Abstract
Identifying and characterizing natural products and synthetic small molecules that inhibit biochemical processes such as ribosomal translation can lead to novel sources of molecular probes and therapeutics. The search for new antibiotics has been invigorated by the increasing burden of drug-resistant bacteria and has identified many clinically essential prokaryote-specific ribosome inhibitors. However, the current cohort of antibiotics is limited with regards to bacterial resistance mechanisms because of structural similarity within classes. From a high-throughput screen for translation inhibitors, we discovered a new compound, T6102, which inhibits bacterial protein synthesis in vitro, inhibits bacterial growth of Bacillus subtilis in vivo, and has a chemical structure that appears to be unique among known classes of translation-inhibiting antibiotics. T6102’s unique structure compared to current clinically-utilized antibiotics makes it an exciting new candidate for the development of next-generation antibiotics.
Keywords: novel antibiotic, translation inhibition, high-throughput screening, drug discovery, adamantane
Introduction
The advent of antibiotic use in the 1940’s dramatically improved infectious disease outcomes, preventing countless complications and deaths over the past seven decades. Nevertheless, the overuse of antibiotics in modern medicine, livestock, and agriculture has allowed for the expansion of drug-resistant strains, limiting the clinical efficacy of many drugs and instigating the augmenting global threat of antibiotic-resistant outbreaks [1].
Resistance to one antibiotic often severely limits the clinical utility of the entire class of structurally similar drugs. Examples include the expression of beta-lactamase in resistance to penicillins, expression of export proteins in resistance to tetracyclines, and mutation of ribosomal proteins in resistance to aminoglycosides like streptomycin [2]. Clinically, antibiotic repertoires are severely bound by the limited number of antibiotic structural classes as well as the limited number of molecular targets like prokaryotic ribosomes and bacterial cell wall enzymes [3]. Finding new structural or target-based classes of antibiotics that act by novel mechanisms is one strategy to combat the persistence of drug-resistance in the clinic.
In this study, we discuss a small molecule with translation-inhibiting properties. It was chosen for further study among other hits from high-throughput screening because of a structure that is unlike any known class of antibiotics. The compound, T6102, has a hydrophobic adamantane group and is reminiscent of the structures of current drugs used for neurological disease, diabetes, and viral infections, such as amantadine and rimantadine, as well as of investigational drugs such as the antitumor compound 2, 2-Bis (4-(4-amino-3-hydroxyphenoxy) phenyl) adamantine [4]. Adamantane structures are increasingly utilized in medicinal chemistry due to their versatility as rigid scaffolds, steric bulk for protecting intramolecular reactions, and lipophilic groups to increase partition coefficients [5]. Adamantane derivatives, due to their hydrophobicity, have also been used as substituents in inhibitor design to bind to hydrophobic clefts in protein targets [6]. T6102, for example, was originally synthesized as a precursor for compounds used for hydrophobic tagging of target proteins to induce proteasomal degradation [7,8].
As far as antibiotics go, there have been reports of a few potential novel antimicrobials with adamantane moieties including antimycobacterial compounds (SQ109, N’-(Adamantan-2-ylidene)thiophene-2-carbohydrazide, 17, 4-(adamantan-1-yl)quinoline, substituted 5-(1-adamantyl)-1,2,4-triazole-3-thiols) [9-12] and antimalarial compounds (adamantyl dihydroartemisinin) [13]. However, unlike T6102, most of these involve derivatization of current antimicrobial structures. The antimycobacterial compound SQ109, for example, was identified in a screen for compounds with activity against tuberculosis from a library of derivatives of ethambutol, an antibiotic that is currently one of the first line treatments against tuberculosis infections [5]. While T6102, like these investigational drugs, also contains an adamantane group, T6102 is unique in that the rest of its structure does not resemble any known class of antibiotics.
There is clearly a dire need for novel, structurally unique antibiotics that interact with a variety of potential targets by diverse mechanisms. Here we report the discovery and synthesis of a new compound, T6102, which inhibits bacterial processes both in vitro and in vivo (in Bacillus subtilis) and may be a useful starting point for the development of structurally novel antibiotics.
