Abstract

Few secondary metabolites have been reported from mammalian microbiome bacteria despite the large numbers of diverse taxa that inhabit warm-blooded higher vertebrates. As a means to investigate natural products from these microorganisms, an opportunistic sampling protocol was developed, which focused on exploring bacteria isolated from roadkill mammals. This initiative was made possible through the establishment of a newly created discovery pipeline, which couples laser ablation electrospray ionization mass spectrometry (LAESIMS) with bioassay testing, to target biologically active metabolites from microbiome-associated bacteria. To illustrate this process, this report focuses on samples obtained from the ear of a roadkill opossum (Dideiphis virginiana) as the source of two bacterial isolates (Pseudomonas sp. and Serratia sp.) that produced several new and known cyclic lipodepsipeptides (viscosin and serrawettins, respectively). These natural products inhibited biofilm formation by the human pathogenic yeast Candida albicans at concentrations well below those required to inhibit yeast viability. Phylogenetic analysis of 16S rRNA gene sequence libraries revealed the presence of diverse microbial communities associated with different sites throughout the opossum carcass. A putative biosynthetic pathway responsible for the production of the new serrawettin analogues was identified by sequencing the genome of the Serratia sp. isolate. This study provides a functional roadmap to carrying out the systematic investigation of the genomic, microbiological, and chemical parameters related to the production of natural products made by bacteria associated with non-anthropoidal mammalian microbiomes. Discoveries emerging from these studies are anticipated to provide a working framework for efforts aimed at augmenting microbiomes to deliver beneficial natural products to a host.
During the past decade, the field of human microbiome science has undergone a revolution. A key component of this transformation is the realization that the human body’s endogenous microbial flora do not live as an isolated community, but instead function within a dynamic human–microbe network.1−4 There is also growing appreciation that the proper function of the human microbial ecosystem plays an important role in forestalling disease and maintaining good health.3 The extent of the interconnectedness between humans and their microbiomes has even led some to refer to it as “our second genome”.5
As the scientific community’s understanding of the human microbiome’s composition has matured, so have the research questions that are being posed concerning the microbiome’s roles in health and disease. Whereas initial inquiries in the field focused on resolving fundamental problems such as, “what is there?” and “what do they do?” microbiome researchers are beginning to ask new questions such as, “how can we control their function?” Emerging efforts to engineer or manipulate human microbiomes offer exciting opportunities to restore and potentially enhance human health and performance.
There are two general approaches that provide feasible options for engineering human microbiomes. The first tactic involves the introduction or elimination of selected microbial taxa from the host.6−8 This approach was successfully tested in animal models of inflammatory bowel disease,9−11 arthritis,12 colon cancer,13 cystic fibrosis,14 and depression,15 resulting in the restoration of disease-free phenotypes. This method was also effectively used in humans to restore resilience against Clostridium difficile colonization.16,17 A second approach that presents a more structured form of microbiome engineering involves genetically modifying selected microorganisms to provide them new functions that may benefit a host.7,18−20 These methods could include the (i) introduction of new metabolic enzymes that are absent in the host and its microbiome consortia or (ii) incorporation of new small-molecule-generating biosynthetic genes that are responsible for the production of biomolecules not currently made in humans.21 One can envision the potential for generating and applying designer microbes that produce compounds capable of warding off selected pathogens or modulating disease processes.
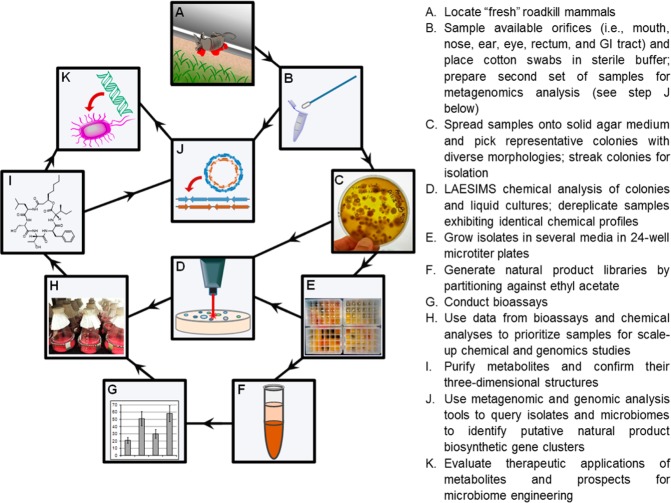
In pursuit of identifying candidate biosynthetic gene clusters whose products might benefit humans, our team investigated natural products made by bacteria from non-anthropoidal mammalian hosts. While several unique metabolites have been previously reported from bacteria inhabiting humans,20,22−25 relatively little is known about the chemicals generated by bacteria colonizing the majority of other mammals.26−28 Accordingly, we developed a bioassay-based pipeline that offers the opportunity to detect natural products with potentially beneficial therapeutic attributes (Figure 1). The rationale behind our approach was that natural products derived from other mammalian species may have arisen through adaptive host-mediated selection, and these compounds might be biomedically useful, as well as relatively safe.
Figure 1.
Overview of the combined microbiological, chemical, and genomic steps employed to investigate microbiome bacteria derived from roadkill mammals for new natural products and their biosynthetic gene clusters. The methodologic approach illustrated here was developed to support the discovery of new bioactive natural products from non-anthropoidal sources, which in turn could be used for the purpose of engineering new biosynthetic pathway functions into the microbiomes of both humans and agriculturally important animals.
To obtain the large number of microbiome bacteria from diverse mammalian sources that we required for our screening process, we used an opportunistic sampling approach to explore roadkill (animals killed as a result of unintentional vehicular collisions), which is an underutilized source of microbiome bacteria.29 In our case, we focused on fresh (recently deceased) roadkill comprising mammals that are native or naturalized to central Oklahoma. Roadkill offers a convenient route to accessing microbiome bacteria since it (i) is abundant in many areas, (ii) presents the opportunity for sampling diverse animals and their associated bacteria across a broad geographical region, (iii) alleviates concerns over the trapping and testing of live animals since only carcasses are sampled, and (iv) offers the possibility to conduct chronologically dependent testing of specific animal populations over extended periods.29,30 In this paper, we present the development and application of our mammalian-microbiome-derived natural product discovery pipeline (Figure 1) and present data for several new and known depsipeptides obtained from opossum-associated bacteria. The compounds were detected based on their abilities to inhibit Candida albicans biofilm formation, which is the single most prevalent cause of human fungal infections.
Results and Discussion
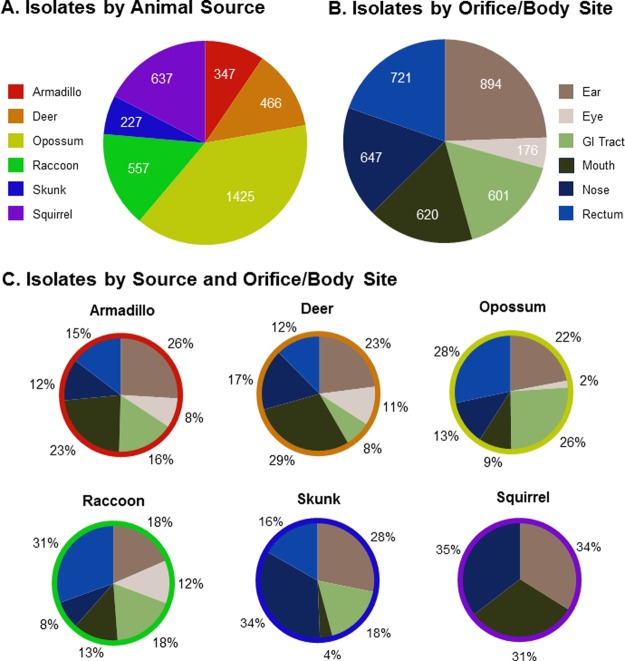
Opportunistic sampling of mammalian roadkill took place over a two-year period near the University of Oklahoma campus (Norman, OK, USA). Carcasses deemed fresh (generally determined to have been struck by motor vehicles no more than 10 h prior to sampling) were selected, and those with one or more intact orifices (i.e., mouth, nose, ear, eye, and rectum) or gastrointestinal tracts were sampled roadside with sterile swabs. The mammalian roadkill carcasses that were sampled included (in order of increasing frequency) the following: skunk (Mephitis mephitis), armadillo (Dasypus novemcinctus), deer (Odocoileus virginianus), raccoon (Procyon lotor), squirrel (Sciurus carolinensis), and opossum (Dideiphis virginiana). Sampling of these carcasses led to the generation of 3659 bacterial isolates (Figure 2). Not only were opossum carcasses the most frequently encountered, they were also a particularly rich source of morphologically unique bacteria, accounting for 39% (1425) of the roadkill-associated isolates collected (Figure 2).
