Abstract
Alterations of microbiota-gut-brain axis have been invoked in the pathogenesis of autism spectrum disorders (ASD). Mouse models could represent an excellent tool to understand how gut dysbiosis and related alterations may contribute to autistic phenotype. In this study we paralleled gut microbiota (GM) profiles, behavioral characteristics, intestinal integrity and immunological features of colon tissues in BTBR T + tf/J (BTBR) inbred mice, a well established animal model of ASD. Sex differences, up to date poorly investigated in animal models, were specifically addressed. Results showed that BTBR mice of both sexes presented a marked intestinal dysbiosis, alterations of behavior, gut permeability and immunological state with respect to prosocial C57BL/6j (C57) strain. Noticeably, sex-related differences were clearly detected. We identified Bacteroides, Parabacteroides, Sutterella, Dehalobacterium and Oscillospira genera as key drivers of sex-specific gut microbiota profiles associated with selected pathological traits. Taken together, our findings indicate that alteration of GM in BTBR mice shows relevant sex-associated differences and supports the use of BTBR mouse model to dissect autism associated microbiota-gut-brain axis alteration.
The ability of the enteric microbes to directly communicate with the central nervous system (CNS), to modulate brain functions and possibly influence behavior is currently under study, and the importance of a microbiota-gut-brain axis has been established1. Communication between the gut microbiota (GM) and brain includes several connections such as vagus nerve, gut hormones, microbial metabolites and immune system2. The observation of high co-morbidity between intestinal inflammatory diseases and psychiatric symptoms such as anxiety and stress3,4 and the frequent occurrence of gastrointestinal dysfunctions in autistic patients5 strongly corroborated the hypothesis of an implication of GM in psychiatric conditions including autistic spectrum disorders (ASD). Pivotal studies revealed improvement in both behavioral and gastrointestinal symptoms of autistic patients upon oral short-term treatment with antibiotics and probiotics6,7, supporting a role of GM in ASD. Although evidence of a deviation of GM composition in ASD patients with respect to normally developing individuals has been reported8,9,10,11,12,13,14, to date no clear-cut conclusion was reached on the association of specific bacterial profiles with ASD. The use of mouse models of ASD may help to clarify the underlying mechanisms involved in communication of enteric microbes with CNS and possibly generating abnormal behaviors. The composition of GM in mice displaying features of ASD was analyzed in models of environmental risk factors such as in utero valproic acid (VPA) exposure and maternal immune activation (MIA)15,16. In utero VPA exposed mice displayed altered GM composition and fatty acid metabolism along with social behaviour abnormalities15. In addition, in the offspring of MIA mouse model, gastrointestinal barrier defects and microbiota alterations were observed, and probiotic treatment was shown to restore microbial composition and ameliorate behavioral alterations16. Given the complexity of autism pathogenesis involving environmental factors and multiple genes, several genetically-based mouse models of autism were developed including mutant and trasgenic mice17. One of the earliest and most studied animal model showing autistic-like phenotype is BTBR T + tf/J (BTBR) inbred mouse strain18,19,20,21,22. These animals are characterized by dysregulation of social communication23,24,25, occurrence of repetitive behaviour21,26,27, reduction in hippocampal neurogenesis28, alterations of BDNF signaling29 in addition to aberrant immune state30, all features that well parallel autistic phenotype. Based on these considerations we performed a comprehensive analysis of fully symptomatic BTBR mice. In particular in this study we paralleled GM profiles, behavioral characteristics such as social interaction, stereotyped and repetitive behavior, intestinal integrity and immunological features of colon tissues in adult male and female BTBR mice. Furthermore because sex differences in the majority of mouse models of psychiatric disorders have been frequently overlooked31, and because sex differences have been reported in gut microbiota composition32, we analyzed sex-related profiles of GM and correlated them with specific pathological traits, separately in male and female BTBR. Interestingly, we studied if specific bacterial taxa drive dysbiosis in BTBR females and males, and their relationship with the aleration of behavior, gut integrity and colon immunological state.
Results and Discussion
Overall structure of GM of male and female BTBR mice
We analyzed sex-related profiles of GM in BTBR mice, a known mouse model of autism to gain insights into relationship between autistic behavior and dysbiosis. Fecal microbiota of fully symptomatic, 12 months old, female and male BTBR (fBTBR and mBTBR, respectively; n = 6 mice each group) and female and male C57 control mice of same age (fC57 and mC57, respectively; n = 6 mice each group;) was analyzed by next generation sequencing (NGS) technology using the Illumina Miseq system. V3–V4 variable regions of the 16S rRNA gene were amplified and sequenced to characterize total bacterial population; 62,009.83 ± 33,665.39 high-quality sequences/sample were obtained from all 24 fecal samples, representing 3,250 operational taxonomic units (OTUs). The results shown were obtained considering a depth of 32,288 sequences/sample clustered in 2,740 OTUs; Good’s coverage > of 99.3% for all sequences in the four groups indicated good sequencing depth for reliable investigation of differences in fecal microbiota between BTBR and control mice. Among the 2,740 OTUs detected across any of the samples, 245 OTUs discriminated between fBTBR and fC57 mice, while 167 discriminated between mBTBR and mC57 mice. Discriminant OTUs were identified using two complementary analyses, LEfSe algorithm and Metastats comparison (Tables S1 and S2).
