Abstract
Objective
In type 2 diabetes (T2D), pancreatic β cells become progressively dysfunctional, leading to a decline in insulin secretion over time. In this study, we aimed to identify key genes involved in pancreatic beta cell dysfunction by analyzing multiple mouse strains in parallel under metabolic stress.
Methods
Male mice from six commonly used non-diabetic mouse strains were fed a high fat or regular chow diet for three months. Pancreatic islets were extracted and phenotypic measurements were recorded at 2 days, 10 days, 30 days, and 90 days to assess diabetes progression. RNA-Seq was performed on islet tissue at each time-point and integrated with the phenotypic data in a network-based analysis.
Results
A module of co-expressed genes was selected for further investigation as it showed the strongest correlation to insulin secretion and oral glucose tolerance phenotypes. One of the predicted network hub genes was Elovl2, encoding Elongase of very long chain fatty acids 2. Elovl2 silencing decreased glucose-stimulated insulin secretion in mouse and human β cell lines.
Conclusion
Our results suggest a role for Elovl2 in ensuring normal insulin secretory responses to glucose. Moreover, the large comprehensive dataset and integrative network-based approach provides a new resource to dissect the molecular etiology of β cell failure under metabolic stress.
Keywords: Diabetes, Pancreas, Beta cell dysfunction, Network analysis, Molecular phenotyping, Metabolic stress
Highlights
-
•
High fat and normal chow fed mice from six common strains were studied over 3 months.
-
•
Islet RNA-Seq and phenotype data were integrated in a network-based analysis.
-
•
Elovl2 was identified as a hub in a network module related to insulin secretion.
-
•
Elovl2 silencing decreased insulin secretion in mouse and human β cell lines.
1. Introduction
An absolute or relative decrease in insulin secretion by pancreatic β-cells underlies the development of type 1 and type 2 diabetes, respectively. These diseases impose a huge burden on welfare systems in both developed and developing countries, affecting ∼8% of the adult population and consuming $160 billion (USD) per annum (International Diabetes Federation, 2015). In particular, a sedentary lifestyle and consumption of highly calorific diets with a substantial fat content play a central role in inducing insulin resistance and type 2 diabetes. The latter develops when insulin secretion by the β-cell becomes insufficient to overcome insulin resistance [1], [2]. Our incomplete knowledge of β-cell biology in health and disease [3] means that only limited therapeutic options presently exist to treat diabetes, and there are none to prevent or cure the disease.
Many recent studies have evaluated the role of specific mechanisms such as the action of reactive oxygen or nitrogen species, endoplasmic reticulum (ER) stress, sphingolipid metabolism, or autophagy in β-cell failure [4], [5], [6]. Although these mechanisms represent normal physiological responses to environmental challenges, if unchecked, they may ultimately lead to cellular dysfunction, de-differentiation [7], [8], or even death [9], particularly when the intensity of the original stimulus exceeds a given threshold and is long-lasting. Importantly, besides the nature of the triggering signal, genetic architecture also dictates cellular sensitivity to these signals. Thus, in man, more than 90 genetic loci are implicated in type 2 diabetes risk, most of these affecting β-cell function [10]. Likewise, C57Bl/6J mice display a defective insulin secretory response to glucose compared to C57Bl/6N mice [11], a difference due to a single mutation in the Nnt gene [12] that alters the susceptibility to develop glucose intolerance and β-cell dysfunction [11]. Dissection of how specific signals lead to different β-cell responses depending on genetic background is thus a critical goal of β-cell research.
Approaches to examining how β-cells respond to pro-diabetic challenges including the fatty acid palmitate have previously used clonal β-cells and human islets and involved transcriptomic analyses by microarray or massive parallel sequencing (RNAseq) [13], [14], [15], [16]. One of the limitations of these earlier studies is the use of in vitro models in which the harmful effects of lipids on β cells are often exaggerated compared to those in vivo. However, in an attempt to overcome this problem, a few studies have been performed using mice fed high fat (HF) diets [17], [18], [19], [20]. Under these conditions, mice usually show significant increases in body weight, as well as increased blood glucose and insulin levels, during the first weeks of exposure to the modified diet. High fat-high sucrose (HFHS) diet-fed mice become severely glucose intolerant and insulin resistance progressively worsens with time [18]. However, the development and severity of the diabetic phenotype depends on several factors, including the choice of genetic background. In addition, the composition of the diets administered, the housing conditions, and the experimental setup for mouse phenotyping and downstream analysis all have an impact on disease severity.
The C57Bl/6J line is established as the strain of choice for the HFHS diet model of diabetes as treated mice develop a more severe diabetic phenotype in response to the diet compared to other strains [18], [21]. This has led to the notion of “resistant” and “susceptible” strains, where the genetic differences in metabolic response are more important for the development of obesity and diabetes than the increased calorific intake itself [21], [22]. Although several previous studies have compared the glycemic and insulinogenic response of different strains on a high fat or a regular diet [23], [24], [25], [26], [27], [28], a systematic evaluation combining phenotypic with islet genomic data is lacking. However, such an analysis could, at least in theory, help to identify new targets associated with β-cell dysfunction induced by metabolic stress in an unbiased manner.
Here, we analyzed in parallel the effect of a HFHS diet on six commonly used laboratory mouse strains using carefully controlled housing and experimental conditions. The strains were chosen as they are amongst those most frequently used in metabolic studies and show marked differences in susceptibility to diet-induced obesity and β-failure [25], [26], [27], [29], [30], [31]. Mice from these six strains were phenotyped in depth for glucose homeostasis, insulin resistance, and islet morphometry after 2, 10, 30, or 90 days of HFHS versus regular chow (RC) diet. This was complemented by deep sequencing of mouse islet mRNA under the same conditions and at the same time-points. After initial evaluation and comparison of the degree of diet-induced dysglycemia in the different strains, we integrated the phenotype measurements with islet gene expression data in a network-based analysis. This multi-parameter systems-based approach led to the identification of a sub-network of islet-expressed genes associated with glucose tolerance and insulin secretory capacity. Using this approach, we provide evidence that Elongase of very long-chain fatty acids 2 (Elovl2), an enzyme involved in very long chain fatty acid synthesis and highlighted by our network analysis, is a key player in regulating glucose-stimulated insulin secretion in the context of β-cell dysfunction.
2. Material and methods
The experimental protocol was approved by the institutional animal care and use committee of the Paris Diderot University (CEEA40).
2.1. Mouse phenotyping
Eight-week old male mice from six different strains: C57Bl/6J, DBA/2J, A/J, AKR/J, 129S2/SvPas, and BALB/cJ were housed on a 12-h light/dark cycle and were fed a standard rodent chow (RC) diet (SAFE A04) or high-fat high-sucrose (HFHS) diet (SAFE 235F, with 46% fat expressed in Kcal/kg), ad libitum. Islets were isolated and mice were phenotyped at 2, 10, 30, and 90 days of HFHS or RC diet. Protocols for islet isolation and phenotyping are described in Supplementary Methods.
