Abstract
As the most common subtype of Leber congenital amaurosis (LCA), LCA10 is a severe retinal dystrophy caused by mutations in the CEP290 gene. The most frequent mutation found in patients with LCA10 is a deep intronic mutation in CEP290 that generates a cryptic splice donor site. The large size of the CEP290 gene prevents its use in adeno-associated virus (AAV)-mediated gene augmentation therapy. Here, we show that targeted genomic deletion using the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system represents a promising therapeutic approach for the treatment of patients with LCA10 bearing the CEP290 splice mutation. We generated a cellular model of LCA10 by introducing the CEP290 splice mutation into 293FT cells and we showed that guide RNA pairs coupled with SpCas9 were highly efficient at removing the intronic splice mutation and restoring the expression of wild-type CEP290. In addition, we demonstrated that a dual AAV system could effectively delete an intronic fragment of the Cep290 gene in the mouse retina. To minimize the immune response to prolonged expression of SpCas9, we developed a self-limiting CRISPR/Cas9 system that minimizes the duration of SpCas9 expression. These results support further studies to determine the therapeutic potential of CRISPR/Cas9-based strategies for the treatment of patients with LCA10.
Keywords: LCA10, CEP290, CRISPR/Cas9
LCA10 is caused by mutations in CEP290, which is too large for the gene therapy vector AAV. Here, Ruan et al. show that gene editing using CRISPR/Cas9 represents a promising therapeutic approach to treat LCA10. They also develop a self-limiting CRISPR/Cas9 system to limit duration of Cas9 expression.
Introduction
Leber congenital amaurosis (LCA) is the most severe form of inherited retinal dystrophy, with patients typically exhibiting the onset of symptoms during their first year of life,1 characterized by a visual acuity that rarely exceeds 20/400.2 The prevalence of LCA in the general population is approximately 1 in 30,000 and accounts for approximately 5% of all inherited retinal dystrophies.3 LCA is a heterogeneous group of diseases that are caused by mutations in one of at least 18 genes. The gene most frequently mutated in patients with LCA is the centrosomal protein 290 kDa (CEP290, MIM610142) gene,4 a condition that is referred to as LCA10. In the retina, CEP290 is mainly located to the connecting cilium of photoreceptors, where it plays an essential role in both cilium assembly and ciliary protein trafficking.5, 6, 7 Of the CEP290 mutations that result in LCA10, the most recurrent mutation, accounting for up to 15% of all LCA cases in many Western countries, is a deep intronic mutation (c.2991+1655A > G) in intron 26 of the CEP290 gene (hereafter referred to as “IVS26 mutation” or “IVS26 splice mutation”).4, 8, 9 This mutation generates a cryptic splice donor site that leads to the inclusion of a cryptic exon bearing a premature stop codon (p.C998X) into approximately one-half of cellular CEP290 transcripts, which results in partial CEP290 activity in patients bearing the IVS26 mutation.4
Proposed therapeutic strategies for the treatment of LCA10 include gene augmentation using viral gene delivery vectors and the correction of the aberrant splice variant using antisense oligonucleotides (AONs). However, because the gene for human CEP290 (encompassing 54 exons and an open reading frame of 7,440 bp) exceeds the typical cargo size (∼4.7 kb) of recombinant adeno-associated viruses (rAAVs), the use of this delivery platform is challenging. While lentiviral vectors can accommodate the large CEP290 cDNA, they reportedly have limited tropism for photoreceptors, which are the affected cells in LCA10. Moreover, precise control of the expression levels of the CEP290 gene is desirable, as previous reports have demonstrated that the overexpression of CEP290 is cytotoxic to photoreceptors.10, 11, 12 Presently, neither rAAV nor lentiviral-mediated gene expression is conducive to modulation. AONs have been proposed as an alternative strategy to correct the aberrant splicing of CEP290.13, 14 However, because AONs have a limited half-life, treatment will likely require repeated (weekly or monthly) subretinal injections by a retinal specialist over the lifetime of the patient. Additionally, a recent study using rAAVs to deliver AONs to mouse photoreceptors showed efficacy, but the efficiency of delivery was lower than that of naked AONs.15 Therefore, the effective treatment of LCA10 will require improved therapeutic strategies.
Recently, the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) protein 9 system (known as CRISPR/Cas9) has been demonstrated as an efficient and simple genome editing tool.16, 17, 18 The CRISPR/Cas9 system consists of two components, a Cas9 nuclease and a single guide RNA (sgRNA). The sgRNA contains a targeting guide sequence that directs the Cas9 nuclease to cleave target DNA and introduce site-specific double-stranded breaks (DSBs). DSBs can be repaired via the non-homologous end joining (NHEJ) pathway or the homology-directed repair (HDR) pathway. Two DSBs introduced by a pair of sgRNAs expressed concomitantly with Cas9 can be used to induce targeted genomic deletion via the NHEJ pathway.19, 20
Presently, there are no approved therapies for patients with LCA10. Here, we describe a CRISPR/Cas9-based strategy for the treatment of patients with LCA10 bearing the IVS26 mutation. We employed a combination of specific pairs of sgRNAs and Cas9 to excise the intronic fragment containing the IVS26 splice mutation in CEP290. This technique led to the sustained generation of increased levels of wild-type CEP290.
Results
Generation of an In Vitro Model of LCA10 Using CRISPR/Cas9
A cellular model of LCA10 was developed by introducing the IVS26 mutation of CEP290 into HEK293FT cells using CRISPR/Cas9-mediated HDR. Single cell clones were isolated from 293FT cells that had been co-transfected with a plasmid expressing both sgRNA and Streptococcus pyogenes Cas9 (SpCas9) along with a single-stranded oligonucleotide (ssODN) that contained 75 nt of homologous arms on either side flanking the IVS26 mutation (Figure 1A). Of the 235 single cell clones isolated, one contained the IVS26 mutation on both CEP290 alleles (hereafter referred to as “mutant cells”). Another clone contained the mutation on one CEP290 allele and an endogenous wild-type sequence on the other allele (hereafter referred to as “heterozygous cells”). Cells that contained two alleles of endogenous wild-type CEP290 DNA are hereafter referred to as “wild-type cells.”
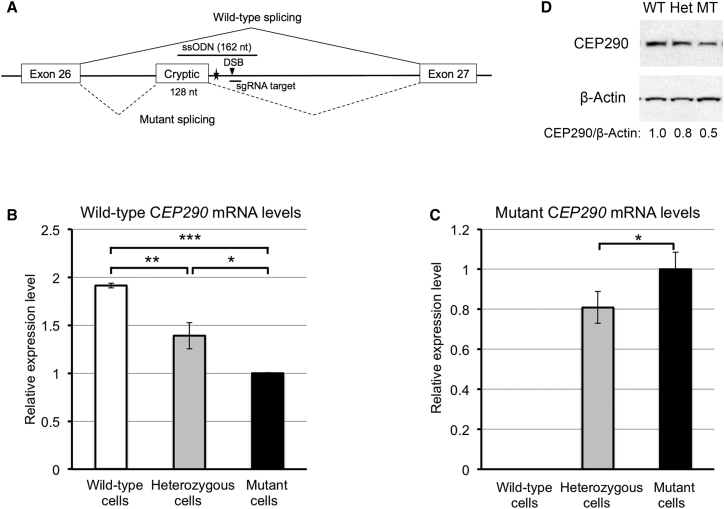
Figure 1.
Generation of an In Vitro Model of LCA10 Using CRISPR/Cas9
(A) A schematic diagram showing the IVS26 mutation (filled star) in the CEP290 gene and the locations of the sgRNA target (antisense strand) and ssODN (sense strand) used for introducing the IVS26 mutation. The DSB site (filled triangle) induced by SpCas9 was located 15 bp downstream of the IVS26 mutation. The splicing patterns for wild-type and mutant are represented by solid and dashed lines, respectively. (B and C) Basal levels of wild-type CEP290 mRNA (B) and mutant CEP290 mRNA (C) in the wild-type (white bar), heterozygous (gray bars), and mutant 293FT cells (black bars), as determined by qRT-PCR. CEP290 mRNA levels were normalized to the levels of Cyclophilin A (PPIA) mRNA. The data are presented as the means ± SD (n = 3). Comparisons were performed using one-way ANOVA followed by the Tukey honest significant difference (HSD) post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001. (D) Immunoblot analysis of lysates prepared from the wild-type (WT), heterozygous (Het), and mutant 293FT cells (MT). The membrane was probed for CEP290 protein (top) and β-actin (bottom; as a loading control). The ratio of CEP290/β-actin is shown at the bottom, with the ratio for the wild-type cells set to 1.0.
