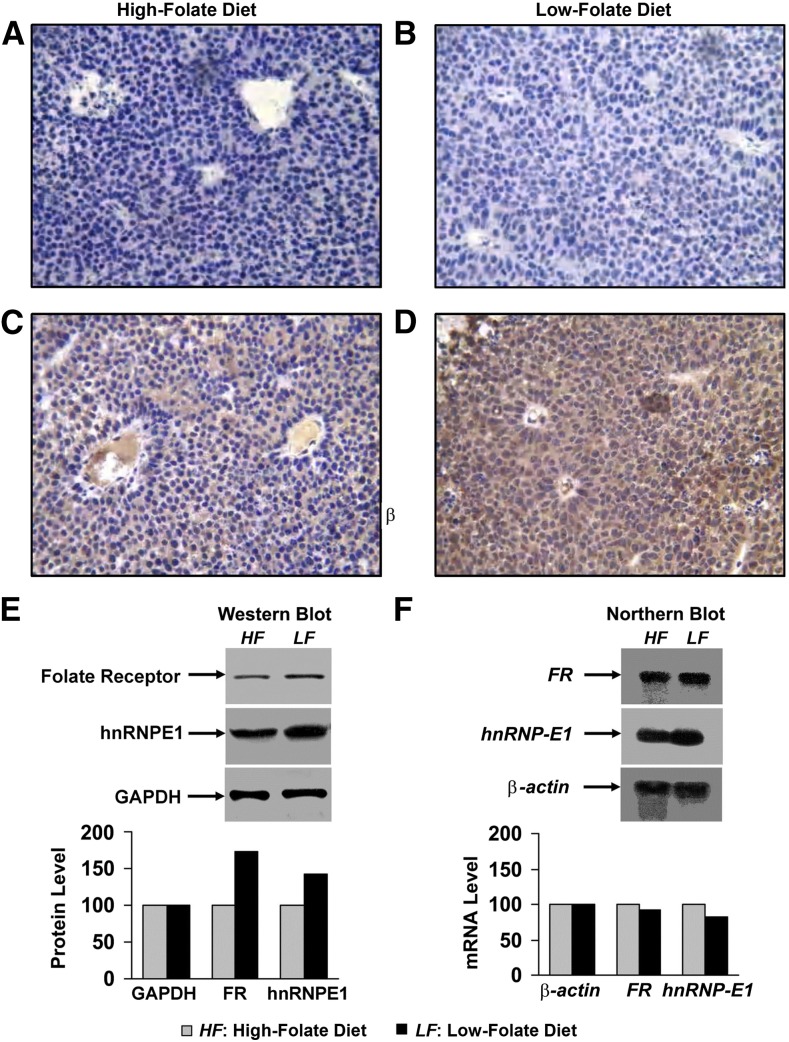
Accordingly, we evaluated the possibility of a dual upregulation of both folate receptor and hnRNP-E1 in HeLa-IU1 cell–derived tumor xenografts in an athymic mouse model (19), in which the tumor was homogeneous and the folate content of the murine diet was controlled (13). Mice fed a folate-restricted diet (120 nmol folate/kg diet) compared with a folate-replete diet (1200 nmol folate/kg diet) throughout the 4 wk (13) before injection of HeLa-IU1 cells into their flanks (19) and over the ensuing 4 wk did not exhibit differences in the amount of food consumed or their weight. However, the folate-restricted diet resulted in a serum homocysteine of 185 μmol/L (consistent with severe folate deficiency), which was nearly 10 times more than the serum homocysteine (19 μmol/L) of control mice (13). Immunohistochemical studies for tumor hnRNP-E1 expression with the use of anti–hnRNP-E1 antiserum revealed enhanced brown staining in folate-deficient xenografts (Figure 7D) compared with folate-replete xenografts (Figure 7C); by contrast, there was little staining with the use of nonimmune serum (Figure 7A, B). Compared with tumors from mice fed a folate-replete diet (Figure 7A, C), tumors that developed in folate-deficient mice (Figure 7B, D) exhibited megaloblastic features with larger nuclei and open chromatin and an abundant cytoplasm (reflecting a high nuclear:cytoplasmic ratio) and consequently fewer cells per 40× magnification field. Western blot analysis with the use of anti-folate receptor antiserum, as well as anti–hnRNP-E1 antiserum, confirmed an increased signal reflecting upregulation of both folate receptors and hnRNP-E1 in tumor xenografts of folate-deficient mice. Moreover, northern blots (Figure 7F) showed no difference in mRNA levels for both folate receptor and hnRNP-E1 mRNA. Thus, there was evidence of upregulation for both folate receptor and hnRNP-E1 protein concentrations within the xenografts of animals that experienced folate deficiency, which likely occurred at the posttranscriptional level. Parenthetically, although immunohistochemistry of the tumors revealed more abundant staining of hnRNP-E1 in cytoplasm and nuclei compared with western blots of tumors (cytosol), the concordance between these distinctly different methods, which confirmed upregulation of hnRNP-E1 and folate receptors in response to folate deficiency, was the important finding. These data were also concordant with those of cultured placental cells propagated in LF medium, as well as the observed effects of homocysteinylated hnRNP-E1 and (HA)-hnRNP-E1(C293S) mutant proteins in triggering the biosynthesis of folate receptors and hnRNP-E1 in these cells.
FIGURE 7.