Figure 2.
Summary of the bacterial isolates collected from roadkill mammals. (A) Breakdown of the 3659 roadkill microbiome isolates based on the source organisms from which they were derived. (B) Distribution of the isolates based on the locations/orifices on the carcasses from which they were derived. (C) Categorization of the isolate data illustrating the percent contribution of each body site to the overall bacterial collection prepared from the different animal species (note that the colors and categories used to construct the slices within each of the pie charts in panel C are the same as those used for the pie chart in panel B).
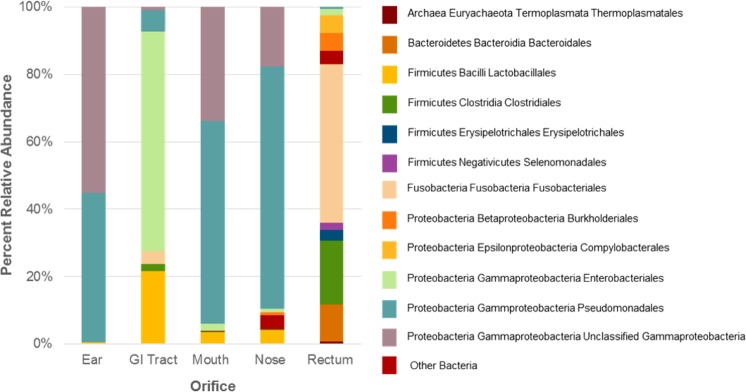
The phylogenetic diversity of the opossum microbiome was further evaluated by sequencing 16S rRNA gene libraries. Swabs taken from five opossum body sites were used to generate 13 994 sequences, which clustered into 73 operational taxonomic units (OTUs) at 97% sequence similarity (Figure 3). Members of the Gammaproteobacteria dominated the communities from the mouth, nose, and ear, constituting greater than 90% of each community. The upper gastrointestinal tract also contained a sizable population of Gammaproteobacteria (72.4%) and Bacilli (21.7%). The rectum was notably more diverse containing large populations of Fusobacteria (47.2%), Clostridia (19.1%), and Bacteroidia (10.8%). The Gammaproteobacteria were largely represented by two abundant operational taxonomic units, one of which was unclassified and another assigned most closely to the genus Pseudomonas.
Figure 3.
Phylogenetic diversity of mammalian microbiome bacteria from different orifices/body sites of a roadkill opossum carcass.
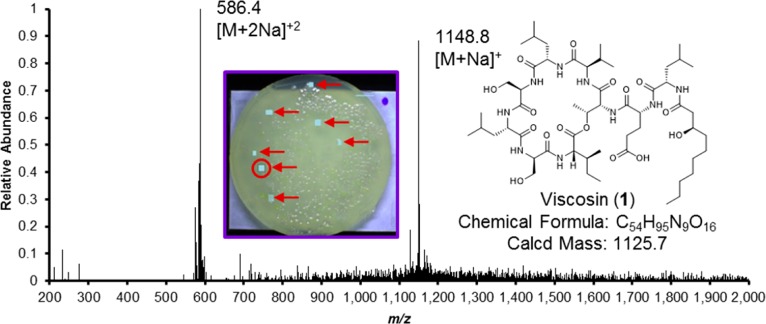
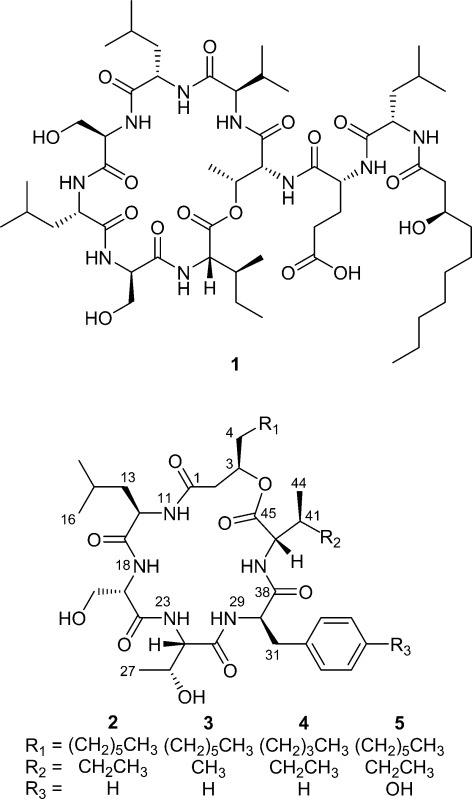
All of the bacterial isolates were individually cultured in two or more broth media, and a library of their ethyl-acetate-soluble natural products was prepared. These samples were subjected to bioassay screening against several medically relevant targets. One of the opossum ear samples showed appreciable activity in an assay designed to identify compounds that inhibited C. albicans biofilm formation. However, upon closer scrutiny, this sample was determined to be derived from a culture containing two bacteria that each produced slightly different-sized white, opaque, mucoidal colonies. When the two microorganisms were obtained in pure culture, the 16S rRNA genes of both isolates were sequenced, and the bacteria were identified as members of the genera Pseudomonas and Serratia. Representative colonies of both isolates were analyzed in situ by laser ablation electrospray ionization mass spectrometry (LAESIMS). A single major metabolite was observed from the Pseudomonas sp. (Figure 4), which was determined to be viscosin (1) based on its MS/MS fragmentation pattern relative to an authentic standard. Its assignment was later confirmed by a single-crystal X-ray diffraction experiment of the metabolite following its scale-up production and purification (Figure 5). LAESIMS analysis of the Serratia sp. isolate led to the purification of serrawettin W2 (2), as well as three co-metabolites that could not be dereplicated (i.e., m/z values and fragmentation patterns did not produce any reasonable matches to reported bacterial natural products). Although the information derived from the LC-MS investigation of the compounds (MS data, LC retention times, and UV–vis PDA profiles) enabled us to postulate that the Serratia sp. metabolites were analogues of 2, we decided to pursue their scale-up purification for structure confirmation and bioactivity testing.
Figure 4.
Targeting microbial natural products using LAESIMS. The inset shows bacterial colonies growing on the surface of an agar plate. The plate was placed inside of the LAESIMS chamber for mass spectrometry profiling. A subset of representative colonies was selected (indicated by red arrows), and a virtual grid was laid over these colonies using the instrument’s software to target where laser ablation would occur. The light blue polygons show where mass data were collected from the colonies within the range of m/z 200–2000. The presented mass data were derived from the circled colony (average of several locations taken from the colony and subtracted from mass data obtained from a blank [uncolonized] portion of the plate), which reveals prominent single and doubly charged sodium adduct ions for viscosin (1).