Assaying the levels of CEP290 mRNAs (both wild-type and mutant) showed that the levels of wild-type CEP290 mRNA in the heterozygous and mutant 293FT cells were reduced by 27% and 48%, respectively, compared with the wild-type cells (Figure 1B). Consistent with the genotype, wild-type cells did not express mutant CEP290 mRNA. Moreover, the levels of mutant CEP290 mRNA were 24% higher in the mutant cells than in the heterozygous cells (Figure 1C). Consistent with the changes in the mRNA levels, the levels of full-length CEP290 protein in the heterozygous and mutant cells were reduced by approximately 20% and 50%, respectively, compared with the wild-type cells (Figure 1D). Therefore, our cellular model accurately recapitulates the CEP290 expression pattern observed in fibroblasts of patients with LCA104 and represents a tool for testing potential therapeutics for patients with LCA10 who harbor the IVS26 splice mutation.
Targeted Deletion of the CEP290 IVS26 Mutation Using Paired sgRNAs and SpCas9
Using the cellular model for LCA10 described above, we tested whether a pair of upstream and downstream sgRNAs could efficiently guide SpCas9 to cleave intron 26 of the CEP290 gene at sites flanking the IVS26 splice mutation, thereby excising an intronic fragment containing the IVS26 mutation (Figure 2A). An upstream sgRNA (U1) was paired with one of three different downstream sgRNAs (D1, D2, and D3) and engineered into plasmids that also encoded SpCas9. All three different sgRNA pairs flanked the IVS26 mutation and were designed to generate genomic deletions of 283 bp (U1D1), 187 bp (U1D2), and 231 bp (U1D3), respectively. As a control, two random sgRNAs were engineered into the same SpCas9 plasmid.
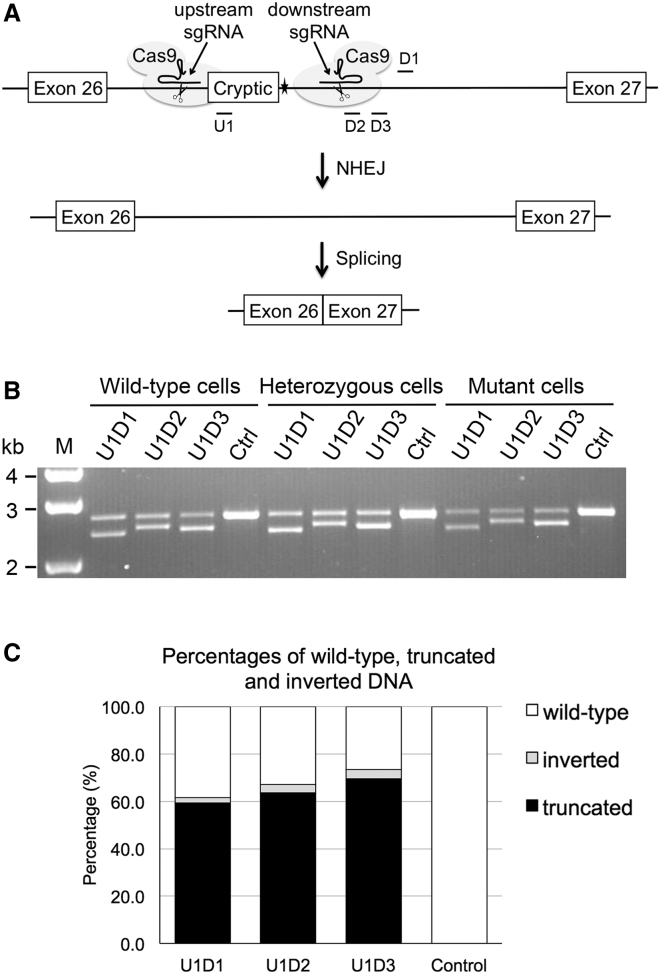
Figure 2.
Targeted Deletion of the IVS26 Mutation with Paired sgRNAs and SpCas9
(A) Schematic diagram depicting the strategy used to remove the IVS26 mutation of CEP290 (filled star). An upstream sgRNA directs the first Cas9 cleavage to a site located upstream of the IVS26 mutation, and a downstream sgRNA directs the second Cas9 cleavage to a site downstream of the mutation. The two cleavage ends are directly ligated through the NHEJ process, resulting in the excision of the intronic fragment flanking the IVS26 mutation. The truncated intron 26 is removed during mRNA processing by the RNA splicing machinery. The locations of the upstream (U1) and downstream (D1, D2, and D3) sgRNA guide sequences are indicated in the diagram. Note that D1 sgRNA targets the sense strand, whereas the other three sgRNAs target the antisense strand. (B) PCR analysis for targeted genomic deletion. 293FT cell lines were transfected with the indicated pairs of sgRNA and SpCas9. Primers were designed to bind outside of the region to be deleted. The upper bands represent PCR products amplified from intron 26 of wild-type CEP290, whereas the lower bands represent PCR products amplified from the CEP290 allele following genomic deletion. M, 1-kb DNA ladder. (C) Percentages of wild-type, truncated, and inverted DNA in the mutant 293FT cells transfected with paired sgRNAs and SpCas9, as determined by next-generation sequencing.
All three test (U1D1, U1D2, and U1D3), but not control, sgRNA pairs induced targeted genomic deletion of the expected sizes when transfected into the three 293FT cell lines (wild-type, heterozygous, and mutant) (Figure 2B). Next-generation sequencing (NGS) analysis of PCR-amplified samples prepared from the mutant cells showed that 59.3%, 63.7%, and 69.5% of the sequences from the U1D1-, U1D2-, and U1D3-transfected cells, respectively, comprised truncated DNA (Figure 2C). Therefore, the three sgRNA pairs were highly efficient at guiding SpCas9 for IVS26 mutation excision. Consistent with previous findings, NGS analysis also indicated that the repair of paired sgRNA-induced genomic deletion was largely (in about 90% of reads for all the three sgRNA pairs) accomplished by precise ligation of blunt-ended DSBs created by SpCas9; each DSB occurred exactly 3 bp upstream of the protospacer-adjacent motif (PAM) sequence.19, 20 The remaining truncated sequences contained small nucleotide insertions or deletions (indels) between the two DSBs. In addition to genomic deletion, we also observed inversion of the intronic sequences between the two DSBs in 2%–4% of total sequences (Figure 2C).
