Abstract
Pseudomonas aeruginosa is the primary cause of chronic airway infections in cystic fibrosis (CF) patients. Persistent infections are seen from the first P. aeruginosa culture in about 75% of young CF patients, and it is important to discover new ways to detect P. aeruginosa at an earlier stage. The P. aeruginosa biomarker hydrogen cyanide (HCN) contains a triple bond, which is utilized in this study because of the resulting characteristic C≡N peak at 2135 cm−1 in a Raman spectrum. The Raman signal was enhanced by surface-enhanced Raman spectroscopy (SERS) on a Au-coated SERS substrate. After long-term infection, a mutation in the patho-adaptive lasR gene can alter the expression of HCN, which is why it is sometimes not possible to detect HCN in the breath of chronically infected patients. Four P. aeruginosa reference strains and 12 clinical P. aeruginosa strains isolated from CF children were evaluated, and HCN was clearly detected from overnight cultures of all wild type-like isolates and half of the later isolates from the same patients. The clinical impact could be that P. aeruginosa infections could be detected at an earlier stage, because daily breath sampling with an immediate output could be possible with a point-of-care SERS device.
Individuals with cystic fibrosis (CF) have an inherited defect in the cystic fibrosis transmembrane conductance regulator (CFTR) gene, causing thickened, dehydrated mucus to form on mucociliary surfaces. This in turn increases the risk of e.g. airway infections. Pseudomonas aeruginosa lung infections are the largest threat to the wellbeing and survival of CF patients, and it has been shown for a cohort of young Danish CF patients that an estimated 75% have a persistent infection already from the first detection of P. aeruginosa1.
If the child is unable to expectorate, today’s methods consist of either a larynx swab, which is insensitive, or induced sputum (naso-laryngeal suction), which is extremely invasive. P. aeruginosa can be difficult to detect, especially in children and young adults without a chronic infection because they have few bacteria in their airways. Therefore more sensitive and non-invasive methods are being investigated utilizing the fact that hydrogen cyanide (HCN) is a biomarker for P. aeruginosa2,3,4. In nature, P. aeruginosa emits the poisonous gas HCN in order to kill competitive microorganisms. This wildtype (WT)-like behavior is also expected to occur in the lungs, at least during the initial stages of infection. To sustain a persistent colonisation and infection, adaptation is necessary. In many cases this involves mutations of different genes, so-called “patho-adaptive” genes. One of the genes that have been suggested as being “patho-adaptive” (i.e. beneficial for P. aeruginosa to mutate) is lasR5,6. The lasR gene is a transcriptional regulator necessary for HCN production7, so a mutation in the lasR gene can imply a change in the bacterial emission of HCN. According to the clinical CF program in Denmark, patients are seen once a month in the outpatient clinic and deliver a sputum sample coughed up from the lungs. Depending on the patient’s age and ability this is either done by expectoration or, as in most young patients, induced by a tube inserted through the nose into the larynx to provoke a cough reflex after which a sample is obtained through the tube (naso-laryngeal suction). Sputum samples are then cultured to identify pathogenic microorganisms.
Several groups have investigated HCN production by P. aeruginosa in vitro3,8,9. The drawbacks of all the applied methods are that they need extensive sample handling before measurements can be obtained, and that it can only be done in solution. To the best of our knowledge, selected ion flow tube – mass spectroscopy (SIFT-MS) is the current state-of-the-art method in terms of sensitivity and specificity10. However, it is also an expensive and complex procedure. The technique was employed in the study by Enderby et al.2, where median values of 13.5 ppb HCN were detected directly in the breath of CF children with a newly discovered P. aeruginosa lung colonization2, whereas in the more recent study by Gilchrist et al.10, HCN concentrations at or below 7.5 ppb were detected from breath samples collected in bags and measured within 24 hours10. An optimal sensor would be a cheap and simple point-of-care device, allowing for measurements to be carried out directly on human breath, without pre-handling of the samples. The so-called surface-enhanced Raman scattering (SERS) technique can potentially fulfill these requirements since hand-held Raman spectrometers are progressively becoming cheaper, better and smaller11.