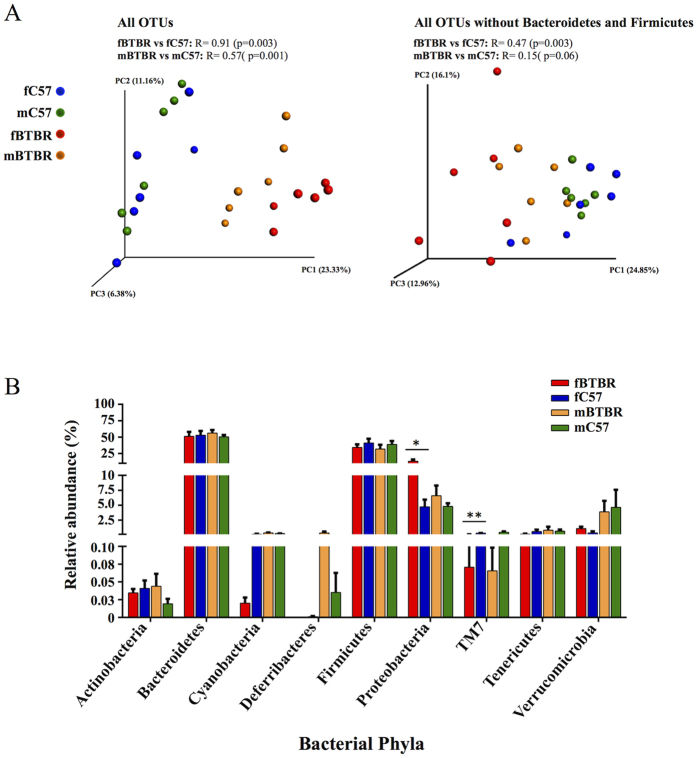
We evaluated ecological features of fecal bacterial communities in fBTBR and mBTBR compared to those of control groups. No significant differences in species richness (number of OTUs) and degree of homogeneity abundance of the species (Shannon index) were observed between groups (data not shown), while strong differences in phylogenetic assortment were detected comparing fBTBR and mBTBR with their respective controls (Fig. 1). Phylogenetic distances among samples were assessed by means of Unweighted Unifrac distance metrics, a qualitative phylogenetic measure that considers the presence/absence of a taxon. ANOSIM R statistic revealed a difference in gut bacterial assortment between BTBR of both sexes and their respective controls, with fBTBR vs fC57 displaying a higher R value compared to mBTBR vs mC57. This effect was evident in the PCoA plot, where fBTBR samples clustered to the extreme right of the plot, while mBTBR samples were positioned midway between fBTBR and control samples of both sexes (Fig. 1A, left plot).
Figure 1. Female and male BTBR mice exhibit an altered gut microbial composition.
(A) Unweighted UniFrac-based 3D PCoA plot constructed on all OTUs (32,288 reads per sample, left) or all OTUs without Bacteroidetes and Firmicutes reads (850 reads per sample, right) of fecal community of BTBR and C57 mice of both sexes. Analysis of similarity (ANOSIM) with 999 permutations was used to detect the statistical significant differences in microbial community composition between fBTBR and mBTBR compared to their controls (fC57 and mC57); on the top of plots are reported both R statistics and p values. (B) Relative abundance of all identified OTUs classified at phylum level. Mean values ± SEM are plotted (n = 6/group). Significant differences are indicated by *p < 0.05 and **p < 0.01 for comparison of fBTBR vs. fC57 and mBTBR vs. mC57. Abbreviations: fBTBR (BTBR female mice); mBTBR (BTBR male mice); fC57 (C57 female mice); mC57 (C57 male mice).
Sequencing data revealed that 89.1% of total reads were taxonomically classified in Bacteroidetes and Firmicutes phyla, and the majority of discriminatory OTUs, both in females and males, were classified in these phyla (Tables S1 and S2). The impact of these taxa was evident when the Unweighted Unifrac analysis was repeated after negative filtering of these phyla from total sequences. After subtraction, ANOSIM analysis on remaining OTUs revealed a weaker grouping level among samples (Fig. 1A, right plot), indicating that Bacteroidetes and Firmicutes were the principal contributors to the BTBR and C57 GM differences both in female and male mice.
GM profiling of BTBR female and male mice
Over the total of 9 bacterial phyla identified, comparison of mean abundances (by nonparametric Kruskal-Wallis test) of primary (Bacteroidetes, Firmicutes) and most of the less abundant phyla showed no significant differences between fBTBR and mBTBR compared to their respective controls (Fig. 1B). Exception was made for Proteobacteria which were found significantly more abundant in fBTBR with respect to fC57 (relative abundance 13.2% ± 2.6% and 4.7% ± 1.2%, respectively), and for TM7 phylum which was found significantly less abundant in fBTBR with respect to fC57 (relative abundance of 0.1% ± 0.04% in fBTBR and 0.3% ± 0.05% in fC57). No significant differences were found for all phyla identified in mBTBR compared to mC57 (Fig. 1B). Gram-negative Proteobacteria include lots of gut commensal species with potential pathogenic features, as the lipopolysaccharide expression. Although comparison between animal model and human disease needs to be under great scrutiny, given the known heterogeneity of ASD in both clinical and basic aspects, it is interesting to note that several families among Proteobacteria were found in the intestine of children with autism and gastrointestinal disturbances9. Even though the role of Proteobacteria in the pathogenesis of autism remains unclear, it is possible that alteration of commensal microbes could activate Proteobacteria as infectious agents for example through their ability to trigger immunological response.
Key phylotypes driving GM profiles of male and female BTBR mice
In order to identify GM key phylotypes responsible for differences between fBTBR and mBTBR compared to controls we applied LEfSe algorithm. 17/30 key genera were found within Bacteroidetes and Firmicutes phyla, mainly within Bacteroidales and Clostridiales orders, even though the comparison of the relative abundances of these phyla did not show significant differences (Fig. 1B). This result confirmed that Bacteroidetes and Firmicutes taxa reassortment mainly marks the differences in the GM between BTBR and C57 mice in both sexes accordingly to ANOSIM results on Unweighted Unifrac analysis (Fig. 1A). Alteration of Clostridial and Bacteroidial OTUs was found to drive the major changes in GM composition also in maternal immune activation (MIA) model of autism16. Among key genera with relative abundance >0.1%, Bacteroides and Parabacteroides (order Bacteroidales) were significantly more abundant in BTBR female and male compared to control mice (Table 1). Conversely, the genus Dehalobacterium (order Clostridiales) was significantly less abundant in BTBR mice of both sexes compared to controls (Table 1). Notably, the identified differences in relative abundance of Bacteroides and Parabacteroides were more pronounced in fBTBR as indicated by fold change (Table 1). Bacteroides and Parabacteroides genera are producers of propionic acid as endproduct of their metabolism and previous studies reported that intraventricular injection of propionate in rats induced pathologic changes characteristic of ASD33. Moreover these microorganisms are lipopolysaccharide producers, and high levels of serum enodotoxin were detected in autistic patients34. Finegold et al.8 also detected high levels of Bacteroides (Bacteroides vulgatus) in fecal samples of severely autistic children8.