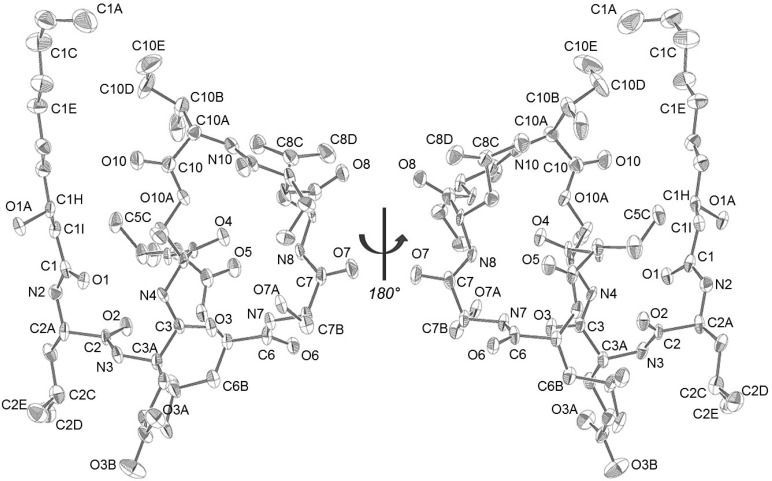
Figure 5.
ORTEP rendering of viscosin (1) illustrating the metabolite’s absolute configuration (determined by refinement of the Flack parameter; the water molecule identified near atom N2 has been removed from the figure for clarity; however, the position of the water can be found in the Supporting Information). The numbering system used for this structure reflects the atom assignments used in the Supporting Information and in the Cambridge Structural Database (CCDC 1511786).
Upon partitioning of the liquid cultures of the Serratia sp. isolate, the combined ethyl acetate layer was determined to retain the putative serrawettin analogues, as well as the biofilm inhibition activity. LAESIMS-guided fractionation of the organic layer by HP20SS VLC, as well as preparative and semipreparative C18 HPLC resulted in the purification of compounds 3–5, along with 2, whose structure was subsequently confirmed by MS experiment, 1H and 13C NMR data (Tables 1 and 2, respectively), and Marfey’s analysis.31,32
Table 1. 1H NMR Data (600 MHz, DMSO-d6) for Serrawettins W2 (2), W4, (3), W5 (4), and W6 (5).
| serrawettin W2 (2) | serrawettin W4 (3) | serrawettin W5 (4) | serrawettin W6 (5) | |
|---|---|---|---|---|
| position | δH, m (J in Hz) | δH, m (J in Hz) | δH, m (J in Hz) | δH, m (J in Hz) |
| Lipid | (3R)-hydroxydecanoic acid | (3R)-hydroxydecanoic acid | (3R)-hydroxyoctanoic acid | (3R)-hydroxydecanoic acid |
| 2a | 2.36, dd (5.9,14.4) | 2.38, dd (6.8, 14.6) | 2.35, dd (6.2, 14.5) | 2.34, dd (5.4, 14.2) |
| 2b | 2.63, dd (3.3, 14.6) | 2.59, dd (3.1, 14.7) | 2.63, dd (3.1, 14.4) | 2.66, m |
| 3 | 4.92, m | 4.96, m | 4.93, d (7.3) | 4.91, m |
| 4 | 1.52, m | 1.53, m | 1.52, m | 1.53, d (7.4) |
| 5 | 1.23, m | 1.22, m | 1.24, m | 1.23, m |
| 6 | 1.23, m | 1.22, m | 1.24, m | 1.23, m |
| 7 | 1.23, m | 1.22, m | 1.24, m | 1.23, m |
| 8 | 1.23, m | 1.22, m | 0.81, m | 1.23, m |
| 9 | 1.23, m | 1.22, m | 1.23, m | |
| 10 | 0.82, m | 0.84, m | 0.82, m | |
| Residue 1 | d-leucine | d-leucine | d-leucine | d-leucine |
| 11 (NH) | 7.81, d (8.7) | 7.86, d (8.5) | 7.82, d (8.6) | 7.86, d (8.8) |
| 12 | 4.35, ddd (3.9, 8.7, 12.0) | 4.39, m | 4.35, ddd (3.8, 8.6, 10.1) | 4.36, m |
| 13a | 1.13, m | 1.22, m | 1.13, m | 1.23, m |
| 13b | 1.42, m | 1.44, m | 1.42, m | 1.45, m |
| 14 | 1.64, m | 1.63, m | 1.64, m | 1.65, dd (5.3, 10.8) |
| 15 | 0.82, m | 0.84, m | 0.81, m | 0.82, m |
| 16 | 0.82, m | 0.84, m | 0.81, m | 0.82, m |
| Residue 2 | l-serine | l-serine | l-serine | l-serine |
| 18 (NH) | 8.07, d (8.5) | 8.09, d (7.9) | 8.18, d (8.6) | 8.24, d (7.4) |
| 19 | 4.41, m | 4.39, m | 4.42, m | 4.43, m |
| 20a | 3.55, m | 3.59, m | 3.58, m | 3.59, m |
| 20b | 3.62, m | |||
| 21 (OH) | 5.02, t (5.5) | 5.04, m | 5.08, s | 5.13, s |
| Residue 3 | l-threonine | l-threonine | l-threonine | l-threonine |
| 23 (NH) | 8.01, d (8.4) | 8.01, d (7.8) | 8.16, d (8.2) | 8.24, d (7.4) |
| 24 | 4.1, dd (3.0, 8.3) | 4.03, dd (3.6, 7.9) | 4.07, dd (3.0, 8.4) | 4.09, dd (2.9, 8.4) |
| 25 | 4.24, m | 4.16, m | 4.20, s | 4.23, s |
| 26 (OH) | 5.09, d (4.7) | 5.09, d (5.0) | 5.18, s | 5.24, d (5.3) |
| 27 | 0.97, d (6.4) | 0.94, d (6.3) | 0.96, d (6.4) | 0.98, d (6.5) |
| Residue 4 | d-phenylalanine | d-phenylalanine | d-phenylalanine | d-tyrosine |
| 29 (NH) | 7.44, d (7.0) | 7.59, d (7.2) | 7.46, d (7.0) | 7.41, d (6.9) |
| 30 | 4.44, dd (4.7, 7.2, 7.3) | 4.39, m | 4.42, m | 4.36, m |
| 31a | 2.88, dd (7.3, 13.8) | 2.88, dd (8.1, 13.8) | 2.88, dd (7.4, 13.8) | 2.79, dd (7.3, 13.8) |
| 31b | 3.12, dd (4.6, 13.9) | 3.15, dd (4.2, 13.9) | 3.11, dd (4.6, 13.9) | 2.98, dd (4.6, 13.8) |
| 33 | 7.14, m | 7.14, m | 7.14, d (6.8) | 6.84, d (8.1) |
| 34 | 7.06, m | 7.14, m | 7.06, d (7.0) | 6.53, d (8.1) |
| 35 | CH, 7.14, m | CH, 7.14, m | CH, 7.14, d (6.8) | OH, 5.32, m |
| 36 | 7.06, m | 7.14, m | 7.06, d (7.0) | 6.53, d (8.1) |
| 37 | 7.14, m | 7.14, m | 7.14, d (6.8) | 6.84, d (8.1) |
| Residue 5 | l-isoleucine | l-valine | l-isoleucine | l-isoleucine |
| 39 (NH) | 8.47, d (6.6) | 8.28, d (7.2) | 8.44, d (6.7) | 8.48, br s |
| 40 | 3.76, m | 3.73, t (7.8) | 3.77, m | 3.76, m |
| 41 | 1.76, m | 1.95, m | 1.76, m | 1.76, m |
| 42a | 1.13, m | (CH3) 0.84, m | 1.13, m | 1.12, m |
| 42b | 1.36, m | 1.35, m | 1.35, m | |
| 43 | 0.82, m | 0.81, m | 0.82, m | |
| 44 | 0.82, m | 0.84, m | 0.81, m | 0.82, m |
Table 2. 13C NMR Data (150 MHz, DMSO-d6) for Serrawettins W2 (2), W4, (3), W5 (4), and W6 (5).
