Abstract
Long non-coding RNAs (lncRNAs) constitute the largest class of non-coding transcripts in the human genome. Results from next-generation sequencing and bioinformatics advances indicate that the human genome contains more non-coding RNA genes than protein-coding genes. Validated functions of lncRNAs suggest that they are master regulators of gene expression and often exert their influences via epigenetic mechanisms by modulating chromatin structure. Specific lncRNAs can regulate transcription in gene clusters. Since the functions of protein-coding genes in clusters are often tied to specific pathways, lncRNAs constitute attractive pharmacological targets. Here we review the current knowledge of lncRNA functions in human cells and their roles in disease processes. We also present forward-looking perspectives on how they might be manipulated pharmacologically for the treatment of a variety of human diseases, in which regulation of gene expression by epigenetic mechanisms plays a major role.
Keywords: lncRNA, epigenetics, pharmacology, gene expression, drug target
Epigenetics: Chromatin-mediated Effects on Gene Regulation
In 1942, Conrad Waddington coined the term ‘epigenetics’ and defined it as “the study of the causal interactions between genes and their products, which leads to phenotype changes during development” [1,2]. Today, epigenetics is a rapidly growing field of research and is now broadly defined as mechanisms leading to changes in gene expression that do not involve changes in DNA sequences per se. At a molecular level, epigenetic mechanisms are primarily mediated by alterations of chromatin structures and changes (especially in DNA methylation and post-translational modifications (PTMs) placed upon nucleosomal histones) can lead to alterations of the expression of genes presented in or near epigenetically modified nucleosomes. Epigenetic regulation is rooted in chromatin structures, which can be divided into two major classes: euchromatin and heterochromatin. Euchromatin is transcriptionally active and consists of DNA that is loosely associated with nucleosomes and thus is accessible to RNA polymerases. Heterochromatin, in contrast, is highly condensed and not readily transcribed, resulting in silenced gene states.
Long Non-coding RNAs
One of the major types of epigenetic regulation utilizes functionally untranslated RNA species. Long non-coding RNAs (lncRNAs) are non-coding RNAs with a length greater than 200 nucleotides. lncRNAs play versatile roles in many aspects of gene regulation, including transcription, mRNA splicing, translation, epigenetic silencing, genomic imprinting, X-chromosome inactivation, and the processing of small ncRNAs [3,4]. Many lncRNAs regulate gene expression by recruiting chromatin complexes by means of DNA methylation and histone PTMs, which are described below.
Roles of lncRNAs in the Regulation of DNA Methylation
The interaction between lncRNAs and DNA methylation enzymes plays a key role in epigenetic regulation. DNA methylation at position 5 within cytosine bases present in CpG dinucleotides plays a critical role in key biological functions, including embryonic development, genomic imprinting, X-chromosome inactivation, the silencing of transposable elements, and many others. Genomic imprinting is an epigenetic mechanism where certain genes are expressed from only one of the two parental chromosomes. Some imprinted genes are maternally expressed (e.g. IGF2R and H19), and others paternally expressed (e.g IGF2) [5]. Xist is transcribed from the X-inactivation center (Xic) and coats the inactive X-chromosome in females in cis, leading to silencing of hundreds of X-linked genes [6]. The Air lncRNA (Antisense Igf2r RNA) has been shown to silence imprinted genes [7]. H19 functions as a lncRNA but it has also been reported by Cai et al. that “H19 functions as a primary microRNA precursor decreasing the post-transcriptional down-regulation of mRNAs during development” [8].
Some lncRNAs are known to regulate DNA methylation through physical interactions with DNA methyltransferases (DNMTs). Ruscio et al. have reported that a lncRNA called Extra-coding CEBPA (ecCEBPA) physically interacts with DNMT1 (the primary maintenance cytosine methyltransferase) and prevents methylation of the CEBPA gene locus in cis [9]. lncRNA Dali physically interacts with DNMT1 protein and modulates DNA methylation of CpG island-associated promoters in trans [10]. The lncRNA Dum causes promoter methylation of developmental pluripotency associated 2 (Dppa2) gene through interactions with DNMT1, and the de novo methyltransferases DNMT3A, and DNMT3B [11]. lncRNAs can thus modulate DNA methylation in cis and trans through interactions with all three DNMTs. Hence, it is possible that dysregulated lncRNAs may be involved in epigenetic changes leading to human diseases.
Histone Modifications
Histone modifications are also known to epigenetically regulate gene transcription as well as DNA repair and replication, chromosome condensation, and alternative splicing [12]. A vertebrate nucleosome consists of an octamer of core nucleosomal histones (two each of histones H2A, H2B, H3, and H4) and a linker histone H1, which binds short stretches of DNA between nucleosomes leading to chromatin compaction. All of the core histones undergo covalent PTMs, which include acetylation, methylation, phosphorylation, ubiquitylation, and sumoylation at specific amino acid residues, mostly within their N-terminal histone tails [13]. Transcriptional activation is mediated by histone acetyltransferases (HATs) and transcriptional repression is mediated by the action of histone deacetylases (HDACs). The polycomb repressive complexes (PRC1 and PRC2) contain both histone methyltransferases leading to H3K27Me3 and histone monoubiquitination, which results in gene silencing, respectively [14]. In contrast, trithorax group (TrxG) complexes methylate H3K4Me3, which facilitates gene transcription [15]. Therefore, histone PTMs are centrally involved in epigenetic regulation at the level of chromatin structure, and this notion is revisited frequently in the remainder of this review.
Epigenetic Mechanisms of Disease
Together, histone PTMs, CpG methylation, and lncRNAs regulate gene expression and are exquisitely involved in a wide range of biological processes, including development and the epigenetic regulation of genes. However, when errors in the maintenance of epigenetic states occur in somatic cells, a variety of human diseases and conditions can occur.
Beckwith-Wiedemann syndrome (BWS), an imprinting disorder of the KCNQ1OT gene, is mediated epigenetically, and at least 50 percent of patients have loss of DNA methylation and loss of histone H3K9Me2 on the maternal KCNQ1OT differentially methylated region (DMR) [16]. He at al. reported that a microdeletion in the human H19 DMR can result in loss of insulin-like growth factor 2 (IGF2) imprinting and BWS [16]. lncRNAs can thus lead to the development of complex diseases through epigenetic mechanisms.
Perturbations in DNA methylation are common in many cancers and global alterations involving DNA hypomethylation are common in cancer, often typified by a 20 to 60 percent reduction in 5-methyl-cytosine content [12]. DNA hypomethylation at the promoter regions of oncogenes can activate their expression, and DNA hypermethylation at the promoters of tumor suppressor genes can lead to their silencing. Loss of imprinting of IGF2 and H19 genes by demethylation can lead to carcinogenesis and tumor progression [17]. The lncRNA LET is repressed by histone deacetylase 3 (HDAC3), which is reported to contribute to hypoxia-mediated cancer cell invasion [18]. The lncRNA ANRASSF1 reduces the expression of Ras association domain family 1 isoform A (RASSF1A) protein by recruiting PRC2 to the RASSF1A promoter region, thereby increasing cellular proliferation and inhibiting cell death [19]. These and other examples too numerous to cite firmly establish altered DNA methylation and histone modification as being intimately involved in carcinogenesis, and often in conjunction with lncRNA actions.