Results
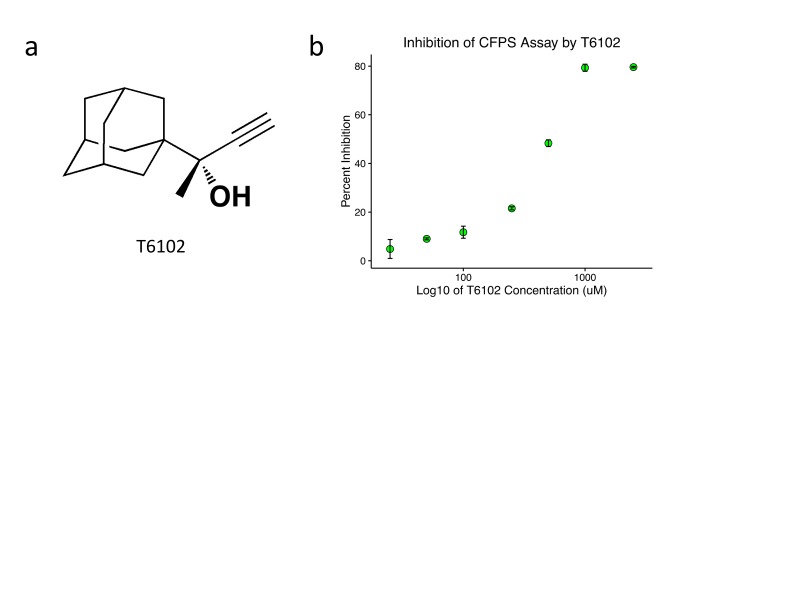
In order to identify small molecule inhibitors of prokaryotic translation, we developed an economical, high-throughput, Escherichia coli lysate-based, cell-free protein synthesis screen for luciferase synthesis inhibition with a kanamycin positive control (Figure 1). 872 crude extracts of natural products from the Yale Rainforest Natural Product Collection [14] and 286 synthetic compounds from the Yale Compound Repository (Yale Center for Molecular Discovery, New Haven, CT) were screened. Among synthetic assay hits, including some that resembled currently utilized antibiotics (Figure 2), T6102 (Figure 3a) was the most structurally unique compared to known classes of antibiotics and was chosen for further analysis. We synthesized T6102 for additional testing of inhibition of the cell-free protein synthesis reaction, and this consistently revealed dose-dependent inhibition as assayed by luciferase activity, with an IC50 of 453.7 +/- 39.3 uM (Figure 3b).
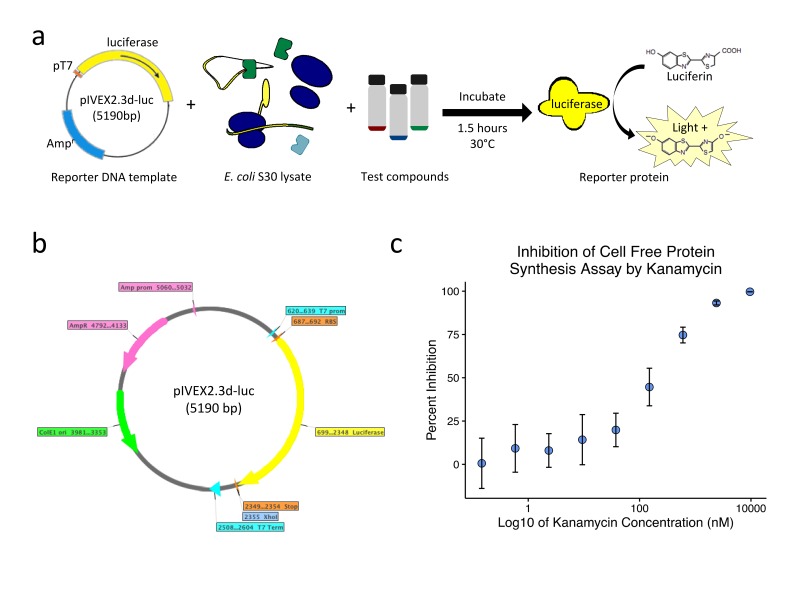
Figure 1.
High-throughput screen to identify small molecule inhibitors of prokaryotic protein synthesis. (a) Overview of the economical, high-throughput bacterial lysate-based cell-free protein synthesis reaction. Shown here is a circular DNA template containing luciferase downstream of a T7 promoter (pT7). T7 RNA polymerase was spiked into bacterial lysate, and the reaction was incubated with test compounds for 1.5 hours before the addition of luciferin for measurement of the luciferase signal. (b) Map of plasmid pIVEX2.3d-luc used for a luciferase template. (c) Dose-dependent sensitivity of the protein synthesis inhibition reaction when incubated with varying concentrations of kanamycin. 1.2 uM of kanamycin was found to sufficiently inhibit the reaction and was used to terminate all subsequent reactions at 1.5 hours. Error bars represent the standard deviation for each condition. Abbreviations: Prom - Promoter, Term - Terminator, RBS - Ribosome binding sequence (AGGAGA), Stop - Tandem stop codons (TAATAA), Ori - Origin, Amp - Ampicillin, R - Resistance
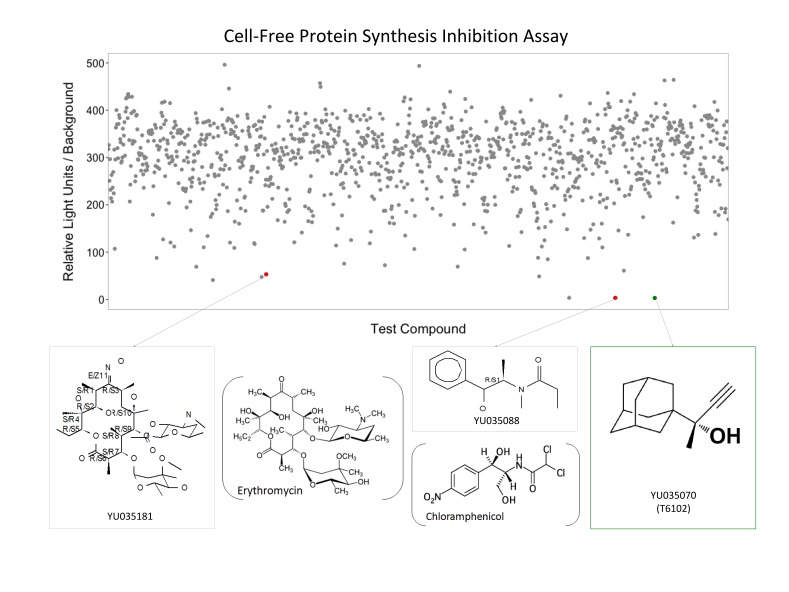
Figure 2.