| serrawettin W2 (2) | serrawettin W4 (3) | serrawettin W5 (4) | serrawettin W6 (5) | |
|---|---|---|---|---|
| position | δC, type | δC, type | δC, type | δC, type |
| Lipid | (3R)-hydroxydecanoic acid | (3R)-hydroxydecanoic acid | (3R)-hydroxyoctanoic acid | (3R)-hydroxydecanoic acid |
| 1 | 169.2, C | 168.8, C | 169.2, C | 168.5, C |
| 2 | 40.6, CH2 | 39.9, CH2 | 40.5, CH2 | 40.6, CH2 |
| 3 | 72.2, CH | 71.6, CH | 72.1, CH | 72.1, CH |
| 4 | 32.4, CH2 | 32.1, CH2 | 32.3, CH2 | 32.4, CH2 |
| 5 | 31.5, CH2 | 31.1, CH2 | 24.8, CH2 | 31.5, CH2 |
| 6 | 29.0, CH2 | 28.4, CH2 | 31.3, CH2 | 29.2, CH2 |
| 7 | 28.8, CH2 | 22.0, CH2 | 22.2, CH2 | 29.2, CH2 |
| 8 | 22.5, CH2 | 24.7, CH2 | 14.2, CH3 | 22.5, CH2 |
| 9 | 21.4, CH2 | 22.0, CH2 | 25.2, CH2 | |
| 10 | 14.4, CH3 | 13.9, CH3 | 14.4, CH3 | |
| Residue 1 | d-leucine | d-leucine | d-leucine | d-leucine |
| 12 | 51.1, CH | 50.6, CH | 51.1, CH | 51.2, CH |
| 13 | 40.8, CH2 | 40.4, CH2 | 40.8, CH2 | 40.6, CH2 |
| 14 | 24.2, CH | 23.8, CH | 24.2, CH | 24.2, CH |
| 15 | 23.8, CH3 | 21.2, CH3 | 21.4, CH3 | 21.4, CH3 |
| 16 | 23.8, CH3 | 23.3, CH3 | 23.8, CH3 | 23.7, CH3 |
| 17 | 171.7, C | 172.2, C | 172.6, C | 170.3, C |
| Residue 2 | l-serine | l-serine | l-serine | l-serine |
| 19 | 57.1, CH | 56.5, CH | 57.2, CH | 57.2, CH |
| 20 | 61.6, CH2 | 61.1, CH2 | 61.7, CH2 | 61.6, CH |
| 22 | 172.5, C | 170.6, C | 171.0, C | 171.8, C |
| Residue 3 | l-threonine | l-threonine | l-threonine | l-threonine |
| 24 | 58.8, CH | 58.8, CH | 59.0, CH | 58.9, CH |
| 25 | 65.8, CH | 65.5, CH | 65.9, CH | 65.9, CH |
| 27 | 20.6, CH3 | 20.1, CH3 | 20.6, CH3 | 20.6, CH3 |
| 28 | 171.0, C | 169.7, C | 170.0, C | 169.2, C |
| Residue 4 | d-phenylalanine | d-phenylalanine | d-phenylalanine | d-tyrosine |
| 30 | 54.0, CH | 54.0, CH | 54.1, CH | 54.3, CH |
| 31 | 37.6, CH2 | 37.0, CH2 | 37.6, CH2 | 36.9, CH2 |
| 32 | 137.2, C | 137.1, C | 139.3, C | 126.6, C |
| 33 | 128.5, CH | 128.0, CH | 128.5, CH | 130.4, CH |
| 34 | 129.5, CH | 129.1, CH | 129.5, CH | 115.4, CH |
| 35 | 126.9, CH | 126.3, CH | 126.8, CH | 156.5, C |
| 36 | 129.5, CH | 129.1, CH | 129.5, CH | 115.4, CH |
| 37 | 128.5, CH | 128.0, CH | 128.5, CH | 130.4, CH |
| 38 | 169.9, C | 171.0, C | 171.6, C | 170.7, C |
| Residue 5 | l-isoleucine | l-valine | l-isoleucine | l-isoleucine |
| 40 | 58.0, CH | 59.4, CH | 58.0, CH | 58.0, CH |
| 41 | 35.0, CH | 28.9, CH | 35.0, CH | 34.9, CH |
| 42 | 25.2, CH2 | 19.1, CH3 | 25.2, CH2 | 25.2, CH2 |
| 43 | 10.6, CH3 | 10.6, CH3 | 10.5, CH3 | |
| 44 | 15.7, CH3 | 18.9, CH3 | 15.7, CH3 | 15.8, CH3 |
| 45 | 171.0, C | 170.6,C | 171.1, C | 171.1, C |
Analysis of HRESIMS data for compound 3 provided a prominent ion with m/z 740.4235 [M + Na]+ that supported a molecular formula of C37H59N5O9. This indicated that compound 3 likely differed from 2 by the loss of a CH2 unit. Comparative analysis of the 1H–1H TOCSY data for 3 versus 2 revealed that the spin system associated with the isoleucine residue was altered. Inspection of the 1H (Table 1) and 1H–1H dqfCOSY NMR data for 3 showed that the amide proton (8.28, d, J = 7.22, 1 H) coupled with the α-proton (3.73, t, J = 7.76, 7.76, 1 H), which coupled to a methine (1.95, m, 2 H) that in turn coupled with methyl protons (0.84, m, 6 H). These results could be accounted for if the isoleucine residue in 2 changed to a valine residue in 3. Subsequent investigation of the metabolite by hydrolysis followed by Marfey’s analysis confirmed that the isoleucine residue was no longer present and instead an l-valine had been incorporated. Thus, metabolite 3 was assigned the trivial name serrawettin W4 in recognition of its structural relationship to metabolite 2.
A similar structure determination strategy was applied to compound 4 after observing that the HRESIMS data contained a prominent ion with m/z 726.4073 [M + Na]+, which corresponded to a molecular formula of C36H57N5O9. This indicated that the metabolite was deficient for two CH2 units relative to compound 2. Inspection of the 1H–1H TOCSY data for 4 provided strong evidence that metabolites 2 and 4 shared identical macrocycles. This was supported by hydrolysis followed by Marfey’s analysis, which confirmed the presence of the same amino acid residues in 2 and 4. Turning our attention to the MSn result, as well as 1H NMR data attributable to the hydrocarbon chain, it was determined that the lipid-derived portion of 4 was missing two CH2 units relative to 2. With the structure of metabolite 4 confirmed, the new compound was given the trivial name serrawettin W5.
An evaluation of metabolite 5 by HRESIMS provided an ion with m/z 770.4346 [M + Na]+ that enabled us to determine its molecular formula was C38H61N5O10. This represented an increase of one oxygen atom in 5 relative to compound 2. Focusing on the 1H NMR data (Table 1) for the amino acid portion of the macrocycle in 5, it was readily apparent that the aromatic protons associated with the phenylalanine residue in 2 were altered and that one hydrogen bonded to a carbon was missing. The new aromatic spin-coupled network appeared as an AA′BB′ system, which led us to propose that 5 incorporated a tyrosine residue in place of the phenylalanine in 2. The assignments of the carbon and hydrogen spins for the tyrosine were subsequently confirmed by HMBC experiment, as well as hydrolysis followed by Marfey’s analysis, which together established the presence of an l-tyrosine in 5. Metabolite 5 was assigned the trivial name serrawettin W6.
With the stereochemical assignments of the amino acid residues completed, we focused on securing the configurations of the C-3 positions in each compound. Applying biosynthetic-based logic to the problem, it appeared reasonable to presume that the stereochemistries of the metabolites’ C-3 positions were set upon incorporation of either 3-hydroxydecanoic acid (2, 3, and 5) or 3-hydroxyoctanoate (4). A survey of the chemical literature concerning the spectroscopic properties of β-hydroxy fatty acids revealed a decisive trend in their chiral-optical data (Table S1). Namely, in chloroform, R-configured β-hydroxy fatty acids displayed consistent levorotatory activities, whereas in ethanol, these molecules exhibited dextrorotatory properties. Returning to the acid hydrolysates prepared from 2–5, we were able to purify milligram quantities of enantiopure β-hydroxy fatty acids from each sample. Specific rotation values were obtained for the products in both ethanol and chloroform, which enabled us to assign R configurations to the C-3 positions in 2–5.