Dual upregulation of hnRNP-E1 and FRs at the posttranscriptional level in HeLa-IU1–derived tumor xenografts of mice fed either a folate-replete diet (A and C) or a folate-deficient diet (B and D) for 1 mo before and after implantation of 1 million HeLa-IU1 cells into their flanks. Because of prohibitive costs, this longitudinal experiment was carried out only once; similar results were obtained from 2 randomly selected tumors each from folate-replete mice (n = 6) and folate-deficient mice (n = 5). Immunohistochemistry for hnRNP-E1 with the use of nonimmune serum (A and B) compared with anti–hnRNP-E1 antiserum (C and D). Magnification was 40×. Western blots (50-μg tumor protein probed with either anti-folate receptor antiserum, anti–hnRNP-E1 antiserum, or anti-GAPDH antibodies) to assess the expression of folate receptor and hnRNP-E1 in the tumor (E). Densitometric evaluation of folate receptor and hnRNP-E1 protein signals in relation to GAPDH is shown below the gels. Northern blots (20 μg total tumor RNA) to assess FR α mRNA and hnRNP-E1 mRNA in tumor xenografts (F). Densitometric scanning of the signals from northern blots was adjusted by a loading control (β-actin) and is shown below the gel. The HF diet contained 1200 nmol folate/kg; the LF diet contained 400 nmol folate/kg. FR, folate receptor; hnRNP-E1, heterogeneous nuclear ribonucleoprotein E1.
Discussion
Perpetuation of hnRNP-E1 biosynthesis during folate deficiency at the translational level.
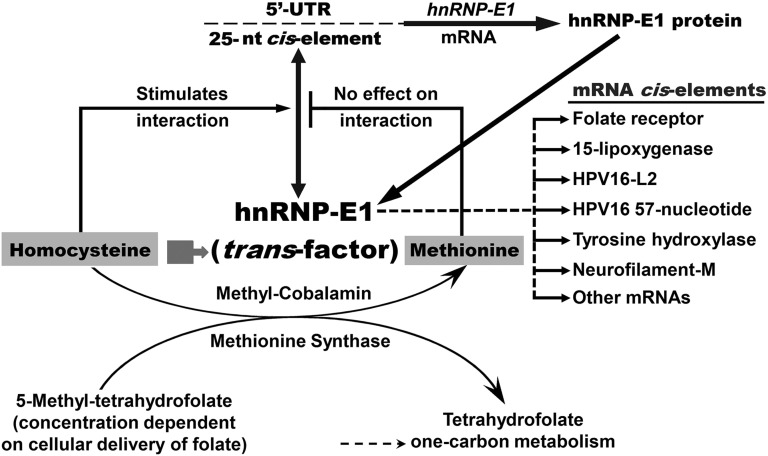
This paper provides support for a model that invokes physiologic auto upregulation of hnRNP-E1 by homocysteinylated hnRNP-E1 during prolonged folate deficiency (Figure 8). Thus, during folate deficiency, with an intracellular accumulation of l-homocysteine, the high affinity interaction of intracellular homocysteinylated hnRNP-E1 with a 25-nucleotide cis element in the 5′-UTR of hnRNP-E1 mRNA triggers an increase in biosynthesis of hnRNP-E1, which results in the upregulation of hnRNP-E1 proteins. With ongoing folate deficiency, newly synthesized hnRNP-E1 would also become homocysteinylated and perpetuate a positive-feedback loop, which generates hnRNP-E1 (and folate receptors) for as long as folate deficiency persists (Figure 8). This amplified and homocysteinylated hnRNP-E1 will also interact (with varying degrees of affinity) with other mRNA members of the nutrition-sensitive folate-responsive posttranscriptional RNA operon during folate deficiency. Because this interaction is sensitive to low concentrations of l-homocysteine, even the small amount of hnRNP-E1 generated during mild folate deficiency could become homocysteinylated and trigger the translation of additional hnRNP-E1, leading to the amplification of this circuit, akin to a “snowball effect” (Figure 8). Conversely, relief of folate deficiency will turn off this positive feedback loop by reactivating one-carbon metabolism and reducing cellular homocysteine concentrations via conversion to methionine (2), which has no effect on the RNA-protein interaction (Figure 8). Meanwhile, the residual homocysteinylated hnRNP-E1 will eventually be degraded (3) while constitutive biosynthesis of hnRNP-E1 is reestablished. This posttranscriptional upregulation of hnRNP-E1 during folate deficiency occurs in a manner similar to that of the physiologic upregulation of folate receptors (3). Therefore, hnRNP-E1 mRNA is yet another member of this posttranscriptional RNA operon that is orchestrated by homocysteinylated hnRNP-E1 during folate deficiency (3).
FIGURE 8.
A model for the interaction between the 25-nucleotide hnRNP-E1 mRNA cis element and homocysteinylated hnRNP-E1, which amplifies hnRNP-E1 (and thereby facilitates ongoing upregulation of folate receptors during prolonged folate deficiency). The cellular accumulation of homocysteine during folate deficiency (2, 22, 23, 28) results in homocysteinylation of hnRNP-E1 (short broad arrow), which unmasks a high affinity mRNA-binding site for target mRNAs (3). In the event of interaction of homocysteinylated hnRNP-E1 with its own 25-nucleotide hnRNP-E1 mRNA cis element, this positive-feedback loop will continue to generate more hnRNP-E1, resulting in the amplification of hnRNP-E1. In this model, only folate replenishment can reduce cellular homocysteine concentrations to a basal state that will turn off this autoregulatory positive feedback loop involving hnRNP-E1 (3). The mRNA-binding site in homocysteinylated hnRNP-E1 also accommodates several diverse mRNAs (some of these are identified as “mRNA cis elements”). The interaction with these and other mRNA members of the nutrition-sensitive posttranscriptional RNA operon (1, 3, 5, 17) with homocysteinylated hnRNP-E1 will result in either up- or downregulation of these mRNA-encoded proteins, many of which may contribute to the cellular features observed in folate-deficient megaloblastic cells. The effect of iron(II) on hnRNP-E1 and other enzyme pathways leading to the intracellular accumulation of l-homocysteine is not shown. hnRNP-E1, heterogeneous nuclear ribonucleoprotein E1; HPV16, human papillomavirus type 16; iron(II), ferrous sulfate heptahydrate; neurofilament-M, neuronal intermediate neurofilament–middle molecular mass; nt, nucleotide; 5′-UTR, 5′-untranslated region.