High throughput screening identifies protein synthesis inhibitors. Raw data of luciferase activity are plotted over the 1,158 compounds tested. Structures of selected hits (colored dots) are shown below. Red dots show compounds (gray boxes) that resemble the antibiotics erythromycin (left) and chloramphenicol (middle). The green box (right) identifies T6102 with a structure unlike any known class of translation inhibitors.
Figure 3.
T6102 exhibits dose-dependent inhibition of protein synthesis. (a) Chemical structure of T6102. (b) Percent inhibition of the Cell Free Protein Synthesis (CFPS) assay, from a circular DNA template and measured by luciferase activity, is plotted against the log (base 10) of the concentration of T6102 incubated in the protein synthesis reaction. Error bars represent the standard deviation for each condition.
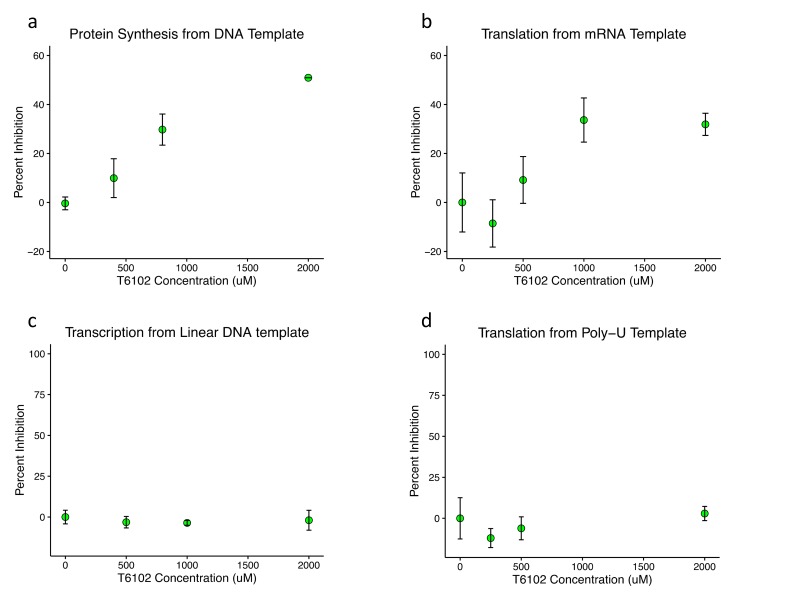
The cell-free protein synthesis system used for this assay was modified from Jewett et al, 2008 [15] and involves four major reactions: transcription, translation, energy regeneration, and aminoacyl-tRNA charging, in addition to luciferase activity. In order to investigate the mechanism of protein synthesis inhibition by T6102, inhibition of the cell free protein synthesis reaction was assayed by incorporation of radiolabeled amino acids with different templates. T6102 exhibited dose-dependent inhibition of protein synthesis from a circular luciferase DNA template (Figure 4a), further validating the initial hit and suggesting that T6102 inhibits protein synthesis rather than primarily inhibiting luciferase activity. When the same reaction was incubated with luciferase mRNA in vitro-transcribed separately by run-off transcription, T6102 again exhibited similar dose-dependent inhibition of protein synthesis (Figure 4b), suggesting that T6102 does not inhibit transcription. The lack of transcription inhibition was confirmed in a transcription inhibition counterscreen (Figure 4c). In order to investigate the remaining three reactions (energy regeneration, translation, and tRNA synthetase activity), T6102 was incubated in a polyuridylic acid [poly(U)] mRNA template-driven elongation reaction with detection of radiolabeled phenylalanine. Curiously, T6102 did not inhibit this elongation reaction at any dose tested (Figure 4d), suggesting that it does not merely nonspecifically disrupt the enzymatic and metabolic components of the protein synthesis reaction, including the energy regeneration system. Thus, T6102 likely inhibits a different step in translation or tRNA charging (except of tRNA-Phenylalanine). To investigate whether T6102 might bind the ribosome, T6102 was soaked into Thermus thermophilis 70S ribosome crystals; however, no obvious additional density was seen on difference (Fo-Fc omit) maps (data not shown). Though this does not provide evidence that T6102 does not bind the ribosome, the lack of additional density in ribosome crystals with T6102 suggests that T6012 may not crystallize with the ribosome. A few possible explanations include low affinity binding, transient interaction, binding in a different conformation of any component, or a completely different molecular target.
Figure 4.
T6102 inhibits translation but does not inhibit transcription, elongation, or energy regeneration. (a) Percent inhibition of the CFPS reaction, from a circular DNA template and measured by incorporation of radioactive amino acids, is plotted against the concentration of T6102 incubated in the protein synthesis reaction. (b) Percent inhibition of the CFPS reaction, from a luciferase RNA template and measured by incorporation of radioactive amino acids, is plotted against the concentration of T6102 incubated in the protein synthesis reaction. (c) Percent inhibition of RNA synthesis by T7 RNA polymerase, from a linearized plasmid and measured by RiboGreen quantification of RNA concentration, is plotted against the concentration of T6102 incubated in the transcription reaction. (d) Percent inhibition of polyphenylalanine [poly(Phe)] synthesis in the CFPS reaction, from a poly(U) RNA template and measured by incorporation of radioactive phenylalanine, is plotted against the concentration of T6102 incubated in the poly(Phe) synthesis reaction. All other reaction components were the same as in (b), suggesting that T6102 does not inhibit elongation or energy regeneration. Error bars represent the standard deviation for each condition.