The genome of the Serratia sp. isolate was sequenced to link the identified natural products to their prospective biosynthetic gene cluster. Over 5.4 million reads were generated and assembled into 35 contiguous sequences after quality control. The annotated 16S rRNA gene was extracted and aligned to OTUs classified as Gammaproteobacteria, which we had observed from the opossum ears. These showed close (98%) homology to an OTU classified as a member of the Enterobacteriaceae. A total of eight potential secondary-metabolite-producing biosynthetic gene clusters were identified by antiSMASH analysis, including four NRPS clusters. One of the candidate clusters presented a near-perfect match to what was required to construct metabolites 2–5 (SI Figure S69).
The purified compounds were evaluated for their abilities to inhibit growth and biofilm production of C. albicans. None of the metabolites were able to kill fungal cells at concentrations up to 100 μM. Compound 1 from the Pseudomonas sp. isolate proved to be the most potent inhibitor of C. albicans biofilm formation, with an IC50 value of 4.6 ± 1.0 μM. The Serratia sp. metabolites exhibited a range of biofilm inhibition capabilities, with 2 being the most potent (IC50 = 7.7 ± 0.7 μM), followed by 4 (IC50 = 13.4 ± 0.2 μM), 5 (IC50 = 29.2 ± 0.4 μM), and 3 (IC50 = 59.8 ± 5.7 μM). The abilities of these bacterial cyclic lipodepsipeptides to limit biofilm formation, while not impacting yeast cell survival, hint at the possibility of alternative chemical options for controlling C. albicans infections by means of biofilm modulation.
The use of opportunistic sampling to humanely investigate the microbiomes of a broad range of animals offers an intriguing method to accelerate the exploration of this resource for new natural products. Notably, the discovery pipeline presented here is an enabling tool for identifying bacterial natural products from nonhominid mammals. Our approach, however, is not without limitations. One potential concern centers on how well our collection strategy accurately reflects the native microbiomes of living mammals since the microbiomes of humans are known to change as the time since death increases.33 While our investigation did not test this point specifically, we did make efforts to sample only recently deceased mammals and took care to avoid orifices that were markedly compromised by collisions with vehicles. Therefore, we remain confident that the isolates obtained in this study are representative members of mammalian microbiomes and not taxa simply derived from the immediate environment or strictly enhanced via decomposition. Further studies are anticipated to provide additional microbiome-associated bacteria, their natural products, and associated biosynthetic gene clusters, which might serve as candidates for future microbiome engineering endeavors. Indeed, colony profiling by LAESIMS suggests the presence of many bacteria-derived peptide-like products that we anticipate will possess potentially useful therapeutic properties.
Experimental Section
Procedures
NMR data were obtained on Varian VNMR spectrometers (Agilent Technologies, Inc., Santa Clara, CA, USA) with broadband and triple resonance probes at 25 ± 0.5 °C unless otherwise noted. Optical rotation measurements were made on a Rudolph Research Autopol III automatic polarimeter (Rudolph Research Analytical Corp., Hackettstown, NJ, USA). UV data were measured with a Hewlett-Packard 8452A diode array spectrophotometer (Agilent Technologies, Inc.). IR spectra were obtained on a Bruker Vector 22 FT-IR spectrometer (Bruker Corp., Billerica, MA, USA). Accurate mass electrospray-ionization mass spectrometry data were collected on an Agilent 6538 High-Mass-Resolution QTOF mass spectrometer (Agilent Technologies, Inc.). HPLC separations were carried out on a Shimadzu system using a SCL-10A VP controller (Shimadzu Scientific Instruments Inc., Columbia, MD, USA) and Gemini 5 μm C18 and hexyl-phenyl columns (110 Å, 250 × 21.2 mm and 250 × 10.0 mm, respectively, Phenomenex Inc., Torrance, CA, USA) with flow rates of 10 or 4 mL/min. All chemicals and solvents used in the study were of research grade quality or better.
Opportunistic Sampling
A 48 km section of Oklahoma State Highway 9 from the University of Oklahoma campus, Norman, OK, to the city of Tecumseh, OK, was used for this study. This route was chosen because it traverses a state park (Lake Thunderbird State Park), a significant quantity of acreage devoted to agriculture, and forested land, which are all prime habitats for many of Oklahoma’s major mammalian species. Additionally, the highway is heavily traveled (a major east–west thoroughfare from Arkansas to Texas) with posted speed limits of 65 mph (105 kph). The juxtaposition of having an active highway that intersects such a variety of mammalian habitats made this a prime location for near daily animal–vehicle collisions.
A sampling permit was obtained from the State of Oklahoma (Scientific Collector Permit #5250), and the University of Oklahoma IACUC was informed of the experiments, resulting in the assignment of an internal case-tracking number (R11-021). When fresh roadkill was encountered (generally less than 10 h old), the intact orifices, which included the mouth, ear, nose, and rectum, were sampled with sterile cotton-tipped swabs and then immediately plunged into sterile phosphate-buffered saline (PBS) buffer. When opportunities presented themselves, samples were also taken by swabbing the inner portions of the lower gastrointestinal tract.
Isolation and Cultivation of Bacteria
Sample swabs were generally processed within 1–2 h after collection. The cotton tips of the swabs were placed into 1.0 mL of PBS and vortexed. These suspensions were diluted 10–3 with PBS, and 50 μL aliquots were spread onto three types of solid agar media (10% tryptic soy agar, 10% brain heart infusion agar, and DM7 agar) and incubated at 30 °C for 3 days. Colonies were targeted for isolation based on their morphological uniqueness. Colony picking was performed using sterile toothpicks and the inoculum used to prepare streak plates on DM7 medium. Single colonies were picked with toothpicks, which were then dipped into tryptic soy broth containing 15% glycerol and incubated at room temperature for 1 week. Cultures of isolates were used to generate samples for cryogenic preservation at −80 °C, as well as inoculate various media for further chemical and bioassay studies. For these experiments, 5 μL aliquots were inoculated into the wells of three 24-well plates containing 50% tryptic soy broth, 50% brain heart infusion broth, and DM7 broth and incubated for 7 days at 30 °C. These cultures were subjected to partitioning three times against equal volumes of ethyl acetate. The organic layers from each sample were combined, and the solvent was removed in vacuo. Extracts were evaluated for bioactivities, and those determined to be active were examined by LCMS and LAESIMS.
Strain Information
The Serratia sp. and Pseudomonas sp. isolates were obtained from swabbing the ear canal of a roadkill opossum encountered on Oklahoma State Highway 9 near Norman, OK. While the original colony selected for the study was thought to be a single isolate, subsequent cultivation of the bacteria led us to determine that the culture contained two taxa, which were separated through isolation on a solid agar medium and analyzed in pure culture.
A single colony of the Serratia sp. was used to inoculate 1/10th strength tryptic soy broth medium, which, after 16 h, was used to obtain genomic DNA by means of an Xpedition Soil/Fecal DNA MiniPrep kit following the manufacturer’s directions (Zymo Research, Irvine, CA, USA). The genomic DNA was quantified using the QuBit BR assay (Life Technologies, Carlsbad, CA, USA), and approximately 1 μg was sheared to a mean insert size of 500 bp and sequenced using PE250 V2 chemistry on the Illumina MiSeq platform (Illumina, San Diego, CA, USA).
The reference strain C. albicans SC5314 was cultured in brain-heart infusion medium (BHI, Becton Dickinson) or RPMI-1640 plus MOPS medium [RPMI-1640 medium (Sigma) buffered to pH 7.0 with 0.17 M MOPS (3-(N-morpholino)propanesulfonic acid, Sigma)] as required.
Sequence Data for the Serratia sp. Isolate
Sequence data used for 16S rRNA gene analysis are available at the NCBI SRA under accession number SRX1601902. The annotated Serratia sp. genome is available under the accession number GCA_001643155.1, and the raw reads used to assemble the genome are available for download at the NCBI SRA under the accession number SRX1585056.
