Abstract
Background: Formate provides one-carbon units for de novo purine and thymidylate (dTMP) synthesis and is produced via both folate-dependent and folate-independent pathways. Folate-independent pathways are mediated by cytosolic alcohol dehydrogenase 5 (ADH5) and mitochondrial aldehyde dehydrogenase 2 (ALDH2), which generate formate by oxidizing formaldehyde. Formate is a potential biomarker of B-vitamin–dependent one-carbon metabolism.
Objective: This study investigated the contributions of ADH5 and ALDH2 to formate production and folate-dependent de novo purine and dTMP synthesis in HepG2 cells.
Methods: ADH5 knockout and ALDH2 knockdown HepG2 cells were cultured in folate-deficient [0 nM (6S) 5-formyltetrahydrofolate] or folate-sufficient [25 nM (6S) 5-formyltetrahydrofolate] medium. Purine biosynthesis was quantified as the ratio of [14C]-formate to [3H]-hypoxanthine incorporated into genomic DNA, which indicates the contribution of the de novo purine synthesis pathway relative to salvage synthesis. dTMP synthesis was quantified as the ratio of [14C]-deoxyuridine to [3H]-thymidine incorporation into genomic DNA, which indicates the capacity of de novo dTMP synthesis relative to salvage synthesis.
Results: The [14C]-formate-to-[3H]-hypoxanthine ratio was greater in ADH5 knockout than in wild-type HepG2 cells, under conditions of both folate deficiency (+30%; P < 0.001) and folate sufficiency (+22%; P = 0.02). These data indicate that ADH5 deficiency increases the use of exogenous formate for de novo purine biosynthesis. The [14C]-deoxyuridine-to-[3H]-thymidine ratio did not differ between ADH5 knockout and wild-type cells, indicating that ADH5 deficiency does not affect de novo dTMP synthesis capacity relative to salvage synthesis. Under folate deficiency, ALDH2 knockdown cells exhibited a 37% lower ratio of [14C]-formate to [3H]-hypoxanthine (P < 0.001) compared with wild-type HepG2 cells, indicating decreased use of exogenous formate, or increased endogenous formate synthesis, for de novo purine biosynthesis.
Conclusions: In HepG2 cells, ADH5 is a source of formate for de novo purine biosynthesis, especially during folate deficiency when folate-dependent formate production is limited. Formate is also shown to be limiting in the growth of HepG2 cells.
Keywords: alcohol dehydrogenase 5, aldehyde dehydrogenase 2, folate, formate, formaldehyde dehydrogenase, purine synthesis, thymidylate synthesis
Introduction
Folate, an essential B-vitamin, serves as a cofactor in the form of tetrahydrofolate (THF)7 polyglutamates to carry and activate one-carbon units for the de novo synthesis of purines and thymidylate (dTMP) and for the remethylation of homocysteine to methionine (Figure 1) (1). Disruptions in folate-mediated one-carbon metabolism (FOCM), which can arise from insufficient intake of folate and other nutrients (e.g., choline and vitamins B-12 and B-6) and/or genetic variants, have been linked to a higher risk of cancer, neurodegenerative diseases, and developmental anomalies (1).
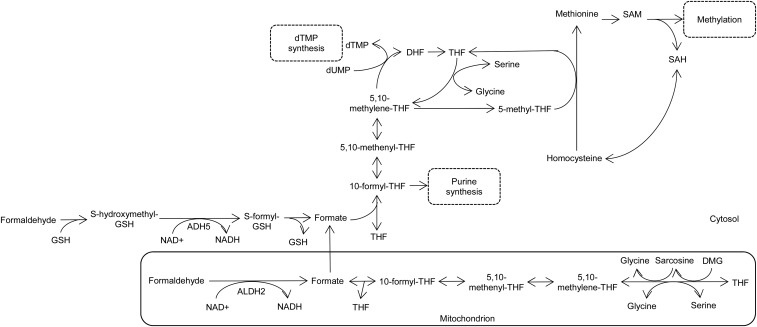
FIGURE 1.
Cytosolic and mitochondrial folate-mediated one-carbon metabolism and formate generation by ADH5 and ALDH2. ADH5, alcohol dehydrogenase 5; ALDH2, aldehyde dehydrogenase 2; DHF, dihydrofolate; DMG, dimethylglycine; dTMP, thymidylate; dUMP, deoxyuridine monophosphate; GSH, reduced glutathione; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; THF, tetrahydrofolate.
FOCM occurs in the cytosol, mitochondria, and nucleus, all of which are interrelated through the exchange of one-carbon units from formate, serine, and glycine (2). Mitochondrial one-carbon metabolism plays a role in the generation of formate from the catabolism of serine, glycine, dimethylglycine, and sarcosine (Figure 1). Mitochondria-derived formate then enters the cytosol or nucleus and is incorporated into 10-formylTHF, which provides the C2 and C8 carbon for de novo purine biosynthesis. Alternatively, cytosolic 10-formylTHF may be reduced to 5,10-methyleneTHF, which can be used for de novo dTMP synthesis or irreversibly reduced to 5-methylTHF for homocysteine remethylation to methionine (Figure 1). Therefore, formate is an intermediate metabolite essential for one-carbon metabolism; its utilization and mitochondrial production are linked to folate status, and hence formate concentrations in serum have the potential to serve as a biomarker of folate status (3). Folate is essential both for formate synthesis and to sequester formate within the cell, because previous studies have reported increased plasma and urinary formate and decreased rates of formate production in rats and mice fed a folate-deficient diet compared with those fed a folate-replete diet (4–6).