lncRNAs in Detail
Recent advances in next generation sequencing and bioinformatics show that much of the mammalian genome is transcribed into RNA, and that much of this RNA is functional. This new view constitutes a paradigm shift in our view of the genome as a whole and the regulation of protein-coding genes. The ENCODE project consortium reported that at least 93 percent of human genomic DNA is transcribed [20]. After completion and analysis of the human genome, it was found that there are only 20,000 to 25,000 protein coding genes in the genome, corresponding to less than 2 percent of the genome [21]. The remaining 98 percent was originally thought to be “junk DNA” [22], but it is now known that much of this DNA encodes functional non-coding RNAs (ncRNAs) [23] and hence, cannot be interpreted simply as ‘transcriptional noise’ [24].
lncRNAs make up the majority of the ncRNA transcripts in the genome [25], and yet the functions of most lncNRAs are unknown. lncRNA lengths can range from 200 nucleotides to over 50 kilobases [26]. Various genomic consortiums and databases including GENCODE [27] have assisted in the task of computational identification, annotation, and interpretation of expansive ncRNA datasets. lncRNAs, such as XIST and H19, were initially identified from cDNA libraries [28]. Iyer et al. curated 7,256 RNA sequencing libraries and identified 58,648 lncRNA genes from the human transcriptome totaling 91,013 expressed genes [29]. lncRNAs have been discovered by a variety of methods including RNA immunoprecipitation (RIP), microarray and tiling array screens, and RNA-sequencing. All these approaches have limitations. For instance, isolating lncRNAs though RIP methods relies upon the specificity of antibodies. Although these studies have provisionally identified a large number of lncRNAs, their functions cannot be fully evaluated until each of these lncRNAs is experimentally validated. Nonetheless, these studies show that the human genome likely contains more lncRNA genes than protein-coding genes.
lncRNAs have been classified into different categories and have been extensively reviewed by Laurent et al. [30]. Briefly, lncRNAs are classified based on their locations in the genome, their lengths, proximity to protein-coding genes, association with DNA elements, mechanisms of action, and sub-cellular localization (nucleus or cytoplasm). lncRNA genes can regulate adjacent protein-coding genes near their sites of synthesis, and these are known as cis-acting lncRNAs. lncRNAs can also regulate genes in distant genomic locations (on other chromosomes), and these are called trans-acting lncRNAs. lncRNA genes often reside near protein-coding genes (often presented in gene clusters) and they can be divided into sense lncRNAs, natural antisense lncRNAs (NATs), and long intronic ncRNAs (linRNAs). Sense lncRNAs are transcribed from the sense strand with respect to regulated protein-coding genes, while NAT lncRNAs are transcribed from the antisense strand with respect to adjacent protein-coding genes (e.g. APOA1-AS) [31]. linRNAs reside within introns of protein-coding genes [32]. Genes for intergenic lncRNAs (located between protein-coding genes) are called long intergenic noncoding RNAs (lincRNAs), and these include the well-studied lncRNAs XIST, H19, and an antisense non-coding RNA in the INK4 locus (ANRIL) [33]. Some lncRNAs are of extraordinary length; these are called very long intergenic noncoding RNAs (vlincRNA) and they consist of transcripts over 50 kilobases (e.g. HELLP) [34]. Based on subcellular localization, lncRNAs can be divided into nuclear or cytoplasmic classes. Most lncRNAs are nuclear (eg. XIST and maternally expressed gene 3; MEG3) but some, like H19, are located in the cytoplasm [23]. Genome-wide physical interactions of lncRNAs with DNA have been determined by biochemical approaches, including Chromatin Isolation by RNA Purification (ChIRP) [35] and Capture Hybridization Analysis of RNA Targets (CHART) [36]. Through these methods, some lncRNAs associate with enhancers (enhancer-associated lncRNA, or elncRNAs) [37], promoters (promoter-associated lncRNAs, or PLARS) [38], or telomeres (telomeric repeat-containing RNA, or TERRA) [39]. Other lncRNAs include transcribed ultraconserved regions (T-UCR) [40], which are so called due to their sequence conservation across species (e.g. PTENP1) [41]. The existing systems of classification have limitations, since individual lncRNAs can fit into multiple classifications. Annotation and classification nomenclature of lncRNAs are still coalescing; hence, new classification strategies that can consider lncRNA properties, functions, and relationships are desirable.
lncRNAs are generally transcribed by RNA polymerase II and undergo post-transcriptional modifications including 5’-capping, polyadenylation, and splicing [23]. RNA polymerase III, which is primarily involved in the transcription of tRNAs and 5S rRNA, also transcribes a neuronal lncRNA, BC200 [42]. More than 25 percent of lncRNAs are alternatively spliced to produce two or more related isoforms [23]. Some lncRNAs, however, are non-polyadenylated [23]. lncRNA promoters exhibit specific histone marks, including methylated H3K4, H3K27, H3K36, and acetylated H3K9 and H3K27, suggesting that they too undergo epigenetic regulation similar to protein-coding genes [23].
The presence of well-defined open reading frames (ORFs) distinguishes protein-coding genes from lncRNAs. The FANTOM consortium assumes that genes containing long ORFs are likely protein-coding genes in the mouse transcriptome [43]. However, some lncRNAs have ORFs consisting of over 100 codons, including Xist, H19, and KCNQ1OT, yet these are not translated into proteins [44]. In a surprising ribosome profiling study by Guttman et al, it was found that the ribosome occupancy on many lincRNAs is detectable, but was similar to small ncRNAs (including small nuclear RNAs, small nucleolar RNAs, microRNA precursors, and lncRNAs) and other non-coding regions, including 5’-untranslated regions (UTRs). This demonstrates that the presence of ribosome occupancy alone is insufficient to determine the coding potential of lncRNAs. However, they defined a method to accurately distinguish protein-coding transcripts from all classes of non-coding transcripts based on the release of translating ribosomes from translated RNAs upon encountering a bona fide stop codon [45].
lncRNAs sequences are generally less conserved across species than protein-coding genes, implying that they undergo evolutionary changes much more rapidly. Even though some lncRNAs are poorly conserved, they are functional, which suggests that recently evolved lncRNAs can still be of functional consequence. Human accelerated regions (HARs) are regions where increased rates of nucleotide substitution occur between the human and chimpanzee. The 118-nucleotide HAR1 region is a part of the HAR1F lncRNA, which is highly expressed in developing human brain. The HAR1 region is folded into an organized RNA secondary structure and mutations in this region in human were shown to stabilize the secondary structure as compared to the chimpanzee [46]. Lack of conservation is a limiting factor for the use of animal models of human diseases, and it is possible that some of the species-specific differences might be due to divergence in lncRNA conservation. However, some lncRNA sequences are conserved across species. lncRNA promoter regions are more conserved than the exonic sequences and exhibit levels of sequence conservation comparable to protein-coding genes in the mouse genome [23,29,47]. lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) is highly conserved in mammals but is not found in non-mammals [48]. Polyadenylated RNA (PAN RNA) is produced and retained in the nucleus during Kaposi’s sarcoma-associated herpesvirus (KSHV) lytic infection. PAN RNA has an expression and nuclear retention element (ENE), which enables the formation of a triple helical structure. The triple helix renders lncRNA stability and also its nuclear retention [49]. The lncRNA thyroid cancer-associated transcript 126 (THCAT126) is presented in nearly all vertebrates [29]. Thus, not all lncRNAs are poorly conserved across species, and Iyer et al. identified 597 intergenic lncRNAs, which contained regions of ultraconserved elements (UCEs) (regions greater than 200 nucleotides that are highly conserved across species) [29].