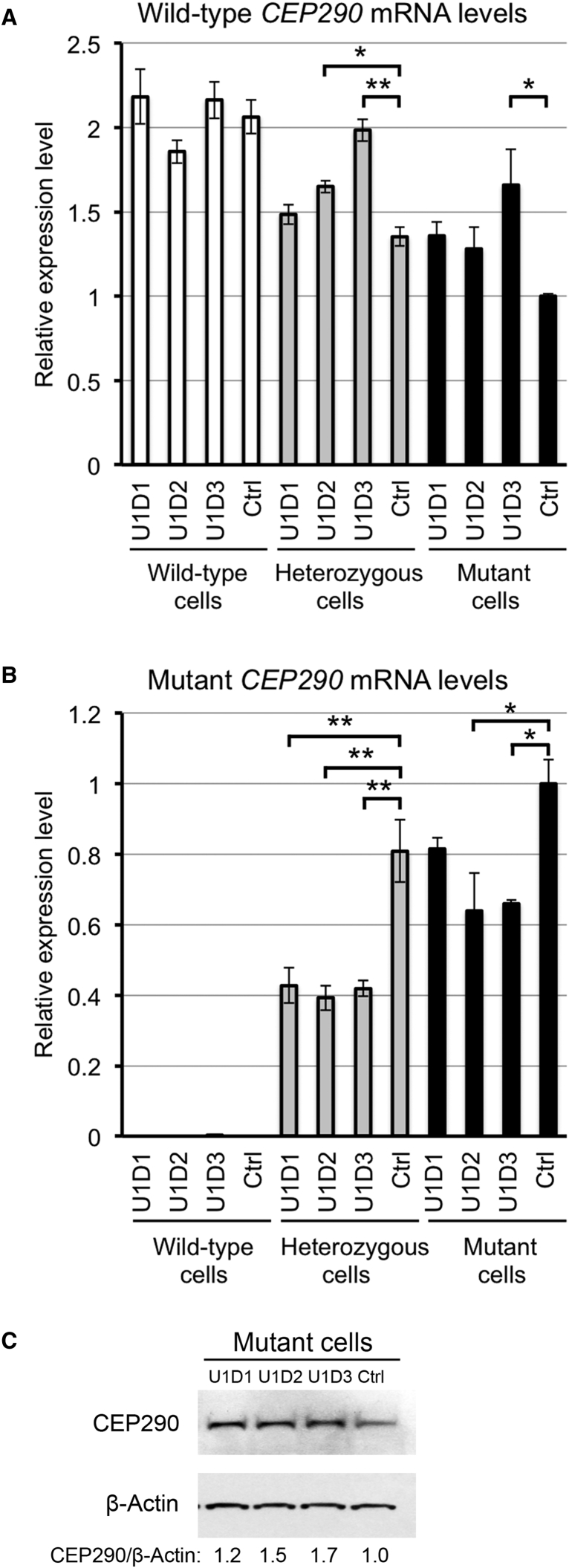
To confirm that the paired sgRNA-induced deletion in the heterozygous and mutant 293FT cells resulted in greater production of wild-type CEP290 mRNA, we assayed the levels of CEP290 mRNAs in these cells. None of the three sgRNA pairs significantly changed the levels of wild-type CEP290 mRNA in the wild-type cells (Figure 3A), which indicated that the targeted genomic deletion did not interfere with the splicing of normal CEP290 transcripts. Compared with the control sgRNA pair, the U1D3 sgRNA pair significantly increased the levels of wild-type CEP290 mRNA and reduced the levels of mutant CEP290 mRNA in the heterozygous and mutant cells (Figures 3A and 3B). The U1D2 sgRNA pair significantly increased the levels of wild-type CEP290 mRNA in the heterozygous cells (Figure 3A) and reduced the levels of mutant CEP290 mRNA in the heterozygous and mutant cells (Figure 3B). The less effective sgRNA pair U1D1 significantly reduced the levels of mutant CEP290 mRNA in the heterozygous cells (Figure 3B). Western blot analysis showed that each of the three test sgRNA pairs resulted in the production of higher levels of full-length CEP290 protein in the mutant cells (Figure 3C). Therefore, the test sgRNA pair U1D3 was highly efficient at circumventing the aberrant splicing of the cryptic exon of CEP290.
Figure 3.
Rescue of the Expression of Wild-Type CEP290 with Paired sgRNAs and SpCas9
(A and B) The levels of wild-type (A) and mutant (B) CEP290 mRNAs in the wild-type (white bars), heterozygous (gray bars), and mutant 293FT (black bars) cells transfected with paired sgRNAs and SpCas9, as measured by RT-qPCR. The data are presented as the means ± SD (n = 2). Comparisons were performed using one-way ANOVA followed by the Tukey HSD post hoc test. *p < 0.05; **p < 0.01. Ctrl, control sgRNA pair. (C) Immunoblot analysis of lysates prepared from the mutant cells transfected with paired sgRNAs and SpCas9. The membrane was probed for CEP290 protein (top) and β-actin (bottom; as a loading control). The ratio of CEP290/β-actin is shown at the bottom, with the ratio for the control sgRNA pair set to 1.0.
To analyze potential off-target mutations for the U1 and D3 sgRNAs, we performed the T7 endonuclease I (T7E1) assay in human 293FT cells to examine the 10 most likely off-target sites for each sgRNA (Table S4) determined by the Benchling CRISPR gRNA Design tool (http://www.benchling.com). None of these sites showed detectable levels of indel mutations (Figure S1).
SaCas9 for the Targeted Deletion of the IVS26 Splice Mutation
The relatively large size of the gene encoding SpCas9 (∼4.1 kb) when combined with the paired sgRNAs is difficult to package into a single adeno-associated virus (AAV) vector. Ran et al.21 recently reported on a shorter version of a Cas9 nuclease from Staphylococcus aureus that they referred to as SaCas9. This observation presents the possibility of assembling both the nuclease and paired sgRNAs into a single AAV vector. Based on this study, we designed three upstream sgRNAs (aU1, aU2, and aU3) and two downstream sgRNAs (aD1 and aD2) for SaCas9. The upstream/downstream sgRNAs pairs were assembled into a single all-in-one AAV expression plasmid that also encoded SaCas9 under the transcriptional control of a minimal cytomegalovirus (minCMV) promoter. In contrast, the SpCas9 gene and the U1D3 sgRNA pair were engineered into two separate AAV expression plasmids (pAAV-SpCas9 + pAAV-U1D3).
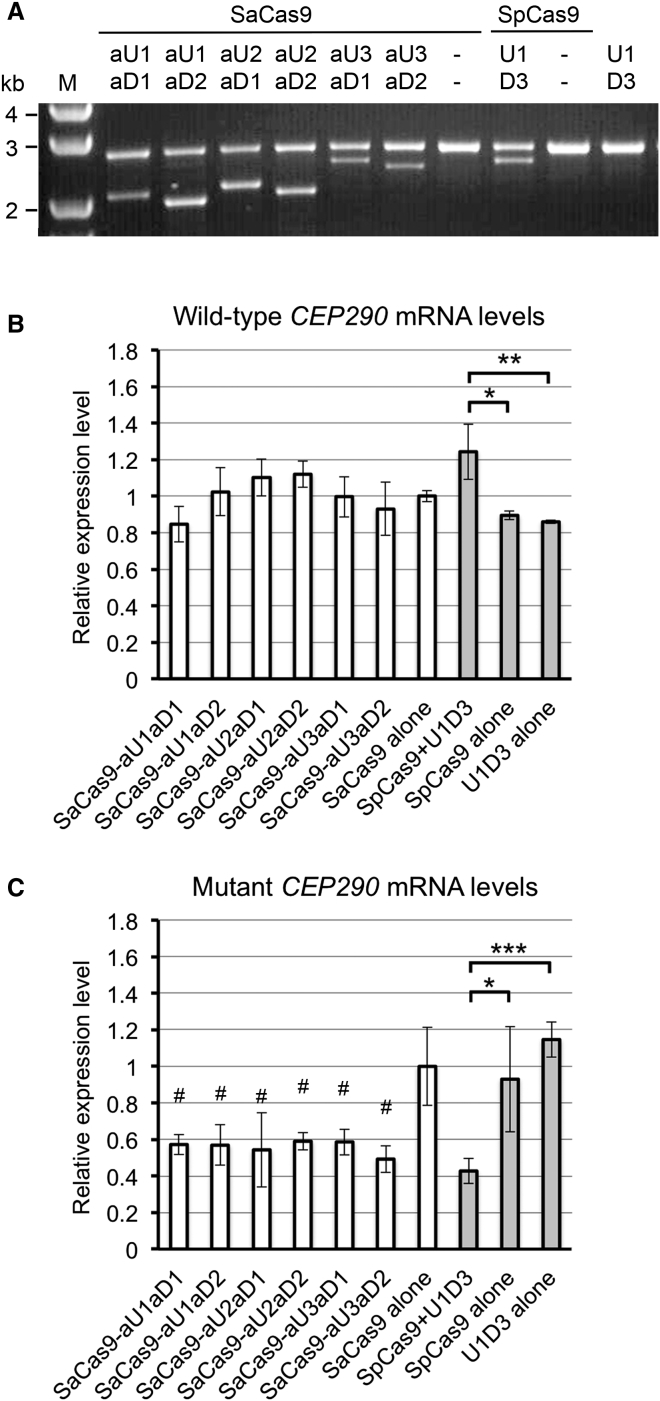
PCR analysis of genomic DNA from the mutant 293FT cells showed that all six test sgRNA pairs co-expressed with SaCas9 induced targeted genomic deletions to remove the IVS26 mutation of CEP290 (Figure 4A). However, surprisingly, none of these six test sgRNA pairs significantly increased the levels of wild-type CEP290 mRNA; the aU2aD1 and aU2aD2 sgRNA pairs showed a slight trend toward higher levels (Figure 4B). In contrast, the dual pAAV-SpCas9 and pAAV-U1D3 plasmids significantly increased the levels of wild-type CEP290 mRNA compared with the individual plasmids (Figure 4B). All of the test sgRNA pairs co-expressed with SaCas9 or SpCas9 significantly reduced the levels of mutant CEP290 mRNA (Figure 4C). Notably, we cannot exclude the possibility that a more potent sgRNA pair than the sgRNA pairs we tested could guide SaCas9 to efficiently rescue the expression of wild-type CEP290 in the mutant cells.