Table 1. Relative abundance of key genera discriminating female and male BTBR from their sex-matched control mice.
| fBTBR | fC57 | p-value; Fold Change | mBTBR | mC57 | p-value; Fold Change | |
|---|---|---|---|---|---|---|
| Prevotella | 4.18 ± 1.70 | 1.50 ± 1.20 | 0.037; 2.8 | 1.95 ± 0.50 | 0.72 ± 0.39 | p > 0.05 |
| Bacteroides | 9.85 ± 3.53 | 0.93 ± 0.45 | 0.006; 10.5 | 6.94 ± 2.47 | 0.80 ± 0.16 | 0.004; 8.7 |
| Parabacteroides | 6.40 ± 1.43 | 0.79 ± 0.30 | 0.004; 8.1 | 4.02 ± 0.84 | 1.08 ± 0.36 | 0.025; 3.7 |
| AF12 (Rikenellaceae) | 0.31 ± 0.07 | 0.76 ± 0.07 | 0.006; 0.4 | 0.57 ± 0.18 | 0.71 ± 0.11 | p > 0.05 |
| Lactobacillus | 0.27 ± 0.08 | 0.11 ± 0.04 | p > 0.05 | 3.48 ± 2.10 | 0.15 ± 0.04 | 0.016; 22.7 |
| Dehalobacterium | 0.06 ± 0.02 | 0.20 ± 0.04 | 0.025; 0.3 | 0.11 ± 0.04 | 0.27 ± 0.04 | 0.025; 0.4 |
| Oscillospira | 5.39 ± 0.68 | 13.71 ± 1.76 | 0.010; 0.4 | 7.16 ± 1.75 | 8.88 ± 0.74 | p > 0.05 |
| Ruminococcus | 0.71 ± 0.17 | 1.11 ± 0.16 | p > 0.05 | 0.68 ± 0.18 | 1.19 ± 0.16 | 0.037; 0.6 |
| Coprobacillus | 0.13 ± 0.07 | 0 | 0.003; 244 | 0.09 ± 0.03 | 0.01 ± 0.01 | 0.014; 9 |
| Sutterella | 4.51 ± 1.05 | 1.07 ± 0.48 | 0.016; 4.2 | 3.01 ± 0.87 | 1.71 ± 0.66 | p > 0.05 |
| U. Desulfovibrionaceae | 2.78 ± 1.24 | 0.16 ± 0.12 | 0.010; 17.3 | 0.55 ± 0.22 | 0.63 ± 0.34 | p > 0.05 |
| Desulfovibrio | 0.39 ± 0.11 | 1.15 ± 0.44 | p > 0.05 | 0.11 ± 0.08 | 1.14 ± 0.34 | 0.006; 0.1 |
| U. Helicobacteraceae | 0.67 ± 0.57 | 0.05 ± 0.01 | p > 0.05 | 0.13 ± 0.03 | 0.03 ± 0.01 | 0.025; 4.2 |
| U. Enterobacteriaceae | 3.38 ± 1.84 | 0.03 ± 0.01 | 0.004; 109 | 1.66 ± 1.11 | 0.13 ± 0.07 | p > 0.05 |
| U. F16 (TM7) | 0.07 ± 0.04 | 0.29 ± 0.05 | 0.004; 0.2 | 0.07 ± 0.03 | 0.44 ± 0.17 | p > 0.05 |
| Akkermansia | 1.08 ± 0.32 | 0.35 ± 0.27 | 0.016; 3.1 | 3.90 ± 1.85 | 4.62 ± 2.96 | p > 0.05 |
Key genera were identified applying the metagenomic biomarker discovery approach of LEfSe and only genera with an LDA significant threshold >2 and relative abundance >0.1% in at least one group of mice, are shown. Fold change was expressed as ratio between the value of mean relative abundance of each genus in fBTBR and mBTBR groups and the value found in the sex-matched controls. Data are shown as average and SEM (n = 6/group).
Abbreviations: fBTBR (BTBR female mice); mBTBR (BTBR male mice); fC57 (C57 female mice); mC57 (C57 male mice).
Among the other key genera, Prevotella, Coprobacillus, Sutterella, Akkermansia (muciniphila) and unclassified members of Desulfovibrionaceae and Enterobacteriaceae significantly increased, while Oscillospira and members of Rikenellaceae and TM7 (AF12 and U. F16, respectively) significantly decreased in fBTBR, possibly driving a female-specific microbial signature in BTBR mice. Key genera specifically altered in mBTBR were Lactobacillus, Ruminococcus, Desulfovibrio and unclassified member of Helicobacteriaceae (Table 1). The alteration in the abundance of key genera that we mainly found in fBTBR was consistent, for a subset of specific taxa, with previously reported data both in human ASD patients and in a different murine model of ASD9,10,13,15. Genus Sutterella, within Proteobacteria, was found to be elevated both in feces of ASD children and in intestinal biopsies from children with autism and intestinal disturbances35,36. Furthermore, high levels of Akkermansia muciniphila were reported in BTBR fed a chow diet, while the ketogenic diet resulted in a normalization of its levels in association with improvement of behavioral symptoms37. Genus Oscillospira accounted for 13.7% in fC57 and was found significantly reduced to 5.4% in fBTBR. Oscillospira, recenlty identified by 16S RNA sequencing, is considered an enigmatic component of the human gut microbiota and, possibly based on butyrate production it was positively associated with human health38. Interestingly butyrate has recently been shown to improve repetitive behavior in BTBR mice39, thus it would be of interest to functionally characterize Oscillospira, and individual species in this genus, to examine their impact on autistic phenotype.
Correlations between GM alterations and behavioral phenotype, gut permeability and cytokine expression in female and male BTBR mice
Along with description of BTBR GM profile we investigated the possible correlation between the levels of specific bacterial taxa and peculiar pathological traits dysregulated in ASD patients such as behavioral abnormalities, gut permeability and immune abnormalities5,40.
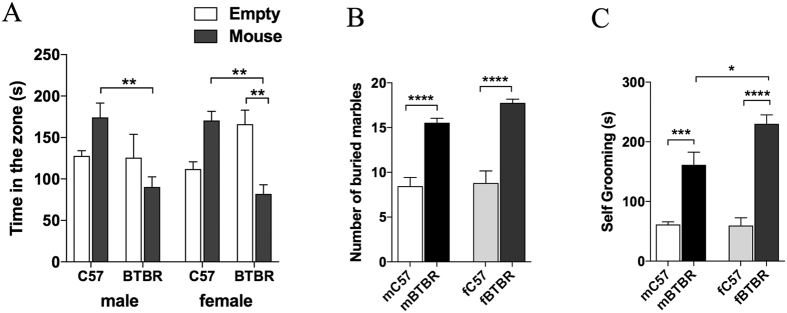
Three chamber paradigm, marble burying and spontaneous self-grooming tests were used to evaluate social, stereotyped and repetitive behavior in control and BTBR mice. For all behavioral tests, both female and male BTBR mice showed deficits compared to controls, however a significant sex-related alteration was observed with fBTBR displaying higher self-grooming scores (Fig. 2A–C).