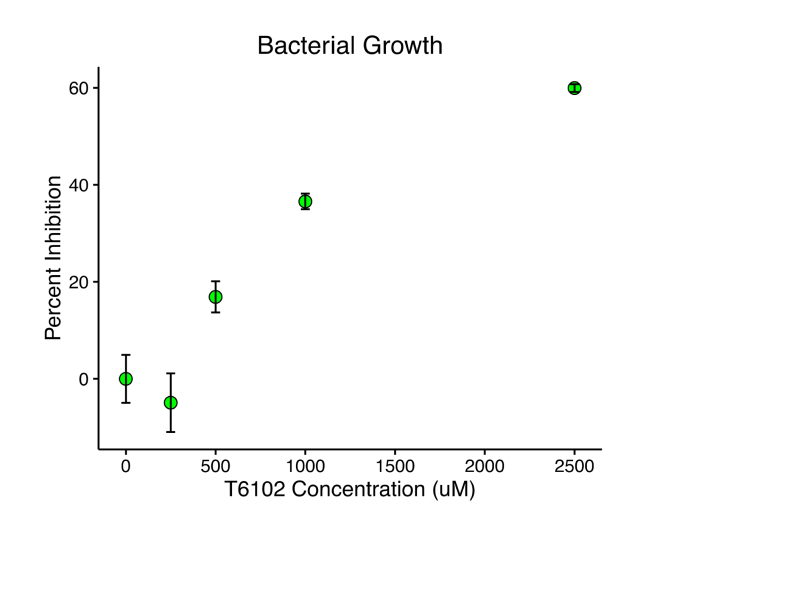
With the newfound mechanistic specificity for protein translation by T6102 in mind, we sought to investigate whether T6102 might also show in vivo antibacterial activity. When B. subtilis cultures were grown in the presence of T6102, T6102 exhibited dose-dependent inhibition of bacterial growth, with an IC50 of 593.6 +/- 245.3 uM (Figure 5), suggesting that it or its analogs may be promising novel antibiotics of a new, unique structural class. Analyzing T6102-resistant bacterial colonies or testing for synergy on bacterial growth inhibition when combined with other known classes of antibiotics may provide additional insights into the mechanism of action of T6102’s antibacterial activity.
Figure 5.
T6102 inhibits bacterial growth. Percent inhibition of B. subtilis growth, measured by optical density at 630 nm, is plotted against the concentration of T6102 incubated in the bacterial culture. Error bars represent the standard deviation for each condition.
Discussion
While the search for new antibiotics has yielded several prokaryote-specific translation inhibitors that have been utilized in the clinic or have provided insight into the structure and function of the ribosome, the current repertoire of antibiotics is insufficient to combat the rise of drug-resistant bacteria. It is well known that bacterial resistance to one antibiotic often renders resistance to the entire class. Thus novel classes of antibiotics are needed to meet current and future therapeutic needs.
Here we describe the discovery and characterization of one potential candidate for a structurally novel class of antibiotics, T6102. Through in vitro assays, we demonstrate that T6012 specifically inhibits bacterial translation, rather than acting through inhibition of transcription, inhibition of energy regeneration, or nonspecific disruption of cellular function. Moreover, through in vivo growth inhibition assays, we show that T6102 inhibits the growth of B. subtilis in culture, making T6102 a promising antibiotic candidate.
One limitation in the study of T6102 was its relative insolubility and precipitation at concentrations higher than 1,500 uM. Along these lines, inhibition of human cell growth could not be reliably assessed due to variability in cell growth and precipitation at high concentrations (data not shown). Nevertheless, chemical analogs of T6102 that may have improved aqueous solubility and potency should be pursued and assessed for eukaryotic cell toxicity to assess potential for clinical antibiotic use. If eukaryotic toxicity is observed, analogs of T6102 can be tested for anticancer activity across various eukaryotic cell lines to determine if any might be specific for growth inhibition of rapidly-dividing cell lines over noncancerous lines. Novel chemotherapeutic agents that do not interfere with DNA replication or transcription, unlike many of the current chemotherapies that act by nonspecific intercalation or transcription inhibition, may be less toxic clinically with fewer mutagenic side effects from action at the RNA, rather than DNA, level [16].
Similar to a few reported investigational antimicrobial agents, T6102 also contains an adamantane group. Nevertheless, the previously reported antimicrobial agents are generally composed of a derivative of the active component of a known antimicrobial drug conjugated to adamantane in order to improve solubility or targeting [5]. For this reason, these drugs likely act in the manner characteristic of the parent drug class rather than through a novel antimicrobial mechanism. T6102, unlike previously reported antimicrobial agents, appears structurally unlike any known class of translation inhibitors and may thus represent a novel class of antibiotics. In this study, T6102 was also shown to specifically inhibit bacterial translation in vitro as well as B. subtilis growth in vivo, making it a promising antibiotic candidate. In the growing context of emerging resistant infectious agents, novel classes of antibiotics, such as one suggested by T6102, are urgently needed to meet current and future therapeutic needs.