Figure 4.
Targeted Deletion of the IVS26 Mutation with Single SaCas9 Plasmids or Dual SpCas9 Plasmids
(A) PCR analysis of mutant 293FT cells transfected with either the single pAAV-SaCas9-paired sgRNAs plasmids or the dual SpCas9 plasmids (pAAV-SpCas9 + pAAV-U1D3) for targeted genomic deletion. Dashes indicate that there is no sgRNA pair in this plasmid. (B and C) The levels of wild-type (B) and mutant (C) CEP290 mRNAs in the mutant cells transfected with either the single pAAV-SaCas9-paired sgRNAs plasmids (white bars) or the dual SpCas9 plasmids (gray bars), as measured by qRT-PCR. The data are presented as the means ± SD (n = 3). Comparisons were performed using one-way ANOVA followed by the Tukey HSD post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001; #p < 0.05 compared with the mutant cells transfected with the pAAV-SaCas9 alone (control).
Targeted Genomic Deletion of an Intronic Fragment of the Mouse Cep290 Gene in the Mouse Retina
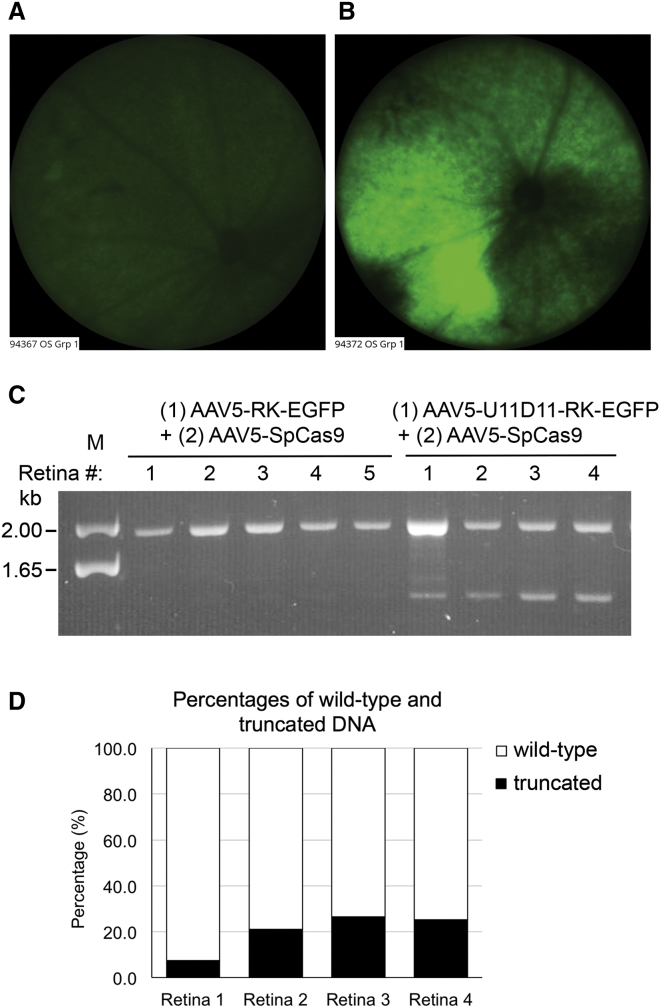
The aforementioned studies using the LCA10 cell model supported the use of select pairs of sgRNAs and SpCas9 to remove the IVS26 mutation and rescue the expression of wild-type CEP290. In subsequent analyses, we sought to determine whether we could translate this CRISPR/Cas9-based approach for treating LCA10 to animals in vivo. However, because there is no animal model that expresses the IVS26 mutation-associated cryptic exon, we elected to perform our validation studies in wild-type mice and test whether a pair of sgRNAs and SpCas9 could induce targeted genomic deletion of intron 25 of the mouse Cep290 gene. Intron 25 in the mouse is homologous to intron 26 of the human CEP290 gene. The 18 sgRNA pairs designed to target intron 25 of the mouse Cep290 gene were first evaluated for their effectiveness in genomic deletion in the mouse Neuro-2a cells. The most efficient sgRNA pair U11D11 induced approximately 50% genomic deletion in Neuro-2a cells (Figure S2) and was used in a subsequent in vivo Cep290 targeting experiment. A dual AAV system was used to package the U11D11 sgRNA pair and SpCas9 separately into two AAV5 vectors. Previous studies have shown that AAV5 serotype vectors efficiently transduce photoreceptors when injected into the eye subretinally.22 The AAV5-U11D11 vector also expressed an EGFP reporter under the transcriptional control of the human rhodopsin kinase (RK) promoter. A rAAV5 vector that expressed EGFP alone was also included as a control. The dual AAV vectors (1 × 109 viral genomes each) were co-injected into the subretinal space of 8- to 10-week-old C57BL/6J mice. EGFP expression in the retina was examined in live animals using a Micron IV retinal microscope at 2 and 4 weeks post-injection (Figures 5A and 5B). The mice were euthanized at 4 weeks post-injection, and genomic DNA was extracted from the neural retinas for PCR analysis. Co-injection of AAV5-SpCas9 with AAV5-U11D11-RK-EGFP, but not with AAV5-RK-EGFP, resulted in a targeted genomic deletion of the expected size (Figure 5C). NGS analysis of four treated retinas revealed that 7.5%, 21.1%, 26.4%, and 25.2% of sequences comprised truncated DNA following U11D11-guided genomic deletion (Figure 5D). The low deletion frequency noted in one of the retinas (7.5%) was consistent with the relatively low EGFP expression observed in this mouse (Figure 5A). These results show that a dual AAV system could be used to delete an intronic fragment in the Cep290 gene of mouse photoreceptors.
Figure 5.
Targeted Deletion of a Fragment in Intron 25 of the Cep290 Gene in the Mouse Retina Using a Dual AAV System
(A and B) Micron IV fluorescence images for treated retina 1 (A) and retina 4 (B) on day 28 after subretinal injection. (C) PCR analysis showing targeted genomic deletion in the mouse retinas that received (1) AAV5-RK-EGFP (control) or AAV5-U11D11-RK-EGFP (treated) and (2) AAV5-SpCas9. The upper bands correspond to PCR products amplified from intron 25 of wild-type Cep290 gene, whereas the lower bands correspond to PCR products amplified from the Cep290 allele following U11D11-guided genomic deletion. (D) Percentages of wild-type and truncated DNA in the four treated retinas, as determined by next-generation sequencing.