2.2. In vitro insulin secretion measurements
MIN6 cells were seeded in 96-well plates and treated for 24 h in the presence of various glucose concentrations. Cells were then pre-incubated in KRBH containing 0.2% fatty-acid free BSA and 2 mM glucose for 30 min. Insulin secretion was measured following a 30 min incubation in KRBH containing 0.2% defatted BSA with 2 mM glucose or 20 mM glucose. The insulin concentration in the medium was determined by Ultra Sensitive Mouse Insulin ELISA kit (Alpco, Salem, USA).
Beta TC-tet cells were washed with PBS and pre-incubated for 2 h in KRBH-BSA (supplemented with 2 mM glucose), then the medium was replaced with fresh KRBH-BSA containing 2 mM glucose or 20 mM glucose + 100 nM Exendin-4 and incubated for 1 h. Secreted and cellular insulin were assessed by radioimmunoassay (RIA) using RIA kit (Millipore, MA, USA) following manufacturer's instructions.
Five days after transfection, EndoC-βH1 cells were starved in 0.5 mM glucose DMEM-based medium. After 24 h starvation, cells were washed twice and then pre-incubated in KRBH containing 0.2% fatty-acid free BSA and 0 mM glucose for 1 h. Insulin secretion was measured following 40 min incubation with KRBH containing 0.2% fatty-acid free BSA and 0 mM or 20 mM glucose. Insulin secretion and intracellular insulin were measured by ELISA as previously described [32].
2.3. Quantitative PCR
Real-time qPCR was performed on total 4 μg RNA isolated from mouse islets using a LightCycler 1.5 detection system (Roche). The housekeeping gene Rpl19 was used to normalize the results. Data are expressed as means ± S.E.M. and significance was assessed by the Student's t test.
2.4. RNA-Seq and downstream bioinformatics analysis
RNA-Seq analysis was performed on RNA isolated from at least 150 islets per mouse, libraries were prepared using Illumina TruSeq protocol and sequencing performed on a HiSeq2000 instrument (50-cycles). 50 nt reads were processed from 341 samples, mapped to mm9 reference genome and summary counts produced per gene. Gene counts were normalized using the trimmed mean method (EdgeR) and differential expression analysis performed using limma (voom method), correcting p-values for multiple testing using the Benjamini Hochberg method [33]. Weighted gene co-expression network analysis (WGCNA) [34] was performed on normalized RNA-Seq data and gene expression modules to phenotypic trait correlations were calculated using the Spearman method. Gene set enrichment analysis (GSEA) was performed for gene co-expression modules against canonical pathways and gene ontology categories in MSigDB. A global network was created with genes, gene co-expression modules, phenotypic traits, and pathways/GO categories represented as nodes and the relationships between them represented as edges. Network visualization was performed using Gephi 0.8.2. For further details see Supplementary Methods.
2.5. Cell transfection of siRNA
MIN6 and Beta TC-tet cells were transfected by diluting 150 nM Stealth RNAi™ siRNA specific for mouse Elovl2 (Invitrogen #MSS285122) or Stealth RNAi™ siRNA Negative Control siRNA duplex (medium GC content, from Stealth RNAi™ siRNA Negative Control Kit, Invitrogen #12935100) in 200 μl serum free medium in a 12 well tissue culture plate. The medium GC content negative siRNA control was used since the GC content of the Elovl2 siRNA is 45%. 3 μl of Lipofectamin RNAiMax (Invitrogen, Zug, Switzerland) was then added and incubated at room temperature for 20 min to form Lipid-siRNA complexes. The transfected cells were incubated at 37 °C in a CO2 and were used for the experiments, 72 h after transfection. Initially, 3 Stealth RNAi™ siRNAs specific for mouse Elovl2 were tested for efficiency of Elovl2 knock down by qPCR. The siRNA corresponding to the most 5′ location in mouse Elovl2 mRNA (NCBI Refseq NM_019423.2) and targeting exon 5 of the Elovl2 gene showed the strongest diminution of transcript (approximately 50%, data not shown) and was selected for the insulin secretion experiment. For EndoC-βH1 cells, 80 nM of Non-Targeting siRNA control pool (Dharmacon #D-001206-13) or ELOVL2 ON-TARGETplus smart pool siRNA (Dharmacon #M-009531-01) containing a pool of 4 ELOVL2 specific siRNAs were transfected using Lipofectamine RNAiMax (Lifetechnologies).
3. Results
3.1. Different mouse strains show distinct susceptibilities to developing a HFHS-induced metabolic phenotype
Male mice from six genetically different mouse strains (C57Bl/6J, DBA/2J, 129S2/SvPas, AKR/J, A/J, and BALB/cJ) were fed a HFHS or RC diet for 2, 10, 30, or 90 days, after which time various phenotypic and molecular measurements were performed. Glucose tolerance (AUC glycemia) and insulin secretion were measured by performing oral glucose tolerance tests (OGTT). Fasting glycemia, mouse weight, and pancreas weight were also recorded. In addition, quantitative histology was performed on pancreatic sections and used to estimate percentage of β and α cell mass (see Supplementary Methods for details). At each time-point, pancreatic islets were isolated and mRNA extracted for RNA-Seq. The experiments were designed to ensure that there were at least six biological replicates for each strain/time-point/diet combination.
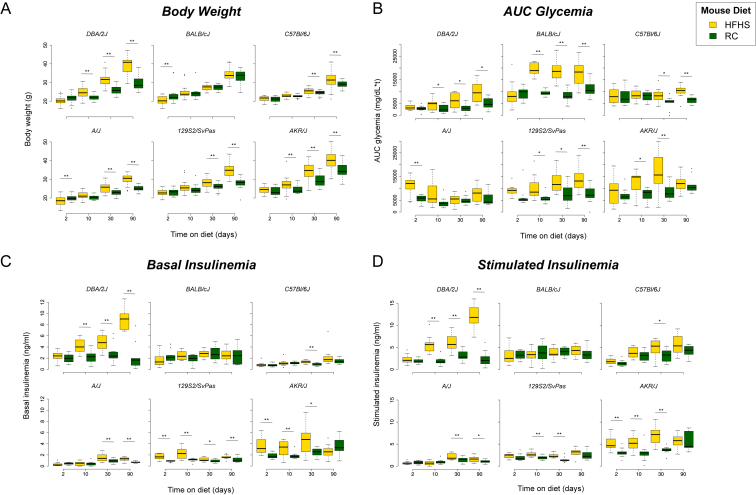
Over the three-month period, the mouse strains showed markedly different responses to HFHS diet. DBA/2J mice were characterized by severe weight gain on HFHS-diet when compared to RC-fed controls (Figure 1A) and this was accompanied by a gradual decrease in glucose tolerance (Figure 1B). In contrast, AKR/J mice showed a transient decrease in glucose tolerance up to day 30, which then started to normalize at day 90. This change was accompanied by a gradual increase in body weight in the HFHS-diet fed mice compared to controls. This was similar for 129S2/SvPas and A/J mice, both of which showed a gradual increase in body weight accompanied by a decrease in glucose tolerance. In contrast, C57Bl/6J mice showed later onset of glucose intolerance starting at day 30, associated with a moderate weight gain, whereas BALB/cJ mice showed massively decreased glucose tolerance at early time-points but showed no difference in body weight gain between HFHS diet and RC-fed mice. Plasma insulin levels (insulinemia), both basal and glucose-stimulated, were dramatically increased in DBA/2J mice on HFHS diet (Figure 1C,D), and this was accompanied by a non-significant tendency towards an increase in β-cell to α cell ratio (Supplementary Figure S1D). No clear differences in β to α cell ratio were observed for the other strains on HFHS versus RC diet, except for AKR/J mice, in which a small but significant increase was observed in HFHS-diet fed mice at day 90.