Although, to our knowledge, the finding that hnRNP-E1 interacts with its own mRNA cis element under physiologic conditions has not been reported, Waggoner and Liebhaber (29) earlier noted that αCP2 (also known as hnRNP-E2) mRNA was associated with specifically immunoprecipitated αCP2/hnRNP-E2. However, despite their prescient suggestion of possible autoregulatory control of αCP2 expression (29), to our knowledge, no formal studies had investigated the physiologic basis for this interaction, particularly as it relates to nutrition. Our data provide a physiologic context for specific interaction between homocysteinylated hnRNP-E1 and its own 25-nucleotide hnRNP-E1 mRNA cis element.
Earlier, we determined that, among the various physiologically relevant thiols, hnRNP-E1 activation and mRNA-binding was optimal with l-homocysteine, but far less so with DL–homocysteine, homocysteine thiolactone, l-cysteine, and glutathione (1, 3, and this paper); however, d-homocysteine and methionine did not have any effect on triggering this RNA-protein interaction involving hnRNP-E1 binding to folate receptor mRNA (1, 5) and hnRNP-E1 mRNA cis element (this paper), as well as to other known target cis elements (17). Based on these considerations, it is biologically plausible that other vitamin deficiencies (of vitamin B-12 and vitamin B-6) and other genetic defects in folate and cobalamin metabolism, which likewise result in accumulation of this thiol intracellularly, would also result in homocysteinylation and activation of hnRNP-E1 to interact with its target RNA cis elements as a collateral, but secondary effect, as discussed earlier (3). This warrants additional study in animal models.
Although in vitro assays that used purified components revealed that RNA-protein interaction can occur with as low as 2.5 μmol l-homocysteine/L, the precise concentration of free intracellular homocysteine that can trigger the interaction of hnRNP-E1 with target mRNA during mild, moderate, and severe folate deficiency is not known. The current state-of-the-art assay for measuring homocysteine in biological specimens detects the sum of all free and protein-bound homocysteine (3), so that fraction of free homocysteine—and other thiols—that can react within hnRNP-E1 in cells remains to be determined. Moreover, distinguishing between that fraction of (activated) homocysteinylated hnRNP-E1 and unmodified hnRNP-E1 in cells at different degrees of folate deficiency awaits additional refinement in the separation of these fractions.
There is some evidence for a potential (albeit minor) role of hnRNP-E1 in the transcription of folate receptor (1, 3, 5). However, because Mayanil’s laboratory has determined that the folate receptor itself is a transcription factor (30), and amplification of hnRNP-E1 during prolonged folate deficiency affects the upregulation of folate receptors (3 and this paper), it will be of interest to identify the entire repertoire of downstream genes that are independently transcribed by hnRNP-E1 and folate receptors during folate deficiency.
Because homocysteine that accumulates in cells leaks out (1) and median values for elevation of serum homocysteine among several cohorts with clinical folate and vitamin B-12 deficiency is ∼50 and 70 μmol/L, respectively (22, 23, 28), such concentrations are capable of homocysteinylation of intracellular hnRNP-E1 and triggering high affinity RNA-protein interactions in vivo. This predicts the upregulation of hnRNP-E1 and posttranscriptional engagement of mRNAs comprising its posttranscriptional RNA operon in both vitamin B-12 and folate deficiency (2, 22, 23, 28). Indeed, the common clinical hematologic manifestations of folate and vitamin B-12 deficiency likely involve contributions from this nutrition-sensitive posttranscriptional RNA operon.
Our data on the upregulation of folate receptors in HeLa-IU1–derived tumors growing in folate-deficient mice are consistent with the studies by Leamon et al. (31). However, our findings that the dual upregulation of folate receptors and hnRNP-E1 likely occurred at the posttranscriptional level in both placental cells and tumor xenografts in response to folate deficiency suggested that endogenous homocysteinylated hnRNP-E1 likely bound to 2 distinct mRNA cis elements in the same tissue. This was corroborated by experiments involving the modulation of hnRNP-E1 within folate-replete placental cells that used siRNA against hnRNP-E1 mRNA and (HA)-hnRNP-E1(C293S) mutant proteins. These results can now explain the constitutive coexpression of these 2 proteins in several human tissues, such as the placenta (4, 5), cervix (1, 6), reticulocytes, and erythroid precursors (7–12), as well as in several murine fetal tissues of dams that experienced gestational folate deficiency (13). Although homogeneous populations of tumor xenografts had uniform upregulation of hnRNP-E1 in response to folate deficiency, there was selective expression of hnRNP-E1 in heterogeneous tissues in different organs from murine fetuses that experienced folate deficiency in utero (Supplemental Figure 5). Future experiments will need to clarify the basis for these selective transcriptional and/or posttranscriptional events that contributed to these observations.
Effect of iron(II) in modulating the RNA-protein interaction.