Development of a Self-Limiting CRISPR/Cas9 System to Limit the Expression of SpCas9
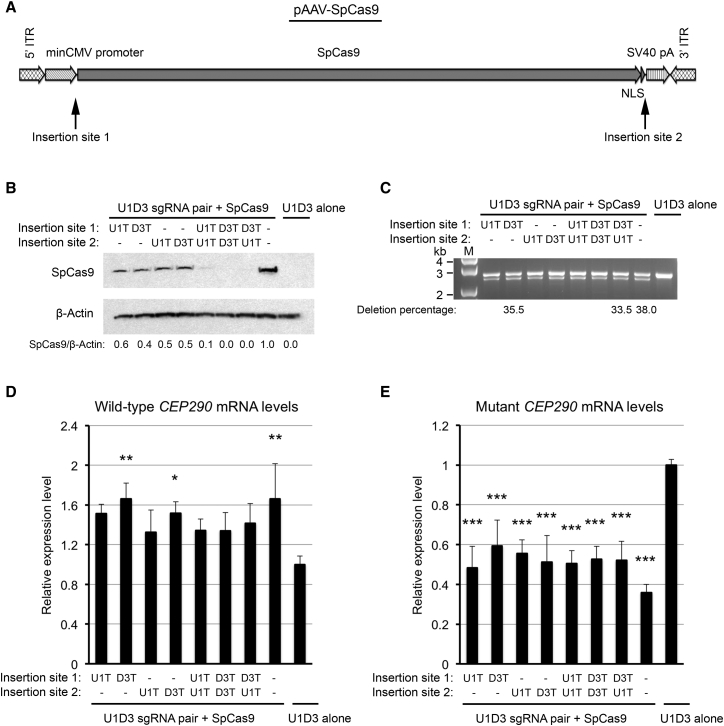
Sustained expression of SpCas9 is not required for gene editing; indeed, prolonged expression of exogenous SpCas9 in the transduced cells may elicit host cellular immunity and engender safety problems.23 Hence, a “hit and go” approach whereby cellular exposure to the SpCas9 protein is limited may be beneficial for in vivo CRISPR/Cas9-based therapies. To this end, we developed a self-limiting CRISPR/Cas9 system by incorporating recognition site(s) for the sgRNA(s) into the SpCas9 plasmid itself, such that the plasmid would be excised and eliminated following the initiation of SpCas9 expression. This system comprises two AAV expression plasmids: a self-limiting pAAV-SpCas9 plasmid and a pAAV-sgRNAs plasmid expressing the U1D3 sgRNA pair. The recognition sequences (sgRNA target sequences plus corresponding PAMs) for the U1 and/or D3 sgRNA (U1T and/or D3T) were incorporated into either one or two of the two insertion sites in the pAAV-SpCas9 plasmid (Figure 6A). Insertion site 1 was located between the minCMV promoter and the SpCas9 coding sequence, whereas insertion site 2 was located between the nuclear localization signal (NLS) sequence and the SV40 polyadenylation (SV40 pA) signal sequence. In this configuration, the U1D3 sgRNA pair would guide the SpCas9 protein for both targeted genomic deletion and cleavage of the SpCas9 plasmid itself and thereby limit the production of the SpCas9 protein.
Figure 6.
Self-Limiting CRISPR/SpCas9 System
(A) Schematic diagram of the pAAV-SpCas9 plasmid. The sgRNA recognition sequences were incorporated into insertion site 1 (between the minCMV promoter and SpCas9) and/or insertion site 2 (between SpCas9-NLS and SV40 pA). ITR, inverted terminal repeat. (B) Immunoblot analysis of lysates prepared from the mutant 293FT cells transfected with the pAAV-U1D3 plasmid and the self-limiting pAAV-SpCas9 plasmids, which contained the U1 sgRNA recognition sequence (U1T) and/or the D3 sgRNA recognition sequence (D3T). The membrane was probed for SpCas9 protein (top) and β-actin (bottom; as a loading control). The ratio of SpCas9/β-actin is shown at the bottom, with the ratio for the control pAAV-SpCas9 plasmid without the sgRNA recognition sequence set to 1.0. Dashes indicate that there is no sgRNA recognition sequence at this site. (C) PCR analysis for targeted genomic deletion in the mutant cells transfected with the pAAV-U1D3 plasmid and the self-limiting pAAV-SpCas9 plasmids. Three selected samples were subjected to NGS analysis, and percentages of truncated DNA were shown below the gel image. (D and E) The levels of wild-type (D) and mutant (E) mRNAs in the mutant cells transfected with the pAAV-U1D3 plasmid and the self-limiting pAAV-SpCas9 plasmids, as measured by qRT-PCR. The data are presented as the means ± SD (n = 3). Comparisons were performed using one-way ANOVA followed by the Tukey HSD post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001 (compared with the mutant cells transfected with the pAAV-U1D3 plasmid alone [control]).
Measurement of SpCas9 protein levels in the mutant 293FT cells transfected with this self-limiting CRISPR/Cas9 system showed that when a single sgRNA recognition sequence (U1T or D3T) was inserted into the self-limiting pAAV-SpCas9 plasmid, the quantities of the SpCas9 protein were reduced by approximately 50% compared with the control pAAV-SpCas9 plasmid that did not contain the sgRNA recognition sequence (Figure 6B). Insertion of two U1Ts, two D3Ts, or D3T and U1T into the two insertion sites reduced the expression of the SpCas9 protein to negligible levels (Figure 6B). Therefore, the self-limiting CRISPR/Cas9 configuration effectively limited the expression of SpCas9.
PCR analysis of genomic DNA isolated from the mutant cells showed that the self-limiting CRISPR/Cas9 system was still able to facilitate targeted deletion, as illustrated by the removal of the IVS26 mutation (Figure 6C). Similar to the control pAAV-SpCas9 plasmid, insertion of D3T into either site significantly increased the levels of wild-type CEP290 mRNA in the mutant cells, and the remaining configurations showed a trend toward higher levels (Figure 6D). All configurations of the self-limiting pAAV-SpCas9 plasmids significantly reduced the levels of mutant CEP290 mRNA (Figure 6E). Therefore, the self-limiting CRISPR/Cas9 system, despite conferring only transient expression of SpCas9 expression, was effective at removing the IVS26 mutation of CEP290. This was illustrated by a reduction in the levels of mutant CEP290 mRNA and a corresponding increase in the levels of wild-type CEP290 mRNA.
Discussion
In the present study, we explored the potential of CRISPR/Cas9-mediated gene editing to correct the IVS26 splice mutation in CEP290. We showed that a combination of specific pairs of sgRNAs and SpCas9 could efficiently correct the aberrant splicing of a common CEP290 variant in a cellular model of LCA10. As a proof of principle, we showed that we could use dual rAAV vectors to induce the deletion of a specific intronic fragment of the mouse Cep290 gene in the photoreceptors of mice. Finally, we developed a “hit and go” approach to limit the expression of SpCas9 in the transfected cells to minimize the potential impact of a host immune response to the exogenous enzyme.
Ocular diseases are particularly conducive to gene therapy because the eye is relatively accessible using established, albeit somewhat invasive, techniques. Correction of the underlying basis for the disease pathology and restoration of visual function can be measured directly and non-invasively in a process that facilitates the determination of the efficacy of treatment. The eye is also relatively immune privileged, and reports of AAV-mediated gene therapy of the retina have shown limited local or systemic immune responses in the form of neutralizing antibodies to the AAV vector or T cell responses to the viral capsid.24, 25 For example, a recent phase 3 trial of subretinal injections of AAV2 vectors encoding RPE65 into patients with LCA2 showed that the procedure is safe (with no serious drug-related adverse events or deleterious immune responses) and, importantly, improved vision and light sensitivity.26 LCA10 represents another attractive ocular target for therapeutic intervention using this technology platform. Previous studies indicated that patients with LCA10 retained a central island of photoreceptors with a normal thickness at the fovea (where the cone photoreceptors are located). Moreover, despite early and severe visual loss, the anatomy of the intracranial visual pathway is preserved for many years, which offers a broad temporal window of opportunity for therapeutic intervention.27, 28
For LCA10 disease, the CRISPR/Cas9-based gene editing strategy may represent an effective approach to correct the IVS26 mutation of CEP290. In contrast to the CEP290 gene, the size of the editing machinery of CRISPR/Cas9 fits within the packaging limits of recombinant AAV vectors. Genome editing using CRISPR/Cas9 also allows for the permanent correction of the IVS26 mutation in patients. Importantly, this approach is not anticipated to alter the transcriptional regulation of the CEP290 gene, which may be pertinent because there are 11 alternative splice isoforms, and their relative contributions to photoreceptor function and activity are unclear.14 In addition, the absolute cellular levels of CEP290 protein, a structural protein, are thought to be important for cell survival.12 Restoring the genetic configuration of CEP290 to a more normal configuration avoids changes in the overall expression levels of CEP290 and the levels of various splicing isoforms of the protein.