Figure 1.
Impact of HFD and age on metabolic parameters. Boxplots showing differences between HFHS (yellow) and RC (green) diet in the 6 mouse strains over time for (A) Body weight (g), (B) AUC glycemia measured during the glucose tolerance test (OGTT), (C) Basal insulinemia (ng/ml) measured at the start of the OGTT, and (D) Stimulated Insulinemia (ng/ml) measured at 15 min following glucose administration. The bottom and top of the boxes represent the first and third quartiles, with the horizontal line representing the median. The upper whiskers represent the third quartile plus 1.5× IQR (interquartile range); the lower whiskers represent the first quartile minus 1.5× IQR. Outlier points beyond this range are indicated above or below the whiskers. Statistical significance between HFHS and RC at each time-point was measured using the two-sided Student's t-test and p-values were corrected for multiple comparisons using the Benjamini Hochberg FDR method [33]. Statistically significant comparisons following FDR correction (FDR ≤ 0.05) are indicated by a double asterisk. Marginally significant comparisons (raw p-value ≤ 0.05) are indicated by a single asterisk.
Interestingly, DBA/2J was the only strain displaying a clear diabetic phenotype from the outset, with significantly higher fasting glycemia in HFHS fed mice at all time-points (Supplementary Figure 1A). These data indicate that DBA/2J and BALB/cJ are the most severely glucose intolerant (with a marked difference in insulin secretion), and along with C57Bl/6J, these three strains appear to show a progressive worsening of disease with time. Of note, whereas DBA/2J and, to a lesser extent, C57Bl/6J mice showed weight gain on HFHS diet, BALB/cJ mice experienced no such weight gain, and indeed weight loss at day 10. In contrast, AKR/J, 129S2/SvPas, and A/J mice show more transient glucose intolerance, which started to improve at three months, indicating that these strains are better able to adapt to metabolic stress. Scatter plots showing the correlations between the phenotypes shown in Figure 1 are provided as Supplementary Figure S2. A table of corresponding Pearson correlation coefficients and associated p-values is provided in Supplementary Table S1.
The mouse strains also showed differential adaptation to HFHS-diet in terms of insulin secretory response to oral glucose. In order to measure the degree of compensatory response to HFHS diet, we calculated an insulinogenic index (AUC insulin/AUC glycemia during OGTT) for each time-point and rank ordered the strains according to the difference in insulinogenic index between HFHS-diet fed and RC-diet averaged over all time-points (Table 1). Overall, DBA/2J mice show the largest insulin adaptation response to HFHS-diet, followed by AKR/J and C57Bl/6J, with 129S2/SvPas, BALB/cJ, and A/J showing the lowest adaptation.
Table 1.
Insulinogenic index difference between HFHS and RC-diet fed strains. The index is calculated as the mean difference over all four time-points.
| Strain | Insulinogenic index difference (HFHS-RC) |
|---|---|
| DBA/2J | 0.0163 |
| AKR/J | 0.0033 |
| C57Bl/6J | 0.0030 |
| A/J | −0.0014 |
| BALB/cJ | −0.0021 |
| 129S2/SvPas | −0.0038 |
3.2. Islet transcript levels are strongly influenced by strain
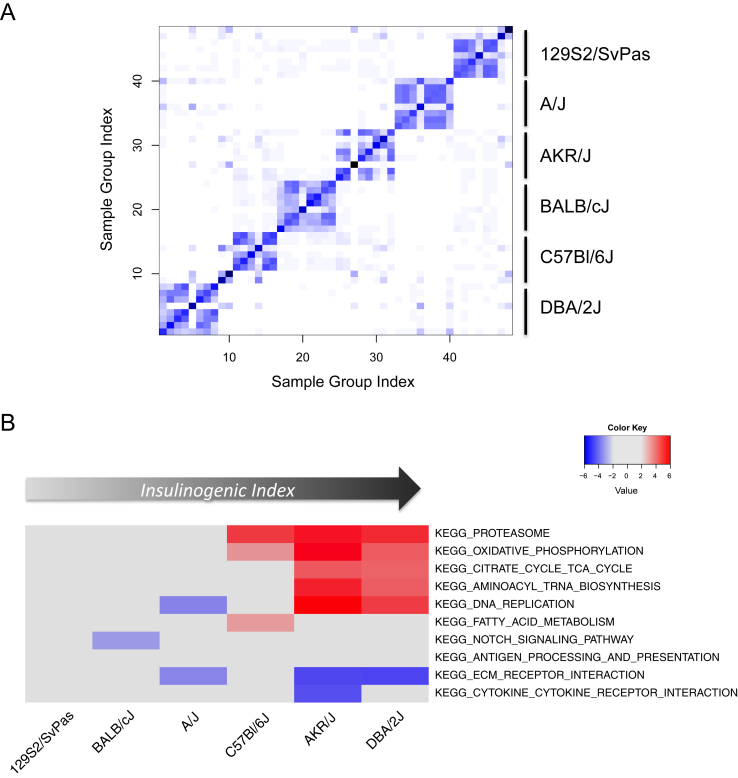
Sample similarity was calculated as previously described [35] using normalized islet transcriptomic data from 338 RNA-Seq samples. The resulting heatmap shows a clear separation of the mouse samples into six clusters (Figure 2A). These sample clusters closely correspond to the different strains used in the study, but the samples do not separate according to type of diet or length of time on the diet, even within the same strain. This indicates that the transcriptional profiles of islets are more closely related to genetic legacy than to environmental factors such as diet.
Figure 2.
Transcriptomic analysis across mouse strains and diets. (A) Sample similarity heatmap generated from gene expression data. Euclidean distances between mean gene expression profiles were calculated for each sample group, then sample similarity calculated using the affinityMatrix function of SNFtool in R [35]. Darker color indicates higher affinity between samples. Strains corresponding to sample clustering are indicated on the right of the heatmap. (B) Heatmap of 10 selected pathways enriched in HFHS vs RC for the 6 mouse strains at day 2. Colors correspond to degree of enrichment: red = positive enrichment; blue = negative enrichment. Enrichment scores were calculated using GSEA against MSigDB V3 canonical pathways (see Methods for details). Only enrichment scores ≥3 with a corresponding FDR ≤ 0.3 are represented as colors in the heatmap.