lncRNAs typically have lower expression levels in tissues as compared to protein-coding genes. Computational analysis of RNA sequencing data from 16 human tissues obtained from the Illumina Human Body Map Project revealed that lncRNAs in general had lower expression in all tissues than protein-coding genes, except in the testis [50]. lncRNAs also have higher tissue-specificity as compared to coding genes. Custom microarray studies on 9,747 lncRNA transcripts from GENCODE version 3c annotation, which assayed lncRNA content from human tissues and cell lines, also suggest that lncRNAs have far lower levels of expression relative to mRNAs [23,50]. lncRNA expression profiles are not identical for polyadenylated and non-polyadenylated RNAs. Djebali et al. reported that lncRNA gene expression ranged over six orders of magnitude (10−2 to 104 reads per kilobase per million reads (RPKM)) for polyadenylated lncRNAs and five orders of magnitude (10−2 to 103 (RPKM) for non-polyadenylated lncRNAs) [51]. It has thus been shown by different studies that most lncRNAs have lower expression and higher tissue specificity of expression than protein-coding genes.
lncRNAs are often more unstable than mRNAs. Two independent studies determined the range of lncRNA stability and reported that their half-lives varied from unstable to highly stable [52,53]. Clark et al. used custom microarray studies in a mouse cell line to identify the half-lives of approximately 800 lncRNAs and 1,200 mRNAs in the mouse Neuro-2a. Most of the lncRNAs were stable, with half-lives greater than 16 hours, and approximately 240 lncRNAs were unstable, with half-lives less than 2 hours. The mean half-life of lncRNAs was found to be 4.8 hours and this was less than mRNAs, which had a mean half-life of 7.7 hours [53]. It has been observed that the nuclear paraspeckle assembly transcript 1 (Neat1) is highly unstable, but is functional and required for the nuclear localization and dynamic regulation of paraspeckles at the Neat1 locus [53]. Several pathways are known to be involved in the degradation of lncRNAs, including nucleolytic degradation by nuclear exosomes and cytoplasmic nonsense-mediated decay [54]. lncRNAs thus regulate key cellular functions irrespective of their stability within the cellular compartment (nucleus or cytoplasm).
Mechanisms of lncRNA Action
lncRNAs play a critical role in many cellular and biological processes, including early embryonic development, embryonic stem cell (ESC) pluripotency and differentiation, cell cycle regulation, proliferation, apoptosis, and senescence [3,4,55]. The molecular mechanisms whereby lncRNAs exert their regulatory impacts on protein-coding genes have been extensively reviewed [56-59]. Individual lncRNAs can operate as a signal, decoy, scaffold, and guide lncRNAs [56,57]. These classes of lncRNAs are discussed below:
Signal lncRNAs: lncRNAs can act as molecular signals to regulate transcription in response to different stimuli. Transcription of some lncRNAs is both tissue-specific and temporally specific. It has been noted by Wang et al. that some signal lncNRAs have regulatory functions while others are triggered by transcriptional events [57]. Wang et al. have reported that “lncRNAs act as signals marking space, time, developmental stage, and expression for gene regulation” [57]. lncRNA KCNQ1OT1 acts as a signal lncRNA and induces transcriptional repression by recruiting G9a histone methyltransferases and PRC2 to the genes both in cis and in trans [60]. Thus, they serve as a marker of transcriptional activity. lncRNAs that are transcribed from the four human homeobox transcription factors (HOX) clusters exhibit specific spatial and temporal patterns of gene expression during development. For example, HOTAIR regulates sequential Hox gene expression during mouse embryogenesis, thus serving as a signal of anatomic position during development [57,61]. lncRNAs also respond to environmental stimuli, such as cold, which triggers their action (e.g., COLDAIR and COOLAIR) [62]. Signal lncRNAs thus serve as sensors that regulate important biological functions.
Decoy (sponge) lncRNAs: Some lncRNAs serve to sequester key cellular components. lncRNAs can bind to transcription factors and microRNAs, where they sequester these factors, thus preventing their action. lncRNA GAS5 acts as a decoy glucocorticoid response element (GRE) and binds to the DNA-binding domains of the glucocorticoid receptor, thereby preventing its association with DNA, downstream effects on the cell cycle and apoptosis [3,63]. lncRNAs also act as decoys (sponges) for miRNAs and splicing factors [4,57]. Pseudogene lncRNA PTENP1 acts as a sponge and sequesters microRNAs that bind the 3’-UTR of the tumor-suppressor gene PTEN, thus indirectly influencing translation of PTEN mRNA [64]. The lncRNA hepatocellular carcinoma upregulated long non-coding RNA (HULC) acts as an endogenous miRNA sponge for miR-372 and downregulates its target gene protein kinase cAMP-activated catalytic subunit beta (PRKACB) [4,65]. Decoy lncRNAs thus inhibit the function of the effector transcription factors and microRNAs by sequestration, and thereby negatively regulate transcription.
Scaffold lncRNAs: Scaffold lncRNAs are structural in nature and provide a framework upon which one or more proteins can simultaneously assemble within different lncRNA domains and regulate chromatin modifications [66]. HOTAIR acts as a scaffold and simultaneously binds to two different protein complexes. The 5' domain of HOTAIR binds to the PRC2 complex (involved in H3K27 methylation and gene silencing) and the 3' domain of HOTAIR binds to LSD1-CoREST complex to mediate H3K4Me2 demethylation [67]. ANRIL binds to both PRC1 and PRC2, leading to repression of the INK4b/ARF/INK4a gene locus [68]. KCNQ1OT1 serves as a scaffold by binding both PRC2 and G9a in order to mediate H3K27 and H3K9 trimethylation and consequent gene silencing [69]. lincRNA functional intergenic repeating RNA element (Firre) acts as a scaffold through its 156-bp repeating RNA domain (RRD). Firre interacts with the nuclear-matrix factor hnRNPU through the Firre RRD for nuclear localization of Firre transcripts and for binding to different chromosomal locations [70]. Thus, scaffold lncRNAs form a binding platform for tethering one or more chromatin-modifying complexes and enzymes to mediate RNA-protein interactions.
Guide lncRNAs: Guide lncRNAs act a guide for the localization of regulatory protein complexes, such as trithorax group proteins, polycomb group proteins, and transcription factors to their target DNA sites in cis or in trans [57]. Xist [71,72] and Air [73] act in cis to regulate syntenic genes that are either subject to dosage compensation or imprinting, respectively. The lncRNA HOXA transcript at the distal tip (HOTTIP) acts functionally as both a signal and guide lncRNA to regulate the HOXA locus, by recruiting the adaptor protein WD repeat domain 5 (WDR5) and mixed lineage in leukemia-1 (MLL-1) protein, which in turn mediate histone methylation and gene transcription in the HOXA locus [74]. Trans-acting lincRNAs like HOTAIR [75] and Jpx also regulate gene expression and chromatin modification upon other chromosomes [76]. Guide lncRNAs can thus regulate gene expression through complex epigenetic mechanisms.