To treat IVS26-associated LCA10, it is likely that a dual AAV system that separately encodes SpCas9 and a pair of sgRNAs will be required. Using this approach, both the AAV-SpCas9 and AAV-sgRNAs vectors will need to transduce the same photoreceptors for therapeutic benefit. Importantly, we do not anticipate this to be a significant hurdle, as the small, enclosed space of the subretinal compartment should facilitate co-transduction, particularly if a high AAV vector-to-cell ratio is used. Indeed, we showed that we could achieve efficient targeted genomic deletion in the photoreceptors of mice following the subretinal injection of a dual AAV system. Other illustrations include the demonstration of highly efficient (∼80%) co-transduction when a pair of AAV1/2-SpCas9 and AAV1/2-sgRNA vectors were co-injected into the hippocampal dentate gyrus of mice.29 A dual AAV system was also successfully used to excise exon 23 of mutant dystrophin in a mouse model of Duchenne muscular dystrophy (DMD), with resultant correction of dystrophin protein expression and skeletal muscle function.30
Testing for efficacy was limited by the absence of an animal model bearing the IVS26 splice mutation in LCA10. Although animal models bearing spontaneous Cep290 mutations (e.g., rd16 mouse and rdAc Abyssinian cat) show some phenotypes that mimic those noted in some patients with LCA10,5, 31 these models cannot be used to test our CRISPR/Cas9-based approach. Garanto et al.32 generated a transgenic knock-in mouse model carrying an IVS26 mutation-bearing intron 26 of the human CEP290 gene, but this model did not recapitulate the disease phenotypes noted in patients with LCA10. This could be due to differences in the ability of the splicing machinery in mice and humans to recognize this splice variant. To assess the therapeutic potential of the CRISPR/Cas9-based approach to treat patients with LCA10 harboring the IVS26 splice mutation, additional efforts will be needed to create a more faithful LCA10 knock-in mouse model.
One safety consideration associated with the use of CRISPR/Cas9-based therapies is the induction of a host immune response to the exogenous bacterial Cas9 protein. Indeed, a recent report showed the induction of humoral immunity against SpCas9 (detected SpCas9-specific cellular immune response) following delivery to animals.23 Therefore, strategies that induce the transient expression of the Cas9 protein could be necessary for in vivo applications. A proposed approach to limit the duration of Cas9 expression involved the use of nanoparticles to deliver either the Cas9 protein or Cas9 mRNA.33, 34 However, the efficiency of delivery of the nanoparticles into the photoreceptors is typically low. In the current study, we developed a “hit and go” self-limiting CRISPR/Cas9 system to minimize the duration of expression of SpCas9. Limiting the exposure of SpCas9 should also reduce the potential for off-target effects, which should improve the safety profile of this system.
In summary, we provide a proof-of-principle study for a CRISPR/Cas9-based therapy that could be used to treat the IVS26 splice mutation associated with patients with LCA10. AAV-mediated gene editing is an attractive strategy for treating inherited ocular diseases, because delivery occurs at a localized site and the genetic correction is permanent for the life of photoreceptors. We also posit that this strategy of targeted genomic deletion may be equally applicable to the treatment of other genetic diseases caused by deep intronic mutations.
Materials and Methods
sgRNA Design
sgRNAs were designed using the MIT CRISPR Design tool (http://crispr.mit.edu/) and the Benchling CRISPR gRNA Design tool (http://www.benchling.com). Both tools find and rank all 20-bp protospacer sequences preceding a NGG protospacer-adjacent motif, and they analyze the potential off-target sites by a bioinformatics blast search with the whole genome DNA sequences.
DNA Constructs
The pSpCas9 plasmid, which was engineered to express SpCas9 under the transcriptional control of a cytomegalovirus (CMV) promoter, was purchased from Sigma. The BbsI restriction site in the bovine growth hormone (BGH) poly(A) was removed using the QuikChange lightning site-directed mutagenesis kit following the manufacturer’s protocol (Agilent Technologies). Paired sgRNAs were subcloned into this plasmid using the Golden Gate cloning method, as previously described.35 Briefly, a DNA fragment containing the U6 promoter-BbsI:BbsI-sgRNA scaffold was synthesized by GeneArt (Thermo Fisher Scientific) and was inserted into the PciI and NheI restriction sites of the pSpCas9-BbsI null plasmid to generate a pSpCas9(BBU) plasmid that was then used to subclone the upstream sgRNA target sequences. A pSpCas9(BBD) plasmid was made compatible for subcloning the downstream sgRNA target sequences. To construct pSpCas9(BBD), PCR was performed with the pSpCas9(BBU) plasmid based on the template and primers listed in Table S2. The PCR product was inserted into the PciI and KpnI sites of the pSpCas9(BBU) plasmid to generate pSpCas9(BBD). The oligos for the upstream and downstream sgRNAs were annealed and subcloned into the two BbsI restriction sites of the pSpCas9(BBU) and pSpCas9(BBD) plasmids, respectively. Finally, the U6 promoter-downstream sgRNA fragment was excised from the pSpCas9(BBD) plasmid using the BsaI restriction enzyme and was subcloned into the two BsaI sites of the pSpCas9(BBU)-upstream sgRNA plasmid. The resultant pSpCas9(BBUD) plasmid expressed two U6 promoter-driven sgRNAs and one CMV promoter-driven SpCas9.
To construct the all-in-one SaCas9 plasmid, a minCMV-SaCas9-NLS-FLAG-BGH pA-U6-BsaI:BsaI-sgRNA scaffold fragment was synthesized by GenScript and subcloned into the PciI and BbsI restriction sites of the pSpCas9 plasmid to replace the CMV-SpCas9-BGH pA cassette. Five sgRNA guide sequences (aU1, aU2, aU3, aD1, and aD2) were subcloned into the two BsaI restriction sites of the pSaCas9 plasmid. To pair the upstream and downstream sgRNAs, the U6 promoter-downstream sgRNA fragment was excised from its plasmid using KpnI and NotI and then was subcloned into the NotI site of the plasmid that expressed the upstream sgRNA. Finally, the entire minCMV-SaCas9-BGH pA-U6-upstream sgRNA-U6-downstream sgRNA fragment was subcloned into an AAV expression plasmid.
To construct the self-limiting SpCas9 plasmid, a DNA fragment containing a minCMV promoter was synthesized by GeneArt and was inserted into the MluI and ApoI sites of the pSpCas9 plasmid to generate a pminCMV-SpCas9-NLS-BGH pA plasmid. Next, a DNA fragment containing the SV40 pA signal was synthesized by GeneArt and was inserted into the XhoI and BbsI restriction sites of the above plasmid to replace the BGH pA signal. The recognition sequences for U1 and/or D3 sgRNAs were subcloned into insertion site 1 (between the minCMV promoter and SpCas9) and/or insertion site 2 (between the NLS and SV40 pA). Finally, the minCMV-SpCas9-NLS-SV40 pA fragment was subcloned into an AAV expression plasmid to generate the self-limiting pAAV-SpCas9 plasmid. To construct an AAV expression plasmid for the U1D3 sgRNA pair, the U6-U1 sgRNA-U6-D3 sgRNA fragment from the pSpCas9(BBUD)-U1D3 plasmid was excised using PciI and KpnI and then was subcloned into the EcoRV and KpnI sites of the AAV expression plasmid.
sgRNA target sequences can be found in Table S1, primer sequences can be found in Table S2, and other DNA sequences used in this study can be found in Table S3.