3.3. Several islet pathways are modified at the transcriptional level in response to HFHS-induced metabolic stress
In order to identify specific metabolic and signaling pathways that were up- or down-regulated by HFHS-diet in islets from different mouse strains, we performed gene set enrichment analysis (GSEA) [36], comparing gene lists ranked by fold change (HFHS vs RC) against Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Figure 2B shows a heatmap of 10 of the most significantly enriched pathways at day 2, representing initial adaptation to HFHS diet (for all time-points see Supplementary Figure S3). The strains are ordered along the x-axis according to insulinogenic index difference (Table 1). DNA replication, oxidative phosphorylation, biosynthesis, and proteasome pathways were up-regulated in the most insulinogenic strains, DBA/2J and AKR/J. Gene expression alterations in C57Bl/6J were less marked but oxidative phosphorylation and proteasome pathways were up-regulated in addition to fatty acid metabolism. The least insulinogenic strains, 129S2/SvPas, BALB/cJ, and A/J, showed no evidence for modulation of these pathways. Interestingly, extracellular matrix (ECM) genes were down-regulated in AKR/J, DBA/2J and to a lesser extent in A/J, suggesting that modulation of ECM could be linked to beta cell adaptation (or maladaptation) to metabolic stress.
3.4. The global network analysis highlights key pathways and processes affected in the different mouse strains
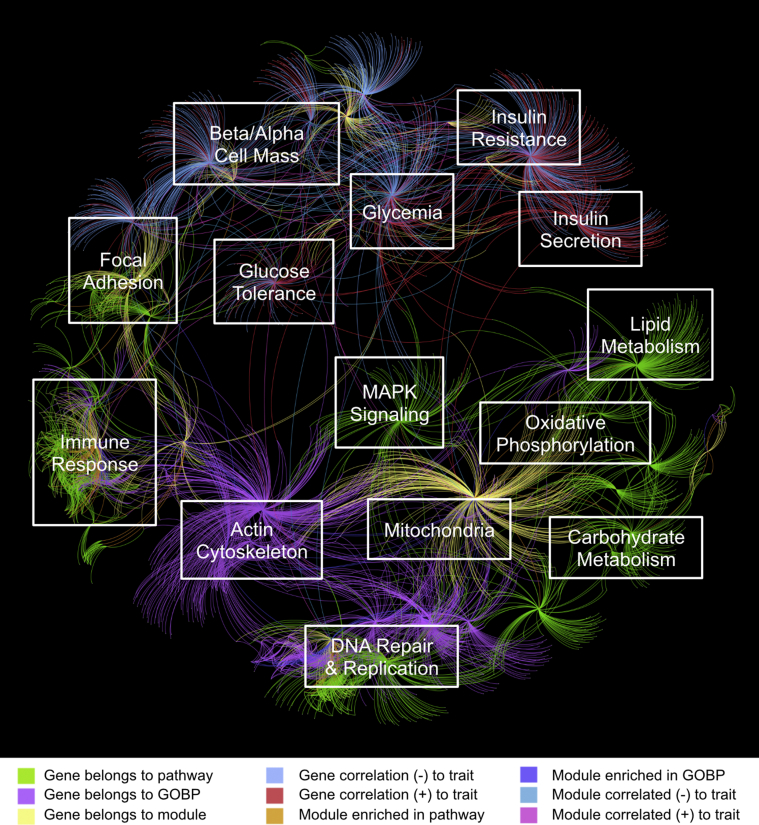
In order to capture the diversity of both islet transcriptomic and phenotypic data collected, we built a transcriptional network integrating the RNA-Seq and phenotypic data together with annotations of known pathways and gene ontology categories (see Supplementary Methods). To perform the integration, we first analyzed the RNA-Seq data to identify gene co-expression modules. Such modules are based on the concept that genes sharing more neighbors in a correlation network are more likely to be functionally related [37]. Each module was then correlated with the phenotypic traits and highly connected ‘hub’ genes were identified in each of the modules. Modules were also tested for enrichment against known pathways and gene ontology categories. The gene expression modules, ‘hub’ genes, enriched pathways, Gene Ontology (GO) categories, and phenotypic traits were integrated together into a single global network. An annotated representation of the network is shown in Figure 3. Manual exploration of the network revealed that several of these node clusters represented distinct sets of pathways, biological processes, or related phenotypic traits. We identified distinct node clusters related to focal adhesion, immune response, actin cytoskeleton, MAPK signaling, lipid metabolism, carbohydrate metabolism, oxidative phosphorylation, DNA replication, cell cycle, insulin secretion, glycemia/glucose tolerance, and β/α cell mass.
Figure 3.
Network representation integrating gene modules, pathways, and phenotypic traits. See Supplementary Methods for details of the network generation. Colors represent different relationship types and are included for illustrative purposes only. The figure is overlaid with manual annotations referring to particular pathways or traits that are approximately represented at that position in the network. Network visualisation was produced using Gephi 0.8.2.
This network captures the integrated genomics and physiological results from our mouse experiment into a single model. The network, which can be explored within an appropriate software tool such as Cytoscape [38], is available as supplementary data.
3.5. A sub-network of islet genes is associated with glucose tolerance
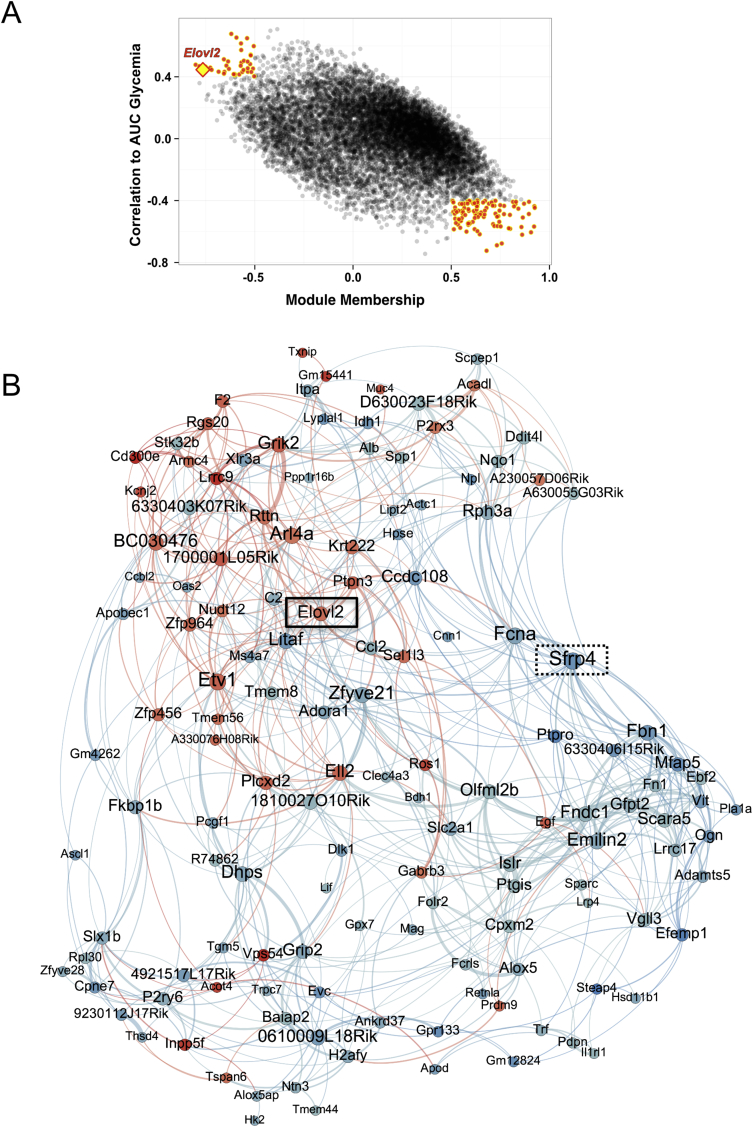
We interrogated the global network described above and identified a gene co-expression module that was significantly correlated to both oral glucose tolerance and insulin secretion. The correlation of this and other modules to the phenotypic traits is shown in Supplementary Figure S4. AUC glycemia is an indicator of glucose intolerance: the higher the AUC, the higher the glucose intolerance. We then searched for genes correlated to both the module and oral glucose tolerance; these may represent genes of key influence that are highly connected within the module and which may also affect the trait. A scatter plot of the correlations of all genes to the module and to oral glucose tolerance is shown in Figure 4A. In the figure, yellow points indicate those that show the highest correlations to both the trait and the module. These genes were then used to create a sub-network of genes related to glucose tolerance. A representation of the sub-network is shown in Figure 4B. Remarkably, one of the prominent genes in this network is Sfrp4 (indicated by a dotted box in Figure 4B), which has recently been identified as a key marker for diabetes in humans [39]. Sfrp4 is a secreted protein modulator of Wnt signaling, which is thought to be linked to islet inflammation and the resulting loss of insulin secretion and reduced tolerance to glucose through regulation by IL1B [39]. Although we did not find evidence for enrichment of inflammatory markers such as interleukin 1B (IL-1B), the sub-network is significantly enriched for secreted and extracellular matrix proteins (GO:0031012 ∼ extracellular matrix, enrichment p-value = 2.97e-7, FDR = 3.54e-4; Supplementary Table S2), indicating that changes occurring in the extracellular space might be linked to islet damage and impaired insulin secretion and glucose tolerance.