DNA Extraction and 16S rRNA Gene Library Preparation
The opossum carcass was sampled with sterile cotton swabs, and the swabs were frozen at −20 °C until DNA extraction was performed. The samples were thawed and then homogenized (BioSpec Products, Bartlesville, OK, USA) for 30 s. Community genomic DNA was extracted using the MoBio Power Biofilm DNA extraction kit (MoBio Laboratories, Inc., Carlsbad, CA, USA). Libraries of bacterial and archaeal 16S rRNA gene fragments were amplified from each DNA extraction by PCR with primers that spanned the V4 region between positions 519 and 802 (E. coli numbering), producing a ∼300 bp fragment. These primers evenly amplify a broad range of both the Bacteria and Archaea. The forward primer (M13L-519F: 5′-GTA AAA CGA CGG CCA GCACMG CCG CGG TAA-3′) contained the M13 forward primer (in bold), followed by the 16S rRNA gene-specific sequence (underlined). The reverse primer (785R: 5′-TAC NVG GGT ATC TAA TCC-3′) was taken directly from “S-D-Bact07850b-A-18” in Klindworth et al.34 Each 50 μL PCR consisted of 3 U of recombinant Taq polymerase (Thermo Fisher Scientific Inc., Grand Island, NY, USA), 1.5 mM MgCl2, 0.1 μM DNTPs (Thermo Fisher Scientific Inc.), 0.2 μM of each primer, and 4 μL of a DNA extraction. Initial amplification was conducted using the following protocol: initial denaturation at 95 °C for 2 min, followed by 30 cycles of denaturation at 95 °C for 45 s, annealing at 52 °C for 45 s, and extension at 72 °C for 45 s, followed by a final extension at 72 °C for 5 min. The amplified 16S rRNA gene fragments in each library were purified using AmpureXP paramagnetic beads according to the manufacturer’s protocols (Beckman Coulter, Inc., Indianapolis, IN, USA). A second, six-cycle PCR was used to add a unique 12 bp barcode35 to each amplicon library using a unique forward primer containing the barcode + M13 forward sequence (5′–3′) and the 785R primer using reaction conditions identical to those listed above. The resulting barcoded PCR products were quantified using the QuBit HS assay (Life Technologies), pooled in equimolar amounts, and concentrated to a final volume of 80 μL using Amicon Ultra-0.5 mL 30K centrifugal filters (Merck Millipore Corp., Billerica, MA, USA). The pooled library was then submitted for sequencing on the MiSeq platform using PE250 V2 chemistry (Illumina).
Sequence Analysis
After sequencing, reads were merged using PEAR,36 demultiplexed in QIIME,37 filtered by quality, and clustered into OTUs using UPARSE.38 The mapping file used to demultiplex the samples is present as a supplementary table. The taxonomy of each OTU was assigned using UCLUST39 and the SILVA database (Release 123).40 Taxonomy summaries were generated within QIIME.
For the genomic sequence, reads were first quality trimmed, and sequence adapters were removed using PEAT41 before assembly within SPAdes42 using kmer values of 21, 33, 55, 77, 99, and 127. After assembly, contiguous sequences less than 300 bp in length were discarded, and contaminating PhiX sequences were removed after visualization of the genome assembly graph in Bandage.43 The assembled genome was annotated using the NCBI PGAP annotation pipeline, and putative metabolite clusters were identified using antiSMASH.44
Defined Medium for Bacterial Growth (DM7)
Single colonies of the Serratia sp. isolate were transferred to DM7 and vortexed. Aliquots of this suspension were inoculated into 40 high-aeration shake flasks (2 L Ultra Yield flask) containing 1 L of DM7. The flasks were shaken at 135 rpm for 7 days at room temperature on an Innova 5000 shaker. The DM7 broth consisted of (per liter) monopotassium phosphate, 2.0 g; ammonium chloride, 1.5 g; magnesium sulfate heptahydrate, 0.5 g; glycerol, 12.6 g; myo-inositol, 0.4 g; monosodium glutamate, 5.0 g; sodium fluoride, 0.084 g; iron(II) sulfate heptahydrate, 0.025 g; zinc(II) sulfate heptahydrate, 0.01 g; cobalt(II) chloride, 0.01 g; calcium carbonate, 0.25 g; and p-aminobenzoate, 0.001 g; with the pH of the final solution adjusted to 7.
Crystal Data for 1
The broth from the liquid culture of the Pseudomonas sp. was partitioned against ethyl acetate. During the partitioning process, crystals of 1 formed on the inner surface of the separatory funnel. The crystals were rinsed from the funnel under a stream of ethyl acetate, and the solvent was removed by leaving the crystals overnight in an open vial at room temperature.
A colorless needle-shaped crystal of 1 of dimensions 0.26 × 0.04 × 0.04 mm was selected for structure analysis. Intensity data for this compound were collected using a diffractometer with a Bruker APEX ccd area detector45,46 and graphite-monochromated Mo Kα radiation (λ = 1.541 78 Å). The sample was cooled to 100(2) K. Cell parameters were determined from a nonlinear least-squares fit of 2010 peaks in the range 4.2° < θ < 68.2°. A total of 39 420 data were measured in the range of 3.626° < θ < 71.353° using φ and ω oscillation frames. The data were corrected for absorption by the empirical method47 giving minimum and maximum transmission factors of 0.835 and 0.972. The data were merged to form a set of 11 566 independent data with R(int) = 0.145 and a coverage of 98.5%.
The orthorhombic space group P212121 was determined by systematic absences and statistical tests and verified by subsequent refinement. The structure was solved by direct methods and refined by full-matrix least-squares methods on F2.48 The positions of hydrogens bonded to carbons were initially determined by geometry and were refined using a riding model. Hydrogens bonded to nitrogens and oxygens were located on a difference map, and their positions were refined independently with X-H restraints. Non-hydrogen atoms were refined with anisotropic displacement parameters. A total of 767 parameters were refined against 51 restraints and 11 566 data to give wR(F2) = 0.3772 and S = 1.238 for weights of w = 1/[σ2(F2) + (0.1800P)2 + 30.0000P], where P = [Fo2 + 2Fc2]/3. The final R(F) was 0.1281 for the 11 016 observed, [F > 4σ(F)] data. The largest shift/s.u. was 0.000 in the final refinement cycle. The final difference map had maxima and minima of 0.783 and −0.891 e/Å3, respectively. The absolute configuration of the structure was determined by refinement of the Flack parameter.49
Purification of 2–5
The broth from each culture flask was pooled and partitioned three times against equal volumes of ethyl acetate. The solvent was removed in vacuo to generate a crude organic residue (10.3 g). The organic material was processed by VLC over HP20SS resin using a step gradient going from water to methanol to generate six fractions. Fraction 4 was further separated by preparative C18 HPLC (gradient elution with MeOH–H2O, 75:25, to 100% organic in 30 min) to yield three subfractions (1–3). Subfraction 2 was further separated by semipreparative C18 HPLC (MeCN–H2O, 55:65) to provide four subfractions (A–D). Subfraction 2C was further purified by semipreparative C18 HPLC (MeCN–H2O, 60:40) to yield 2 (46.7 mg, 4.53% yield). Subfraction 2B was further purified by semipreparative C18 HPLC (MeCN–H2O, 65:35) to yield 3 (47.1 mg, 4.57% yield). Subfraction 2A was further purified by semipreparative hexyl-phenyl HPLC (MeCN–0.1% formic acid in H2O, 65:35) to yield 4 (2.8 mg, 0.271% yield) and 5 (3.7 mg, 0.359% yield).