lncRNAs can regulate mRNA stability both positively and negatively [77]. Antisense lncRNA BACE1-AS is known to increase the stability of BACE1 mRNA. BACE1-AS forms a RNA-RNA duplex with BACE1 mRNA. BACE1-AS competes with miR 485-5p for the same binding site in the BACE1 mRNA. This abrogates miRNA-induced repression and thus stabilizes the BACE1 mRNA. BACE1-AS expression is increased in the brains of Alzheimer’s patients [78]. Alu elements are primate-specific repeat elements. lncRNAs containing Alu repeats can destabilize mRNA through Staufen mediated decay (SMD). SMD is a process by which mRNA degradation is mediated by the binding of Staufen 1 (STAU1) (a protein that binds to double-stranded RNA) to STAU1-binding sites (SBS) within the 3'-UTR of the target mRNA. Alu elements within the 3’-UTR of lncRNAs imperfectly base-pair with the mRNA 3’-UTR Alu elements, creating a double stranded SBS. STAU-1 binds to this site and destabilizes the target mRNA. Gong et al. have shown that lncRNA-AF087999 base-pairs with the SERPINE1 mRNA at its 3'-UTR Alu element sequence and facilitates the binding of STAU1 protein to mRNAs leading to SMD [79]. In summary, lncRNAs are involved in various aspects of mRNA stability.
lncRNAs in Human Disease
Dysregulated or mutated lncRNAs play a critical role in the etiology and pathogenesis of many diseases (Table 1). Many different types of cancers and syndromes are associated with mutations in lncRNA genes [3]. Genome-wide array studies show differential lncRNA expression patterns in comparisons between normal and tumor cells [80-82]. Transcribed ultraconserved regions (T-UCRs) are highly conserved sequences between orthologous regions of human, rat, and mouse genomes located in intra- and intergenic regions. Genome-wide microarray profiling studies of T-UCRs indicate that they are differentially expressed in human leukemias and carcinomas and are regulated by miRNAs both in vitro and in vivo [83]. HOTAIR is associated with multiple types of cancer (reviewed by Hajjari et al.), [84] where it interacts with PRC2 and LSD1 to repress target gene transcription [85]. ANRIL binds to chromobox 7 (CBX7), which is a component of the PRC1 complex, to induce gene silencing, and both ANRIL and CBX7 are upregulated in prostate cancer [86]. lincRNA-p21 is a p53 repressor and is associated with the development and progression of prostate cancer, chronic lymphocytic leukemia, atherosclerosis, and rheumatoid arthritis [87,88]. A chromosomal translocation involving the lncRNA gene DISC2 on chromosome 1 (1;11) (q42.1;q14.3) is associated with schizophrenia and psychiatric disorders in a large Scottish family [89]. Genome-wide association studies (GWAS) have shown that germline deletion (403 kb) of INK4b/ARF locus including the ANRIL gene is associated with hereditary cutaneous malignant melanoma (CMM) and neural system tumors (NST) syndrome [90]. Expansion of CTG trinucleotides in the primary sequence of lncRNA gene ATXN8OS produces a toxic RNA that alters RNA splicing of spinocerebellar ataxia type 8 (SCA8) mRNA [91]. BWS is an imprinting disorder associated with abnormal imprinting of the KCNQ1OT and IGF2 genes resulting in congenital malformation and tumor predisposition [16]. Thus, as more and more lncRNAs are being functionally validated, there is increasing evidence for the role of dysregulated or mutated lncRNAs in the etiology and prognosis of various neurodegenerative, cardiovascular, metabolic diseases, and cancer of various organs.
Table 1. lncRNAs associated with human diseases.
| Affected organ system | Disease | Associated lncRNAs | References |
| Autoimmune Diseases | Psoriasis | PRINS | [122] |
| Rheumatoid arthritis | HOTAIR | [123] | |
| Cardiovascular diseases | Cardiac hypertrophy | 7SK; CHRF | [124,125] |
| Myocardial infarction | MIAT; KCNQ1OT1; ANRIL | [126,127] | |
| Digestive system disorders | Barrett's | AFAP1-AS1 | [128] |
| Crohn's | DQ786243 | [129] | |
| Endocrine & metabolic disorders | Pseudohypoparathyroidism type Ib | GNAS-AS1 | [130] |
| Genetic disorder | Fragile X syndrome | FMR4; FMR5; FMR6 | [131,132] |
| HELLP syndrome | HELLPAR | [34] | |
| Infectious diseases | Leishmania | 7SL | [133] |
| Musculoskeletal system disorders | Duchenne muscular dystrophy | KUCG1; linc-MD1 | [134,135] |
| Facioscapulohumeral muscular dystrophy | D4Z4; DBE-T | [136] | |
| Neurological diseases | Angelman syndrome | UBE3A-AS1 | [137] |
| Parkinson's | naPINK1 | [138] | |
| West syndrome | BX118339 | [139] | |
| Reproductive system diseases | Mullerian aplasia | H19 | [140] |
| Cancer | Adenocarcinoma | HNF1A-AS1; ZXF1 | [141,142] |
| Breast cancer | GAS5 | [143,144] | |
| Colorectal cancer | MALAT1; H19; HOTAIR | [145-147] | |
| Esophageal squamous cell cancer | ANRIL; SPRY4-IT1 | [148,149] | |
| Gastric cancer | GHET1 | [150] | |
| Kaposi's | PAN | [151] | |
| Liver | HULC | [152,153] | |
| Lung | MALAT1; LincRNA-p21 | [154,155] | |
| Testicular cancer | BOK-AS1 | [156] |
lncRNAs as Drug Targets: Future Strategies
Existing pharmaceutical agents to treat diseases through epigenetic mechanisms are nonspecific. Epigenetic drugs, such as the DNA methyltransferase (DNMT) inhibitor 5-aza-2’-deoxycytidine, and histone deacetylase (HDAC) inhibitors, such as sodium valproate, depsipeptide, tetrapeptide suberoylanilide hydroxamic acid (SAHA), and CI-994 (N-acetyldinaline) [92], have all been used to treat various types of cancer; however, their modes of action alter chromatin structure throughout the genome. Histone- or DNA-modifying enzymes attached to gene-specific zinc finger proteins may in the future lead to targeted treatments that will specifically bind to targeted epimutation sites [93]. However, since these are proteins, they must be delivered by venous injection. Since lncRNAs normally regulate relatively small sets of genes, often with related functions, the ability to target lncRNAs pharmacologically should result in improved specificity and lowered incidence of side effects. Gene clusters often contain genes with similar functions (i.e. they function in a shared pathway), and these gene clusters are typically regulated by one or few specific lncRNAs. Therefore, lncRNAs constitute attractive and potentially highly specific drug targets.
A number of opportunities and challenges exist for the future pharmacological manipulation of lncRNAs. RNA therapeutics can take advantage of various lncRNA cellular functions and target those pathways through gene silencing and structure disruption mechanisms. lncRNAs are functional molecules that can be detected in the body fluids; hence, they can serve as diagnostic biomarkers for various diseases. Occasionally, a single lncRNA can target several mRNAs, and in such situations, manipulating the lncRNA can help to modulate multiple genes and their functions. Extensive secondary structures and long lncRNA size may hinder the design of effective small interfering RNAs (siRNAs) and small molecule inhibitors. Toxicity might be observed with siRNA or antisense oligonucleotide (ASO)-mediated knockdown strategies designed to disrupt lncRNA functions. Single-stranded ASOs are highly unstable in cells and subject to nucleases. Other difficulties encountered with ASOs include low target affinity and low potency, which may require the use of higher concentrations that in turn could lead to off-target effects. Chemical modifications to ASOs, such as phosphorothioate modifications, heterocyclic modifications, 2’-O-methyl modifications, and 5′-, 3′-end-locked nucleic acid (LNA) modifications, are known to increase affinity and cellular uptake and decrease toxicity [94]. The low expression of lncRNAs may permit the use of lower doses, which may alleviate some toxicities. Off-target hybridization effects can be minimized by careful bioinformatics selection of ASO sequences. Some of the possible pharmacological approaches to target lncRNAs are discussed below and are summarized in Figure 1.