Raman spectroscopy is a fingerprinting technique that measures the inelastic scattering of monochromatic light when interacting with matter. The frequency shift (Raman shift) of the inelastically scattered light corresponds to the energies of the molecular vibrations12. HCN contains a triple bond between C and N, which will result in a Raman band around 2135 wavenumbers (cm−1). But since the inelastically scattered light is only a tiny part of light scattered from a sample (<10−8), Raman spectroscopy in general has a weak signal-to-noise ratio, and there is a need for signal enhancement. Using SERS, the Raman signal of small molecules is enhanced by utilizing the collective oscillations of conducting electrons (surface plasmons) that take place in the vicinity of clustered noble-metal nanoparticles13,14. SERS has been used in a wide array of medical applications15,16,17. An example hereof is using label free Ag colloid SERS to detect single-nucleotide mismatch in short DNA sequences to discover even small nucleotide changes18. We have demonstrated previously that the SERS technique is suitable for cyanide quantification with the estimated lower limit of detection between 1.8 and 18 ppb using highly sensitive gold-coated silicon (Si) nanopillar SERS substrates19. These substrates consist of vertically standing Si nanopillars with gold (Au) caps which can lean against each other for enhanced SERS detection in liquid or gas phase20. In this study we utilize identical SERS substrates and investigate the capability of the method to detect the biomarker HCN(g) in emissions from clinical early and late P. aeruginosa isolates. In order to determine its applicability as a non-invasive diagnostic tool for detection of P. aeruginosa we qualitatively evaluate results which indicate HCN presence in gases from early P. aeruginosa isolates, and since our SERS substrate is able to detect ppb values of HCN, this novel method is expected to be able to discover P. aeruginosa from breath at an earlier stage than by using today’s clinical practice.
Materials and Methods
Preparation of the SERS substrate
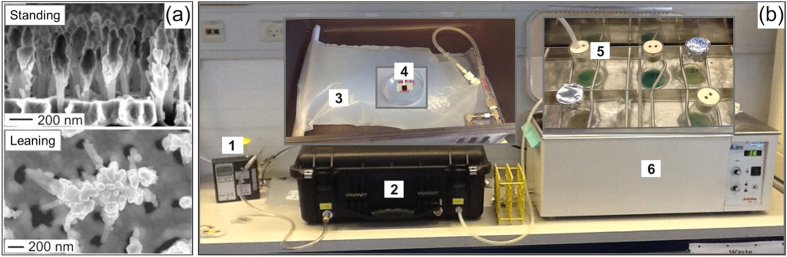
The disposable SERS substrate was etched in a single-side polished Si wafer using 3 min Reactive Ion (plasma) Etching with alternating SF6 bombardment and O2 protection of the emerging nanopillars, followed by a 1 min O2 cleaning step. Before use, Au was evaporated onto the Si nanopillars, forming 225 nm caps used for SERS detection. See Fig. 1(a). On the day of exposure the wafer was cut into 6 × 6 mm chips using a diamond cutter and tweezers. The chips were cleaned by immersion into ethanol (Absolute grade, CHROMASOLV R, Sigma-Aldrich) for 3 min followed by H2O (Molecular Biology Reagent grade, Sigma-Aldrich) for 3 min and left to dry on a tissue whereby the pillars would lean to enable SERS detection. For easy handling and transportation, the SERS substrate was mounted inside a small Petri dish, using double sided adhesive tape (Scotch), with a Post-It on top, the tacky side facing up for holding the substrate (Fig. 1(b. The wall of the Petri dish protected the delicate SERS substrate so it would not get in contact with the bag and be scratched (See section on SERS substrate exposure to bacterial emissions).
Figure 1. Setup for SERS measurements on bacterial volatiles.
(a) SEM images of the SERS substrate before and after leaning for formation of electromagnetic “hot spots” for SERS enhancement. (Courtesy of Kaiyu Wu. Reprinted with permission from editor of ref. 19). (b) The pump (1) induces a vacuum inside the vacuum chamber (2), leading to inflation of the bag (3) and exposure of the SERS substrate (4) to volatiles created by the bacterial culture (5) in the water bath (6).
P. aeruginosa reference strains
PAO1 is a reference strain commonly used to benchmark various P. aeruginosa strains against. PA-SD2 is a PAO1 isolate with a knockout mutation in lasR, as described in ref. 20. DK02 is a P. aeruginosa lineage that spread among Danish CF patients for over 40 years. The earliest isolate is called DK02-1973, and its lasR mutated strain DK02-197921. All four reference strains were evaluated in the study.