Figure 2. Analysis of social interaction, stereotyped and repetitive behavior in female and male BTBR mice.
(A) Three-chamber social interaction test showing time spent in each chamber by BTBR and C57 mice of both sexes (n = 12/group; p = 0.7424 for chamber, p = 0.0077 for strain, p = 0.7761 for sex, p < 0.0001 for chamber x strain and p = 0.1671 for chamber x strain x sex, by three-way ANOVA). (B) Number of buried marble by BTBR and C57 mice of both sexes after a 30 min testing session (n = 12/group; p < 0.0001 for strain, p = 0.1322 for sex and p = 0.2689 for strain x sex, by two-way ANOVA). (C) Seconds spent in repetitive grooming measured for BTBR and C57 mice of both sexes during 10 min test session (n = 12/group; p < 0.0001 for strain, p = 0.0372 for sex, and p = 0.0269 for strain x sex, by two-way ANOVA). Significant differences are indicated by *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 using Bonferroni post-hoc tests following three-way ANOVA with chamber, strain and sex as factors (A) or two-way ANOVA with strain and sex as factors (B and C). Abbreviations: fBTBR (BTBR female mice); mBTBR (BTBR male mice); fC57 (C57 female mice); mC57 (C57 male mice). Data are shown as mean values ± SEM.
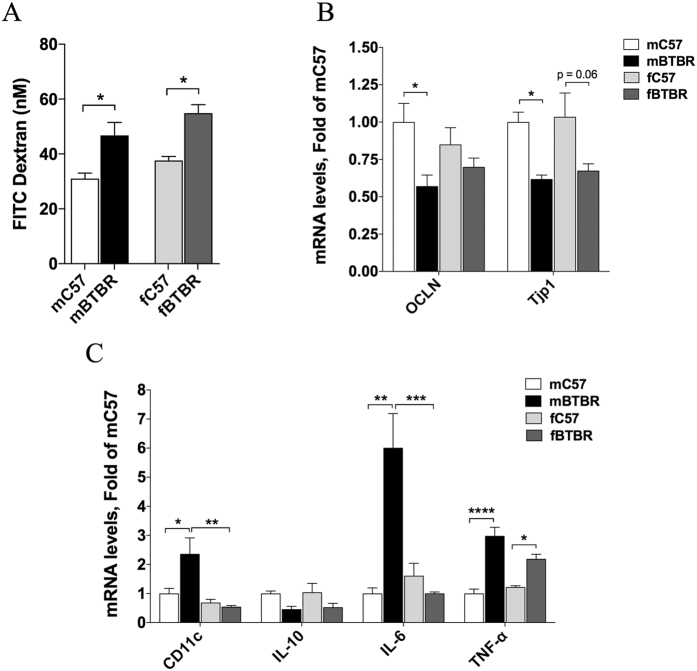
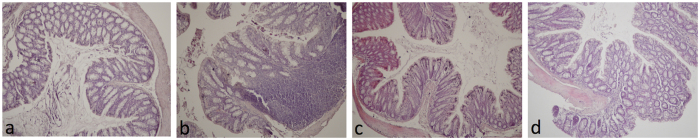
Alteration of gut permeability, as evidenced by increased FITC-dextran translocation across the intestinal epithelium into blood, was observed both in female and male BTBR mice (Fig. 3A). Consistent with deficit in intestinal barrier integrity, a significant reduction of occludin and zonuline mRNA levels was detected in colon of male BTBR mice and a similar trend was observed in females (Fig. 3B). In addition, expression of a subset of cytokines (TNF alpha, IL-6 and IL-10) and CD11c integrin was determined in colon tissue of male and female BTBR and C57 mice. Increased expression of TNF alpha was observed in BTBR mice of both sexes (Fig. 3C). Significant increase of IL-6 and CD11c was observed in mBTBR compared to both sex-mached controls and fBTBR (Fig. 3C). These results indicate that, although with different marks, BTBR mice of both sexes present an increased gut permeability and altered cytokines pattern in colon tissue. In addition, histological evaluation of colon tissues showed tissue damage and evident inflammatory cells infiltration in both mBTBR and fBTBR (Fig. 4).
Figure 3. Analysis of intestinal integrity and inflammation levels in female and male BTBR mice.
(A) Intestinal epithelial permeability to fluorescein isothiocyanate (FITC)-dextran 4 kDa of BTBR and C57 mice of both sexes. Data are represented as plasma concentration of FITC dextran (nM) (n = 5/group; p = 0.0002 for strain, p = 0.0469 for sex and p = 0.8187 for strain x sex, by two-way ANOVA). (B) Colon occludin (OCLN) and zonuline-1 (Tjp1), gene expression normalized to GAPDH gene in BTBR and C57 mice of both sexes. Data were normalized to mC57 control (n = 6/group; OCLN: p = 0.0067 for strain, p = 0.9105 for sex and p = 0.1623 for strain x sex; Tjp1: p = 0.0004 for strain, p = 0.6202 for sex and p = 0.9105 for strain x sex, by two-way ANOVA). (C) Colon mRNA levels of inflammatory markers (CD11c, IL-10, IL-6 and TNF-alpha) normalized to GAPDH in BTBR and C57 mice of both sexes. Data for each gene were normalized to mC57 controls (n = 6/group; CD11c: p = 0.0529 for strain, p = 0.0018 for sex and p = 0.019 for strain x sex; IL-10: p = 0.0041 for strain, p = 0.7827 for sex and p = 0.9739 for strain x sex; IL-6: p = 0.0106 for strain, p = 0.0107 for sex and p = 0.002 for strain x sex; TNF-alpha: p < 0.0001 for strain, p = 0.2196 for sex and p = 0.0354 for strain x sex, by two-way ANOVA). In bar charts all data are expressed as mean values ± SEM; significant differences are indicated by *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001; near-significant differences are also reported (Bonferroni post-hoc tests following two-way ANOVA with strain and sex as factors). Abbreviations: fBTBR (BTBR female mice); mBTBR (BTBR male mice); fC57 (C57 female mice); mC57 (C57 male mice).