Marfey’s Analysis
Samples of the metabolites (∼300 μg) were incubated overnight in 6 M HCl (500 μL) at 110 °C. The hydrosylates were dried under nitrogen gas, and 20 μL of 1 M NaHCO3 was added to each sample to facilitate neutralization. Next, 100 μL of 1% 1-fluoro-2,4-dinitrophenyl 5-l-alanine amide was added to the samples, and the resulting solutions were heated at 45 °C for 1 h. The reaction mixtures were neutralized with 20 μL of 1 M HCl and diluted with 500 μL of MeCN. The samples were centrifuged and analyzed by LC-MS (100 Å, C18 Kinetex 2.6 μm, 75 × 3.0 mm column with gradient elution using MeCN with 0.1% formic acid in H2O, 10:90, to 100% organic over 15 min.). Standards eluted as l-isoleucine at 7.96 min, d-isoleucine at 8.19 min, l-valine at 7.68 min, d-valine at 7.68 min, l-leucine at 7.78 min, d-leucine at 8.26 min, l-phenylalanine at 7.80 min, d-phenylalanine at 8.13 min, l-serine at 7.57 min, d-serine at 8.13 min, l-tyrosine at 8.57 min, d-tyrosine at 8.88 min, l-threonine at 8.53 min, l-allo-threonine at 5.73 min, and d-allo-threonine at 5.90 min.
Examination of 3-Hydroxy Fatty Acids by Polarimetry
The hydrosylates were partitioned with equal portions of hexanes (×3). The hexanes were removed in vacuo, the resultant compound was suspended in 2 mL of chloroform or ethanol, and their specific rotation values were determined.
Assays for Growth Inhibition and Biofilm Formation
The effects of compounds on the growth of C. albicans were tested using the method described in the NCCLS 2008 CLSI M27-A3 guidelines24 with the following modifications. Cells of C. albicans SC5314 were cultured in BHI medium (Becton Dickinson Co., Franklin Lakes, NJ, USA) at 37 °C overnight. The cells were pelleted by centrifugation, washed with sterile PBS (pH 7.4), and resuspended in RPMI-1640 plus MOPS medium. Test compounds were prepared in DMSO at stock concentrations of 2 mM before being diluted in RPMI-1640 plus MOPS for testing. Aliquots of yeast suspension (100 μL containing 2.5 × 103 cells mL–1) were added to the medium containing the diluted compounds or DMSO [final concentrations did not exceed 2% (v/v)] before being transferred to 96-well plates (Corning). After 48 h of incubation at 37 °C, the viability of the yeast was measured using the XTT assay. In brief, yeast cells were treated with 0.1 mg mL–1 XTT at 37 °C for 1 h. Absorbance measurements were taken at 492 nm using a microplate reader (Infinite M200). The minimum inhibitor concentrations (MIC) for growth were defined as the lowest antifungal concentrations that caused ≥85% reduction in metabolic activity.
For measuring biofilm formation, the medium was aspirated and the wells were washed twice with sterile PBS to remove nonadherent cells. Fresh medium (100 μL of RPMI-1640 plus MOPS) was then added back to each well. The formation of biofilms was measured using the XTT assay. All experiments were performed in triplicate on three separate occasions. The 50% inhibitory concentration (IC50) values for biofilm inhibition were calculated using GraphPad Prism 5.
Serrawettin W2 (2):
white, powdery solid; [α]D20 = −7.3 (c 0.164, CHCl3); UV (MeOH) λmax (log ε) 206 (3.52); IR (film) νmax 3600, 3516, 3412, 3003, 2965, 2926, 1715, 1425, 1362, 1221, 1092, 903, 783, 527; 1H NMR (DMSO-d6, 600 MHz) refer to Table 1; 13C NMR (DMSO-d6, 151 MHz) refer to Table 2; HRESIMS m/z 730.4402 [M – H]− (calcd for C38H60N5O9, 730.4391).
Serrawettin W4 (3):
white, powdery solid; [α]D20 = −7.4 (c 0.270, CHCl3); UV (MeOH) λmax (log ε) 206 (3.17); IR (film) νmax 3607, 3524, 3412, 3003, 2967, 2925, 1713, 1707, 1427, 1361, 1221, 1094, 903, 783, 527; 1H NMR (DMSO-d6, 600 MHz) refer to Table 1. 13C NMR (DMSO-d6, 151 MHz) refer to Table 2; HRESIMS m/z 740.4235 [M + Na]+ (calcd for C37H59N5O9Na, 740.4210).
Serrawettin W5 (4):
white, powdery solid; [α]D20 = −7.6 (c 0.245, CHCl3); UV (MeOH) λmax (log ε) 204 (3.05); IR (film) νmax 3603, 3520, 3412, 3003, 2968, 2926, 1714, 1427, 1362, 1221, 1094, 905, 783, 523; 1H NMR (DMSO-d6, 600 MHz) refer to Table 1. 13C NMR (DMSO-d6, 151 MHz) refer to Table 2; HRESIMS m/z 726.4073 [M + Na]+ (calcd for C36H57N5O9Na, 726.4054).
Serrawettin W6 (5):
white, powdery solid; [α]D20 = −7.4 (c 0.140, CHCl3); UV (MeOH) λmax (log ε) 206 (3.11); IR (film) νmax 3609, 3528, 3412, 3003, 2695, 2924, 1746, 1712, 1422, 1362, 1222, 1092, 903, 785, 530; 1H NMR (DMSO-d6, 600 MHz) refer to Table 1. 13C NMR (DMSO-d6, 151 MHz) refer to Table 2; HRESIMS m/z 770.4346 [M + Na]+ (calcd for C38H61N5O10Na, 770.4316).
Acknowledgments
Research reported in this publication was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (1R21AI101487) (R.H.C. and B.S.S.). X-ray data were collected by L. Thomas in the OU Macromolecular Crystallography Laboratory, which is supported, in part, by an Institutional Development Award from the National Institutes of Health, General Medical Sciences (P20GM103640). The C. albicans SC5314 culture was kindly provided by A. Dongari-Bagtzoglou, University of Connecticut Health Center. The LC-ESIMS instrument used for this project was funded in part by a Challenge Grant from the Office of the Vice President for Research, University of Oklahoma, Norman Campus, and an award through the Shimadzu Equipment Grant Program (R.H.C.).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jnatprod.6b00772.
X-ray crystal data for 1, NMR data for 2–5 (1H and 13C NMR, HMBC, HSQC, dqfCOSY, TOCSY, and 1H–13C HSQCTOCSY), Marfey’s analysis data, specific rotation data for β-hydroxy fatty acids, C. albicans biofilm growth inhibition data, and putative serrawettin biosynthetic genes (PDF)
Serratia sequence mapping file (TXT)
Crystallographic data (CIF)
Author Contributions
⊥ J. L. Motley and B. W. Stamps contributed equally.
The authors declare no competing financial interest.
Dedication
Dedicated to Professor Phil Crews, of the University of California, Santa Cruz, for his pioneering work on bioactive natural products.