In addition to formate synthesis through the folate-dependent mitochondrial pathways, formate can be produced through folate-independent pathways, one of which involves the oxidation of formaldehyde to formate (3). This can occur via the reaction mediated by the cytosolic glutathione and NAD+-dependent enzyme alcohol dehydrogenase 5 [ADH5, class III (alternative abbreviation, ADH3); also known as formaldehyde dehydrogenase]. Specifically, ADH5 oxidizes S-hydroxymethylglutathione, a molecule formed spontaneously from formaldehyde and glutathione, to S-formylglutathionine, which is further converted to formate (Figure 1) (7). Formaldehyde can also be oxidized to formate by a mitochondrial NAD+-dependent aldehyde dehydrogenase class II (ALDH2; Figure 1) (8). ALDH2 and ADH5 are ubiquitously expressed in various tissues, including liver, kidney, and brain, and are most abundant in the liver (9–12).
Although there has been increasing interest in the role of mitochondria-derived formate in the functioning of FOCM and as a biomarker of nutrient status, very little is known about the contribution of folate-independent sources of formate to one-carbon metabolism. The objective of the current study was to investigate the effects of ADH5 and ALDH2 activity on de novo purine and dTMP biosynthesis by inhibiting ADH5 or ALDH2 expression in human hepatocarcinoma (HepG2) cells.
Methods
Cell culture.
HepG2 cells were maintained in DMEM (Corning) with 10% (vol:vol) FBS (HyClone), 1% penicillin/streptomycin (Corning), and 4 mM l-glutamine (Corning) at 37°C and 5% CO2. For all experiments, modified DMEM lacking glycine, serine, methionine, folate, choline, and all nucleosides and nucleotides was used with 10% dialyzed and charcoal-treated FBS, 1% penicillin/streptomycin, and 4 mM l-glutamine.
Generation of ADH5 knockout HepG2 cells by CRISPR/Cas9.
The CRISPR single guide RNA (5′-TGAACATGGCGAACGAGGTA-3′) targeting exon 1 of human ADH5 (NM_000671) was cloned into the pSpCas9(BB)-2A-Puro CRISPR/Cas9 vector as previously described (13). Cells were transfected for 48 h by using the FuGene 6 transfection reagent (Promega) following the manufacturer’s instructions. The transfected cells were selected in the presence of 2 μg puromycin/mL (RPI). The efficiency of ADH5 knockout was verified by immunoblotting.
Gene knockdown by small interfering RNA transfection.
Cells were transfected with either negative control small interfering RNA (siRNA; Qiagen) or FlexiTube GeneSolution (GS217) siRNA for ALDH2 (Qiagen) by using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s instructions. Cells were harvested 72 h after transfection. The efficiency of ALDH2 knockdown was verified by immunoblotting.
Cellular total folate measurement.
Total folate concentrations in cells were quantified by using a Lactobacillus casei microbiological assay as previously described (14).
Immunoblotting.
Cellular proteins were extracted and quantified as previously described (15). Proteins were resolved on 4–15% (vol:vol) gradient SDS-PAGE gels (Bio-Rad) and transferred to Immobilon-P PVDF membrane (Millipore). The membrane was blocked for 1 h at room temperature in 5% BSA in PBS with 0.2% Tween. Primary antibodies were diluted in 5% BSA in PBS with 0.2% Tween and incubated overnight at 4°C. Secondary antibodies were diluted in 5% nonfat dry milk in PBS with 1% Nonidet P-40 (US Biologicals) and added to the membrane for 1 h at room temperature. ADH5 and ALDH2 were detected with a 1:1000 rabbit anti-ADH5 antibody and a 1:2000 rabbit anti-ALDH2 antibody, respectively (Proteintech Group), followed by a 1:5000 dilution of HRP-conjugated donkey anti-rabbit secondary antibody (Pierce). As loading controls, 1:1000 mouse anti-α-Tubulin antibody (Active Motif) and a 1:3000 mouse anti-α-Calpain antibody (Affinity BioReagents) were used followed by a 1:5000 dilution of HRP-conjugated goat anti-mouse secondary antibody (Pierce). The membrane was visualized by autoradiography after the addition of SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Purine biosynthesis assay.
Cells were seeded on 100-mm plates in modified DMEM lacking glycine, serine, and all nucleosides and nucleotides but supplemented with 200 μM methionine and 30 μM choline, with 25 nM (6S) 5-formylTHF (folate sufficiency) or without (6S) 5-formylTHF (folate deficiency). After 2 doublings, cells were plated in triplicate on 6-well plates and allowed to grow for another doubling in the same media but supplemented with 10 μM [14C]-formate and 1 nM [3H]-hypoxanthine (Moravek Biochemicals). Cells were harvested, and genomic DNA was isolated by using a High Pure PCR template preparation kit (Roche) with RNase A treatment according to the manufacturer’s instructions. Isotope incorporation into genomic DNA was quantified by using a Beckman LS6500 scintillation counter in dual disintegrations/minute mode (16). Data are shown as the ratio of [14C]-formate to [3H]-hypoxanthine, which indicates the incorporation of formate into DNA via the folate-dependent de novo purine synthesis pathway relative to the incorporation of hypoxanthine into DNA via the folate-independent purine salvage pathway.
dTMP biosynthesis assay.