Methods
Cell-Free Protein Synthesis Reaction. An Escherichia coli in-house lysate-based cell-free protein synthesis system similar to Seidelt et al, 2009 [17] was optimized for reporter protein, DNA or RNA template, time, temperature, Mg2+ concentration, volume, T7 RNA polymerase concentration, and organic solvent tolerance. Cell-free protein synthesis reactions were set up according to [17] with the following exceptions: Reactions were performed in 25 uL final volumes with 15 mM magnesium glutamate (Sigma), 60 mM sodium pyruvate (Sigma), 8 mM sodium oxalate (Sigma), 170.6 ug/mL E. coli MRE-600 tRNA (uncharged) in water (Sigma), 15 mg/mL circular pIVEX-2.3d-luc (Figure 1b) DNA, 1 mg/mL T7 RNA polymerase (in house), and 2 percent dimethylsulfoxide (DMSO) (Sigma). Reactions were incubated for 1.5 hours at 30°C after which kanamycin (Sigma) was added to a final concentration of 1.2 uM to stop the reaction.
Luciferase Assay. 180 uL of room-temperature luciferase stabilization buffer [18] was added to each well of a black, opaque-bottom 96-well plate (Corning). 20 uL of each completed 25 uL cell-free protein synthesis reaction was added to individual wells. On a Synergy 4 Plate Reader (Biotek) at 25°C, background luminescence was measured, 15 uL of Steady-Glo Luciferase Assay reagent (Promega) at 25°C was added to each well, the plate was shaken for 10 seconds, and luminescence was measured. Final luminescence per sample was calculated by subtracting the sample’s background luminescence units from the sample’s measured luminescence units.
Protein Synthesis with Radiolabeled Amino Acids. 25 uL cell-free protein synthesis reactions were performed as described above with the following exceptions: the 2 mM L-amino acids alanine, arginine, aspartic acid, glutamic acid, histidine, isoleucine, leucine, lysine, phenylalanine, serine, threonine, tryptophan, and valine were replaced with their 14C-labeled counterparts (PerkinElmer NEC445E) at 0.1 mCi/mL. Completed reactions were assayed for protein incorporation of radioactivity by trichloroacetic acid precipitation as previously described [19].
Poly-U Translation. Poly-U translation was performed according to Boddeker et al, 2002 [20] with the following modification: reactions contained a final concentration of 2 percent DMSO.
B. subtilis growth. Aliquots of overnight culture of B. subtilis were diluted one hundred-fold into fresh LB medium. The diluted culture was incubated with 4 uL of DMSO or antibiotic in DMSO to a final volume of 200 uL for 12 hours at 37°C alongside non-inoculated LB medium. The optical density at 630 nm was measured on a Synergy 4 Plate Reader (Biotek).
High Throughput Screening. Cell free protein synthesis reactions were performed as described above with the following exceptions: 10 uL final volumes were used and the reactions were incubated at 28°C. For high-throughput screens, liquid handling was performed by hand or by Multidrop (Thermo) in Corning(C) white, non-binding surface 384 well plates (Sigma). Compounds in DMSO were added as 20 nL additions using a robotic pintool PlateMate2x2 (Matrix) to 20 nM final concentrations against 1.2 uM kanamycin positive controls in 10 uL cell-free protein synthesis reactions. Plates were sealed and incubated at 28°C for 1.5 hours, after which 35 uL of room-temperature Steady-Glo Luciferase Assay Reagent (Promega) diluted two hundred fold in 70 mM HEPES pH 7.7, 7 mM MgSO4, 3 mM dithiotreitol, and 1 percent Bovine Serum Albumin was added to the reactions followed by brief orbital shaking, a 5 minute incubation at room temperature, and luminescence measurement on Envision (PerkinElmer) plate reader. A luciferase inhibition counterscreen was performed exactly as above except 20 nL additions of compounds were performed after the 1.5 hour incubation. Greater than 98 percent inhibition of luciferase readout as compared to a 20 nL DMSO control was considered luciferase inhibition, and these hits were not pursued in downstream analysis. Signal analysis and significance of hits were analyzed with the assistance of Janie Merkel of the Yale Center for Molecular Discovery.
Cloning of pIVEX2.3d-luc. The firefly luciferase gene, luc, was polymerase chain amplified using Phusion High-Fidelity DNA Polymerase (NEB) according to the manufacturer’s protocol with pBESTluc plasmid template (Promega) and primers (5’- TATATACCATGGAAGACGCCAAAAACATAAAGAAAGG, 5’- TATATCTCGAGTTATTACAATTTGGACTTTCCGCCC, W.M. Keck Foundation). The product and vector pIVEX2.3d (5Prime) were digested with NcoI (NEB) and XhoI (NEB) and ligated after treatment of the vector with Calf Intestinal Phosphatase (NEB) using T7 DNA Ligase (NEB) all according to the manufacturers’ protocols. Plasmids were transformed into E. coli XL10Gold cells (Agilent Technologies, Inc.), and DNA was isolated and purified using QIAGEN plasmid isolation kits according to the manufacturers’ protocols. DNA was visualized and purified by agarose gel electrophoresis performed in 0.8 percent agarose gels containing 1X SYBR Gold Stain (Invitrogen). Gels were submerged in 1X TAE Buffer (40 mM Tris-acetate, 1 mM EDTA pH 8.3) and run against 1 kb or 100 bp DNA ladders (Invitrogen).