Although hnRNP-E1 (also known as PCBP1) is a cellular sensor of folate deficiency (3), it is also an iron-binding protein that is believed to chaperone cytosolic iron to ferritin (24, 25). Although both cysteine and glutathione are candidate ligands for the cytosolic labile iron pool, and bind iron(II) with high affinity (26, 27), the much higher physiologic concentration of glutathione (10 mmol/L) than that of cysteine (<0.5 mmol/L) will dominate the binding of iron. Indeed, Hider and Kong (26, 27) suggested that the major component of the labile iron pool in the cytosol is the simple glutathione-Fe(II) complex, and that this complex will dominate the speciation of iron(II) over cellular iron(II) concentrations of 10−7 to 10−5. We observed that, in the presence of a fixed concentration of l-homocysteine that triggered homocysteinylated hnRNP-E1 binding to hnRNP-E1 mRNA cis element, there was a specific dose-dependent effect of iron(II) in quenching this interaction. Although iron(III) alone had no effect in modulating RNA-protein complex formation, when iron(III) was rapidly reduced to iron(II) by 10 mmol glutathione/L (26, 27), there was a similar specific effect [as iron(II)] in reducing the interaction of homocysteinylated hnRNP-E1 with hnRNP-E1 mRNA cis element. The molecular basis for the effect of iron(II) and glutathione-Fe(II) complex on hnRNP-E1 remains to be clarified.
Despite these findings, it remains unclear whether these effects of the addition of iron(II) evoked experimentally with the use of purified components are biologically relevant in iron-replete cells. This is because all our previous studies involving hnRNP-E1 that documented significant posttranscriptional upregulation of folate receptors in cells (3), and upregulation of hnRNP-E1 in murine fetuses of dams that experienced gestational folate deficiency (13), as well as in cultured placental cells and xenografts in mice (this paper), were conducted under conditions with sufficient iron(II) present. However, a more plausible and clinically relevant scenario in which a role of iron(II) can be better appreciated is during combined iron and folate deficiency, which is found in well over one-half of pregnant women in resource-limited countries (2, 22, 32, 33). In such a scenario, the reduced intracellular availability of iron(II) to interact with hnRNP-E1 would be permissive in facilitating the capacity of l-homocysteine to covalently bind to hnRNP-E1 and unmask its mRNA-binding site, leading to an increased binding affinity for target mRNA (Figure 8). Therefore, the net effects of iron(II) deficiency—when superimposed on a given degree of folate deficiency—could be to further enhance the engagement of member mRNAs (of the nutrition-sensitive posttranscriptional RNA operon) by homocysteinylated hnRNP-E1; this could lead to even more adverse effects than the effects of pure folate deficiency (i.e., iron deficiency has the potential to magnify the adverse effects of folate deficiency). This hypothesis warrants further testing in vivo in animals with combined folate and iron deficiency, particularly because of its potential relevance to fetal neurodevelopment. For example, we documented a dose-dependent interaction of homocysteinylated hnRNP-E1 with tyrosine hydroxylase and neuronal intermediate neurofilament–middle molecular mass (neurofilament M) mRNAs, which was associated with overexpression of tyrosine hydroxylase and neurofilament M proteins, respectively, in the brains of murine fetuses that experienced folate deficiency (3). Therefore, in a fetus that experiences combined iron and folate deficiency, the augmented interaction of homocysteinylated hnRNP-E1 with tyrosine hydroxylase mRNA (3) could lead to an even greater overexpression of tyrosine hydroxylase (34, 35) and predisposition to increased biosynthesis of neurotransmitters (dopamine and norepinephrine). Likewise, the augmented interaction of homocysteinylated hnRNP-E1 with neurofilament M mRNA, leading to greater overexpression of neurofilament M (3), could further perturb its delicately balanced synthesis with neurofilament low- and high-molecular-mass triplet proteins, resulting in abnormal intermediate filaments and disturbed formation of neurons (36–42). Thus, assessing how the superimposition of iron deficiency to folate deficiency can further worsen the already disordered histopathology in the brains of mouse fetuses that experienced folate deficiency in utero (13), and studying how these dual deficiencies can further modulate their postnatal anxiety phenotype (43), warrant further investigation. This is particularly relevant because of 1) the burgeoning literature pointing to clinical correlates of our murine studies (3, 13, 43), in which women with dietary insufficiency of folate during pregnancy appear to have a higher risk of bearing children with neuropsychiatric and/or behavioral problems (44–48), and 2) the widespread prevalence of iron deficiency during pregnancy and acknowledgment of the critical role of iron during human neurodevelopment (49–58).
The spectrum of mRNAs bound by hnRNP-E1 during folate deficiency.
Although the precise number of mRNAs that can interact with homocysteinylated hnRNP-E1 is unclear, earlier studies demonstrated 160 mRNAs that interact with the closely related protein, αCP2 (hnRNP-E2), in a human hematopoietic cell line (28). These mRNAs encoded a plethora of cellular proteins, including those related to components of the cytoskeleton, transcription factors, proto-oncogenes, factors involved in cell signaling, constituents of hemoglobin, and cell proliferation, differentiation, and apoptosis. Because of significant homology in the K-homology domains of hnRNP-E2 and hnRNP-E1 (3), such data predict that the mRNA-binding domain of hnRNP-E1 will also interact with a comparably large number of mRNAs. Indeed, several other studies indicate that hnRNP-E1 interacts with mRNAs involved in cell differentiation, viral mRNAs, neurotransmitters, nerve and neuronal cell components, and cell cycle proteins (1, 3, 5, 8, 9, 14, 15, 17, 18, 21, 36, 59–71). Parenthetically, in every instance in which hnRNP-E1 binds these various mRNAs in the presence of nonphysiologic reducing agents such as DTT and β-mercaptoethanol, there is also an interaction with the physiologically relevant reducing agent, l-homocysteine (1, 3, 5, 18). So it is clearly important to identify the entire spectrum of mRNA members of this posttranscriptional RNA operon that interact with homocysteinylated hnRNP-E1. Another challenge from a pathobiological standpoint is to identify the rank-order and temporal sequence of binding of these diverse mRNAs to hnRNP-E1 as it is progressively homocysteinylated during mild, moderate, and severe clinical folate deficiency. This is because the net effects of up- and downregulation of the proteins encoded by the many different mRNAs belonging to this operon are likely to influence several aspects of the cellular pathobiology of evolving megaloblastic changes during progressive folate deficiency.