Cells were plated and grown in modified DMEM lacking glycine, serine, and all nucleosides and nucleotides but supplemented with 200 μM methionine and 30 μM choline, with 25 nM (6S) 5-formylTHF (folate sufficiency) or without (6S) 5-formylTHF (folate deficiency). After 2 doublings, cells were plated in triplicate on 6-well plates and allowed to grow for another doubling in the same media but supplemented with 2 μM [14C]-deoxyuridine and 25 nM [3H]-thymidine (American Radiolabeled Chemicals). [14C]-Deoxyuridine is incorporated into DNA via the folate-dependent de novo pathway, whereas [3H]-thymidine is incorporated into DNA via the salvage pathway. Total genomic DNA was isolated from the harvested cells, and the isotope incorporation was quantified as described above. Data are shown as the ratio of [14C]-deoxyuridine to [3H]-thymidine (17).
Cell growth assay.
Cell proliferation was determined by using a colorimetric MTT [3-(4,5)-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay (18). Cells were plated in 96-well plates and grown in modified DMEM lacking glycine, serine, and all nucleosides and nucleotides but supplemented with 200 μM methionine, 30 μM choline, and 25 nM (6S) 5-formylTHF. The effect of formate supplementation on cell proliferation was determined by using the same media further supplemented with either 30 μM or 90 μM sodium formate. From 24 to ≤72 h, cell growth was measured by adding 20 μL of 2.5 g MTT reagent/L to each well followed by 4 h of incubation at 37°C in 5% CO2. The insoluble formazan product was resuspended in 100 μL DMSO, and A570 was measured on a microplate reader (Epoch; BioTek).
Statistical analysis.
Histograms and scatterplots of the residuals were used to assess normality and variance homogeneity. The effects of gene expression (wild-type compared with ADH5 knockout or wild-type compared with ALDH2 knockdown) were assessed by using t tests. To examine the effects of gene expression and folate status as well as their interaction, a 2-factor ANOVA was used with post hoc Bonferroni corrections. Linear mixed models with Bonferroni corrections were used to assess the effect of gene expression on cell growth over time. Data are shown as means ± SDs of 3–5 biological replicates/condition. All statistical tests were performed with IBM SPSS Statistics (version 20), and significance was defined as P < 0.05.
Results
ADH5 deficiency increases use of exogenous formate for de novo purine biosynthesis.
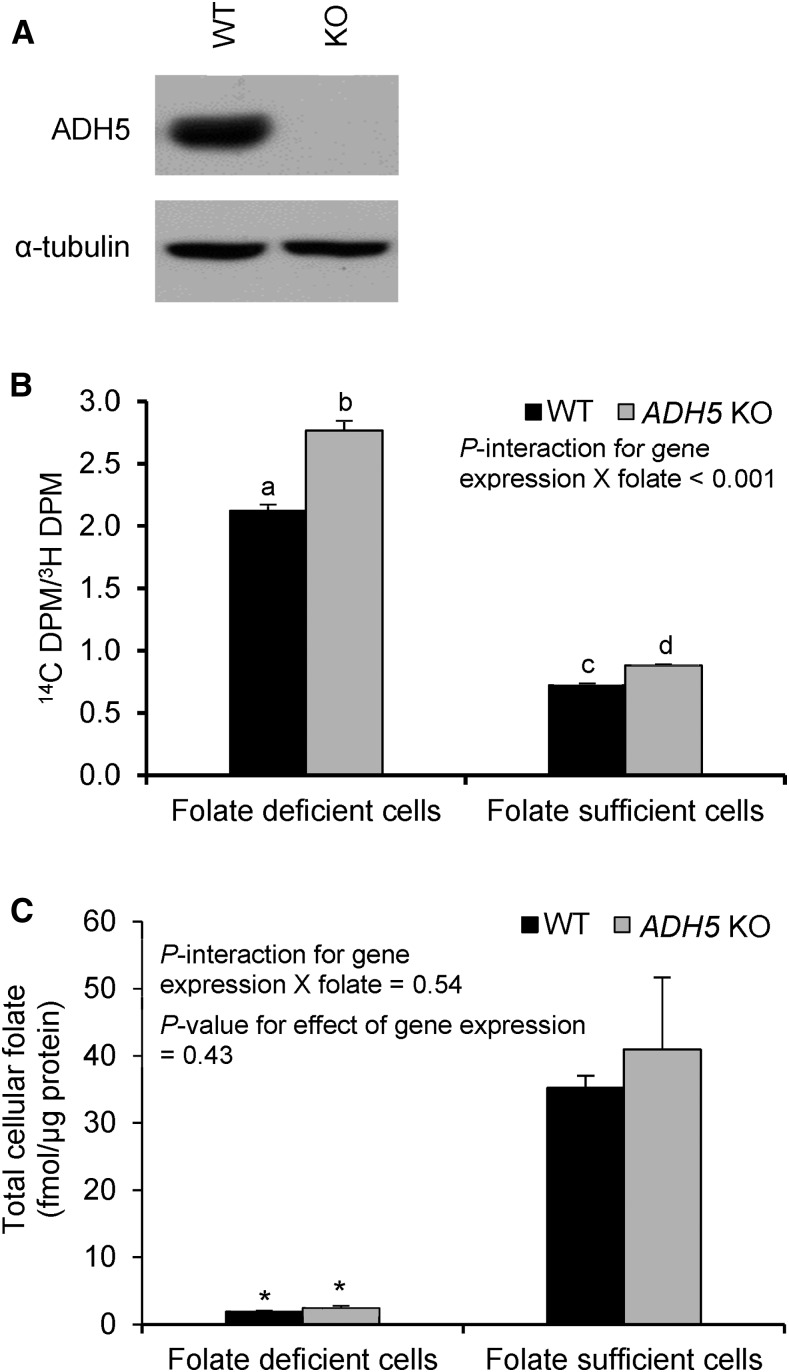
HepG2 cells lacking ADH5 were generated by using CRISPR/Cas9 genome editing (13). Ablation of ADH5 protein expression was confirmed by immunoblotting (Figure 2A). The wild-type and ADH5 knockout HepG2 cells were cultured in folate-deficient or folate-sufficient culture medium containing [14C]-formate and [3H]-hypoxanthine. The ratio of [14C]-formate to [3H]-hypoxanthine in DNA serves as a measure of de novo purine synthesis efficiency relative to salvage purine synthesis.