Template mRNA Preparation and Analysis. pIVEX-2.3d-luc was digested with XhoI (NEB) according to the manufacturer’s protocol for the generation of an mRNA template. Transcription was performed by run-off transcription as described above except for the addition of 1 U/mL SUPERase·In (Ambion) to reactions. After transcription reactions were performed, reactions were spun for 30 minutes at 13,000 rpm on a table-top centrifuge at 4°C to pellet pyrophosphate and precipitated with 7.5 M LiCl, 50 mM EDTA pH 8.0 as described in the manufacturer’s protocol (Ambion) and visualized by formaldehyde agarose gel electrophoresis (FAGE) in 1.2 percent agarose gels containing 20 mM MOPS, 5 mM sodium acetate, 1 mM EDTA, 1X SYBR Gold Stain (Invitrogen) and 5 percent formaldehyde (v/v) pre-equilibrated in 20 mM 3-(N-morpholino)propanesulfonic acid (MOPS), 5 mM sodium acetate and 1 mM EDTA running buffer for 30 minutes at 4°C. Samples were mixed with 5 mM MOPS, 0.8 mM sodium acetate, 0.25 mM EDTA, 6 percent formaldehyde (v/v), 16 percent formamide (v/v), and xylene cyanol, incubated at 65°C for 10 minutes, and loaded and run on FAGE at 60 V and 4°C for 1.5 hours.
In vitro transcription. Run-off transcription reactions were performed using 100 ug/mL linear template DNA, 50 ug/mL T7 RNA Polymerase P266L mutant (in-house), and 5 mM of each rNTP pH 7.5 in 80 mM Tris-HCl pH 7.6, 24 mM MgCl2, 2.5 mM spermidine, and 50 mM DTT in DEPC-treated water and incubated at 37°C for 2.0 hours.
Transcription Inhibition Counterscreen. A plasmid containing the Vibrio cholera VC1422 glycine riboswitch [21] for use as a test transcription product was linearized by restriction digest and in vitro transcribed by run-off transcription as described above but with the following modification: compounds dissolved in DMSO were added to a final concentration of 2 percent DMSO. Transcription products for the Transcription Inhibition Counterscreen were mixed with 12.5 mM EDTA pH 8.0, 50 percent formamide (v/v), xylene cyanol and bromophenol blue and incubated at 65°C for 10 minutes before visualization on 6 percent denaturing polyacrylimide gels containing 7 M urea in 1X TBE buffer (10 mM Tris, 90 mM Boric Acid, 1 mM EDTA) run at 30 W for 40 minutes. Gels were stained for qualitative visualization of RNA with Toluidine Blue, 0.01 percent Toluidine Blue (w/v) and 0.1 percent sodium tetraborate (w/v), and destained with water. RNA was quantified by Quant-iT RiboGreen RNA Reagent and Kit (Invitrogen) assays according to the manufacturer’s protocol. Briefly, 1 U per ug of template DNA of RQ1 RNase-free DNase (Promega) was added to each reaction and incubated at 37°C for 20 minutes. 100 uL of 1:1000 dilutions of DNase-treated transcription reactions in RNase/DNase-free 1X TE Buffer (10 mM Tris-HCl, 1 mM EDTA pH 7.5) were prepared in black, opaque bottom 96 well plates. Background fluorescence was read using Excitation: 485/20, Emission: 530/25 filter wheels. 100 uL of freshly made 2X RiboGreen solution was added to each well, the plate was shaken for 2 minutes and incubated for another 3 minutes at room temperature, and fluorescence was reread on a Synergy 4 Plate Reader (Biotek). RNA was quantified against serial dilutions of an E. coli 16S and 23S ribosomal RNA standard (Invitrogen) and compared as a percentage of the uninhibited control.
Crystal Soaks. 70S ribosomes from T. thermophilus HB8 were prepared and crystallized as previously described [22]. Briefly, sitting drop vapor diffusion trays were set up with a 500 uL reservoir solution of 2.9 percent poly(ethylene glycol) 20k (PEG20k), 9 percent 2-methyl-2,4-pentanediol (MPD), 175 mM arginine and 100 mM Tris-HCl pH 7.6. 5 uL sitting drops contained ~2.5 uL of 12-13 mg/mL 70S T. thermophilus ribosomes (from D. Bulkley) in 5 mM HEPES pH 7.5, 10 mM ammonium chloride, 10 mM magnesium acetate, 50 mM KCl and ~2.5 uL of well solution before equilibration. Crystals were cryoprotected by increasing the MPD concentration from 25 percent to 30 percent to 35 percent in 2.9 percent PEG20k, 100 mM Tris-HCl pH 7.6, 10 mM magnesium acetate, 10 mM ammonium chloride, 50 mM KCl. Antibiotics were soaked into the cryo-stabilized crystals for several hours to overnight at concentrations around 100 uM in 40 percent MPD. Data was collected at Brookhaven National Laboratory on beamline X29 and the Advanced Photon Source on beamlines 24-ID-E and 24-ID-C and processed using XDS [23]. Structures were solved by molecular replacement using the apo T. thermophilus 70S as a model (from Y. Polikanov) and refined on PHENIX [24].