The children strain collection
At the CF clinic at Rigshospitalet in Copenhagen a special collection of bacterial strains has been sampled from CF children and young adults since 2004, resulting in a unique collection for genomics and other evolutional studies to be made5,22. From this collection early (lasR WT) and late (lasR mutant) isolates from five patients have been selected, encompassing three different clone types, for comparison of adaptations between and within patients. The selected strains originate from CF patients who today are all chronically infected, and the late isolates represent various locations of lasR mutations. Selected strains include the first P. aeruginosa isolate and at least one later isolate from the same patient after onset of chronicity and with detected mutations in the lasR gene. From the children strain collection, 12 P. aeruginosa isolates were evaluated.
Ethics
The local ethics committee at the Capital Region of Denmark Region Hovedstaden approved the use of the samples: registration number H-4-2015-FSP. All patients have given informed consent. For patients below 18 years of age, informed consent was obtained from their parents. The study was carried out in accordance with the approved guidelines and the University Hospital Rigshospitalet approved the experimental protocol.
Identification of mutations in lasR
Using a modified version of the pipeline previously described in Andersen et al.23, we identified mutations in the CF isolates using the sequences of isolates from young CF patients, previously published by Marvig et al. (2015)5. Briefly: Reads were mapped to the lasR reference sequence (NP_250121.1) from the reference strain Pseudomonas aeruginosa PAO1, using the Burrows-Wheeler alignment tool24 (BWA version 0.7.12), with the paired-end reads setting. Alignments were filtered to remove unmapped reads, sorted, and indexed using SAMtools25. Each isolate was assigned to a read group using Picard Tools (open-source software version 1.14, http://pickard.sourceforge.net) and differences between isolates and the reference lasR sequence were identified with SAMtools. Mutations were manually checked using the Integrative Genomics Viewer26 (IGV, version 2.3.68). The functional impact (missense/nonsense/silent) was determined using the translated sequence of lasR in the IGV. The full pipeline can be found in Supplementary material.
Preparation of bacterial cultures
P. aeruginosa strain PAO1, its engineered lasR mutant PA-SD2 and two isolates from the classic clinical lineage DK02 from 1973 and 197927, as well as P. aeruginosa strains from five paediatric CF patients were stored as pure cultures at −80 °C, wherefrom they were streaked onto LB agar plates and incubated at 37 °C over night (ON). One colony was then inoculated in 10 mL LB growth medium (4% salt concentration) and incubated shaking at an angle of ~45°, 225 rpm overnight ON. One mL of the ON culture was added to 9 mL of fresh LB in a 100 mL Erlenmeyer flask, plugged with a two-holed rubber plug, covered with aluminum foil and placed in a 37 °C water bath at ~200 rpm ON, wherefrom SERS substrates were exposed (Fig. 1). One flask was prepared for each measurement needed.
SERS substrate exposure to bacterial emissions
In the setup we used an SK 224-PCMTX4 Universal air sampling pump (SKC Inc., PA, US), connected via a silicone tube to a large Vac-U-Chamber, 231–939 (SKC Inc., PA, US), Fig. 1(b. Inside the vacuum chamber a 10 L reusable 60 μm PTFE (Teflon) sample bag (Scentroid, ON, Canada), equipped with a 1/2” compression fitting was connected to the sample inlet (3). The bag was specially designed with one end open for mounting the SERS substrate, and closed by a clamp. The sample inlet of the vacuum chamber was connected via a silicone tube to the 100 mL Erlenmeyer culture flask (5), plugged with a rubber plug with two holes, one for the tube and one for air inlet. The pump was operating at ~500 mL/min for 15 min to fill the bag, followed by 5 min holding time for further substrate exposure. Before pumping out bacterial gases, all fittings were tested by closing the vacuum chamber and turning on the pump to let in air from the lab to verify that the bag would inflate and no emissions would be lost when the bacterial culture was connected to the system.
OD and SERS measurements
The Optical density (OD) was measured at 600 nm in a UV-1800 spectrophotometer (SHIMADZU, Kyoto, Japan), giving the absorbance as a measure of cell density. SERS measurements were recorded using a FT-Raman instrument (Bruker VERTEX 70, Bruker Optik, Ettlingen, Germany), equipped with a 1064 nm laser and an InGaAs detector. Samples were measured in a 180 degrees backscattering geometry using 32 scans at a resolution of 4 cm−1. Each sample was measured at 5 different positions on the substrate and the 5 spectra were averaged before subsequent data analysis.