Figure 4.
A gene co-expression module correlated to insulin secretion and oral glucose tolerance. (A) Scatter plot of AUC glucose correlation against module membership (correlation to module) for all genes. Genes with the strongest correlations to both the module (Spearman's |r| ≥ 0.5) and to AUC glycemia (Spearman's |r| ≥ 0.4) are highlighted by red points outlined in yellow. These genes were used to generate the network in (B). Elovl2 is indicated by a yellow diamond outlined in red. (B) Network generated from selected module genes. Node size is proportional to degree and node color indicates correlation to AUC glycemia (blue: negative correlation; red: positive correlation). Edges (connections) between nodes indicate correlation between genes (blue: negative; red: positive). Elovl2 (solid box) and Sfrp4 (dotted box) are indicated in the network. Full details of network generation are described in Supplementary Methods.
3.6. Elovl2 expression positively correlates with glucose intolerance and insulin secretion
Genes involved in fatty acid metabolism were amongst the top pathways and biological processes enriched in the sub-network (GO:0006631 ∼ fatty acid metabolic process, enrichment p-value = 0.001, FDR = 2; Supplementary Table S2). There were several genes related to fatty acid metabolism in the sub-network, notably Elovl2 (Elongase of very long-chain fatty acids 2), Acadl (Acyl-CoA Dehydrogenase, Long Chain, which catalyzes the initial step of mitochondrial beta oxidation to straight fatty acid), and Acot4 (Acyl-CoA Thioesterase 4, which regulates intracellular levels of Acyl-CoA). The expression levels of all three genes were positively correlated with insulin secretion and AUC glycemia, but of the three genes, Elovl2 was also amongst the most highly connected genes in the network (indicated by a solid rectangle in Figure 4B) and was also amongst the top genes correlating with AUC glycemia (Figure 4A). These two observations led us to hypothesize that Elovl2 could be a key gene involved in regulating insulin secretion.
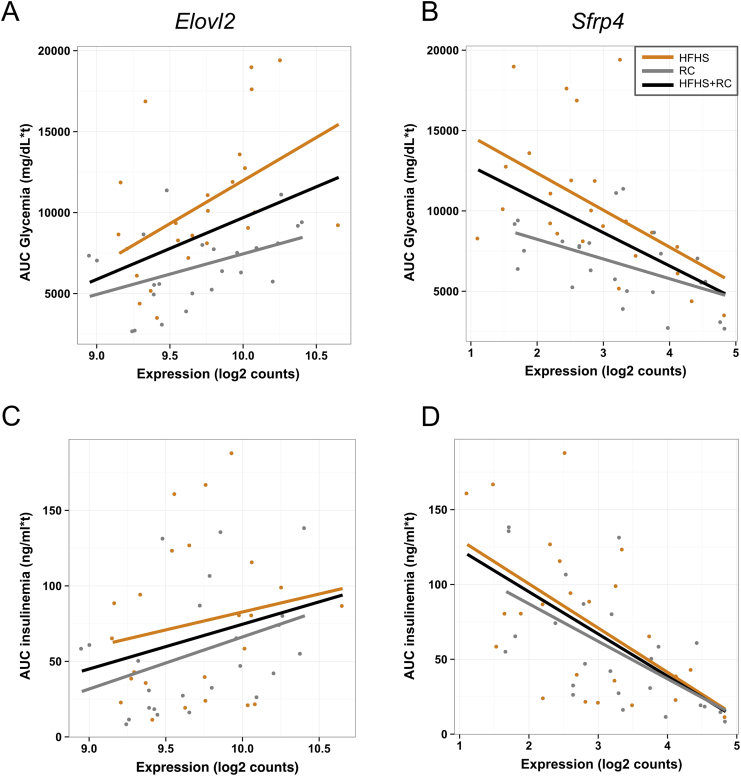
Scatter plots of Elovl2 gene expression against AUC glycemia and insulinemia are shown in Figure 5. Correlations of Sfrp4 with the same traits are shown for comparison. Elovl2 expression is positively correlated with both AUC glycemia (glucose intolerance) and AUC insulinemia (Figure 5A,C) in both HFHS and RC fed mice. The positive correlation of Elovl2 expression with increased glucose intolerance may appear counterintuitive since Elovl2 is also positively correlated with insulin secretion. However, we observed a significant correlation between glucose intolerance and insulin secretion across the mouse models (Figure S5). This suggests that in our experiment, we captured the initial adaptation of islets to metabolic challenge, where insulin secretion was boosted to try to counteract decreasing tolerance to glucose. These data suggest that Elovl2 may have a regulatory role in β cell function such that declining Elovl2 expression could contribute to impaired insulin secretion.
Figure 5.
Scatter plots showing correlations between Elovl2 and Sfrp4 islet gene expression with AUC glycemia and insulinemia. (A) Elovl2 is positively correlated with AUC glycemia: Pearson's r = 0.39, p-value = 8.82e-05 (all samples); r = 0.48, p-value = 0.018 (HFHS samples); r = 0.45, p-value = 0.028 (RC samples). (B) Sfrp4 is negatively correlated with AUC glycemia: Pearson's r = −0.52, p-value = 6.16e-08 (all samples); r = −0.51, p-value = 0.01 (HFHS samples); r = 0.51, p-value = 0.01 (RC samples). (C) Elovl2 is weakly positively correlated with AUC insulinemia: Pearson's r = 0.26, p-value = 0.01 (all samples); r = 0.18, p-value = 0.4 (HFHS samples); r = 0.37, p-value = 0.07 (RC samples). (D) Sfrp4 is negatively correlated with AUC insulinemia: Pearson's r = −0.6, p-value = 1.057e-10 (all samples); r = −0.56, p-value = 0.004 (HFHS samples); r = −0.62, p-value = 0.001 (RC samples).