Nucleofection and Single Cell Clone Screening
Nucleofection was used to generate the cellular model for LCA10, whereas Lipofectamine 3000 (Thermo Fisher Scientific) was used for all other 293FT transfections. For nucleofection, 2.5 μg pSpCas9(BBU)-sgRNA plasmid DNA and 5 μL ssODN (10 μM) were co-transfected into 1 × 106 293FT cells using the Amaxa SF cell line 4D-Nucleofector X Kit L and the program CM-130 in a 4D-Nucleofector System (Lonza). To identify single cell clones bearing the IVS26 mutation, cells were dissociated into single cells at 48 hr after co-transfection and were serially diluted to a final concentration of 0.5 cells per 100 μL. Approximately 100 μL of diluted cells was plated into each well of nine 96-well plates. The cells were expanded in a 5% CO2, 37°C incubator for 2 weeks. Then, 235 single cell clones were identified and subjected to screening for the IVS26 mutation. Genomic DNA was extracted using the QuickExtract DNA extraction solution (Epicenter) and amplified using GoTaq Hot Start Master Mix (Promega). Sanger sequencing was performed to determine the presence of the IVS26 mutation.
qRT-PCR
mRNAs were extracted using the RNeasy Plus Mini Kit (QIAGEN). Then, 0.5 or 1 μg total RNA was used to synthesize cDNA based on the iScript cDNA synthesis kit (Bio-Rad). cDNAs were subjected to real-time qPCR amplification using Fast Plus EvaGreen qPCR Master Mix (Biotium) and primers that specifically detect wild-type and mutant CEP290 mRNAs on an ABI Prism 7500 Real-Time PCR System (Applied Biosystems). The specificity of amplification products was determined from melting curve analysis performed at the end of each run. Data were analyzed using SDS 2.3 software (Applied Biosystems). The levels of CEP290 mRNA were normalized to the levels of PPIA mRNA.
Western Blot Analysis
Cells were lysed in RIPA lysis buffer (Cell Signaling Technology) supplemented with 1 mM PMSF and 1× protease inhibitor cocktail on ice. Protein samples were separated using the NuPAGE Electrophoresis System (Thermo Fisher Scientific), after which the proteins were transferred to polyvinylidene fluoride (PVDF) membranes. Membranes were blocked by Pierce TBST (Tris-buffered saline with Tween 20 detergent) buffer containing 1% non-fat dry milk for 1 hr at room temperature and then were incubated in the indicated primary antibody overnight at 4°C. The primary antibodies used were a rabbit polyclonal anti-CEP290 antibody (a kind gift from Professor Hemant Khanna at University of Massachusetts, Worcester) and a mouse monoclonal anti-SpCas9 antibody (clone 7A9; Millipore). The membranes were then incubated with the appropriate secondary antibody (Alexa Fluor 647-conjugated anti-rabbit or anti-mouse IgG; Cell Signaling Technology) for 1 hr at room temperature. To re-probe membranes for horseradish peroxidase (HRP)-conjugated rabbit monoclonal anti-β-actin antibody (clone 13E5; Cell Signaling Technology), the PVDF membranes were stripped by incubating them at 37°C for 30 min in Restore western blot stripping buffer (Thermo Fisher Scientific). The immunoblot data were quantified by densitometric analysis using NIH ImageJ software. The blotting data shown in this study are representative of at least three independent experiments.
Next-Generation Sequencing
NGS analysis was performed at ACGT. The DNA samples were fragmented to an average target fragment size of 350 bp by ultrasonication and were used to construct a sequencing library using the Illumina TruSeq DNA PCR-free sample preparation kit. The library was quantified and assessed using a Qubit and 2100 Bioanalyzer, after which the samples were loaded onto an Illumina platform to generate PE150 reads. Approximately 150,000 reads (±20%) per sample were generated. Raw Illumina data were de-multiplexed and converted into fastq format, and low-quality (Q < 20) and short reads (N < 50) were filtered out. The filtered reads were aligned to the reference sequences using Bowtie2. For quantification, BEDTools was used to calculate the numbers of reads mapped to a 40-bp sequence flanking the cleavage site that are unique to either the wild-type DNA or the truncated DNA; these reads were used to determine the percentages of wild-type and truncated DNA in each sample.
T7E1 Assay
293FT cells were transfected with either pSpCas9(BBU)-U1 sgRNA or pSpCas9(BBD)-D3 sgRNA plasmid DNA. Genomic DNA was extracted 48 hr post-transfection using the QuickExtract DNA extraction solution and was subjected to T7E1 assay with the EnGen mutation detection kit following the manufacturer’s instructions (New England Lab). All primers used for the PCR amplifications are listed in Table S5. Resulting PCR products ranged in size from 450 bp to 700 bp.
AAV Production
Recombinant AAV vectors were produced by triple transfection of 293FT cells, as previously described.36 Briefly, an AAV expression plasmid, a plasmid containing the rep gene from serotype 2 and a capsid gene from serotype 5, and a helper adenoviral plasmid (Agilent Technologies) were used. Virus was collected at 72 hr post-transfection and was purified by AVB Sepharose affinity chromatography (GE Healthcare). Genome copy (GC) titers of AAV vectors were determined by TaqMan-based qPCR analysis (Thermo Fisher Scientific).
Animals
Eight- to 10-week-old female C57BL/6J mice were purchased from Jackson Laboratories and were maintained at the Sanofi vivarium. The animals were given free access to food and water for the duration of the study. All animal procedures were conducted in compliance with the Animal Welfare Act, and the Guide for the Care and Use of Laboratory Animals, the Office of Laboratory Animal Welfare and in accordance with the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Vision Research.
Subretinal AAV Injection
Mice were sedated using 3.5% isoflurane in 800 mL/min of oxygen delivered to the animal via a nose cone. Mydriasis and cycloplegia were induced in mice with the topical application of Tropicamide (Alcon). A pilot incision was made in the cornea, and a 33-gauge blunt-tipped needle was directed through the incision and advanced posteriorly between the iris and the lens capsule until the tip penetrated the posterior neurosensory retina. A total of 1 × 109 viral genomes (VGs) of AAV5-SpCas9 along with 1 × 109 VGs of AAV5-U11D11-RK-EGFP or the control AAV5-RK-EGFP were delivered to the left eye (OS) of each mouse in a total volume of 1 μL at a rate of 300 nL/second. The needle was held in position for approximately 5 seconds before withdrawal. The animal was allowed to recover from anesthesia prior to returning to its cage. EGFP expression in the retina was evaluated in live animals using a Micron IV retinal microscope (Phoenix Research Labs) at 2 and 4 weeks post-injection, and mice lacking EGFP expression were excluded from the study. All animals were euthanized at 4 weeks post-injection.
Genomic DNA Extraction from Mouse Retinas
Mouse eyes were enucleated and placed in PBS. Retinas were isolated using micro-dissecting scissors under a dissection microscope. The retinal pigment epithelium (RPE) layer was carefully removed. Retinas were homogenized in QuickExtract DNA extraction solution with pestles powered by a cordless motor. Genomic DNA was extracted following the manufacturer’s instructions, diluted to 10 ng/μL, and amplified using GoTaq Hot Start Master Mix and PCR primers flanking the deleted region.
Author Contributions
Conceptualization, G.-X.R. and A.S.; Methodology, G.-X.R., E.B., and D.Y.; Investigation, G.-X.R., E.B., D.Y., and M.L.; Writing – Original Draft, G.-X.R.; Writing – Review & Editing, G.-X.R., E.B., M.L., S.H.C., and A.S.; Supervision, S.H.C. and A.S.
Conflicts of Interest
All authors are employees of Sanofi Genzyme.
Acknowledgments
We thank the Gene Therapy Vector Core group at Sanofi Genzyme for assistance with AAV production, Dongyu Liu for assistance with NGS analysis, and Hemant Khanna at the University of Massachusetts for providing us with the anti-CEP290 antibody.
Footnotes
Supplemental Information includes two figures and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2016.12.006.