Results and Discussion
SERS on PAO1 and DK02 ON culture emissions
The clinical strains were growing much slower than PAO1, and ON cultures were prepared for SERS exposure and measurements. In Fig. 2(a,b) it is seen that PAO1 and the isolate from 1973 both emitted substantial amounts of HCN, as seen in the intense C≡N stretching peak at 2135 cm−1, whereas the lasR mutated PAO1 and the late (1979) isolate did not. This was as expected since the late DK2 and the lasR mutated PAO1 should not have a functional LasR protein.
Figure 2. SERS on overnight cultures of PAO1, the DK02 lineage and their lasR mutated strains.
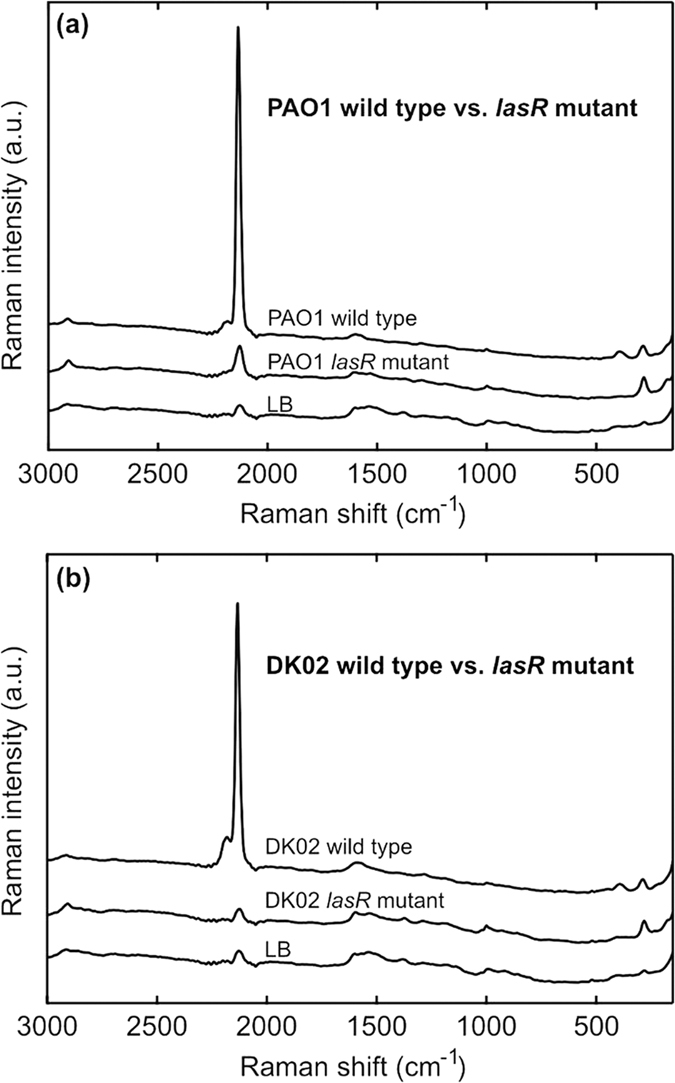
(a) SERS on emissions from overnight cultures of P. aeruginosa PAO1, wild type vs. its engineered lasR mutated strain PA-SD2. (b) SERS on emissions from overnight cultures of the P. aeruginosa DK02 lineage, wild type from 1973 vs. the lasR mutated strain from 1979. Both wild types show intense CN signals at 2135 cm−1 and also a small peak at 2189 cm−1 due to formation of the [Au(CN)2]− complex, whereas the lasR mutated strains emit no HCN. Spectra of LB emissions are included as reference.
HCN emission from PAO1 during growth
As seen in the growth curve of Fig. 3(a), PAO1 has a lag phase of about 2 hours before onset of exponential growth, with stationary growth, and expected HCN production, beginning after 4–6 hours. According to previous studies HCN production starts at the end of exponential/beginning of stationary phase8,28. This is due to the fact that HCN production is quorum sensing (QS) dependent. After a few hours on the substrate much of the C≡N peak had shifted to 2189 cm−1 because of sufficient amounts and the fact that it had had some time to interact with the Au-coated SERS substrate, forming the stabile [Au(CN)2]− complex, as explained in ref. 19. Figure 3(b) shows the SERS intensity of the cumulated cyanide peaks at 2135 and 2189 cm−1 from 2 until 20 h (ON). It is observed that HCN production begins after 4 h growth and is still detectable in the overnight (20 h) culture, whereas the 2 and 3 h samples only had the same background as the LB reference. The results in Fig. 3(b) and the shape of the HCN emission curve with time are very similar to previous findings8,29.