Supplementary Material
References
- Lee M. S.; Oh S.; Tang H. Biomed Mater. Eng. 2014, 24, 3737. [DOI] [PubMed] [Google Scholar]
- Faust K.; Sathirapongsasuti J. F.; Izard J.; Segata N.; Gevers D.; Raes J.; Huttenhower C. PLoS Comput. Biol. 2012, 8, e1002606. 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen U. P.; Nielsen H. B.; Hildebrand F.; Raes J.; Sicheritz-Ponten T.; Kouskoumvekaki I.; Panagiotou G. ISME J. 2013, 7, 730. 10.1038/ismej.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel N.; Balskus E. P. Cell Chem. Biol. 2016, 23, 18. 10.1016/j.chembiol.2015.12.008. [DOI] [PubMed] [Google Scholar]
- Grice E. A.; Segre J. A. Annu. Rev. Genomics Hum. Genet. 2012, 13, 151. 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimee M.; Citorik R. J.; Lu T. K. Adv. Drug Delivery Rev. 2016, 105, 44. 10.1016/j.addr.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaung S. J.; Church G. M.; Wang H. H. Methods Mol. Biol. 2014, 1151, 3. 10.1007/978-1-4939-0554-6_1. [DOI] [PubMed] [Google Scholar]
- Sheth R. U.; Cabral V.; Chen S. P.; Wang H. H. Trends Genet. 2016, 32, 189. 10.1016/j.tig.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emge J. R.; Huynh K.; Miller E. N.; Kaur M.; Reardon C.; Barrett K. E.; Gareau M. G. Am. J. Physiol Gastrointest Liver Physiol 2016, 310, G989. 10.1152/ajpgi.00086.2016. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Wu Y.; Wang J.; Wu G.; Long W.; Xue Z.; Wang L.; Zhang X.; Pang X.; Zhao Y.; Zhao L.; Zhang C. Sci. Rep. 2016, 6, 27572. 10.1038/srep27572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks M. G.; Veiga P.; Wardwell-Scott L. H.; Tickle T.; Segata N.; Michaud M.; Gallini C. A.; Beal C.; van Hylckama-Vlieg J. E.; Ballal S. A.; Morgan X. C.; Glickman J. N.; Gevers D.; Huttenhower C.; Garrett W. S. ISME J. 2014, 8, 1403. 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marietta E. V.; Murray J. A.; Luckey D. H.; Jeraldo P. R.; Lamba A.; Patel R.; Luthra H. S.; Mangalam A.; Taneja V. Arthritis Rheumatol. 2016, 10.1002/art.39785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano Shields C. E.; Van Meerbeke S. W.; Housseau F.; Wang H.; Huso D. L.; Casero R. A. Jr.; O’Hagan H. M.; Sears C. L. J. Infect. Dis. 2016, 214, 122. 10.1093/infdis/jiw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazett M.; Bergeron M. E.; Haston C. K. Sci. Rep. 2016, 6, 19189. 10.1038/srep19189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P.; Zeng B.; Zhou C.; Liu M.; Fang Z.; Xu X.; Zeng L.; Chen J.; Fan S.; Du X.; Zhang X.; Yang D.; Yang Y.; Meng H.; Li W.; Melgiri N. D.; Licinio J.; Wei H.; Xie P. Mol. Psychiatry 2016, 21, 786. 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- Ross C. L.; Spinler J. K.; Savidge T. C. Anaerobe 2016, 41, 37. 10.1016/j.anaerobe.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nale J. Y.; Spencer J.; Hargreaves K. R.; Buckley A. M.; Trzepinski P.; Douce G. R.; Clokie M. R. Antimicrob. Agents Chemother. 2016, 60, 968. 10.1128/AAC.01774-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite A. J.; Shou W. Methods Mol. Biol. 2014, 1151, 27. 10.1007/978-1-4939-0554-6_2. [DOI] [PubMed] [Google Scholar]
- Claesen J.; Fischbach M. A. ACS Synth. Biol. 2015, 4, 358. 10.1021/sb500258b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipperer A.; Konnerth M. C.; Laux C.; Berscheid A.; Janek D.; Weidenmaier C.; Burian M.; Schilling N. A.; Slavetinsky C.; Marschal M.; Willmann M.; Kalbacher H.; Schittek B.; Brotz-Oesterhelt H.; Grond S.; Peschel A.; Krismer B. Nature 2016, 535, 511. 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- Wong C. K.; Tan M. H.; Rasouliha B. H.; Hwang I. Y.; Ling H.; Poh C. L.; Chang M. W. Methods Mol. Biol. 2014, 1151, 117. 10.1007/978-1-4939-0554-6_9. [DOI] [PubMed] [Google Scholar]
- Donia M. S.; Cimermancic P.; Schulze C. J.; Wieland Brown L. C.; Martin J.; Mitreva M.; Clardy J.; Linington R. G.; Fischbach M. A. Cell 2014, 158, 1402. 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner P. M.; Liu J.; Zhang Z.; Merritt J.; Qi F.; Cichewicz R. H. Org. Biomol. Chem. 2010, 8, 5486. 10.1039/c0ob00579g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Du L.; You J.; King J. B.; Cichewicz R. H. Org. Biomol. Chem. 2012, 10, 2044. 10.1039/c2ob06856g. [DOI] [PubMed] [Google Scholar]
- Donia M. S.; Fischbach M. A. Science 2015, 349, 1254766. 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham S.; Wong W. R.; Linington R. G.; Spadafora C.; Iturrado L.; Arnold A. E. PLoS One 2014, 9, e84549. 10.1371/journal.pone.0084549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez L. M.; Wong W. R.; Riener R. M.; Schulze C. J.; Linington R. G. PLoS One 2012, 7, e35398. 10.1371/journal.pone.0035398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodore C. M.; Stamps B. W.; King J. B.; Price L. S.; Powell D. R.; Stevenson B. S.; Cichewicz R. H. PLoS One 2014, 9, e90124. 10.1371/journal.pone.0090124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessup D. A. ILAR J. 2003, 44, 277. 10.1093/ilar.44.4.277. [DOI] [PubMed] [Google Scholar]
- Smith-Patten B. D.; Patten M. A. Environ. Manage. 2008, 41, 844. 10.1007/s00267-008-9089-3. [DOI] [PubMed] [Google Scholar]
- Bhushan R.; Bruckner H. Amino Acids 2004, 27, 231. 10.1007/s00726-004-0118-0. [DOI] [PubMed] [Google Scholar]
- Bhushan R.; Bruckner H. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2011, 879, 3148. 10.1016/j.jchromb.2011.05.058. [DOI] [PubMed] [Google Scholar]
- Can I.; Javan G. T.; Pozhitkov A. E.; Noble P. A. J. Microbiol. Methods 2014, 106, 1. 10.1016/j.mimet.2014.07.026. [DOI] [PubMed] [Google Scholar]
- Klindworth A.; Pruesse E.; Schweer T.; Peplies J.; Quast C.; Horn M.; Glockner F. O. Nucleic Acids Res. 2013, 41, e1. 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M.; Walker J. J.; Harris J. K.; Gold N. J.; Knight R. Nat. Methods 2008, 5, 235. 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Kobert K.; Flouri T.; Stamatakis A. Bioinformatics 2014, 30, 614. 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G.; Kuczynski J.; Stombaugh J.; Bittinger K.; Bushman F. D.; Costello E. K.; Fierer N.; Pena A. G.; Goodrich J. K.; Gordon J. I.; Huttley G. A.; Kelley S. T.; Knights D.; Koenig J. E.; Ley R. E.; Lozupone C. A.; McDonald D.; Muegge B. D.; Pirrung M.; Reeder J.; Sevinsky J. R.; Turnbaugh P. J.; Walters W. A.; Widmann J.; Yatsunenko T.; Zaneveld J.; Knight R. Nat. Methods 2010, 7, 335. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Nat. Methods 2013, 10, 996. 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. Bioinformatics 2010, 26, 2460. 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Pruesse E.; Quast C.; Knittel K.; Fuchs B. M.; Ludwig W.; Peplies J.; Glockner F. O. Nucleic Acids Res. 2007, 35, 7188. 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. L.; Weng J. C.; Hsiao C. C.; Chou M. T.; Tseng C. W.; Hung J. H. BMC Bioinf. 2015, 16, S2. 10.1186/1471-2105-16-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A.; Nurk S.; Antipov D.; Gurevich A. A.; Dvorkin M.; Kulikov A. S.; Lesin V. M.; Nikolenko S. I.; Pham S.; Prjibelski A. D.; Pyshkin A. V.; Sirotkin A. V.; Vyahhi N.; Tesler G.; Alekseyev M. A.; Pevzner P. A. J. Comput. Biol. 2012, 19, 455. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R.; Schultz M. B.; Zobel J.; Holt K. E.. Bandage; 2015. [DOI] [PMC free article] [PubMed]
- Blin K.; Medema M. H.; Kazempour D.; Fischbach M. A.; Breitling R.; Takano E.; Weber T. Nucleic Acids Res. 2013, 41, W204. 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APEX2; Bruker AXS Inc.: Madison, WI, USA, 2007.
- SAINT; Bruker AXS Inc.: Madison, WI, USA, 2007.
- Sheldrick G. M.SADABS; Bruker AXS Inc.: Madison, WI, USA, 2002.
- Sheldrick G. M.SHELXL; Georg-August Universitat: Gottingen, Germany, 2014.
- Parsons S.; Flack H. D.; Wagner T. Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2013, 69, 249. 10.1107/S2052519213010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.