Figure 4. Histological evaluation of colon inflammatory cell infiltration in female and male BTBR mice.
Representative hematoxylin and eosin–stained sections from colon tissues of mice. (a) Colon tissue from mC57 mice showing absence of inflammatory cells. (b) Colon tissue of mBTBR group showing leukocyte infiltration in the mucosa and submucosa. (c) Colon tissue of fC57 group, showing absence of inflammatory cells. (d) Colon tissue from fBTBR group showing moderate leukocyte infiltration in the mucosa. Original magnification 10x. Histological evaluation of inflammatory cells infiltration was scored along the entire colon length, inspecting the colon mucosa, submucosa and transmural areas considering the following parametres: leukocyte density (mC57 = 0.33 ± 0.58, mBTBR = 2 ± 1, fC57 = 0, fBTBR = 2.67 ± 0.58) and expansion of leukocyte infiltration (mC57 = 0.33 ± 0.58, mBTBR = 1.33 ± 0.58, fC57 = 0, fBTBR = 1.67 ± 0.58). The histologic scoring system is reported in Material and Methods section. Data reported as mean ± SD, n = 3/group.
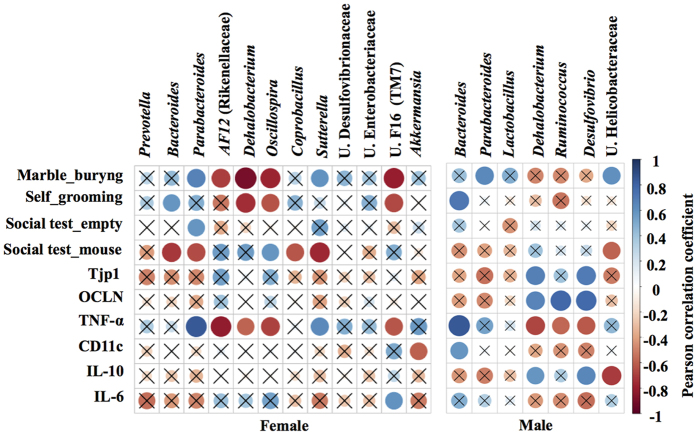
Pearson correlation was applied to correlate the abundance of key genera, that discriminated fBTBR and mBTBR from sex-matched controls, with behavioral tests, colon mRNA expression of occludin, zonuline and immune-markers (Fig. 5). In particular, increase of Parabacteroides and Sutterella, together with decrease of Dehalobacterium, Oscillospira and unclassified member of TM7 were strongly associated to altered behavior and TNF-alpha expression in fBTBR. Onore and collegues (2013) already reported a relationship between the increase in inflammation levels and repetitive grooming behavior in BTBR mice41. Here we add the observation that remodeling of gut microbiota composition by reassortment of specific bacterial genera, may contribute to the immune dysregulation and possibly to the altered behavior of BTBR mice. Unclassified members of Helicobacteriaceae associated with abnormal behavior and low IL-10 expression in mBTBR. Finally, in mBTBR mice lower levels of Dehalobacterium, Ruminococcus and Desulfovibrio were associated with increased gut permeability (Fig. 5).
Figure 5. Correlogram showing the Pearson’s correlation between key genera and behavioral scores, gut integrity and immune-markers in BTBR and C57 mice of both sexes.
Blue circles designate a positive correlation while red ones designate a negative correlation. With X are barred no significant results according to the significance level of 0.05.
Conclusions
Results showed that BTBR mice of both sexes presented a marked intestinal dysbiosis with respect to prosocial C57 strain. Bacteroides, Parabacteroides, Sutterella, Dehalobacterium and Oscillospira genera were identified as key drivers of sex-specific GM profiles associated with altered behavior, increased gut permeability and colon proinflammatory state. Our analysis put in evidence that definition of sex specific signatures of GM should be considered in assessing the role of GM in gut-brain axis connection. In addition, our data indicate that, despite the difficulty and limits in the use of animal models to translate human disease, BTBR mouse model is a mimic, in some relevant aspects, of autistic phenotype. Noteworthy, BTBR mice, as example of idiopathic autism, could be considered an animal model useful to investigate whether restoring GM balance may ameliorate pathological traits. A description that takes into account sex differences in GM profiles and relevant pathological traits characterizing the autistic-like phenotype, could be important for future studies aimed to translate animal findings to human diseases.
Material and Methods
Animals
All procedures involving animals and their care were conducted in conformity with international and national law and policies (EU Directive 2010/63/EU for animal experiments, ARRIVE guidelines and the Basel declaration including the 3R concept). The procedures reported here were approved by the Institutional Committee on the Ethics of Animal Experiments (CSV) of the University of Naples “Federico II” and by the Ministero della Salute under protocol no. 0022569-P-20/12/2010. Before killing and prior to serum and sample collection, animals, kept overnight fasted, were anesthetized by enflurane and euthanized by an intraperitoneal injection of a cocktail of ketamine/xylazine. As suggested by the animal welfare protocol, all efforts were made to minimize animal suffering and to use only the number of animals necessary to produce reliable scientific data. C57Bl/6J (C57) and BTBR T + tf/J (BTBR) inbred strains of mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and a colony was established and maintained. For the study, 24 fully symptomatic BTBR mice (12 months of age, 12 for gender) and 24 C57 control mice (12 months of age, 12 for gender) were housed in the same room under standard 12-h light/12-h dark cycle with free access to water and standard laboratory chow diet. Mice were from different litters and housed by gender separately 2/3 per cage.
Marble burying test
Male and female BTBR and C57 (n = 12 each group) were individually placed in a plastic container (46 cm long by 24 cm wide by 21 cm deep) with 3 cm of clean woodchip bedding (Northeastern Products, NY) where 20 glass marbles (1.5 cm in diameter) were placed on top of clean bedding, arranged in five rows of four. Once a mouse was gently allocated into the test container, a wire lid was placed on top and mouse is allowed to freely explore the cage. After 30 minutes, each mouse was removed from the testing container and replaced to their home cage. When a threshold of 75% coverage for each marble was observed, it was considered buried and recorded. After the test, the marbles were thoroughly cleaned and new bedding was used for each mouse.