Figure 3. PAO1 HCN emission during growth.
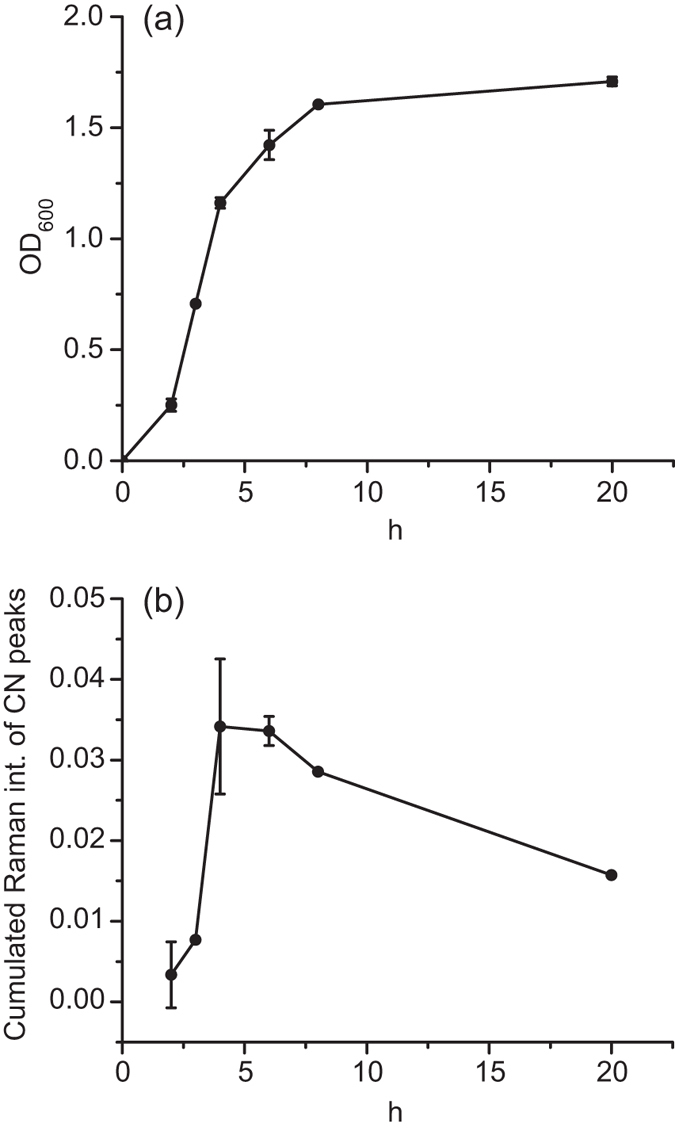
(a) Optical density (OD600) growth curve for PAO1 in LB medium at 37 °C. The lag phase takes about 2 hours, followed by the log phase until about 4–6 hours’ growth after which the stationary state occurs. Final OD absorbance is about 1.7 at 600 nm. (b) Intensity of cumulated cyanide peaks from PAO1 during growth. The cumulated Raman intensity is the (2135 + 2189) cm−1 SERS intensity on the PAO1 exposed substrate minus the background signal of the substrate exposed to LB emissions. It is seen that PAO1 starts producing HCN after 4 hours’ growth, which is at the end of its exponential/beginning of stationary growth phase. After 20 hours HCN is still detectable, although not as intense as after 4 hours. The shape of the CN curve resembles previous studies on P. aeruginosa HCN emission during growth8,28.
HCN emission from P. aeruginosa strains isolated from CF children
A total of 12 isolates from five CF children were measured with the SERS sensor in the developed setup. In Fig. 4 the intensity of the 2135 cm−1 C≡N stretching band shows that all early (WT-like) strains and also some of the lasR mutated strains isolated from paediatric CF patients emitted HCN. Thereby, the principle had been proven that the SERS substrate was able to detect HCN from cultures of early P. aeruginosa colonizations, which was the aim of the study. Since it was possible to detect HCN from emissions of all early ON cultures, it was plausible that HCN could also be detected from breath of a patient with an early P. aeruginosa colonization. Genome sequencing of the lasR mutated isolates5 showed that isolates with lasR mutations located towards the 3′ terminal of the gene did not produce HCN, and it was an interesting finding that only lasR mutated cultures with the mutation located upstream or at the central part of the gene emitted detectable HCN. The continued production of HCN could be explained by mutations or changes in regulation of other genes that are also important for QS and HCN production circumventing a malfunctioning lasR.