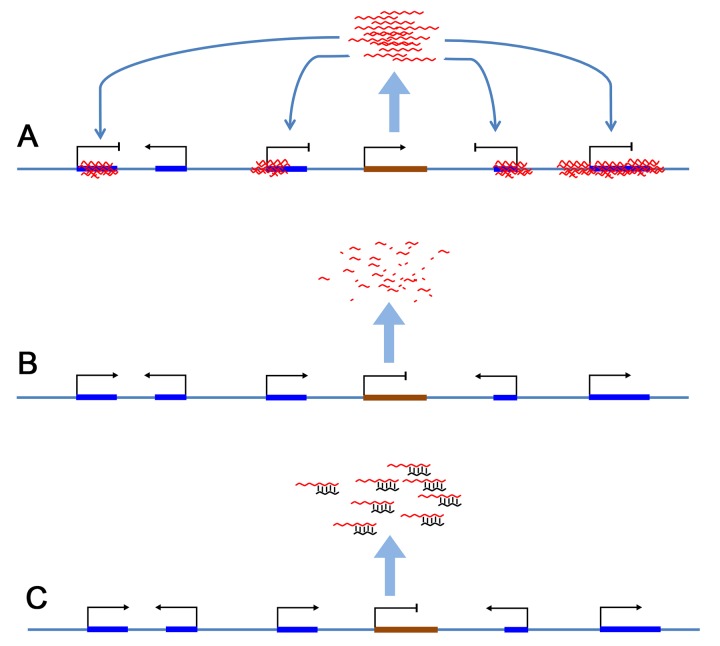
Figure 1.
An Overview of pharmacological strategies to modulate lncRNA functions. A. A diagram of a typical gene cluster regulated by a lncRNA. lncRNA genes produce a pool of lncRNAs, which then interact with nearby genes. Since a gene cluster often contains genes that function in the same pathways, a single lncRNA can coordinately regulate the locus and consequently a pathway of interest. B. Pharmacological targeting strategies can be designed whose mode of action is the direct destruction of lncRNAs. Examples include antisense oligonucleotides, siRNA approaches, and ribozymes. C. Pharmacological strategies can also be devised whose mode of action is the competitive inhibition of interactions between lncRNAs and their target genes or interacting ribonucleoproteins. Examples include small molecule inhibitors and synthetic stabilized oligonucleotides.
SINEUPs: Short interspersed nuclear elements (SINEs) are short (< 500 bp) non-coding repetitive sequences that can transpose into new parts of the genome by means of an RNA intermediate and reverse transcriptase. SINEUPs are a new class of natural antisense lncRNAs that contain an embedded SINEB2 (SINE of B2 family element), and these can Up-regulate translation of target mRNAs. SINEUPs are known to enhance protein synthesis of their target mRNA and function post-transcriptionally [95,96]. SINEUPs contain two functional domains: a Binding Domain (BD), which provides specificity (via base-pairing) to the targeted mRNA, and an Effector Domain (ED) that acts as an activator of translation. The BD consists of 72 nucleotides, which include sequences that are complimentary to the 5’-UTR, the translation initiation codon and initial codons. The ED contains repetitive SINEB2 sequences. The SINEUP lncRNA base-pairs with the mRNA, and the SINEB2 element facilitates association of the mRNA with polysomes, thus increasing the rate of translational initiation [96].
The use of synthetic SINEUPs to increase protein synthesis has been demonstrated both by in vitro and in vivo studies. Uchl1 (ubiquitin carboxyterminal hydrolase L1) is a gene associated with Parkinson’s disease and other neurodegenerative diseases. AS Uchl1 is a natural anti-sense lncNRA that targets Uchl1 mRNA. Under conditions of cellular stress, AS Uchl1 shuttles to the cytoplasm, where it induces Uchl1 mRNA to become highly loaded with ribosomes, thereby increasing its translation [97]. Mutations in subunit 7B of cytochrome C oxidase (cox7B) result in microphthalmia with linear skin defects (MLS) syndrome. In an in vivo study conducted on medakafish (Oryzias latipes), MLS (microphthalmia and microcephaly) was induced by downregulating cox7B using morpholinos that targets cox7B. A synthetic (SINEUP-cox7B) reversed the microphthalmia and microcephaly in MLS Cox7B morphants in a dose-dependent manner, without any increase in the Cox7B mRNA levels [98]. More research into efficient in vivo delivery systems may enable the use of SINEUPs as a therapeutic agent to treat a wide variety of diseases caused by reduced mRNA translation.
RNA interference (RNAi): RNA interference is a mechanism for inducing gene silencing by double stranded RNA. siRNAs are 21-23 nucleotide long RNAs with 3'-dinucleotide overhangs produced by cleavage of double-stranded RNAs by the enzyme Dicer. siRNAs are incorporated into a protein-RNA complex, the RNA-induced Silencing Complex (RISC). The siRNA then binds to the target mRNA and degrades it through perfect sequence complementarity, leading to the recruitment of ribonucleases. siRNAs are highly potent and generally do not require any chemical modifications for activity. siRNA drugs designed to disrupt mRNAs involved in cancer are currently in clinical trials [99]. Similar siRNA strategies can be applied to target lncRNAs. Efficient delivery of siRNAs using nanoparticles and lipid-encapsulation will increase uptake and pharmacokinetic duration of drug delivery [100]. A siRNA directed against MALAT-1 lncRNA in prostate cancer cells resulted in down-regulation of MALAT-1, inhibited cell growth, invasion, migration, and induced cell cycle arrest [101]. siRNA-mediated knockdown of HOTAIR lncRNA inhibited matrix invasion in breast cancer cell lines [85], and injection of siRNA-transfected cells inhibited xenograft efficiency of gastric tumors and metastasis in peritoneal and non-small cell lung cancer [102,103]. Thus, RNAi is a robust and effective strategy to downregulate pathogenic lncRNAs.
Antisense Oligonucleotides (ASO): Antisense oligonucleotides and antisense drugs have been explored as RNA inhibitors in the past to treat various diseases and are currently in different phases of clinical trials. The use of ASO technology to target lncRNAs is a logical next step [104]. ASOs are single-stranded DNA sequences and can be made complementary to the target lncRNAs [105]. In the nucleus, they hybridize with targeted lncRNAs to form RNA:DNA heteroduplexes, which trigger cleavage of the RNA moiety by endogenous RNase H1 activity [106]. ASO blocking of MALAT-1 was shown to prevent lung cancer metastasis [107] and ASO blocking of lncRNA APOA1-AS upregulated high-density lipoprotein particles (HDL) [31]. lncRNA lengths and complex secondary structures complicate ASO design, but systematic evolution of ligands by exponential enrichment (SELEX)-based approaches can be used to identify the best target RNA sequences [108,109]. However, lncRNAs must fold properly in vitro in order for SELEX-based approaches to correctly identify target sequences. ASOs can be delivered in vivo and chemical modifications such as phosphorothioate and LNA modifications enable its endocytotic uptake by cell surface receptors. Thus, ASOs serve as an important platform to modulate gene expression, but more research is required to optimize the synthesis of oligonucleotides to efficiently target lncRNAs, hopefully with high-potency and limited toxicity.