3.7. Elovl2 knock down decreases glucose-stimulated insulin secretion in mouse and human β cell lines
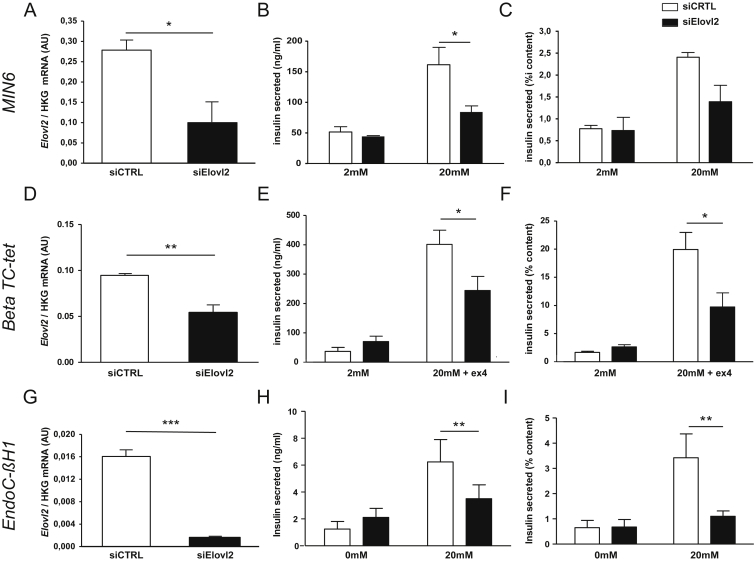
In order to gain further insight into the potential role of Elovl2 in β cell function, we tested the effect of reducing the expression of Elovl2 mRNA on glucose-induced insulin secretion in cells grown in vitro. Initially we treated two mouse β-cell lines (MIN6 and Beta TC-tet) with specific small interfering RNAs (siRNA) against Elovl2 (Figure 6, A-F) under conditions of 2 mM and 20 mM glucose. The effects of siRNA directed towards Elovl2 or a random sequence (CTRL siRNA) are shown in Figure 6A, D: Elovl2 mRNA expression was significantly decreased compared to controls. For Beta TC-tet cells, 100 nM exendin was added to increase glucose-induced insulin secretion. Whilst we saw no difference at 2 mM glucose, we observed a significant reduction in insulin secreted from Elovl2 siRNA-treated cells compared to controls in both cell lines at 20 mM glucose (Figure 6B, E). The reduction of insulin secretion in siRNA-treated cells was not due to reduction of total insulin content (Figure 6C, F displays insulin secreted normalized on insulin content), suggesting that the effect of the siRNA was specifically acting on insulin secretion. This prompted us to test the effect of Elovl2 knock down in human cells. For this, we used the recently described human β-cell line, EndoC-βH1, which displays many properties of functional human β cells [32], [40], [41]. In human EndoC-βH1 cells, a pool of Elovl2 siRNA dramatically decreased Elovl2 expression (Figure 6G) compared to a CTRL siRNA pool. Insulin secretion was significantly decreased at 20 mM but not at 0 mM glucose (Figure 6H). This effect was even more marked when normalizing for total insulin content (Figure 6I), suggesting that Elovl2 expression is required to ensure a correct insulin secretory response to glucose. We thus demonstrate impaired insulin secretion with Elovl2 knock down in three different cell systems. This result is very unlikely to be due to off-target effects since different siRNAs were used in mouse compared to human cells, with the latter comprising a pool of four Elovl2 siRNA to increase targeting efficiency and specificity. In addition, using an adenoviral approach, we over-expressed Elovl2 in MIN6 cells and determined insulin secretion in response to glucose (Supplementary Figure S6). We found that Ad-Elovl2 had no effect on basal insulin secretion but significantly potentiated insulin secretion induced by high glucose, supporting the idea that Elovl2 is a new player in insulin secretion. Moreover, we found that Ad-Elovl2 significantly increased DHA levels in MIN6 cells suggesting that intracellular DHA could play a role in the regulation of glucose-induced insulin secretion (Supplementary Figure S6D).
Figure 6.
Effects of Elovl2 loss of function in glucose-stimulated insulin secretion, in mouse β-cell lines and human β-cell line. (A) Elovl2 mRNA levels expressed as the ratio between the signal of Elovl2 and the signal of the housekeeping gene rpL19 (ribosomal protein L19), (B) Insulin secreted in ng/ml and (C) insulin secreted (% of content) in MIN6 cells. (D) Elovl2 mRNA levels expressed as the ratio between the signal of Elovl2 and the signal of the housekeeping gene gusb (glucuronidase), (E) Insulin secreted in ng/ml and (F) insulin secreted (% of content) in Beta TC-tet cells. (G) Elovl2 mRNA levels expressed as the ratio between the signal of Elovl2 and the signal of rpL19, (H) Insulin secreted in ng/ml and (I) insulin secreted (% of content) as mean values (±SE) of three independent experiments. ***p < 0.001; **p < 0.01; *p < 0.05.
4. Discussion
4.1. A novel mouse resource for the study of islet dysfunction
The resource we present here integrates islet expression data with deep phenotypic measurements from six mouse strains under controlled diet and housing conditions. Since non-biological variability is minimized due to controlled conditions and rigorous experimental design, this enables direct comparison of molecular and physiological phenotypes between the different mouse strains. We also performed a systems level analysis, integrating expression and phenotypic information with known pathways and biological functions, thus capturing in a single network model the knowledge gained from the entire experiment. We interrogated this model and built a co-expression sub-network of genes related to whole body glucose tolerance and plasma insulin levels following oral glucose stimulation. From this sub-network, we identified a highly connected gene, Elovl2, which was correlated to both glucose tolerance and plasma insulin. Elovl2 expression was subsequently confirmed to be important for glucose-induced insulin secretion by siRNA knock down in both mouse and human β cell lines. Although we describe the importance of only a single identified gene, Elovl2, here, the integrated database contains a wealth of evidence for further potential candidates. Indeed, using these data, nucleus-encoded mitochondrial genes Ndufb8 and Atp5a1 are also shown to be key regulators of insulin secretion (Parnis et al., submitted).
4.2. Strain phenotypes are mirrored by distinct islet gene expression profiles
Phenotypically, we observe that mice from the different strains show a distinct response to HFHS diet. This is in accordance with a previous study from Gauguier and colleagues [24] showing strain-specific patterns of adaptation to HFHS diet [23], [24], [25], [27], [30], [31]. Intriguingly, we found that BALB/cJ mice fed a HFHS diet did not develop obesity or increased body weight compared to chow-fed controls although they displayed a dramatic intolerance to glucose as early as day 10. However, the percentage of fat mass (measured with an EchoMRI) was strongly increased in HFHS-fed BALB/cJ mice after 90 days of diet (1.6 fold, data not shown) suggesting that this is likely due to early ectopic fat storage in insulin target tissues. In comparison, DBA/2J mice, which present early signs of obesity compared to mice fed with regular chow, have a 3.6 fold increase in the percentage of fat mass at three months, largely reflecting storage in classical adipose tissue depots (e.g. subcutaneous, inguinal). These data underline the important phenotypic differences between the mouse strains and suggest that they may be at least in part due to differential underlying response of the β-cells. Additional studies would be required in female mice to address potential sex dependent differences.