FIGURE 2.
Purine biosynthesis in WT and ADH5 KO HepG2 cells. (A) Silencing of ADH5 was confirmed by immunoblotting. (B) The ratio of [14C]-formate to [3H]-hypoxanthine indicates the incorporation of formate into DNA via the de novo purine synthesis pathway relative to the incorporation of hypoxanthine into DNA via the purine salvage pathway. Labeled means without a common letter differ, P < 0.05. (C) Total cellular folate concentrations. *Different from folate-sufficient cells, P < 0.05. Data were analyzed by using 2-factor ANOVA with Bonferroni corrections. Values are means ± SDs of 3 biological replicates/condition. ADH5, alcohol dehydrogenase 5; DPM, decays per minute; KO, knockout; WT, wild-type.
Culturing cells in folate-deficient medium increased the ratio of [14C]-formate to [3H]-hypoxanthine in both ADH5 knockout and wild-type HepG2 cells (P < 0.001; Figure 2B). This effect was driven by increased incorporation of [14C]-formate into DNA (Supplemental Figure 1). Given that folate is required for mitochondria-derived formate production, we hypothesized that loss of ADH5 protein expression may lead to greater incorporation of exogenous formate into DNA under folate-deficient culture conditions than under folate-sufficient culture conditions. Notably, there was a significant interaction (P < 0.001) between the gene expression (comparing wild-type with ADH5 knockout) and folate in the culture medium (folate deficiency compared with folate sufficiency) on the ratio of [14C]-formate to [3H]-hypoxanthine (Figure 2B). Specifically, the ratio of [14C]-formate to [3H]-hypoxanthine was significantly greater in ADH5 knockout than in wild-type HepG2 cells in both culture conditions, but with a greater increase under conditions of folate deficiency (+30%; P < 0.001) than with folate sufficiency (+22%; P = 0.02). In the folate-deficient condition, [14C]-formate incorporation normalized to DNA content was 30% greater in ADH5 knockout than in wild-type HepG2 cells (P < 0.001), whereas the incorporation of [3H]-hypoxanthine into DNA did not differ (P = 0.89) between the cell lines (Supplemental Figure 1). In folate sufficiency, the incorporation of [14C]-formate into DNA did not differ between ADH5 knockout and wild-type HepG2 cells (P = 0.52), whereas [3H]-hypoxanthine incorporation into DNA decreased by 12% in ADH5 knockout (compared with wild-type) HepG2 cells (P = 0.008). Overall, these data suggest that ADH5-mediated formate production contributes to de novo purine biosynthesis, especially during folate deficiency when mitochondrial formate production is limited.
The effect of both ADH5 expression and exogenous folate availability on intracellular folate concentrations was determined. Intracellular folate concentrations were ∼94% lower in folate-deficient than in folate-sufficient medium in both ADH5 knockout and wild-type HepG2 cells (P ≤ 0.004; Figure 2C). However, no effect of ADH5 gene expression was observed on intracellular folate concentrations (P = 0.43; P-interaction between gene expression and folate in culture medium = 0.54).
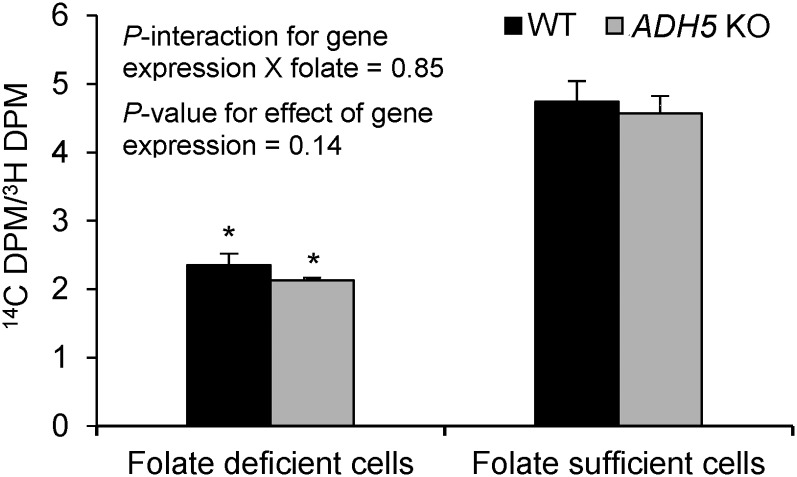
ADH5 deficiency does not affect de novo dTMP synthesis.