Ribozymes: Hammerhead ribozymes are approximately 30-nucleotide long, self-cleaving, and nuclease-resistant catalytic RNA oligonucleotides that can bind to and attack the 2′-OH that is 5′- to the scissile bond (a covalent bond, which can be broken by enzymes) in specific RNA targets, resulting in destabilization of the phosphodiester backbone of the targeted RNA molecule [110-112]. Once cleaved, the ribozymes dissociate from the products and may cleave other target RNAs. The cleavage is highly sequence-specific and is sensitive to single nucleotide mismatches; hence, toxicity due to off-target effects can be minimal [113]. Stability of short ribozyme sequences in the presence of endogenous ribonucleases poses a problem, but this has been addressed by the chemical modification of the phosphate and sugar moieties. Incorporation of four phosphorothioate linkages at the 5′- end of the ribozymes and 2′-O-Me nucleotides in place of the 2′- hydroxyl group has been shown to stabilize ribozymes without altering their catalytic activity [114,115]. Targeted delivery of ribozymes to specific cellular compartments where lncRNAs are located using liposomes or peptide-based delivery systems provides an alternative to RNAi [116]. Synthetic ribozymes against VEGF mRNA administered in vivo decreased growth and metastasis of solid tumors, and the same methodology can be adapted to target specific lncRNAs [112,117]. Thus, ribozymes are an exciting future therapeutic tool that can be used to treat various lncRNA-associated diseases.
Small molecule inhibitors: Small molecule inhibitors can alter gene expression by perturbing the interactions of lncRNAs with chromatin modifying proteins. lncRNAs are folded into secondary structures that often change conformation upon interaction with ribonucleoproteins [118]. PAN RNAs produced by KSHV and MALAT-1 both have an ENE in their structure, which stabilizes these lncRNAs and retains them in the nucleus. ENEs are activated when the 3′-poly (A) tail hybridizes to the U-rich internal loop in the ENE to form a triple helical structure. This triple helix confers stability upon the lncRNA from rapid deadenylation-dependent decay [119]. Future small molecule inhibitors that can disrupt stable lncRNA secondary structure or inhibit lncRNA association with accessory proteins or target genes may provide other avenues to target dysregulated lncRNAs. Fatemi et al. used Amplified Luminescent Proximity Homogeneous Assay (ALPHA screen assay) to analyze the RNA-protein interactions and identified small molecules using high-throughput compound screening methods. They reported the specific and quantifiable binding of brain-derived neurotrophic factor antisense (BDNF-AS) lncRNA to protein EZH2 (component of PCR2) and also identified a small-molecule inhibitor Ellipticine that upregulated its downstream target genes [120,121]. In-depth studies involving lncRNA secondary structures are necessary in the future to allow the possibility of identifying new and efficient small molecule inhibitors that can specifically bind to the target lncRNAs.
Conclusions
An emerging paradigm in epigenetics research indicates that lncRNAs regulate many genes in clusters and this is an active area of research, which needs more focus in the future. lncRNA dysregulation has recently been observed as a common feature in a wide range of human diseases and disorders. lncRNAs function by a variety of mechanisms, but typically they function to regulate the expression of protein-coding genes through epigenetic mechanisms, often by recruiting chromatin remodeling enzymes to gene clusters. Therefore, lncRNAs can be viewed as master regulators of gene expression and constitute attractive targets for specific epigenetic pharmacological therapy. Tractable pharmacological approaches are now available that can either degrade overexpressed lncRNAs or competitively inhibit their function. Now, for the first time, it may be possible to develop drugs that target specific lncRNAs, thus offering the ability to epigenetically modulate specific biochemical pathways. If so, lncRNA drugs will offer significant improvements to the very low specificity of existing drugs that function as epigenetic modifiers.
Acknowledgments
This work was supported in the part by the National Institutes of Health National Institute of General Medical Sciences (Grants R01GM118367 and R01GM087376) to XBZ.
Glossary
- ASO
Antisense oligonucleotide
- BWS
Beckwith-Wiedemann syndrome
- DMR
differentially methylated region
- DNMT
DNA methyltransferase
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- lincRNA
long intergenic noncoding RNA
- lncRNA
long non-coding RNA
- MBP
methyl-CpG-binding protein
- NAT
natural antisense lncRNA
- ORF
open reading frame
- PTM
post-translational modification
- PRC
Polycomb repressive complex
- RPKM
reads per kilobase per million base pairs
- SINE
short interspersed nuclear element SINEUP, to up-regulate translation
- siRNA
small interfering RNA
- TrxG
trithorax group
- T-UCR
transcribed ultraconserved regions
- UTR
untranslated region
References
- Waddington CH. The Epigenotype. Int J Epidemiol. 2012;41(1):10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128(4):635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Karapetyan AR, Buiting C, Kuiper RA. et al. Regulatory roles for long ncRNA and mRNA. Cancers (Basel) 2013;5(2):462–490. doi: 10.3390/cancers5020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner MV, Pauler MV, Huang R. et al. The function of non-coding RNAs in genomic imprinting. Development. 2009;136(11):1771–1783. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autuoro JM, Pirnie SP, Carmichael GG. Long noncoding RNAs in imprinting and X chromosome inactivation. Biomolecules. 2014;4(1):76–100. doi: 10.3390/biom4010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeulle C, Heard E. Antisense RNA in imprinting: Spreading silence through air. Trends Genet. 2002;18(9):434–437. doi: 10.1016/s0168-9525(02)02749-x. [DOI] [PubMed] [Google Scholar]
- Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13(3):313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T. et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503(7476):371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalei V, Sansom SN, Kong L. et al. The long non-coding RNA dali is an epigenetic regulator of neural differentiation. Elife. 2014;3:e04530. doi: 10.7554/eLife.04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhao Y, Bao X. et al. LncRNA dum interacts with dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015;25(3):335–350. doi: 10.1038/cr.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T. et al. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012;8(7):e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128(4):635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- He JH, Han ZP, Li YG. Association between long non-coding RNA and human rare diseases (review). Biomed Rep. 2014;2(1):19–23. doi: 10.3892/br.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1(2):239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Huo XS, Yuan SX. et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49(6):1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Beckedorff FC, Ayupe AC, Crocci-Souza R. et al. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genet. 2013;9(8):e1003705. doi: 10.1371/journal.pgen.1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Birney E, Stamatoyannopoulos E, Dutta A. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Genome Sequencing Consortium I. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Ohno S. So much “junk” DNA in our genome. Brookhaven Symp Biol. 1972;23:366–370. [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G. et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Non-coding RNA. Hum Mol Genet. 2006;15(90001):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM. et al. GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL. et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015;31(5):239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley P, Kadakkuzha BM, Faghihi MA. et al. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014;6(1):222–230. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: Expression noise or expression choice? Genomics. 2009;93(4):291–298. doi: 10.1016/j.ygeno.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–237. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk M, Thulluru HK, Mulders J. et al. HELLP babies link a novel lincRNA to the trophoblast cell cycle. J Clin Invest. 2012;122(11):4003–4011. doi: 10.1172/JCI65171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL. et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD. Capture hybridization analysis of RNA targets (CHART). Curr Protoc Mol Biol. 2013;Chapter 21(Unit 21):25. doi: 10.1002/0471142727.mb2125s101. [DOI] [PubMed] [Google Scholar]