Calculations. Percent inhibition was calculated by dividing each condition’s measurement by the average of the negative controls’ measurement (DMSO only), and then subtracting this number from 1. All experiments in Figures 3, 4, and 5 were performed in triplicate. The average of percent inhibition was taken of all of the replicates for each condition. IC50 values were calculated using the IC50 Toolkit (http://www.ic50.tk/).
Spectroscopic Analysis. 1H and 13C spectra were recorded on Bruker Avance DPX-500 Nuclear Magnetic Resonance (NMR) spectrometers. 1H NMR spectra are represented as follows: chemical shift, multiplicity (s = singlet, d = doublet, m = multiplet, br = broad), integration, and coupling constant (J) in Hertz (Hz). 1H NMR chemical shifts were reported relative to CDCl3 (7.26 ppm). 13C NMR were recorded relative to the central line of CDCl3 (77.00 ppm).
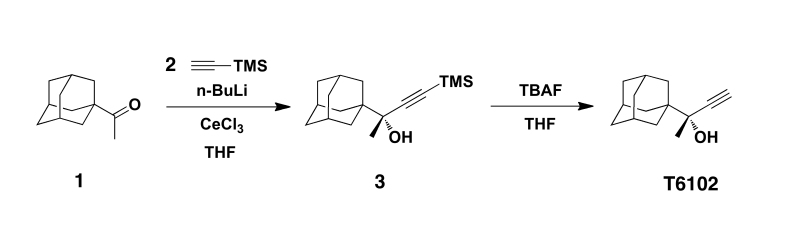
Materials and Purification. The synthesis of T6102 is shown in Figure 6. 1-Adamantyl methyl ketone, ethynyltrimethylsilane, n-butyllithium (n-BuLi, 2.5 M solution in hexanes), cerium (III) chloride, and tetrabutylammonium fluoride (TBAF, 1.0 M solution in tetrahydrofuran) were purchased from Sigma-Aldrich. Tetrahydrofuran (THF) was distilled from sodium/benzophenone. Thin layer chromatography (TLC) was performed using glass plates precoated with silica gel (0.25 mm). TLC plates were stained by submersion into aqueous ceric ammonium molybdate (CAM) followed by brief heating on a hot plate. Flash column chromatography was performed using silica gel 60 (230-400 mesh, Merck) with the indicated solvents.
Figure 6.
Synthesis of T6102. Compound labels and reaction scheme are described in Methods under Materials and Purification.
To a solution of ethynyltrimethylsilane 2 (206 mg, 2.1 mmol) in anhydrous THF (1.5 mL) at -78°C under N2 was added dropwise n-butyllithium (2.5 M solution in hexanes, 0.8 mL, 2.0 mmol). The mixture was allowed to warm to room temperature for 1.0 hour. This mixture was added to a stirred suspension of cerium (III) chloride (493 mg, 2.0 mmol) in anhydrous THF (2.5 mL) at -78°C via cannula. After stirring at -78°C for 1.0 hour, a solution of 1-adamantyl methyl ketone 1 (178 mg, 1.0 mmol) in anhydrous THF (1.5 mL) was added to the mixture. The resulting mixture was stirred at -78°C for 0.5 hours, quenched with saturated aqueous NH4Cl solution (10 mL), and extracted twice with diethyl ether. The extracts were washed with brine, dried over anhydrous Na2SO4, filtered, and concentrated. The crude product 3 was used without further purification for the next reaction.
TBAF (1.0 M solution in THF, 2 mL, 2.0 mmol) was added dropwise to a solution of the crude residue 3 in THF (4 mL) at 0°C. The reaction mixture was stirred at 0°C for 3.5 hours, quenched with saturated aqueous NH4Cl solution, and extracted twice with diethyl ether. The extracts were washed with brine, dried over anhydrous Na2SO4, filtered, and concentrated. The residue was chromatographed (eluting with 100 percent hexanes initially, grading to 10 percent ethylacetate in hexanes) on silica gel to afford T6102 (137 mg, 67 percent) as a white solid. 1H NMR (500 MHz, CDCl3) d 2.43 (s, 1H), 2.03 (brs, 3H), 1.82 (s, 1H), 1.72 (d, J = 2.9 Hz, 6H), 1.72-1.68 (m, 3H), 1.65-1.62 (m, 3H), 1.42 (s, 3H). 13C NMR (125 MHz, CDCl3) d 87.1, 73.8, 72.3, 39.0, 36.9, 36.1, 28.4, 23.4. TLC (20 percent EtOAc in Hexanes), Rf 0.68 (CAM).
Glossary
- CFPS
Cell Free Protein Synthesis
- poly(U)
Polyuridylic acid
- poly(Phe)
Polyphenylalanine
- DMSO
Dimethylsulfoxide
- FAGE
Formaldehyde agarose gel electrophoresis
- MOPS
3-(N-morpholino)propanesulfonic acid
- PEG20k
Poly(ethylene glycol) 20k
- MPD
2-methyl-2,4-pentanediol
- n-BuLi
n-butyllithium
- THF
Tetrahydrofuran
- TBAF
Tetrabutylammonium fluoride
- TLC
Thin layer chromatography
- CAM
Ceric ammonium molybdate
- NMR
Nuclear magnetic resonance
- s
Singlet
- d
Doublet
- m
Multiplet
- br
Broad
- J
Coupling constant
- Hz
Hertz
- pT7
T7 promoter
- prom
Promoter
- term
Terminator
- RBS
Ribosome binding sequence (AGGAGA)
- Stop
Tandem stop codons (TAATAA)
- ori
Origin
- amp
Ampicillin
- R
Resistance
Author Contributions
DT performed all of the experiments detailed here and wrote the manuscript. DT was supported by the Barry M. Goldwater Foundation, the Arnold & Mabel Beckman Foundation, and the MD/PhD Summer to Advance Research Training @ Yale Program. HST performed the synthesis of T6102.