Figure 4. Cyanide production of clinical P. aeruginosa overnight cultures from 5 CF patients.
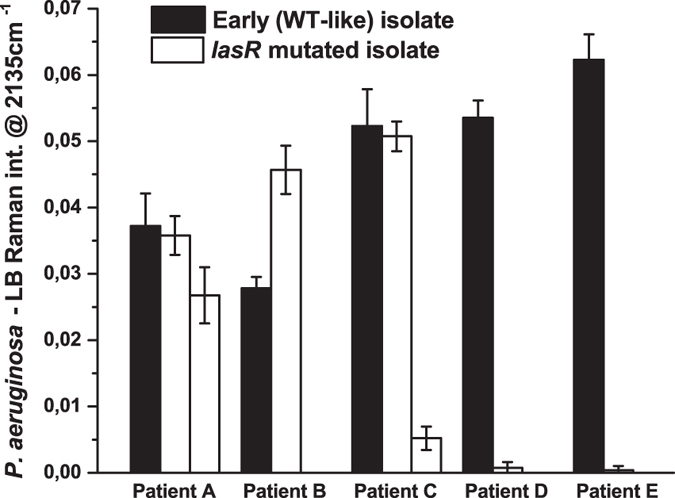
The peak intensity is the SERS signal at 2135 cm−1 minus the background signal of the LB reference samples. It is seen that all WT-like isolates emitted clearly detectable HCN, and so did also some of the lasR mutated isolates. The latest isolates from patients C, D and E did not emit HCN.
Clinical control strains
To make sure HCN could be used as a specific marker for P. aeruginosa colonisation, we also tested other clinically relevant species. Figure 5 shows examples of two other bacteria colonising the lungs of CF patients, and known to be able to cause chronic infections: Stenotrophomonas maltophilia30 and Achromobacter xylosoxidans31. Clinical isolates of these control strains were tested in the same setup as P. aeruginosa, but no HCN could be detected. The findings were in accordance with Bumunang & Babalola32.
Figure 5. SERS on overnight cultures of Stenotrophomonas maltophilia and Achromobacter xylosoxidans.
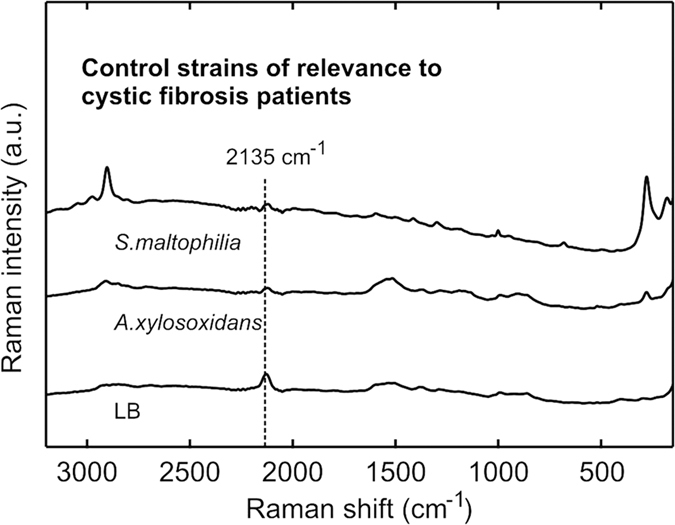
SERS on emissions from overnight cultures of S. maltophilia and A. xylosoxidans. None of the bacteria emitted any detectable HCN, as seen in the low intensity of the triple bond peak at 2135 cm−1. Especially the S. maltophilia spectrum has different peaks than P. aeruginosa. A spectrum of LB emissions is included as reference.
Conclusions and Outlook
We have developed a SERS-based method for detection of HCN from P. aeruginosa cultures, and a setup for exposure of the SERS substrate to bacterial volatiles. The purpose was to see whether the wild type like strains were producing HCN in sufficient amounts for detection by the SERS substrate and to check whether the lasR mutated strains were still producing HCN. For the reference strain PAO1 HCN production started at the end of stationary phase and was still detectable in overnight cultures. In all clinical wild type-like strains isolated from the airways of cystic fibrosis patients there was a clear HCN SERS signal proving the principle that we are able to detect gases from early P. aeruginosa isolates. HCN could only be detected from half of the strains with mutation in the lasR transcriptional regulator gene, which may explain why HCN cannot be detected in the breath of all patients with a chronic P. aeruginosa airway infection.