The effects of ADH5 silencing on dTMP synthesis in folate-deficient and folate-sufficient conditions were investigated. The ratio of [14C]-deoxyuridine (an indicator of de novo dTMP synthesis) to [3H]-thymidine (an indicator of salvage dTMP synthesis) in DNA did not differ between the wild-type and ADH5 knockout HepG2 cells (P = 0.14) independent of folate conditions (P-interaction = 0.85) (Figure 3). When grown under folate-deficient compared with folate-sufficient conditions, both wild-type and ADH5 knockout HepG2 cells had a significantly lower ratio of [14C]-deoxyuridine to [3H]-thymidine (P < 0.001), indicating that folate deficiency may upregulate the salvage pathway to meet cellular dTMP requirements.
FIGURE 3.
dTMP biosynthesis in WT and ADH5 KO HepG2 cells. The ratio of [14C]-deoxyuridine to [3H]-thymidine indicates the relative contribution of the de novo pathway to the salvage pathway for dTMP synthesis. Data were analyzed by using 2-factor ANOVA with Bonferroni corrections. Values are means ± SDs of 3 biological replicates/condition. *Different from folate-sufficient cells, P < 0.05. ADH5, alcohol dehydrogenase 5; DPM, decays per minute; dTMP, thymidylate; KO, knockout; WT, wild-type.
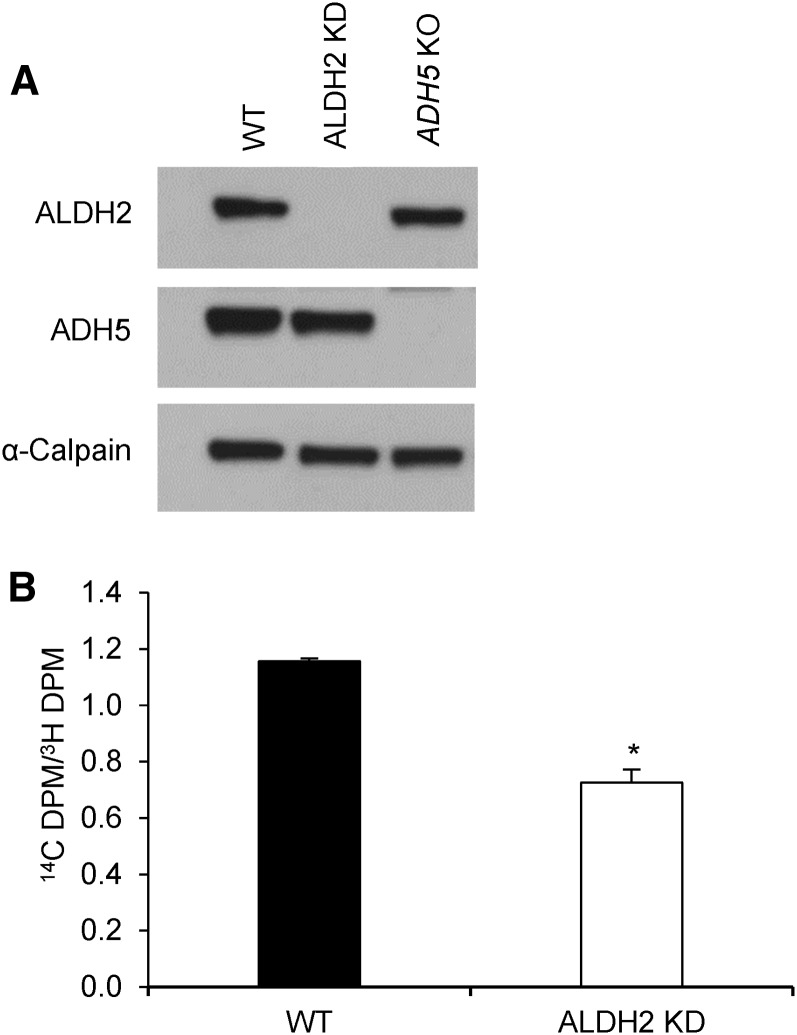
Reduced expression of ALDH2 decreases use of exogenous formate for de novo purine biosynthesis.
To determine the effect of ALDH2 on purine biosynthesis, ALDH2 expression was reduced by using ALDH2-targeting siRNA transfection and verified by immunoblotting (Figure 4A). The wild-type and ALDH2 knockdown HepG2 cells were cultured in folate-deficient medium with [14C]-formate and [3H]-hypoxanthine. The ratio of [14C]-formate to [3H]-hypoxanthine was 37% lower in ALDH2 knockdown HepG2 cells than in wild-type cells (P < 0.001; Figure 4B). ALDH2 knockdown HepG2 cells exhibited decreases in the incorporation of both [14C]-formate (−82%; P < 0.001) and [3H]-hypoxanthine (−72%; P = 0.002) into DNA (Supplemental Figure 2). Overall, these data suggest that reduced ALDH2 expression decreases the use of exogenous formate for de novo purine biosynthesis, indicating increased endogenous production of formate.
FIGURE 4.