References
- Lowell AN. et al. Microscale Adaptation of In Vitro Transcription/Translation for High-Throughput Screening of Natural Product Extract Libraries. Chem Biol Drug Des. 2015;86(6):1331–1338. doi: 10.1111/cbdd.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox SK, Cavey GS, Pearson JD. et al. Single ribosomal protein mutations in antibiotic-resistant bacteria analyzed by mass spectrometry. Antimicrob Agents Chemother. 2001;45(11):3046–3055. doi: 10.1128/AAC.45.11.3046-3055.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov YS. et al. Amicoumacin a inhibits translationby stabilizing mRNA interaction with the ribosome. Mol Cell. 2014;56(4):531–540. doi: 10.1016/j.molcel.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PS. et al. Anticolorectal cancer effects and pharmacokinetic application of 2, 2-Bis [4-(4-amino-3-hydroxyphenoxy) phenyl] adamantane. Int J Clin Exp Med. 2015;8(9):14805–14815. [PMC free article] [PubMed] [Google Scholar]
- Liu J. et al. The many faces of the adamantyl group in drug design. Eur J Med Chem. 2011;46(6):1949–1963. doi: 10.1016/j.ejmech.2011.01.047. [DOI] [PubMed] [Google Scholar]
- Imig JD, Hammock BD. Soluble epoxide hydrolase as a therapeutic target for cardiovascular diseases. Nat Rev Drug Discov. 2009;8(10):794–805. doi: 10.1038/nrd2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa TK. et al. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat Chem Biol. 2011;7(8):538–543. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson JL. et al. Small-molecule-mediated degradation of the androgen receptor through hydrophobic tagging. Angew Chem Int Ed Engl. 2015;54(33):9659–9662. doi: 10.1002/anie.201503720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacksteder KA. et al. Discovery and development of SQ109: a new antitubercular drug with a novel mechanism of action. Future Microbiol. 2012;7(7):823–837. doi: 10.2217/fmb.12.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladkov LL. et al. Vibrational spectroscopy of N'-(Adamantan-2-ylidene)thiophene-2-carbohydrazide, a potential antibacterial agent. Spectrochim Acta A Mol Biomol Spectrosc. 2014;128:874–879. doi: 10.1016/j.saa.2014.02.071. [DOI] [PubMed] [Google Scholar]
- Patel SR. et al. Synthesis, biological evaluation and 3D-QSAR study of hydrazide, semicarbazide and thiosemicarbazide derivatives of 4-(adamantan-1-yl)quinoline as anti-tuberculosis agents. Eur J Med Chem. 2014;85:255–267. doi: 10.1016/j.ejmech.2014.07.100. [DOI] [PubMed] [Google Scholar]
- Al-Abdullah ES. et al. Synthesis, antimicrobial, and anti-inflammatory activity, of novel S-substituted and N-substituted 5-(1-adamantyl)-1,2,4-triazole-3-thiols. Drug Des Devel Ther. 2014;8:505–518. doi: 10.2147/DDDT.S62465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C, Chaudhary S, Puri SK. Orally active esters of dihydroartemisinin: Synthesis and antimalarial activity against multidrug-resistant Plasmodium yoelii in mice. Bioorg Med Chem Lett. 2008;18(4):1436–1441. doi: 10.1016/j.bmcl.2007.12.074. [DOI] [PubMed] [Google Scholar]
- Strobel SA, Strobel GA. Plant endophytes as a platform for discovery-based undergraduate science education. Nat Chem Biol. 2007;3(7):356–359. doi: 10.1038/nchembio0707-356. [DOI] [PubMed] [Google Scholar]
- Jewett MC. et al. An integrated cell-free metabolic platform for protein production and synthetic biology. Mol Syst Biol. 2008;4:220. doi: 10.1038/msb.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szikriszt B, Póti Á, Pipek O. et al. A comprehensive survey of the mutagenic impact of common cancer cytotoxics. Genome Biol. 2016;17:99. doi: 10.1186/s13059-016-0963-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidelt B. et al. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326(5958):1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novac O, Guenier AS, Pelletier J. et al. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 2004;32(3):902–915. doi: 10.1093/nar/gkh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Soll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci U S A. 2010;107(9):4028–4033. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddeker N. et al. Characterization of a novel antibacterial agent that inhibits bacterial translation. Rna. 2002;8(9):1120–1128. doi: 10.1017/s1355838202024020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erion TV, Strobel SA. Identification of a tertiary interaction important for cooperative ligand binding by the glycine riboswitch. Rna. 2011;17(1):74–84. doi: 10.1261/rna.2271511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulkley D. et al. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc Natl Acad Sci U S A. 2010;107(40):17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]