Potential clinical consequences of perturbed RNA-protein interactions.
Subtle mutations in several members of the hnRNP family have given rise to diverse clinical syndromes. This includes fragile X syndrome arising from mutations in the fragile X mental retardation 1 (FMR1) gene on the X chromosome, results in a failure to express the fragile X mental retardation protein that is required for normal neural development. Other paraneoplastic neurologic disorders involving antibodies directed against one member of the hnRNP family, Nova-1, trigger the clinical syndrome of paraneoplastic opsoclonus myoclonus ataxia (72, 73), whereas antibodies to another member, Hu neuronal antigen, induce subacute sensory neuropathy/encephalomyelopathy syndrome (74). Thus, in analogy, our data suggest that it is possible for single nucleotide mutations in either the hnRNP-E1 mRNA cis element or the hnRNP-E1 protein to profoundly perturb physiologic RNA-protein interaction and result in novel clinical syndromes. For example, mutations in the first and/or third CUCC motif of the 25-nucleotide hnRNP-E1 mRNA cis element would reduce or abolish interaction with hnRNP-E1 (Figure 4). Conversely, the mutation of a single cysteine among several potential cysteine residues in hnRNP-E1 (Supplemental Figure 2) that leads to an (HA)-hnRNP-E1(C293S)–like mutant has the potential to markedly increase interaction with operon-associated mRNAs. Such mutations could give rise to profoundly distinct clinical syndromes that arise from quantitatively different (and/or opposing) expression of key proteins encoded by mRNAs that belong to the posttranscriptional RNA operon controlled by hnRNP-E1 (1, 3, 5, 8, 9, 14, 15, 17, 18, 21, 36, 59–71). Therefore experimental studies in animals bearing such mutations could provide clues to clinical syndromes in humans for which the genetic basis has remained obscure.
Finally, the putative homocysteinylation of other members of the hnRNP family, which are closely related to hnRNP-E1 [such as hnRNP-E2 (3, 75), and Nova-1 (76), among others], could result in the activation of a coordinated network of posttranscriptional RNA operons that together comprise a higher-order, nutrition-sensitive, folate-responsive, posttranscriptional RNA regulon (3, 16, 17, 77). In such a construct, the degree to which the mRNA-binding site in each related hnRNP protein is unmasked by homocysteinylation will dictate the extent of its participation in such a putative regulon. In this context, the profound pathophysiologic changes observed in the murine fetal brain that experienced folate deficiency (3, 13), as well as the postnatal behavioral features of anxiety (43), could well have arisen from the activation >1 of these putatively homocysteinylated hnRNP family members in utero.
Acknowledgments
This paper is dedicated to the sacred memory of Hiremagalur N Jayaram, a wonderful colleague and very dear friend. Y-ST conducted the research (hands-on execution of the experiments and data collection), analyzed the data, and performed the statistical analysis in the experiments leading to Supplemental Table 1, Figures 1–8, and Tables 1–9; RAK and SX conducted the research and analyzed the data in the experiments leading to Figures 2 and 7; RAK conducted the research and analyzed the data in the experiments leading to Supplemental Figures 2 and 3; SX conducted the research and analyzed the data in the experiments leading to Supplemental Figures 3–5; DKH conducted the research for the experiments leading to Supplemental Figures 4 and 5; PK and HNJ conducted the research and analyzed the data in the experiments leading to Figure 7; SPS measured thiols for the experiments leading to Figures 3 and 7 and Supplemental Figures 4 and 5; and ACA designed the research (project conception, development of overall research plan, and study oversight), analyzed the data, wrote the paper, and is primarily responsible for the final content of the paper. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: CAT, chloramphenicol acetyltransferase; FAM, 5′6-carboxyfluorescein; GST, glutathione S-transferase; HA, highest affinity; HF, high-folate; hnRNP-E1, heterogeneous nuclear ribonucleoprotein E1; HPV16, human papillomavirus type 16; iron(II), ferrous sulfate heptahydrate; iron(III), ferric chloride hexahydrate; JOE, 6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein; KD, dissociation constant; LF, low-folate; neurofilament M, neuronal intermediate neurofilament–middle molecular mass; siRNA, small interfering RNA; 5′-UTR, 5′-untranslated region.