Purine biosynthesis in WT and ALDH2 KD HepG2 cells cultured in folate-deficient medium. (A) ALDH2 KD was confirmed by immunoblotting. (B) The ratio of [14C]-formate to [3H]-hypoxanthine indicates the incorporation of formate into DNA via the de novo purine synthesis pathway relative to the incorporation of hypoxanthine into DNA via the purine salvage pathway. Data were analyzed by using a t test. Values are means ± SDs of 3 biological replicates/condition. *Different from WT, P < 0.05. ADH5, alcohol dehydrogenase 5; ALDH2, aldehyde dehydrogenase 2; DPM, decays per minute; KD, knockdown; KO, knockout; WT, wild-type.
Formate is limiting for HepG2 cell growth.
The growth of wild-type, ALDH2 knockdown, and ADH5 knockout HepG2 cells in the presence of exogenous formate was investigated by supplementing culture medium with sodium formate. No significant interaction between gene expression (wild-type compared with ADH5 knockout) and time was detected (P = 0.3), indicating that the growth rate of ADH5 knockout HepG2 cells did not differ from the wild-type cells (Supplemental Figure 3). However, ALDH2 knockdown HepG2 cells exhibited significantly lower growth rates relative to wild-type cells over 72 h [P-interaction between gene expression (wild-type compared with ALDH2 knockdown) and time < 0.001], indicating that reduced ALDH2 expression inhibits cell proliferation. The ALDH2 knockdown HepG2 cells exhibited 23% and 31% lower cell viability at 48 and 72 h, respectively, compared with wild-type HepG2 cells (P < 0.001).
There was a significant interaction between gene expression (wild-type HepG2 cells compared with ADH5 knockout or ALDH2 knockdown HepG2 cells) and formate supplementation on cell proliferation (P ≤ 0.004; Table 1). Specifically, the addition of formate (30 or 90 μM) in culture medium increased the growth of wild-type HepG2 cells at 48 and 72 h (P < 0.001); at 72 h, the wild-type cells supplemented with 90 μM formate exhibited a 30% increase in their growth compared with those without formate supplementation. However, the addition of formate did not affect the proliferation of ALDH2 knockdown or ADH5 knockout HepG2 cells (P ≥ 0.82).
TABLE 1.
Effect of formate supplementation on the growth of WT, ALDH2 knockdown, and ADH5 knockout HepG2 cells1
| Formate, μM |
|||||||||
| 24 h |
48 h |
72 h |
|||||||
| 0 | 30 | 90 | 0 | 30 | 90 | 0 | 30 | 90 | |
| WT | 1.00 ± 0.02 | 1.03 ± 0.02 | 1.04 ± 0.04 | 1.55a ± 0.02 | 1.67b ± 0.04 | 1.70b ± 0.03 | 1.96a ± 0.01 | 2.09b ± 0.05 | 2.55c ± 0.06 |
| ALDH2 knockdown | 0.85 ± 0.02 | 0.81 ± 0.00 | 0.83 ± 0.03 | 1.19 ± 0.05 | 1.20 ± 0.08 | 1.20 ± 0.05 | 1.35 ± 0.23 | 1.31 ± 0.19 | 1.23 ± 0.28 |
| ADH5 knockout | 1.43 ± 0.04 | 1.51 ± 0.02 | 1.51 ± 0.03 | 2.10 ± 0.09 | 2.15 ± 0.07 | 2.21 ± 0.10 | 2.54 ± 0.22 | 2.41 ± 0.32 | 2.22 ± 0.45 |
Values are means ± SDs of 5 biological replicates/condition. Values were normalized to the A570 value of WT HepG2 cells cultured in the medium without formate at 24 h. Data were analyzed by using a linear mixed model with Bonferroni corrections. Data from the formate 0-μM condition were presented in Supplemental Figure 3 to show the difference in the growth rates between the cell lines. Within each time point, labeled means in a row without a common superscript letter differ, P < 0.05. ADH5, alcohol dehydrogenase 5; ALDH2, aldehyde dehydrogenase 2; WT, wild-type.
Discussion
Formate provides one-carbon units for the de novo synthesis of purines and dTMP and for the remethylation of homocysteine to methionine. It also plays an important role in embryonic development, as reported by previous studies showing that formate has a protective effect on neural tube closure defects in a mouse model (3, 19, 20). Formate can be produced via both folate-dependent mitochondrial pathways and folate-independent pathways, but the relative contributions of these pathways to formate production and utilization are unknown (3). One of the folate-independent formate-generating pathways is mediated by NAD+-dependent cytosolic ADH5 and mitochondrial ALDH2, which function in the oxidation of formaldehyde to formate (Figure 1). The current study investigated whether ADH5 and ALDH2 enzymatic reactions contribute to the generation of endogenous formate for FOCM.