Spontaneous self-grooming behavior
The same subjects tested in marble burying test were also examined for grooming behavior. Mice were individually allocated in an empty plastic cage (28 cm wide × 17 cm long × 12 cm high) in a separate room for a total of 20 minutes where animals were allowed to freely explore for 20 minutes. The first 10 min served as a habituation period while during the second phase of 10 min, the cumulative period that mice spent in grooming was manual scored by a trained observer. Grooming behavior included head washing, body grooming, genital/tail grooming and paw and leg licking. After the test the cage was thoroughly cleaned.
Social approach testing
The same subjects tested in marble burying and self-grooming tests were also examined for social approach behavior. Social approach behavior was tested in a three-chambered apparatus, using methods previously described18,19. The apparatus (60 × 40 cm) has two doorways that divide it into a three chambers apparatus (20 × 40 cm each). Number of entries and time spent in each chamber were automatically detected by a videocamera coupled with a video-tracking software (Any-maze, Stoelting). The sociability test was preceded by 5-min habituation session where each mouse is restrained in the center of the middle chamber. After these phases, a novel sex, strain and age matched mouse (not used in later testing and previously habituated) is placed in one side of the chamber under an enclose cup while the other side contained an empty cup. During this sociability phase, walls between the compartments are removed and the tendency to approach a novel mouse is compared with tendency to approach a novel object. Each mouse was free to explore all three chambers for 10 minutes and both sides were alternated between the left and right chambers across subjects.
In vivo intestinal permeability assay
In vivo intestinal permeability assay was performed for a subset of mice using fluorescein isothiocyanate-labeled dextran (FITC-dextran) method, as previously described42. Briefly, food and water were withdrawn for 6 h and BTBR T + tf/J and C57Bl/6 J female and male mice (n = 5, each group) were administrated by gavage with FITC labeled dextran 4000 (Sigma-Aldrich, Milan, Italy), as permeability tracer (60 mg/100 g body weight). After 24 hours blood of all animals was collected by intracardiac puncture and centrifuged (3000 rpm for 15 min at RT). Then plasma FITC-dextran concentration was determined (excitation, 485 nm; emission, 535 nm; HTS-7000 Plus-plate-reader; Perkin Elmer, Wellesley, MA, USA), using a standard curve generated by serial dilution of the tracer.
Histological Analysis
Samples of colon from male and female mice of both strains (n = 3) were removed, washed and then fixed in paraformaldehyde (4% v/v; Carlo Erba, Italy) for 12 hours. Samples were dehydrated, embedded in paraffin and cut into 5 μm thick sections before being stained with hematoxylin-eosin (H&E; Carlo Erba, Italy). Images were obtained by a Leica DFC320 video camera (Leica, Milan, Italy) connected to a Leica DM RB microscope using the Leica Application Suite software V2.4.0. Sections were examined and scored using a 0-to-4 scale in a blinded fashion. The histologic scoring system was as follows: a) The severity of inflammatory cell infiltration was evaluated by percentage of leukocyte density in lamina propria area and estimated in a high-power field (HPF) representative of the section (0 for no signs of inflammation, 1 for minimal <10%, 2 for mild 10–25% with scattered neutrophils, 3 for moderate 26–50%, 4 for marked >51% with dense infiltrate); b) The extent of the inflammation was estimated as expansion of leukocyte infiltration (0 for none, 1 for mucosal, 2 for mucosal and submucosal and 3 for mucosal, submucosal and transmural level).
Quantification of gene expression using RT-PCR
Colon samples were collected and immediately frozen at −80 °C until use for RNA extraction. Total RNA was extracted from colon tissues using TRIzol Reagent (Bio-Rad Laboratories), according to the manufacturer’s instructions. cDNA was synthesized using a reverse transcription kit (NucleoSpin®, MACHEREY-NAGEL GmbH & Co, Düren, Germany) from 4 μg total RNA. PCRs were performed with a Bio-Rad CFX96 Connect Real-time PCR System instrument and software (Bio-Rad Laboratories). The PCR conditions were 15 min at 95 °C followed by 40 cycles of two-step PCR denaturation at 94 °C for 15 s, annealing at 55 °C for 30 s and extension at 72 °C for 30 s. Each sample contained 20 ng cDNA in 2X QuantiTect SYBRGreen PCR Master Mix and primers pairs to amplify zonuline-1 (Tjp1), occludin (Ocln), TNF-α (Tnf), interleukin-6 (Il6), interleukin-10 (Il10), CD11c (Itgax) (Qiagen, Hilden, Germany), in a final volume of 50 μl. The relative amount of each studied mRNA was normalized to GAPDH as housekeeping gene, and data were analyzed according to the 2−ΔΔCT method.
Microbial DNA extraction, 16S ribosomal DNA (rDNA) library preparation and sequencing
Freshly evacuated fecal pellets were kept directly in a sterile microtube one day before the sacrifice of mice and stored at −80 °C until assayed. Bacterial genomic DNA was extracted from frozen fecal samples using the QIAamp DNA Stool Mini Kit (Qiagen) according to manufacturer’s instructions. DNA concentration was measured fluorometrically using Qubit dsDNA BR assay kit (Invitrogen) and quality was assessed by spectrophotometric measurements with NanoDrop (ThermoFisher Scientific Inc). Samples were stored at −20 °C until processed for amplification. It is well documented that various compartments of the gastro-intestinal tract harbour different bacterial populations. We chose to analyze readly accessible fecal samples for gut microbiome analyses mainly because fairly representative of the whole gastro-intestinal tract, with exception of some surface-adherent bacterial species. Sequencing samples were prepared according to the protocol 16S Metagenomic Sequencing Library Preparation for Illumina Miseq System with some modifications. The V3–V4 regions of the 16S rDNA gene were amplified starting from 200 ng of DNA template in a reaction volume of 50 μL containg 1x Fast start High Fidelity Reaction Buffer, 5 μM of each primer, 0.2 nM of dNTPs, 3 mM MgCl2, and 2 U FastStart High Fidelity PCR System (Roche Applied Science). PCR was performed using the following cycles conditions: an initial denaturation step at 95 °C for 2 min, followed by 30 cycles of 95 °C for 30 s, 55 °C for 45 s, 72 °C for 55 s and ended with an extension step at 72 °C for 5 minutes; products were visualized by electrophoresis on 1.2% agarose gel. After a purification step with Agencourt AMPure XP (Beckman Coulter Inc), the amplicons were indexed with 10 subsequent cycles of PCR using the Nextera XT Index Kit (Illumina). Each PCR reaction contained 10 μL of amplicons from first PCR, 5 μl index 1 primer (N7xx), 5 μl index 2 primer (S5xx), 5 μl 1x Fast start High Fidelity Reaction Buffer, 6 μL MgCl2 (3 mM), 1 μL dNTPs (0.2 nM), 0.4 μL FastStart High Fidelity PCR System (2U) and 17.6 μl PCR grade water. PCRs were carried out, visualized using gel electrophoresis and subsequently cleaned as described above. Library sizes were assessed using a Bioanalyzer DNA 1000 chip (Agilent technologies) and quantified with Qubit. Normalized libraries were pooled, denatured with NaOH, then diluted to 10pM and combined with 25% (v/v) denatured 10pM PhiX, according to Illumina guidelines. Sequencing run was performed on an Illumina Miseq system using v3 reangents for 2 × 281 cycles.