We acknowledge the possible limitations to this SERS-based breath device, which could have a slightly lower sensitivity than SIFT-MS, which seems to be state-of-the-art for breath detection of HCN. General limitations to ppb level breath detection could be that the child has recently been coughing, whereby the low amounts of bacterial volatiles could be lost, or that the child is too young and unable to exhale properly. Another issue could be if the infection is already chronic and P. aeruginosa has stopped to emit HCN. Still, our results suggest that it is indeed possible to use SERS detection of HCN as an early indicator of P. aeruginosa infection. The method offers the possibility of a simple, non-invasive point-of-care monitoring device, which could be used in the patients’ home or at the general practitioner for more frequent breath measurements, hopefully leading to an earlier detection of P. aeruginosa lung infections in patients with CF.
Additional Information
How to cite this article: Lauridsen, R. K. et al. SERS detection of the biomarker hydrogen cyanide from Pseudomonas aeruginosa cultures isolated from cystic fibrosis patients. Sci. Rep. 7, 45264; doi: 10.1038/srep45264 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors would like to thank The Danish Council for Independent Research for supporting the Sapere Aude project “NAPLAS”, which this research is part of. The research was also funded by the Danish National Research Foundation (DNRF122) and Villum Fonden (Grant No. 9301). We would like to thank Flemming Larsen for seeing the opportunity of forming this CF project. Michael Stenbæk Schmidt is regarded for inventing the first nanopillar SERS substrate; and Susanne Knøchel, Marina Kryger Bjørklund, Susanne (Søs) Koefoed and Alicia Jiménez Fernández are acknowledged for their help in realizing the bacterial studies. Kinga Zór is acknowledged for valuable feed-back on the paper. HKJ was funded by a clinical research stipend from The Novo Nordisk Foundation and Rigshospitalets Rammebevilling 2015–17 and Lundbeckfonden Grant R167-2013-15229.
Footnotes
The authors declare no competing financial interests.
Author Contributions R.L. performed the experiments and wrote the majority of the paper. L.S. wrote the parts on genomics and gave feed-back on the remainder. H.J. identified the bacterial strains, supervised on the experiments and gave feed-back on the paper. T.R. wrote the parts on SERS sensitivity and finished the figures. S.M. supervised on the experiments. L.J. gave feed-back on the paper. S.E. supervised on the Raman part and gave feed-back on the paper. A.B. supervised on the experiments and gave feed-back on the paper.
References
- Johansen H. et al. What makes Pseudomonas aeruginosa persist in the lungs of CF patients? Pediatr. Pulmonol 50(41), 77–107 (2015). [Google Scholar]
- Enderby B., Smith D., Carroll W. & Lenney W. Hydrogen cyanide as a biomarker for Pseudomonas aeruginosa in the breath of children with cystic fibrosis. Pediatr. Pulm. 44, 142–147 (2009). [DOI] [PubMed] [Google Scholar]
- Ryall B., Davies J., Wilson R., Shoemark A. & Williams H. Pseudomonas aeruginosa, cyanide accumulation and lung function in CF and non-CF bronchiectasis patients. Eur. Resp. J 32(3), 740–747 (2008). [DOI] [PubMed] [Google Scholar]
- Sanderson K., Wescombe L., Kirov S., Champion A. & Reid D. Bacterial cyanogenesis occurs in the cystic fibrosis lung. Eur. Resp. J 32, 329–333 (2008). [DOI] [PubMed] [Google Scholar]
- Marvig R., Sommer L., Molin S. & Johansen H. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet. 47(1), 57–64 (2014). [DOI] [PubMed] [Google Scholar]
- Smith E. E. et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 103, 8487–8492 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G. & Haas D. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the Quorum-Sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182(24), 6940–6949 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blier A. et al. Quantification of Pseudomonas aeruginosa hydrogen cyanide production by a polarographic approach. J. Microbiol. Meth. 90, 20–24 (2012). [DOI] [PubMed] [Google Scholar]
- Broderick K. et al. Cyanide Produced by Human Isolates of Pseudomonas aeruginosa Contributes to Lethality in Drosophila melanogaster. J. Inf. Dis. 197, 457–464 (2008). [DOI] [PubMed] [Google Scholar]