Similarly to the marked phenotypic differences we observed, our data show that islet transcriptional profiles are very different between strains. This indicates that strain-specific/strain-dependent changes occurring over time in both RC and HFHS diet fed mice may be important to link transcriptomic signatures to islet function. In order to further investigate this link, we exploited the differences in islet gene expression signatures by measuring correlations both between islet genes, and also between gene co-expression modules and phenotypic traits. The result is a global network that captures the strain-specific/strain-dependent changes over time under metabolic stress, linking the mouse phenotypes with islet transcriptomics in one model. Many of the phenotypic changes we observe are a result of the interplay between many different tissues. In this regard, it would be important to extend the model in the future to include insulin sensitive tissues such as liver, muscle, and adipose tissue.
4.3. Elovl2 is a novel player in insulin secretion
We identified a sub-network of genes correlated to glucose tolerance and insulinemia and found that Elovl2 gene expression was amongst the most strongly associated with both glucose intolerance and insulin secretion within the sub-network. Elovl2 encodes the enzyme that synthetizes DHA (C22:6), an omega-3 series very long chain poly-unsaturated fatty-acid (PUFA) [42]. Expression of Elovl2 has been found in various tissues such as liver, testis, uterus, placenta, mammary gland, retina, and certain areas of the brain, all of which are tissues that are documented as having significant levels of DHA [43], [44]. DHA is also known to play a critical role in brain development and has neuroprotective effects [45].
Several studies have shown that saturated fatty acids decrease insulin secretion and worsen insulin sensitivity, whereas polyunsaturated fatty acids (PUFAs) such as DHA preserve or improve insulin secretion and sensitivity [46], [47]. However, most of these earlier studies are based on exogenous PUFAs originating from fish oil [48], and the role of endogenously synthesized PUFAs as opposed to PUFAs taken up through diet is relatively unknown. Using mice, a previous study showed a role of Elovl2 in sperm maturation [49]. More recently, the same group demonstrated that Elovl2 positively influenced de novo lipogenesis by modulating hepatic levels of key lipogenic genes [50]. However, Elovl2−/− mice were resistant to hepatic steatosis and diet-induced weight gain, implying that hepatic DHA synthesis via Elovl2 could also regulate lipid storage and fat mass expansion.
To date, the expression and role of Elovl2 have not been examined in pancreatic islets or β cells. Our study reveals that Elovl2 is expressed in the pancreatic β cell lines MIN6 and Beta TC-tet and in the human β cell line EndoC-βH1 [51]. Correspondingly, recently published transcriptomic data [52], [53] also demonstrate detectable, although low, levels of Elovl2 mRNA in adult human β cells. Interestingly, Elovl2 expression was also up-regulated ∼2-fold in mouse islets in response to deletion of the tumor promoter Liver Kinase B1 (LKB1) [54], a condition in which glucose-stimulated insulin secretion is also enhanced [55]. Interestingly, we also found that over-expression of Elovl2 in β cells potentiated glucose-induced insulin secretion and was associated with an increase of DHA levels.
We demonstrate that Elovl2 loss of function leads to decreased insulin secretion in two different mouse beta cell lines: MIN6 cells known for their strong responsiveness to glucose, and Beta TC-tet cells that are responsive to glucose plus Exendin-4, a GLP1 receptor agonist. In addition, we show the same defect in glucose-induced insulin secretion after Elovl2 silencing in human EndoC-βH1 cells, a unique and novel tool to study functional human beta cells in standardized assays [32]. To our knowledge, we provide the first demonstration that Elovl2 is involved in the control of glucose-stimulated insulin secretion. Since both in vitro and in vivo exogenous DHA has been shown to amplify (either directly or indirectly) insulin secretion [48], we suggest that a decrease of Elovl2 expression and consequently ELOVL2 activity may lead to an endogenous DHA production and thus to blunted glucose-induced insulin secretion. Previous studies have shown that PUFAs including DHA may play a protective role against the development of metabolic diseases mainly by regulating insulin sensitivity [56], [57]. Our results suggest for the first time that endogenously produced DHA, controlled by Elovl2, may also mediate its effect by acting directly on pancreatic β cells.
Availability of data
All RNA-Seq data has been deposited in NCBI GEO under accession code GSE78183. The data have been managed according to FAIR guidelines [58]. This has been performed by encoding both raw and normalized RNA-Seq expression data, together with all measurements and detailed phenotypic data down to the level of the individual mouse in Resource Description Format (RDF), using existing ontologies to encode the metadata. The RDF formatted data as well as a description of all metadata is available on request.
Author contributions
BT, CM, and MI conceived the study. JDe, NF, PN-L, NK, CR, A-LL, IW, and JDa performed mouse phenotyping and prepared tissues and RNA samples under the supervision of CC-G, CM, CB, and AK. TH, WR and IU generated RNA-Seq libraries and performed deep sequencing. TG, DS, and AM performed histological analysis and generate β and α cell mass data. XB and LB performed siRNA knock down experiments and insulin secretion tests on mouse cell lines under the supervision of BT and HLS. MO performed siRNA knock down experiments using human cell line material under the supervision of RS. MI, RL, DM, LW, and FB stored, structured and integrated the data, and performed systems biology and statistical analyses under guidance from IX. CC-G, MI, LB, BT, and CM wrote the manuscript with contributions from HLS, MO, XB, and GAR.
Acknowledgements
The work leading to this publication has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement n° 155005 (IMIDIA), resources of which are composed of a financial contribution from the European Union's Seventh Framework Programme (FP7/2007-2013) (CM, BT, MI, GAR) and EFPIA companies' in kind contribution (C.M., G.A.R.). BT received additional support from Swiss National Science Foundation grant ((3100A0B-128657) and a European Research Council Advanced Grant (INSIGHT). GAR thanks the MRC (UK) for Programme grant MR/J0003042/1, the BBSRC (UK) for a Project grant (BB/J015873/1), the Royal Society for a Wolfson Research Merit Award and the Wellcome Trust for a Senior Investigator Award (WT098424AIA). MI, DK, RL, FB, LW, and IX would like to thank Jerven Bolleman for initial help with RDF formatting of the data and to Anne Niknejad for manual curation work on gene candidates. The authors declare no financial conflict of interest.
We would like to dedicate this work to the memory of Alain Ktorza, whose enthusiasm, guidance, support and friendship will be sorely missed by all who knew and worked with him.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2017.01.009.
Contributor Information
Christophe Magnan, Email: christophe.magnan@univ-paris-diderot.fr.
Mark Ibberson, Email: mark.ibberson@sib.swiss.