Sequencing data analysis
V3-V4 16S rDNA FASTQ paired-end reads were quality filtered and assembled using PEAR43. Only sequences showing average PHRED score ≥30, read length between 400 and 500 bp and overlapping regions between mate-pair end of at least 40 nucleotides were retained in this step. Passing filter sequences were then processed with PRINSEQ44 in order to obtain FASTA and quality files for further analyses. Metagenomic analyses on the resulting data were conducted using Quantitative Insights Into Microbial Ecology (QIIME, version 1.8.0)45. 16S sequences were used to pick OTUs at 97% of sequences similarity from Greengenes 16S gene database (GG, may 2013 version)46 with a closed reference-based OTU picking method. The GG database was used to taxonomically classify the identified OTUs and to compute their distribution across different taxonomic levels. To avoid sample size biases in subsequent alpha and beta diversity analyses, a sequence rarefaction procedure was applied using a maximum depth of 32,228 sequences/sample.
To assess sampling depth coverage and species heterogeneity in each sample, alpha diversity metrics were employed on rarefied OTU table using Good’s coverage, Observed species and Shannon’s diversity index. A two-sample permutation t-test, using 999 Monte Carlo permutations to compute p-value, was performed to compare the alpha diversities between sample groups. OTUs diversity among sample communities (beta diversity) was assessed by applying unweighted Unifrac distances. Statistical significance of beta diversities was assessed on unweighted UniFrac distances matrixes using ANOSIM method47 with 999 permutations. Statistical differences in OTUs frequencies across sample groups at different taxonomic levels were assessed using nonparametric Kruskal-Wallis test.
Next, two analyses were applied on OTU tables generated by QIIME to identify key OTUs that discriminate female and male BTBR mice from their respective controls: Metastats comparison using the online interfaces48 and LDA Effect Size analysis (LEfSe)49. Only those OTUs reported by both methods to be significantly different between the two groups (p < 0.05 for Metastats, LDA > 2 and p < 0.05 for LEfSe) have been considered as key discriminatory OTUs. Key genera that discriminate female and male BTBR mice from their respective controls were identified applying only LEfSe.
Other statistical methods
Marble buring, self-grooming, plasma FITC-dextran, gene expression data were analyzed by two-way ANOVA with strain and sex as factors; social approach behavior results were analyzed by three-way ANOVA with chamber, strain and sex as factors. Multiple comparisons were performed using Bonferroni’s post-hoc test. Pearson correlation test was used to assess the eventual relationship between the amount of key genera and behavioral scores, intestinal permeability and inflammation.
In this study results were considered statistically significant at p-value <0.05. Significant differences were indicated in figures by *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ANOSIM and permutation t-test were performed using QIIME scripts, all other analyses were performed using R 3.2.050. Bar plots were created by using GraphPad Prism 6.0.
Additional Information
How to cite this article: Coretti, L. et al. Sex-related alterations of gut microbiota composition in the BTBR mouse model of autism spectrum disorder. Sci. Rep. 7, 45356; doi: 10.1038/srep45356 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was partially funded by Epigen Flagship Project C.N.R. and by Regione Campania, l.5 to LCh. We wish to thank Prof. Francesca Borrelli and Dr Giuseppe Piegari for their help in the interpretation of histological analyses.
Footnotes
The authors declare no competing financial interests.
Author Contributions L.Co. performed the sequencing experiments, analyzed metagenomic data and wrote the manuscript. C.C. performed behavioral tests, permeability study and histological experiments, collected samples and analyzed the data. A.L. performed the mRNA expression experiments. E.F., G.S., S.K. and M.C. participated to the experiments and the analysis of data. O.P. analyzed and interpreted histologic experiments. R.R., R.P., G.M.R., R.M., S.C., and A.C. contributed to the data analysis and interpretation. L.Ch. and F.L. analyzed data and wrote the main manuscript text. All authors reviewed the manuscript.