- Gilchrist F. et al. Exhaled breath hydrogen cyanide as a marker of early Pseudomonas aeruginosa infection in children with cystic fibrosis. Eur. Resp. J 1(00044), 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Pang S., Labuza T. & He L. Evaluation of surface-enhanced Raman scattering detection using a handheld and a bench-top Raman spectrometer: A comparative study. Talanta. 129, 79–85 (2014). [DOI] [PubMed] [Google Scholar]
- Kneipp K., Kneipp H., Itzkan I., Dasari R. & Feld M. Ultrasensitive chemical analysis by Raman spectroscopy. Chem. Rev. 99, 2957–2975 (1999). [DOI] [PubMed] [Google Scholar]
- Kneipp K., Kneipp H., Itzkan I., Dasari R. & Feld M. Surface-enhanced Raman scattering and biophysics. Topical review, J. Phys.: Condens. Matter 14, R597–R624 (2002). [DOI] [PubMed] [Google Scholar]
- Nie L., Liu F., Ma P. & Xiao X. Applications of gold nanoparticles in optical biosensors. J. Biomed. Nanotechnol. 10(10), 2700–2721 (2014). [DOI] [PubMed] [Google Scholar]
- Xie W. & Schluecker S. Medical applications of surface enhanced Raman scattering. Phys. Chem. Chem. Phys. 15(15), 5329–5344 (2013). [DOI] [PubMed] [Google Scholar]
- McNay G., Eustace D., Smith W., Faulds K. & Graham D. Surface-enhanced Raman scattering (SERS) and surface-enhanced resonance Raman scattering (SERRS): a review of applications. Appl. Spectrosc. B(8), 825–837 (2011). [DOI] [PubMed] [Google Scholar]
- Das R. & Agrawal Y. Raman spectroscopy: recent advancements, techniques and applications. Vib. Spectrosc. 57(2), 163–176 (2011). [Google Scholar]
- Papadopoulou E. & Bell S. Label-Free Detection of Single-Base Mismatches in DNA by Surface-Enhanced Raman Spectroscopy. Angew. Chem. Int. Ed. 50, 9058–9061 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauridsen R. et al. Towards quantitative SERS detection of hydrogen cyanide at ppb level for human breath analysis. Sens. Biosens. Res. 5, 84–89 (2015). [Google Scholar]
- Schmidt M., Hübner J. & Boisen A. Large area fabrication of leaning silicon nanopillars for surface enhanced Raman spectroscopy. Adv. Mat. 24, OP11–OP18 (2012). [DOI] [PubMed] [Google Scholar]
- Damkiær S., Yang L., Molin S. & Jelsbak L. Evolutionary remodeling of global regulatory networks during long-term bacterial adaptation to human hosts. PNAS 110(19), 7766–7771 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S., Marvig R., Molin S., Johansen H. & Griffin A. Long-term social dynamics drive loss of function in pathogenic bacteria. Proc. Natl. Acad. Sci. USA 112(34), 10756–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S., Marvig R., Molin S., Johansen H. & Griffin A. Long-term social dynamics drive loss of function in pathogenic bacteria. PNAS 112(34), 10756–10761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 25, 1754–60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 25, 2078–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. et al. Integrative Genomics Viewer. Nat. Biotech. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvig R., Johansen H., Molin S. & Jelsbak L. Genome analysis of a transmissible lineage of Pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLOS Gen. 9(9), 1–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric P., Ebert R. & Castric K. The relationship between growth phase and cyanogenesis in Pseudomonas aeruginosa. Curr. Microbiol. 2, 287–292 (1979). [Google Scholar]
- Chen W. et al. Detection of hydrogen cyanide from oral anaerobes by cavity ring down spectroscopy. Sci. Rep. 6(22577), 1–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbøge C., Hansen C., Pressler T., Høiby N. & Johansen H. Chronic pulmonary infection with Stenotrophomonas maltophilia and lung function in patients with cystic fibrosis. J. Cyst. Fibros. 10(5), 318–325 (2011). [DOI] [PubMed] [Google Scholar]
- Hansen C., Pressler T., Ridderberg W., Johansen H. & Skov M. Achromobacter species in cystic fibrosis: cross-infection caused by indirect patient-to-patient contact. J. Cyst. Fibros. 12(6), 609–615 (2013). [DOI] [PubMed] [Google Scholar]
- Bumunang E. W. & Babalola O. O. Characterization of Rhizobacteria from Field Grown Genetically Modified (GM) and Non-GM Maizes. Braz. Arch. Biol. Technol. 57(1), 1–8 (2014). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.