Supplemental Information
References
- 1.Leber T. Ueber Retinitis pigmentosa und angeborene Amaurose. Graefes Arch. Clin. Exp. Ophthalmol. 1869;15:1–25. [Google Scholar]
- 2.Cremers F.P., van den Hurk J.A., den Hollander A.I. Molecular genetics of Leber congenital amaurosis. Hum. Mol. Genet. 2002;11:1169–1176. doi: 10.1093/hmg/11.10.1169. [DOI] [PubMed] [Google Scholar]
- 3.Koenekoop R.K. An overview of Leber congenital amaurosis: a model to understand human retinal development. Surv. Ophthalmol. 2004;49:379–398. doi: 10.1016/j.survophthal.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 4.den Hollander A.I., Koenekoop R.K., Yzer S., Lopez I., Arends M.L., Voesenek K.E., Zonneveld M.N., Strom T.M., Meitinger T., Brunner H.G. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S.K., Cheng H., Scott A., Hurd R.E. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbelanne M., Song J., Ahmadzai M., Tsang W.Y. Pathogenic NPHP5 mutations impair protein interaction with Cep290, a prerequisite for ciliogenesis. Hum. Mol. Genet. 2013;22:2482–2494. doi: 10.1093/hmg/ddt100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craige B., Tsao C.C., Diener D.R., Hou Y., Lechtreck K.F., Rosenbaum J.L., Witman G.B. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 2010;190:927–940. doi: 10.1083/jcb.201006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perrault I., Delphin N., Hanein S., Gerber S., Dufier J.L., Roche O., Defoort-Dhellemmes S., Dollfus H., Fazzi E., Munnich A. Spectrum of NPHP6/CEP290 mutations in Leber congenital amaurosis and delineation of the associated phenotype. Hum. Mutat. 2007;28:416. doi: 10.1002/humu.9485. [DOI] [PubMed] [Google Scholar]
- 9.Vallespin E., Lopez-Martinez M.A., Cantalapiedra D., Riveiro-Alvarez R., Aguirre-Lamban J., Avila-Fernandez A., Villaverde C., Trujillo-Tiebas M.J., Ayuso C. Frequency of CEP290 c.2991_1655A>G mutation in 175 Spanish families affected with Leber congenital amaurosis and early-onset retinitis pigmentosa. Mol. Vis. 2007;13:2160–2162. [PubMed] [Google Scholar]
- 10.Tan E., Wang Q., Quiambao A.B., Xu X., Qtaishat N.M., Peachey N.S., Lem J., Fliesler S.J., Pepperberg D.R., Naash M.I., Al-Ubaidi M.R. The relationship between opsin overexpression and photoreceptor degeneration. Invest. Ophthalmol. Vis. Sci. 2001;42:589–600. [PubMed] [Google Scholar]
- 11.Seo S., Mullins R.F., Dumitrescu A.V., Bhattarai S., Gratie D., Wang K., Stone E.M., Sheffield V., Drack A.V. Subretinal gene therapy of mice with Bardet-Biedl syndrome type 1. Invest. Ophthalmol. Vis. Sci. 2013;54:6118–6132. doi: 10.1167/iovs.13-11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burnight E.R., Wiley L.A., Drack A.V., Braun T.A., Anfinson K.R., Kaalberg E.E., Halder J.A., Affatigato L.M., Mullins R.F., Stone E.M., Tucker B.A. CEP290 gene transfer rescues Leber congenital amaurosis cellular phenotype. Gene Ther. 2014;21:662–672. doi: 10.1038/gt.2014.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collin R.W., den Hollander A.I., van der Velde-Visser S.D., Bennicelli J., Bennett J., Cremers F.P. Antisense oligonucleotide (AON)-based therapy for Leber congenital amaurosis caused by a frequent mutation in CEP290. Mol. Ther. Nucleic Acids. 2012;1:e14. doi: 10.1038/mtna.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerard X., Perrault I., Hanein S., Silva E., Bigot K., Defoort-Delhemmes S., Rio M., Munnich A., Scherman D., Kaplan J. AON-mediated exon skipping restores ciliation in fibroblasts harboring the common Leber congenital amaurosis CEP290 mutation. Mol. Ther. Nucleic Acids. 2012;1:e29. doi: 10.1038/mtna.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garanto A., Chung D.C., Duijkers L., Corral-Serrano J.C., Messchaert M., Xiao R., Bennett J., Vandenberghe L.H., Collin R.W. In vitro and in vivo rescue of aberrant splicing in CEP290-associated LCA by antisense oligonucleotide delivery. Hum. Mol. Genet. 2016;25:2552–2563. doi: 10.1093/hmg/ddw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandl C., Ortiz O., Röttig B., Wefers B., Wurst W., Kühn R. Creation of targeted genomic deletions using TALEN or CRISPR/Cas nuclease pairs in one-cell mouse embryos. FEBS Open Bio. 2014;5:26–35. doi: 10.1016/j.fob.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Q., Cai X., Tan M.H., Schaffert S., Arnold C.P., Gong X., Chen C.Z., Huang S. Precise gene deletion and replacement using the CRISPR/Cas9 system in human cells. Biotechniques. 2014;57:115–124. doi: 10.2144/000114196. [DOI] [PubMed] [Google Scholar]
- 21.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boye S.E., Alexander J.J., Boye S.L., Witherspoon C.D., Sandefer K.J., Conlon T.J., Erger K., Sun J., Ryals R., Chiodo V.A. The human rhodopsin kinase promoter in an AAV5 vector confers rod- and cone-specific expression in the primate retina. Hum. Gene Ther. 2012;23:1101–1115. doi: 10.1089/hum.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D., Mou H., Li S., Li Y., Hough S., Tran K., Li J., Yin H., Anderson D.G., Sontheimer E.J. Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum. Gene Ther. 2015;26:432–442. doi: 10.1089/hum.2015.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett J. Immune response following intraocular delivery of recombinant viral vectors. Gene Ther. 2003;10:977–982. doi: 10.1038/sj.gt.3302030. [DOI] [PubMed] [Google Scholar]
- 25.Maguire A.M., Simonelli F., Pierce E.A., Pugh E.N., Jr., Mingozzi F., Bennicelli J., Banfi S., Marshall K.A., Testa F., Surace E.M. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schimmer J., Breazzano S. Investor outlook: focus on upcoming LCA2 gene therapy phase III results. Hum. Gene Ther. Clin. Dev. 2015;26:144–149. doi: 10.1089/humc.2015.29001.sch. [DOI] [PubMed] [Google Scholar]
- 27.Cideciyan A.V., Aleman T.S., Jacobson S.G., Khanna H., Sumaroka A., Aguirre G.K., Schwartz S.B., Windsor E.A., He S., Chang B. Centrosomal-ciliary gene CEP290/NPHP6 mutations result in blindness with unexpected sparing of photoreceptors and visual brain: implications for therapy of Leber congenital amaurosis. Hum. Mutat. 2007;28:1074–1083. doi: 10.1002/humu.20565. [DOI] [PubMed] [Google Scholar]
- 28.Boye S.E., Huang W.C., Roman A.J., Sumaroka A., Boye S.L., Ryals R.C., Olivares M.B., Ruan Q., Tucker B.A., Stone E.M. Natural history of cone disease in the murine model of Leber congenital amaurosis due to CEP290 mutation: determining the timing and expectation of therapy. PLoS ONE. 2014;9:e92928. doi: 10.1371/journal.pone.0092928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swiech L., Heidenreich M., Banerjee A., Habib N., Li Y., Trombetta J., Sur M., Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menotti-Raymond M., David V.A., Schäffer A.A., Stephens R., Wells D., Kumar-Singh R., O’Brien S.J., Narfström K. Mutation in CEP290 discovered for cat model of human retinal degeneration. J. Hered. 2007;98:211–220. doi: 10.1093/jhered/esm019. [DOI] [PubMed] [Google Scholar]
- 32.Garanto A., van Beersum S.E., Peters T.A., Roepman R., Cremers F.P., Collin R.W. Unexpected CEP290 mRNA splicing in a humanized knock-in mouse model for Leber congenital amaurosis. PLoS ONE. 2013;8:e79369. doi: 10.1371/journal.pone.0079369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuris J.A., Thompson D.B., Shu Y., Guilinger J.P., Bessen J.L., Hu J.H., Maeder M.L., Joung J.K., Chen Z.Y., Liu D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakuma T., Nishikawa A., Kume S., Chayama K., Yamamoto T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 2014;4:5400. doi: 10.1038/srep05400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao X., Li J., Samulski R.J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.