References
- 1.Antony A, Tang YS, Khan RA, Biju MP, Xiao X, Li QJ, Sun XL, Jayaram HN, Stabler SP. Translational upregulation of folate receptors is mediated by homocysteine via RNA-heterogeneous nuclear ribonucleoprotein E1 interactions. J Clin Invest 2004;113:285–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antony AC. Hematology: basic principles and practice. 6th ed Hoffman R, Benz EJ Jr., Silberstein LE, Heslop HE, Weitz JI, Anastasi J, editors. Philadelphia (PA): Elsevier Saunders; 2013. p. 473–504. [Google Scholar]
- 3.Tang YS, Khan RA, Zhang Y, Xiao S, Wang M, Hansen DK, Jayaram HN, Antony AC. Incrimination of heterogeneous nuclear ribonucleoprotein E1 (hnRNP-E1) as a candidate sensor of physiological folate deficiency. J Biol Chem 2011;286:39100–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antony AC, Utley C, Van Horne KC, Kolhouse JF. Isolation and characterization of a folate receptor from human placenta. J Biol Chem 1981;256:9684–92. [PubMed] [Google Scholar]
- 5.Xiao X, Tang YS, Mackins JY, Sun XL, Jayaram HN, Hansen DK, Antony AC. Isolation and characterization of a folate receptor mRNA-binding trans-factor from human placenta. Evidence favoring identity with heterogeneous nuclear ribonucleoprotein E1. J Biol Chem 2001;276:41510–7. [DOI] [PubMed] [Google Scholar]
- 6.Pillai MR, Chacko P, Kesari LA, Jayaprakash PG, Jayaram HN, Antony AC. Expression of folate receptors and heterogeneous nuclear ribonucleoprotein E1 in women with human papillomavirus mediated transformation of cervical tissue to cancer. J Clin Pathol 2003;56:569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antony AC, Kincade RS, Verma RS, Krishnan SR. Identification of high affinity folate binding proteins in human erythrocyte membranes. J Clin Invest 1987;80:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ostareck-Lederer A, Ostareck DH, Standart N, Thiele BJ. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J 1994;13:1476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell 1997;89:597–606. [DOI] [PubMed] [Google Scholar]
- 10.Antony AC, Bruno E, Briddell RA, Brandt JE, Verma RS, Hoffman R. Effect of perturbation of specific folate receptors during in vitro erythropoiesis. J Clin Invest 1987;80:1618–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chkheidze AN, Lyakhov DL, Makeyev AV, Morales J, Kong J, Liebhaber SA. Assembly of the alpha-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein alphaCP. Mol Cell Biol 1999;19:4572–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiledjian M, Wang X, Liebhaber SA. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J 1995;14:4357–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao S, Hansen DK, Horsley ET, Tang YS, Khan RA, Stabler SP, Jayaram HN, Antony AC. Maternal folate deficiency results in selective upregulation of folate receptors and heterogeneous nuclear ribonucleoprotein-E1 associated with multiple subtle aberrations in fetal tissues. Birth Defects Res A Clin Mol Teratol 2005;73:6–28. [DOI] [PubMed] [Google Scholar]
- 14.Ostareck-Lederer A, Ostareck DH, Hentze MW. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem Sci 1998;23:409–11. [DOI] [PubMed] [Google Scholar]
- 15.Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 2002;8:265–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1′s multifunctional regulatory roles. RNA 2010;16:1449–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao S, Tang YS, Khan RA, Zhang Y, Kusumanchi P, Stabler SP, Jayaram HN, Antony AC. Influence of physiologic folate deficiency on human papillomavirus type 16 (HPV16)-harboring human keratinocytes in vitro and in vivo. J Biol Chem 2012;287:12559–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun XL, Antony AC. Evidence that a specific interaction between an 18-base cis-element in the 5′-untranslated region of human folate receptor-alpha mRNA and a 46-kDa cytosolic trans-factor is critical for translation. J Biol Chem 1996;271:25539–47. [PubMed] [Google Scholar]
- 19.Sun XL, Murphy BR, Li QJ, Gullapalli S, Mackins J, Jayaram HN, Srivastava A, Antony AC. Transduction of folate receptor cDNA into cervical carcinoma cells using recombinant adeno-associated virions delays cell proliferation in vitro and in vivo. J Clin Invest 1995;96:1535–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufman MH. The atlas of mouse development. San Diego (CA): Academic Press; 1992. [Google Scholar]
- 21.Wang X, Kiledjian M, Weiss IM, Liebhaber SA. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human alpha-globin mRNA stability. Mol Cell Biol 1995;15:1769–77. Erratum in: Mol Cell Biol 1995 Apr;15(4):2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antony AC. Goldman-Cecil medicine, (Cecil's textbook of medicine) 25th ed Goldman L, Schafer AI, editors. New York: Elsevier Saunders; 2015. p. 1104–14. [Google Scholar]
- 23.Stabler SP, Marcell PD, Podell ER, Allen RH, Savage DG, Lindenbaum J. Elevation of total homocysteine in the serum of patients with cobalamin or folate deficiency detected by capillary gas chromatography-mass spectrometry. J Clin Invest 1988;81:466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi H, Bencze KZ, Stemmler TL, Philpott CC. A cytosolic iron chaperone that delivers iron to ferritin. Science 2008;320:1207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philpott CC. Coming into view: eukaryotic iron chaperones and intracellular iron delivery. J Biol Chem 2012;287:13518–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hider RC, Kong X. Iron speciation in the cytosol: an overview. Dalton Trans 2013;42:3220–9. [DOI] [PubMed] [Google Scholar]
- 27.Hider RC, Kong XL. Glutathione: a key component of the cytoplasmic labile iron pool. Biometals 2011;24:1179–87. [DOI] [PubMed] [Google Scholar]
- 28.Stabler SP. Clinical practice. Vitamin B12 deficiency. N Engl J Med 2013;368:149–60. [DOI] [PubMed] [Google Scholar]
- 29.Waggoner SA, Liebhaber SA. Identification of mRNAs associated with alphaCP2-containing RNP complexes. Mol Cell Biol 2003;23:7055–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boshnjaku V, Shim KW, Tsurubuchi T, Ichi S, Szany EV, Xi G, Mania-Farnell B, McLone DG, Tomita T, Mayanil CS. Nuclear localization of folate receptor alpha: a new role as a transcription factor. Sci Rep 2012;2:980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leamon CP, Reddy JA, Dorton R, Bloomfield A, Emsweller K, Parker N, Westrick E. Impact of high and low folate diets on tissue folate receptor levels and antitumor responses toward folate-drug conjugates. J Pharmacol Exp Ther 2008;327:918–25. [DOI] [PubMed] [Google Scholar]
- 32.Torheim LE, Ferguson EL, Penrose K, Arimond M. Women in resource-poor settings are at risk of inadequate intakes of multiple micronutrients. J Nutr 2010;140:2051S–8S. [DOI] [PubMed] [Google Scholar]
- 33.WHO. Micronutrient deficiencies: iron deficiency anaemia [Internet]. [cited 2010 Sep 10]. Available from: http://www.who.int/nutrition/topics/ida/en/index.html.