References
- Cryan J. F. & Dinan T. G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 13, 701–12 (2012). [DOI] [PubMed] [Google Scholar]
- El Aidy S., Stilling R., Dinan T. G. & Cryan J. F. Microbiome to Brain: Unravelling the Multidirectional Axes of Communication. Adv Exp Med Biol. 874, 301–36 (2016). [DOI] [PubMed] [Google Scholar]
- Cámara R. J. A., Ziegler R., Begré S., Schoepfer A. M. & Von Känel R. The role of psychological stress in inflammatory bowel disease: Quality assessment of methods of 18 prospective studies and suggestions for future research. Digestion. 80, 129–139 (2009). [DOI] [PubMed] [Google Scholar]
- Mawdsley J. E. & Rampton D. S. The role of psychological stress in inflammatory bowel disease. Neuroimmunomodulation. 13, 327–36 (2006). [DOI] [PubMed] [Google Scholar]
- de Magistris L. et al. Alterations of the intestinal barrier in patients with autism spectrum disorders and in their first-degree relatives. J Pediatr Gastroenterol Nutr. 51, 418–424 (2010). [DOI] [PubMed] [Google Scholar]
- Sandler R. H. et al. Short-term benefit from oral vancomycin treatment of regressive-onset autism. J Child Neurol. 15, 429–435 (2000). [DOI] [PubMed] [Google Scholar]
- Parracho H. M. R. T. et al. A double-blind, placebo-controlled, crossover-designed probiotic feeding study in children diagnosed with autistic spectrum disorders. Int J Probiotics Prebiotics. 5, 69–74 (2000). [Google Scholar]
- Finegold S. M. et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 16, 444–453 (2010). [DOI] [PubMed] [Google Scholar]
- Williams B. L. et al. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One. 6(9), e24585 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis M. et al. Fecal Microbiota and Metabolome of Children with Autism and Pervasive Developmental Disorder Not Otherwise Specified. PLoS One. 8(10), e76993 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parracho H. M., Bingham M. O., Gibson G. R. & McCartney A. L. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J Med Microbiol. 54, 987–991(2005). [DOI] [PubMed] [Google Scholar]
- Song Y., Liu C. & Finegold S. M. Real-time PCR quantitation of clostridia in feces of autistic children. Appl Environ Microbiol. 70, 6459–6465 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl Environ Microbiol. 77, 6718–6721 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D. W. et al. Reduced Incidence of Prevotella and Other Fermenters in Intestinal Microflora of Autistic Children. PLoS One. 8(7), e68322 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Theije C. G. M. et al. Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain Behav Immun. 37, 197–206 (2014). [DOI] [PubMed] [Google Scholar]
- Hsiao E. Y. et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 155, 1451–1463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley J. N. Translational animal models of autism and neurodevelopmental disorders. Dialogues Clin Neurosci. 14(3), 293–305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy S. S. et al. Mouse behavioral tasks relevant to autism: Phenotypes of 10 inbred strains. Behav Brain Res. 176, 4–20 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane H. G. et al. Autism-like behavioral phenotypes in BTBR T + tf/J mice. Genes, Brain Behav. 7, 152–163 (2008). [DOI] [PubMed] [Google Scholar]
- Scattoni M. L., Gandhy S. U., Ricceri L. & Crawley J. N. Unusual repertoire of vocalizations in the BTBR T + tf/J mouse model of autism. PLoS One. 3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. L. et al. Motor and cognitive stereotypies in the BTBR T + tf/J mouse model of autism. Genes, Brain Behav. 10, 228–235 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino G. et al. Peroxisome proliferator-activated receptor alpha plays a crucial role in behavioral repetition and cognitive flexibility in mice. Mol Metab. 4, 528–536 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobbe R. L. et al. Expression of social behaviors of C57BL/6J versus BTBR inbred mouse strains in the visible burrow system. Behav Brain Res. 214(2), 443–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbrod A. et al. Automated long-term tracking and social behavioural phenotyping of animal colonies within a semi-natural environment. Nat Commun. 4, 2018 (2013). [DOI] [PubMed] [Google Scholar]
- Karvat G. & Kimchi T. Acetylcholine elevation relieves cognitive rigidity and social deficiency in a mouse model of autism. Neuropsychopharmacology. 39(4), 831–40 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodeo D. A., Jones J. H., Sweeney J. A. & Ragozzino M. E. Differences in BTBR T + tf/J and C57BL/6J mice on probabilistic reversal learning and stereotyped behaviors. Behav Brain Res. 227(1), 64–72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvat G. & Kimchi T. Systematic autistic-like behavioral phenotyping of 4 mouse strains using a novel wheel-running assay. Behav Brain Res. 233(2), 405–14 (2012). [DOI] [PubMed] [Google Scholar]
- Stephenson D. T. et al. Histopathologic characterization of the BTBR mouse model of autistic-like behavior reveals selective changes in neurodevelopmental proteins and adult hippocampal neurogenesis. Mol Autism. 2(1), 7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scattoni M. L., Martire A., Cartocci G., Ferrante A. & Ricceri L. Reduced social interaction, behavioural flexibility and BDNF signalling in the BTBR T + tf/J strain, a mouse model of autism. Behav Brain Res. 251, 35–40 (2013). [DOI] [PubMed] [Google Scholar]
- Heo Y., Zhang Y., Gao D., Miller V. M. & Lawrence D. A. Aberrant immune responses in a mouse with behavioral disorders. PLoS One. 6(7), e20912 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokras N. & Dalla C. Sex differences in animal models of psychiatric disorders. Br J Pharmacol. 171, 4595–4619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jašarević E., Morrison K. E. & Bale T. L. Sex differences in the gut microbiome-brain axis across the lifespan. Philos Trans R Soc Lond B Biol Sci. 371(1688), 20150122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFabe D. F. et al. Neurobiological effects of intraventricular propionic acid in rats: Possible role of short chain fatty acids on the pathogenesis and characteristics of autism spectrum disorders. Behav Brain Res. 176, 149–169 (2007). [DOI] [PubMed] [Google Scholar]
- Emanuele E. Low-grade endotoxemia in patients with severe autism. Neurosci Lett 471, 162–165 (2010). [DOI] [PubMed] [Google Scholar]
- Wang L. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol Autism. 4, 42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. L., Hornig M., Parekh T. & Ian Lipkin W. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. MBio. 3, 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell C. et al. Ketogenic diet modifies the gut microbiota in a murine model of autism spectrum disorder. Mol Autism. 7(1), 37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konikoff T. & Gophna U. Oscillospira: a Central, Enigmatic Component of the Human Gut Microbiota. Trends Microbiol. 24(7), 523–4 (2016). [DOI] [PubMed] [Google Scholar]
- Kratsman N., Getselter D. & Elliott E. Sodium butyrate attenuates social behavior deficits and modifies the transcription of inhibitory/excitatory genes in the frontal cortex of an autism model. Neuropharmacology. 102, 136–45 (2016) [DOI] [PubMed] [Google Scholar]
- Estes M. L. & McAllister A. K. Immune mediators in the brain and peripheral tissues in autism spectrum disorder. Nat Rev Neurosci. 16, 469–486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onore C. E. et al. Inflammatory macrophage phenotype in BTBR T + tf/J mice. Front Neurosci. 7, 158 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani P. D. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 58, 1091–1103 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kobert K., Flouri T. & Stamatakis A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 30, 614–620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R. & Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 27, 863–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 7, 335–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 72, 5069–5072 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K. R. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 18, 117–143 (1993). [Google Scholar]
- White J. R., Nagarajan N. & Pop M. Statistical Methods for Detecting Differentially Abundant Features in Clinical Metagenomic Samples. PLoS Comput Biol 5(4), e1000352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N. Metagenomic biomarker discovery and explanation. Genome Biol. 12(6), R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- R. Core Team R: A Language and Environment for Statistical Computing. R Found Stat Comput Vienna Austria 0:{ISBN} 3–900051–07–0 (2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.