- Lam MT, Li W, Rosenfeld MG. et al. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39(4):170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa R. Promoter-associated long noncoding RNAs repress transcription through a RNA binding protein TLS. Adv Exp Med Biol. 2011;722:196–208. doi: 10.1007/978-1-4614-0332-6_12. [DOI] [PubMed] [Google Scholar]
- Luke B, Lingner J. TERRA: Telomeric repeat-containing RNA. EMBO J. 2009;28(17):2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I. et al. Ultraconserved elements in the human genome. Science. 2004;304(5675):1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Milligan MJ, Lipovich L. Pseudogene-derived lncRNAs: Emerging regulators of gene expression. Front Genet. 2015;5:476. doi: 10.3389/fgene.2014.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M. et al. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23(12):614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T. et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR. et al. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput Biol. 2008;4(11):e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT. et al. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154(1):240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson P, Lipovich L, Grander D. et al. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014;1840(3):1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–237. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM. et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto CC, Pari GS. PAN’s labyrinth: Molecular biology of kaposi’s sarcoma-associated herpesvirus (KSHV) PAN RNA, a multifunctional long noncoding RNA. Viruses. 2014;6(11):4212–4226. doi: 10.3390/v6114212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A. et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Mizutani R, Salam KA. et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22(5):947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Johnston RL, Inostroza-Ponta M. et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22(5):885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Sacco L, Baldassarre A, Masotti A. Bioinformatics tools and novel challenges in long non-coding RNAs (lncRNAs) functional analysis. Int J Mol Sci. 2012;13(1):97–114. doi: 10.3390/ijms13010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F. et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32(2):232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Questa JI, Sun Q. et al. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci U S A. 2014;111(45):16160–16165. doi: 10.1073/pnas.1419030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T. et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J. et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Wu H. et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: Long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6(5):539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K. et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Guseva N. et al. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137(15):2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C. et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA firre. Nat Struct Mol Biol. 2014;21(2):198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Lessons from X-chromosome inactivation: Long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23(16):1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Mlynarczyk-Evans S, Nusinow DA. et al. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA. et al. The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322(5908):1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT. The long noncoding RNA, jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143(3):390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM. et al. Expression of a noncoding RNA is elevated in alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14(7):723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via alu elements. Nature. 2011;470(7333):284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Sun W, Ye G. et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11(225):5876. doi: 10.1186/1479-5876-11-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qian CY, Li XP. et al. Genome-scale long noncoding RNA expression pattern in squamous cell lung cancer. Sci Rep. 2015;5:11671. doi: 10.1038/srep11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Sun Q, Cheng X. et al. Genome-wide analysis of long noncoding RNA (lncRNA) expression in colorectal cancer tissues from patients with liver metastasis. Cancer Med. 2016;5(7):1629–1639. doi: 10.1002/cam4.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Ferracin M. et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- Hajjari M, Salavaty A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12(1):1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM. et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SS, Zheng BY, Xiong XD. LincRNA-p21: Implications in human diseases. Int J Mol Sci. 2015;16(8):18732–18740. doi: 10.3390/ijms160818732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S. et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9(9):1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Pasmant E, Laurendeau I, Heron D. et al. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67(8):3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- Mosemiller AK, Dalton JC, Day JW. et al. Molecular genetics of spinocerebellar ataxia type 8 (SCA8). Cytogenet Genome Res. 2003;100(1-4):175–183. doi: 10.1159/000072852. [DOI] [PubMed] [Google Scholar]
- Ptak C, Petronis A. Epigenetics and complex disease: From etiology to new therapeutics. Annu Rev Pharmacol Toxicol. 2008;48:257–276. doi: 10.1146/annurev.pharmtox.48.113006.094731. [DOI] [PubMed] [Google Scholar]
- Jamieson AC, Miller JC, Pabo CO. Drug discovery with engineered zinc-finger proteins. Nat Rev Drug Discov. 2003;2(5):361–368. doi: 10.1038/nrd1087. [DOI] [PubMed] [Google Scholar]
- Bennett CF, Swayze EE. RNA targeting therapeutics: Molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50(1):259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- Zucchelli S, Cotella D, Takahashi H. et al. SINEUPs: A new class of natural and synthetic antisense long non-coding RNAs that activate translation. RNA Biol. 2015;12(8):771–779. doi: 10.1080/15476286.2015.1060395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchelli S, Fasolo F, Russo R. et al. SINEUPs are modular antisense long non-coding RNAs that increase synthesis of target proteins in cells. Front Cell Neurosci. 2015;9:174. doi: 10.3389/fncel.2015.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M. et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Indrieri A, Grimaldi C, Zucchelli S. et al. Synthetic long non-coding RNAs [SINEUPs] rescue defective gene expression in vivo. Sci Rep. 2016;6:27315. doi: 10.1038/srep27315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam JK, Chow MY, Zhang Y. et al. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L. et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23(8):1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Ren S, Liu Y, Xu W. et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013;190(6):2278–2287. doi: 10.1016/j.juro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Okugawa Y, Toiyama Y, Hur K. et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35(12):2731–2739. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XH, Liu ZL, Sun M. et al. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastelein JJ, Wedel MK, Baker BF. et al. Potent reduction of apolipoprotein B and low-density lipoprotein cholesterol by short-term administration of an antisense inhibitor of apolipoprotein B. Circulation. 2006;114(16):1729–1735. doi: 10.1161/CIRCULATIONAHA.105.606442. [DOI] [PubMed] [Google Scholar]
- Chan JH, Lim S, Wong WF. Antisense oligonucleotides: From design to therapeutic application. Clinical and Experimental Pharmacology and Physiology. 2006;33(5-6):533–540. doi: 10.1111/j.1440-1681.2006.04403.x. [DOI] [PubMed] [Google Scholar]
- Zong X, Huang L, Tripathi V. et al. Knockdown of nuclear-retained long noncoding RNAs using modified DNA antisense oligonucleotides. Methods Mol Biol. 2015;1262:321–331. doi: 10.1007/978-1-4939-2253-6_20. [DOI] [PubMed] [Google Scholar]
- Gutschner T, Hammerle M, Eissmann M. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17(1):106–116. doi: 10.1093/bib/bbv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: New links in cancer progression. Cancer Res. 2011;71(1):3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Gerlach WL. Simple RNA enzymes with new and highly specific endoribonuclease activities. Biotechnology. 1992;24:264–269. [PubMed] [Google Scholar]