- 34.Czyzyk-Krzeska MF, Paulding WR, Beresh JE, Kroll SL. Post-transcriptional regulation of tyrosine hydroxylase gene expression by oxygen in PC12 cells. Kidney Int 1997;51:585–90. [DOI] [PubMed] [Google Scholar]
- 35.Paulding WR, Czyzyk-Krzeska MF. Regulation of tyrosine hydroxylase mRNA stability by protein-binding, pyrimidine-rich sequence in the 3′-untranslated region. J Biol Chem 1999;274:2532–8. [DOI] [PubMed] [Google Scholar]
- 36.Thyagarajan A, Szaro BG. Phylogenetically conserved binding of specific K homology domain proteins to the 3′-untranslated region of the vertebrate middle neurofilament mRNA. J Biol Chem 2004;279:49680–8. [DOI] [PubMed] [Google Scholar]
- 37.Thyagarajan A, Strong MJ, Szaro BG. Post-transcriptional control of neurofilaments in development and disease. Exp Cell Res 2007;313:2088–97. [DOI] [PubMed] [Google Scholar]
- 38.Thyagarajan A, Szaro BG. Dynamic endogenous association of neurofilament mRNAs with K-homology domain ribonucleoproteins in developing cerebral cortex. Brain Res 2008;1189:33–42. [DOI] [PubMed] [Google Scholar]
- 39.Kong J, Tung VW, Aghajanian J, Xu Z. Antagonistic roles of neurofilament subunits NF-H and NF-M against NF-L in shaping dendritic arborization in spinal motor neurons. J Cell Biol 1998;140:1167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong PC, Marszalek J, Crawford TO, Xu Z, Hsieh ST, Griffin JW, Cleveland DW. Increasing neurofilament subunit NF-M expression reduces axonal NF-H, inhibits radial growth, and results in neurofilamentous accumulation in motor neurons. J Cell Biol 1995;130:1413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Z, Marszalek JR, Lee MK, Wong PC, Folmer J, Crawford TO, Hsieh ST, Griffin JW, Cleveland DW. Subunit composition of neurofilaments specifies axonal diameter. J Cell Biol 1996;133:1061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Z, Tung VW. Overexpression of neurofilament subunit M accelerates axonal transport of neurofilaments. Brain Res 2000;866:326–32. [DOI] [PubMed] [Google Scholar]
- 43.Ferguson SA, Berry KJ, Hansen DK, Wall KS, White G, Antony AC. Behavioral effects of prenatal folate deficiency in mice. Birth Defects Res A Clin Mol Teratol 2005;73:249–52. [DOI] [PubMed] [Google Scholar]
- 44.Schlotz W, Jones A, Phillips DI, Gale CR, Robinson SM, Godfrey KM. Lower maternal folate status in early pregnancy is associated with childhood hyperactivity and peer problems in offspring. J Child Psychol Psychiatry 2010;51:594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steenweg-de Graaff J, Roza SJ, Steegers EA, Hofman A, Verhulst FC, Jaddoe VW, Tiemeier H. Maternal folate status in early pregnancy and child emotional and behavioral problems: the Generation R Study. Am J Clin Nutr 2012;95:1413–21. [DOI] [PubMed] [Google Scholar]
- 46.Surén P, Roth C, Bresnahan M, Haugen M, Hornig M, Hirtz D, Lie KK, Lipkin WI, Magnus P, Reichborn-Kjennerud T, et al. Association between maternal use of folic acid supplements and risk of autism spectrum disorders in children. JAMA 2013;309:570–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strand TA, Taneja S, Ueland PM, Refsum H, Bahl R, Schneede J, Sommerfelt H, Bhandari N. Cobalamin and folate status predicts mental development scores in North Indian children 12–18 mo of age. Am J Clin Nutr 2013;97:310–7. [DOI] [PubMed] [Google Scholar]
- 48.Veena SR, Krishnaveni GV, Srinivasan K, Wills AK, Muthayya S, Kurpad AV, Yajnik CS, Fall CH. Higher maternal plasma folate but not vitamin B-12 concentrations during pregnancy are associated with better cognitive function scores in 9- to 10- year-old children in South India. J Nutr 2010;140:1014–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lozoff B, Jimenez E, Wolf AW. Long-term developmental outcome of infants with iron deficiency. N Engl J Med 1991;325:687–94. [DOI] [PubMed] [Google Scholar]
- 50.Lozoff B, Jimenez E, Hagen J, Mollen E, Wolf AW. Poorer behavioral and developmental outcome more than 10 years after treatment for iron deficiency in infancy. Pediatrics 2000;105:E51. [DOI] [PubMed] [Google Scholar]
- 51.Lozoff B, De Andraca I, Castillo M, Smith JB, Walter T, Pino P. Behavioral and developmental effects of preventing iron-deficiency anemia in healthy full-term infants. Pediatrics 2003;112:846–54. [PubMed] [Google Scholar]
- 52.Chang S, Wang L, Wang Y, Brouwer ID, Kok FJ, Lozoff B, Chen C. Iron-deficiency anemia in infancy and social emotional development in preschool-aged Chinese children. Pediatrics 2011;127:e927–33. [DOI] [PubMed] [Google Scholar]