This study provides evidence that ADH5 is a meaningful source of formate for de novo purine biosynthesis. We found a higher ratio of [14C]-formate to [3H]-hypoxanthine in ADH5 knockout than in wild-type HepG2 cells, indicating that ADH5 knockout HepG2 cells incorporated higher amounts of exogenous formate into de novo purine biosynthesis than did wild-type cells. In addition, the use of exogenous formate for de novo purine synthesis in ADH5 knockout HepG2 cells increased during folate deficiency, presumably due to decreased mitochondria-derived formate generation. These findings are consistent with a previous study that showed that rats fed a folate-deficient diet exhibited a 44% reduction in the rate of endogenous formate production compared with those fed a folate-replete diet (5). Moreover, in liver mitochondria isolated from folate-deficient rats, formate production from choline metabolites (dimethylglycine and sarcosine) increased, which may be attributable to formaldehyde production, as suggested by the authors (5). Specifically, in folate deficiency, dimethylglycine and sarcosine are sources of one-carbon units through the reactions mediated by dimethylglycine dehydrogenase and sarcosine dehydrogenase, thereby generating formaldehyde (21). However, HepG2 cells do not metabolize sarcosine or dimethylglycine (data not shown). Alternatively, endogenous formaldehyde can be generated as a byproduct of the enzymatic demethylation reactions, including histone, RNA, and DNA demethylation (22–24). These findings, as well as the results of our study, support a role for ADH5 in formate production through the oxidation of formaldehyde during folate deficiency. ADH5 also makes contributions to the formate pool in states of folate sufficiency. In this study, the ratio of [14C]-formate to [3H]-hypoxanthine was higher in the ADH5 knockout than in the wild-type HepG2 cells when they were cultured in the presence of 25 nM (6S) 5-formylTHF. Overall, our findings show a role for the folate-dependent and folate-independent production of formate for de novo purine biosynthesis.
Notably, the effect of ADH5 deficiency was different between de novo purine and dTMP synthesis. The ratio of [14C]-deoxyuridine to [3H]-thymidine in DNA did not differ between ADH5 knockout and wild-type HepG2 cells, indicating that dTMP synthesis is not compromised by ADH5 deficiency. De novo dTMP synthesis occurs at the sites of DNA synthesis in the nucleus (25), whereas de novo purine biosynthesis occurs in the cytoplasm, where it requires the formation of a multienzyme complex referred to as a purinosome (26). Although the underlying mechanisms need further elucidation, the differences in the effect of ADH5 deficiency between purine and dTMP synthesis suggest that the source of the nuclear formate pool may be different from that of the cytoplasmic formate pool. Given that ADH5 is localized in the cytosol, it may contribute to the formate pool in the cytosol, but not in the nucleus, for de novo purine biosynthesis.
ALDH2 knockdown HepG2 cells exhibited a 37% reduction in the ratio of [14C]-formate to [3H]-hypoxanthine compared with wild-type cells, indicating that reduced ALDH2 expression decreases the use of exogenous formate for de novo purine biosynthesis. These results indicate that ALDH2 knockdown HepG2 cells may upregulate endogenous formate production from the other sources as a compensatory response to ALDH2 deficiency. Alternatively, ALDH2 deficiency may enhance the conversion of formaldehyde to formate. However, given that the mitochondrial ratio of NAD+ to NAD(H) ranges between 7 and 8 (27), the latter scenario that ALDH2 reduces formate seems unlikely. Overall, the differential effects on de novo purine biosynthesis between ADH5 knockout (increased exogenous formate use) and ALDH2 knockdown (decreased exogenous formate use) cells suggest that cytosolic ADH5 and mitochondrial ALDH2 may have distinct roles in providing formate for de novo purine biosynthesis.
ALDH2 knockdown HepG2 cells exhibited significantly slower growth rates relative to wild-type cells, indicating that reduced ALDH2 expression inhibits cell proliferation. Supplementation with formate in culture medium did not rescue the growth of these cells, suggesting that the observed growth inhibition is unlikely to be associated with formate production and/or utilization. As evidenced by a previous study (28), decreased proliferation of ALDH2 knockdown cells may be due to cell cycle arrest and enhanced apoptosis caused by elevated concentrations of reactive oxygen species and toxic aldehyde due to reduced ALDH2 activity. Interestingly, formate supplementation stimulated the growth of wild-type HepG2 cells, indicating that formate is limiting for the growth of these cells. This finding is consistent with a previous study that showed that methyleneTHF dehydrogenase 1 is essential for cell growth (29). Taken together, the results suggest that formate availability may be limiting for cell growth in some cells. In conclusion, this study shows that endogenous formate produced by ADH5 is used in de novo purine biosynthesis and its contribution to the formate pool is enhanced in folate deficiency.
Acknowledgments
SB, MSF, and PJS designed the research and wrote the manuscript; SB and JC conducted the research; SB analyzed the data; and PJS had primary responsibility for the final content. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: ADH5, alcohol dehydrogenase 5; ALDH2, aldehyde dehydrogenase 2; dTMP, thymidylate; FOCM, folate-mediated one-carbon metabolism; siRNA, small interfering RNA; THF, tetrahydrofolate.