- Scott WG, Murray JB, Arnold JR. et al. Capturing the structure of a catalytic RNA intermediate: The hammerhead ribozyme. Science. 1996;274(5295):2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- Citti L, Rainaldi G. Synthetic hammerhead ribozymes as therapeutic tools to control disease genes. Curr Gene Ther. 2005;5(1):11–24. doi: 10.2174/1566523052997541. [DOI] [PubMed] [Google Scholar]
- Phylactou LA, Tsipouras P, Kilpatrick MW. Hammerhead ribozymes targeted to the FBN1 mRNA can discriminate a single base mismatch between ribozyme and target. Biochem Biophys Res Commun. 1998;249(3):804–810. doi: 10.1006/bbrc.1998.9241. [DOI] [PubMed] [Google Scholar]
- Beigelman L, McSwiggen JA, Draper KG. et al. Chemical modification of hammerhead ribozymes. catalytic activity and nuclease resistance. J Biol Chem. 1995;270(43):25702–25708. doi: 10.1074/jbc.270.43.25702. [DOI] [PubMed] [Google Scholar]
- Heidenreich O, Benseler F, Fahrenholz A. et al. High activity and stability of hammerhead ribozymes containing 2’-modified pyrimidine nucleosides and phosphorothioates. J Biol Chem. 1994;269(3):2131–2138. [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: Regulation of gene expression without proteins. Nat Rev Genet. 2007;8(10):776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavco PA, Bouhana KS, Gallegos AM. et al. Antitumor and antimetastatic activity of ribozymes targeting the messenger RNA of vascular endothelial growth factor receptors. Clin Cancer Res. 2000;6(5):2094–2103. [PubMed] [Google Scholar]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- Brown JA, Bulkley D, Wang J. et al. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol. 2014;21(7):633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram Fatemi R, Salah-Uddin S, Modarresi F. et al. Screening for small-molecule modulators of long noncoding RNA-protein interactions using AlphaScreen. J Biomol Screen. 2015;20(9):1132–1141. doi: 10.1177/1087057115594187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi RP, Velmeshev D, Faghihi MA. De-repressing LncRNA-targeted genes to upregulate gene expression: Focus on small molecule therapeutics. Mol Ther Nucleic Acids. 2014;3:e196. doi: 10.1038/mtna.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegedi K, Sonkoly E, Nagy N. et al. The anti-apoptotic protein G1P3 is overexpressed in psoriasis and regulated by the non-coding RNA, PRINS. Exp Dermatol. 2010;19(3):269–278. doi: 10.1111/j.1600-0625.2010.01066.x. [DOI] [PubMed] [Google Scholar]
- Song J, Kim D, Han J. et al. PBMC and exosome-derived hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin Exp Med. 2015;15(1):121–126. doi: 10.1007/s10238-013-0271-4. [DOI] [PubMed] [Google Scholar]
- Muniz L, Kiss T, Egloff S. Misregulation of P-TEFb activity: Pathological consequences. Med Sci (Paris) 2012;28(2):200–205. doi: 10.1051/medsci/2012282019. [DOI] [PubMed] [Google Scholar]
- Wang K, Liu F, Zhou LY. et al. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. 2014;114(9):1377–1388. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- Ishii N, Ozaki K, Sato H. et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51(12):1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res. 2014;115(7):668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- Wu W, Bhagat TD, Yang X. et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in barrett’s esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144(5):956–966. doi: 10.1053/j.gastro.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao YQ, Huang ML, Xu AT. et al. LncRNA DQ786243 affects treg related CREB and Foxp3 expression in crohn’s disease. J Biomed Sci. 2013;20:87. doi: 10.1186/1423-0127-20-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillambhi S, Turan S, Hwang DY. et al. Deletion of the noncoding GNAS antisense transcript causes pseudohypoparathyroidism type ib and biparental defects of GNAS methylation in cis. J Clin Endocrinol Metab. 2010;95(8):3993–4002. doi: 10.1210/jc.2009-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Faghihi MA, Modarresi F. et al. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One. 2008;3(1):e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori C, Peschansky VJ, Barbouth D. et al. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in fragile X syndrome and fragile X-associated tremor/ataxia syndrome. Hum Genet. 2014;133(1):59–67. doi: 10.1007/s00439-013-1356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Tripathi MK, Chaudhuri G. Down-regulation of 7SL RNA expression and impairment of vesicular protein transport pathways by leishmania infection of macrophages. J Biol Chem. 2005;280(32):29364–29373. doi: 10.1074/jbc.M504162200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I. et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TH, Zhang Z, Yagi M. et al. Molecular characterization of an X(p21.2;q28) chromosomal inversion in a duchenne muscular dystrophy patient with mental retardation reveals a novel long non-coding gene on Xq28. J Hum Genet. 2013;58(1):33–39. doi: 10.1038/jhg.2012.131. [DOI] [PubMed] [Google Scholar]
- Cabianca DS, Casa V, Bodega B. et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149(4):819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause angelman syndrome. Nat Genet. 1997;15(1):70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Scheele C, Petrovic N, Faghihi MA. et al. The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genomics. 2007;8:74. doi: 10.1186/1471-2164-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeweyer G, Van der Aa N, Ceulemans B. et al. A de novo balanced t(2;6)(p15;p22.3) in a patient with west syndrome disrupts a lnc-RNA. Epilepsy Res. 2012;99(3):346–349. doi: 10.1016/j.eplepsyres.2011.12.009. [DOI] [PubMed] [Google Scholar]
- Sandbacka M, Bruce S, Halttunen M. et al. Methylation of H19 and its imprinted control region (H19 ICR1) in mullerian aplasia. Fertil Steril. 2011;95(8):2703–2706. doi: 10.1016/j.fertnstert.2011.03.019. [DOI] [PubMed] [Google Scholar]
- Yang X, Song JH, Cheng Y. et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63(6):881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou XF, Pan GF. et al. Enhanced expression of long non-coding RNA ZXF1 promoted the invasion and metastasis in lung adenocarcinoma. Biomed Pharmacother. 2017;68(4):401–407. doi: 10.1016/j.biopha.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Pickard MR, Williams GT. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: Implications for chemotherapy. Breast Cancer Res Treat. 2014;145(2):359–370. doi: 10.1007/s10549-014-2974-y. [DOI] [PubMed] [Google Scholar]
- Sarver AL, Murray CD, Temiz NA. et al. MYC and PVT1 synergize to regulate RSPO1 levels in breast cancer. Cell Cycle. 2016;15(7):881–885. doi: 10.1080/15384101.2016.1149660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Zhang L, Liu X. et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111(4):736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WC, Fu WM, Wong CW. et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZH, Wang XL, Tang HM. et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32(1):395–402. doi: 10.3892/or.2014.3186. [DOI] [PubMed] [Google Scholar]
- Chen D, Zhang Z, Mao C. et al. ANRIL inhibits p15(INK4b) through the TGFbeta1 signaling pathway in human esophageal squamous cell carcinoma. Cell Immunol. 2014;289(1-2):91–96. doi: 10.1016/j.cellimm.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Xie HW, Wu QQ, Zhu B. et al. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumour Biol. 2014;35(8):7743–7754. doi: 10.1007/s13277-014-2013-y. [DOI] [PubMed] [Google Scholar]
- Yang F, Xue X, Zheng L. et al. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-myc mRNA stability. FEBS J. 2014;281(3):802–813. doi: 10.1111/febs.12625. [DOI] [PubMed] [Google Scholar]
- Conrad NK. New insights into the expression and functions of the kaposi’s sarcoma-associated herpesvirus long noncoding PAN RNA. Virus Res. 2016;212:53–63. doi: 10.1016/j.virusres.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Kong G, You X. et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287(31):26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braconi C, Valeri N, Kogure T. et al. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2011;108(2):786–791. doi: 10.1073/pnas.1011098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Hammerle M, Eissmann M. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JJ, Navarro A, Vinolas N. et al. LincRNA-p21 impacts prognosis in resected non-small-cell lung cancer patients through angiogenesis regulation. J Thorac Oncol. 2016 doi: 10.1016/j.jtho.2016.07.015. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gao S, De Geyter C. A natural antisense transcript, BOKAS, regulates the pro-apoptotic activity of human bok. Int J Oncol. 2009;34(4):1135–1138. doi: 10.3892/ijo_00000241. [DOI] [PubMed] [Google Scholar]