- 53.Monk C, Georgieff MK, Osterholm EA. Research review: maternal prenatal distress and poor nutrition - mutually influencing risk factors affecting infant neurocognitive development. J Child Psychol Psychiatry 2013;54:115–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brunette KE, Tran PV, Wobken JD, Carlson ES, Georgieff MK. Gestational and neonatal iron deficiency alters apical dendrite structure of CA1 pyramidal neurons in adult rat hippocampus. Dev Neurosci 2010;32:238–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Georgieff MK. Long-term brain and behavioral consequences of early iron deficiency. Nutr Rev 2011;69 Suppl 1:S43–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tran PV, Dakoji S, Reise KH, Storey KK, Georgieff MK. Fetal iron deficiency alters the proteome of adult rat hippocampal synaptosomes. Am J Physiol Regul Integr Comp Physiol 2013;305:R1297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baker RD, Greer FR. Diagnosis and prevention of iron deficiency and iron-deficiency anemia in infants and young children (0–3 years of age). Pediatrics 2010;126:1040–50. [DOI] [PubMed] [Google Scholar]
- 58.Christian P, Murray-Kolb LE, Khatry SK, Katz J, Schaefer BA, Cole PM, Leclerq SC, Tielsch JM. Prenatal micronutrient supplementation and intellectual and motor function in early school-aged children in Nepal. JAMA 2010;304:2716–23. [DOI] [PubMed] [Google Scholar]
- 59.Meng Q, Rayala SK, Gururaj AE, Talukder AH, O’Malley BW, Kumar R. Signaling-dependent and coordinated regulation of transcription, splicing, and translation resides in a single coregulator, PCBP1. Proc Natl Acad Sci USA 2007;104:5866–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobbyn HC, Hill K, Hamilton TL, Spriggs KA, Pickering BM, Coldwell MJ, de Moor CH, Bushell M, Willis AE. Regulation of BAG-1 IRES-mediated translation following chemotoxic stress. Oncogene 2008;27:1167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis SM, Veyrier A, Hosszu Ungureanu N, Bonnal S, Vagner S, Holcik M. Subcellular relocalization of a trans-acting factor regulates XIAP IRES-dependent translation. Mol Biol Cell 2007;18:1302–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lewis SM, Holcik M. For IRES trans-acting factors, it is all about location. Oncogene 2008;27:1033–5. [DOI] [PubMed] [Google Scholar]
- 63.Jiang Y, Xu XS, Russell JE. A nucleolin-binding 3′ untranslated region element stabilizes beta-globin mRNA in vivo. Mol Cell Biol 2006;26:2419–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Czyzyk-Krzeska MF, Bendixen AC. Identification of the poly(C) binding protein in the complex associated with the 3′ untranslated region of erythropoietin messenger RNA. Blood 1999;93:2111–20. [PubMed] [Google Scholar]
- 65.Pickering BM, Mitchell SA, Evans JR, Willis AE. Polypyrimidine tract binding protein and poly(rC)- binding protein 1 interact with the BAG-1 IRES and stimulate its activity in vitro and in vivo. Nucleic Acids Res 2003;31:639–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holcik M, Korneluk RG. XIAP, the guardian angel. Nat Rev Mol Cell Biol 2001;2:550–6. [DOI] [PubMed] [Google Scholar]
- 67.Chappell SA, LeQuesne JP, Paulin FE, deSchoolmeester ML, Stoneley M, Soutar RL, Ralston SH, Helfrich MH, Willis AE. A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: a novel mechanism of oncogene de-regulation. Oncogene 2000;19:4437–40. [DOI] [PubMed] [Google Scholar]
- 68.Rondon IJ, MacMillan LA, Beckman BS, Goldberg MA, Schneider T, Bunn HF, Malter JS. Hypoxia up-regulates the activity of a novel erythropoietin mRNA binding protein. J Biol Chem 1991;266:16594–8. [PubMed] [Google Scholar]
- 69.Giles KM, Daly JM, Beveridge DJ, Thomson AM, Voon DC, Furneaux HM, Jazayeri JA, Leedman PJ. The 3′-untranslated region of p21WAF1 mRNA is a composite cis-acting sequence bound by RNA-binding proteins from breast cancer cells, including HuR and poly(C)-binding protein. J Biol Chem 2003;278:2937–46. [DOI] [PubMed] [Google Scholar]
- 70.Collier B, Goobar-Larsson L, Sokolowski M, Schwartz S. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J Biol Chem 1998;273:22648–56. [DOI] [PubMed] [Google Scholar]
- 71.Czyzyk-Krzeska MF, Beresh JE. Characterization of the hypoxia-inducible protein binding site within the pyrimidine-rich tract in the 3′-untranslated region of the tyrosine hydroxylase mRNA. J Biol Chem 1996;271:3293–9. [DOI] [PubMed] [Google Scholar]
- 72.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science 2003;302:1212–5. [DOI] [PubMed] [Google Scholar]
- 73.Darnell RB, Posner JB. Paraneoplastic syndromes affecting the nervous system. Semin Oncol 2006;33:270–98. [DOI] [PubMed] [Google Scholar]
- 74.Darnell RB. Onconeural antigens and the paraneoplastic neurologic disorders: at the intersection of cancer, immunity, and the brain. Proc Natl Acad Sci USA 1996;93:4529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Waggoner SA, Johannes GJ, Liebhaber SA. Depletion of the poly(C)-binding proteins alphaCP1 and alphaCP2 from K562 cells leads to p53-independent induction of cyclin-dependent kinase inhibitor (CDKN1A) and G1 arrest. J Biol Chem 2009;284:9039–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Musunuru K, Darnell RB. Determination and augmentation of RNA sequence specificity of the Nova K-homology domains. Nucleic Acids Res 2004;32:4852–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 2007;8:533–43. [DOI] [PubMed] [Google Scholar]