References
- 1.Fox JT, Stover PJ. Folate-mediated one-carbon metabolism. Vitam Horm 2008;79:1–44. [DOI] [PubMed] [Google Scholar]
- 2.Field MS, Kamynina E, Stover PJ. MTHFD1 regulates nuclear de novo thymidylate biosynthesis and genome stability. Biochimie 2016;126:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosnan ME, Brosnan JT. Formate: the neglected member of one-carbon metabolism. Annu Rev Nutr 2016;36:369–88. [DOI] [PubMed] [Google Scholar]
- 4.Lamarre SG, Molloy AM, Reinke SN, Sykes BD, Brosnan ME, Brosnan JT. Formate can differentiate between hyperhomocysteinemia due to impaired remethylation and impaired transsulfuration. Am J Physiol Endocrinol Metab 2012;302:E61–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrow GP, MacMillan L, Lamarre SG, Young SK, MacFarlane AJ, Brosnan ME, Brosnan JT. In vivo kinetics of formate metabolism in folate-deficient and folate-replete rats. J Biol Chem 2015;290:2244–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field MS, Kamynina E, Agunloye OC, Liebenthal RP, Lamarre SG, Brosnan ME, Brosnan JT, Stover PJ. Nuclear enrichment of folate cofactors and methylenetetrahydrofolate dehydrogenase 1 (MTHFD1) protect de novo thymidylate biosynthesis during folate deficiency. J Biol Chem 2014;289:29642–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duester G, Farrés J, Felder MR, Holmes RS, Höög JO, Parés X, Plapp BV, Yin SJ, Jörnvall H. Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem Pharmacol 1999;58:389–95. [DOI] [PubMed] [Google Scholar]
- 8.Teng S, Beard K, Pourahmad J, Moridani M, Easson E, Poon R, O’Brien PJ. The formaldehyde metabolic detoxification enzyme systems and molecular cytotoxic mechanism in isolated rat hepatocytes. Chem Biol Interact 2001;130–132:285–96. [DOI] [PubMed] [Google Scholar]
- 9.Chen CH, Ferreira JC, Gross ER, Mochly-Rosen D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev 2014;94:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galter D, Carmine A, Buervenich S, Duester G, Olson L. Distribution of class I, III and IV alcohol dehydrogenase mRNAs in the adult rat, mouse and human brain. Eur J Biochem 2003;270:1316–26. [DOI] [PubMed] [Google Scholar]
- 11.Estonius M, Svensson S, Höög JO. Alcohol dehydrogenase in human tissues: localisation of transcripts coding for five classes of the enzyme. FEBS Lett 1996;397:338–42. [DOI] [PubMed] [Google Scholar]
- 12.Keller DA, Heck HD, Randall HW, Morgan KT. Histochemical localization of formaldehyde dehydrogenase in the rat. Toxicol Appl Pharmacol 1990;106:311–26. [DOI] [PubMed] [Google Scholar]
- 13.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suh JR, Oppenheim EW, Girgis S, Stover PJ. Purification and properties of a folate-catabolizing enzyme. J Biol Chem 2000;275:35646–55. [DOI] [PubMed] [Google Scholar]
- 15.Bensadoun A, Weinstein D. Assay of proteins in the presence of interfering materials. Anal Biochem 1976;70:241–50. [DOI] [PubMed] [Google Scholar]
- 16.Field MS, Szebenyi DM, Stover PJ. Regulation of de novo purine biosynthesis by methenyltetrahydrofolate synthetase in neuroblastoma. J Biol Chem 2006;281:4215–21. [DOI] [PubMed] [Google Scholar]
- 17.Field MS, Kamynina E, Watkins D, Rosenblatt DS, Stover PJ. Human mutations in methylenetetrahydrofolate dehydrogenase 1 impair nuclear de novo thymidylate biosynthesis. Proc Natl Acad Sci USA 2015;112:400–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anguera MC, Field MS, Perry C, Ghandour H, Chiang EP, Selhub J, Shane B, Stover PJ. Regulation of folate-mediated one-carbon metabolism by 10-formyltetrahydrofolate dehydrogenase. J Biol Chem 2006;281:18335–42. [DOI] [PubMed] [Google Scholar]
- 19.Momb J, Lewandowski JP, Bryant JD, Fitch R, Surman DR, Vokes SA, Appling DR. Deletion of Mthfd1l causes embryonic lethality and neural tube and craniofacial defects in mice. Proc Natl Acad Sci USA 2013;110:549–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudiwala S, De Castro SC, Leung KY, Brosnan JT, Brosnan ME, Mills K, Copp AJ, Greene ND. Formate supplementation enhances folate-dependent nucleotide biosynthesis and prevents spina bifida in a mouse model of folic acid-resistant neural tube defects. Biochimie 2016;126:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porter DH, Cook RJ, Wagner C. Enzymatic properties of dimethylglycine dehydrogenase and sarcosine dehydrogenase from rat liver. Arch Biochem Biophys 1985;243:396–407. [DOI] [PubMed] [Google Scholar]
- 22.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem 2010;79:155–79. [DOI] [PubMed] [Google Scholar]
- 23.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004;119:941–53. [DOI] [PubMed] [Google Scholar]
- 24.Walport LJ, Hopkinson RJ, Schofield CJ. Mechanisms of human histone and nucleic acid demethylases. Curr Opin Chem Biol 2012;16:525–34. [DOI] [PubMed] [Google Scholar]
- 25.Anderson DD, Woeller CF, Chiang EP, Shane B, Stover PJ. Serine hydroxymethyltransferase anchors de novo thymidylate synthesis pathway to nuclear lamina for DNA synthesis. J Biol Chem 2012;287:7051–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 2008;320:103–6. [DOI] [PubMed] [Google Scholar]
- 27.Stein LR, Imai S. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol Metab 2012;23:420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebert AD, Kodo K, Liang P, Wu H, Huber BC, Riegler J, Churko J, Lee J, de Almeida P, Lan F, et al. Characterization of the molecular mechanisms underlying increased ischemic damage in the aldehyde dehydrogenase 2 genetic polymorphism using a human induced pluripotent stem cell model system. Sci Transl Med 2014;6:255ra130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen KE, Patel H, Kuzmanov U, Mejia NR, MacKenzie RE. Disruption of the Mthfd1 gene reveals a monofunctional 10-formyltetrahydrofolate synthetase in mammalian mitochondria. J Biol Chem 2005;280:7597–602. [DOI] [PubMed] [Google Scholar]