Conflict of interest
The authors confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Unger R. Lipotoxicity in the pathogenesis of obesity-dependent NIDDM. Genetic and clinical implications. Diabetes. 1995;44:863–870. doi: 10.2337/diab.44.8.863. [DOI] [PubMed] [Google Scholar]
- 2.Forouhi N.G., Wareham N.J. Epidemiology of diabetes. Medicine (Abingdon) 2014;42(12):698–702. doi: 10.1016/j.mpmed.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rutter G.A., Pullen T.J., Hodson D.J., Martinez-Sanchez A. Pancreatic beta-cell identity, glucose sensing and the control of insulin secretion. The Biochemical Journal. 2015;466(2):203–218. doi: 10.1042/BJ20141384. [DOI] [PubMed] [Google Scholar]
- 4.Unger R.H., Clark G.O., Scherer P.E., Orci L. Lipid homeostasis, lipotoxicity and the metabolic syndrome. Biochimica et Biophysica Acta. 2010;1801(3):209–214. doi: 10.1016/j.bbalip.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Poitout V., Robertson R.P. Minireview: secondary beta-cell failure in type 2 diabetes–a convergence of glucotoxicity and lipotoxicity. Endocrinology. 2002;143(2):339–342. doi: 10.1210/endo.143.2.8623. [DOI] [PubMed] [Google Scholar]
- 6.Bellini L., Campana M., Mahfouz R., Carlier A., Veret J., Magnan C. Targeting sphingolipid metabolism in the treatment of obesity/type 2 diabetes. Expert Opinion on Therapeutic Targets. 2015;19(8):1037–1050. doi: 10.1517/14728222.2015.1028359. [DOI] [PubMed] [Google Scholar]
- 7.Talchai C., Xuan S., Lin H.V., Sussel L., Accili D. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dor Y., Glaser B. Beta-cell dedifferentiation and type 2 diabetes. The New England Journal of Medicine. 2013;368(6):572–573. doi: 10.1056/NEJMcibr1214034. [DOI] [PubMed] [Google Scholar]
- 9.Butler A.E., Janson J., Bonner-Weir S., Ritzel R., Rizza R.A., Butler P.C. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52(1):102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 10.Marullo L., El-Sayed Moustafa J.S., Prokopenko I. Insights into the genetic susceptibility to type 2 diabetes from genome-wide association studies of glycaemic traits. Current Diabetes Reports. 2014;14(11):551. doi: 10.1007/s11892-014-0551-8. [DOI] [PubMed] [Google Scholar]
- 11.Fergusson G., Ethier M., Guevremont M., Chretien C., Attane C., Joly E. Defective insulin secretory response to intravenous glucose in C57Bl/6J compared to C57Bl/6N mice. Molecular Metabolism. 2014;3(9):848–854. doi: 10.1016/j.molmet.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toye A.A., Lippiat J.D., Proks P., Shimomura K., Bentley L., Hugill A. A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia. 2005;48(4):675–686. doi: 10.1007/s00125-005-1680-z. [DOI] [PubMed] [Google Scholar]
- 13.Cnop M., Abdulkarim B., Bottu G., Cunha D.A., Igoillo-Esteve M., Masini M. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes. 2014;63(6):1978–1993. doi: 10.2337/db13-1383. [DOI] [PubMed] [Google Scholar]
- 14.Busch A.K., Cordery D., Denyer G.S., Biden T.J. Expression profiling of palmitate- and oleate-regulated genes provides novel insights into the effects of chronic lipid exposure on pancreatic beta-cell function. Diabetes. 2002;51(4):977–987. doi: 10.2337/diabetes.51.4.977. [DOI] [PubMed] [Google Scholar]
- 15.Malmgren S., Spegel P., Danielsson A.P., Nagorny C.L., Andersson L.E., Nitert M.D. Coordinate changes in histone modifications, mRNA levels, and metabolite profiles in clonal INS-1 832/13 beta-cells accompany functional adaptations to lipotoxicity. The Journal of Biological Chemistrty. 2013;288(17):11973–11987. doi: 10.1074/jbc.M112.422527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao J., Gregersen S., Kruhoffer M., Pedersen S.B., Orntoft T.F., Hermansen K. The effect of chronic exposure to fatty acids on gene expression in clonal insulin-producing cells: studies using high density oligonucleotide microarray. Endocrinology. 2001;142(11):4777–4784. doi: 10.1210/endo.142.11.8483. [DOI] [PubMed] [Google Scholar]
- 17.Roat R., Rao V., Doliba N.M., Matschinsky F.M., Tobias J.W., Garcia E. Alterations of pancreatic islet structure, metabolism and gene expression in diet-induced obese C57BL/6J mice. PLoS One. 2014;9(2):e86815. doi: 10.1371/journal.pone.0086815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winzell M.S., Ahren B. The high-fat diet-fed mouse: a model for studying mechanisms and treatment of impaired glucose tolerance and type 2 diabetes. Diabetes. 2004;53(Suppl. 3):S215–S219. doi: 10.2337/diabetes.53.suppl_3.s215. [DOI] [PubMed] [Google Scholar]
- 19.Peyot M.L., Pepin E., Lamontagne J., Latour M.G., Zarrouki B., Lussier R. Beta-cell failure in diet-induced obese mice stratified according to body weight gain: secretory dysfunction and altered islet lipid metabolism without steatosis or reduced beta-cell mass. Diabetes. 2010;59(9):2178–2187. doi: 10.2337/db09-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montgomery M.K., Hallahan N.L., Brown S.H., Liu M., Mitchell T.W., Cooney G.J. Mouse strain-dependent variation in obesity and glucose homeostasis in response to high-fat feeding. Diabetologia. 2013;56(5):1129–1139. doi: 10.1007/s00125-013-2846-8. [DOI] [PubMed] [Google Scholar]
- 21.Surwit R.S., Feinglos M.N., Rodin J., Sutherland A., Petro A.E., Opara E.C. Differential effects of fat and sucrose on the development of obesity and diabetes in C57BL/6J and A/J mice. Metabolism. 1995;44(5):645–651. doi: 10.1016/0026-0495(95)90123-x. [DOI] [PubMed] [Google Scholar]
- 22.Poussin C., Ibberson M., Hall D., Ding J., Soto J., Abel E.D. Oxidative phosphorylation flexibility in the liver of mice resistant to high-fat diet-induced hepatic steatosis. Diabetes. 2011;60(9):2216–2224. doi: 10.2337/db11-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Funkat A., Massa C.M., Jovanovska V., Proietto J., Andrikopoulos S. Metabolic adaptations of three inbred strains of mice (C57BL/6, DBA/2, and 129T2) in response to a high-fat diet. The Journal of Nutrition. 2004;134(12):3264–3269. doi: 10.1093/jn/134.12.3264. [DOI] [PubMed] [Google Scholar]
- 24.Fearnside J.F., Dumas M.E., Rothwell A.R., Wilder S.P., Cloarec O., Toye A. Phylometabonomic patterns of adaptation to high fat diet feeding in inbred mice. PLoS One. 2008;3(2):e1668. doi: 10.1371/journal.pone.0001668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexander J., Chang G.Q., Dourmashkin J.T., Leibowitz S.F. Distinct phenotypes of obesity-prone AKR/J, DBA2J and C57BL/6J mice compared to control strains. International Journal of Obesity (London) 2006;30(1):50–59. doi: 10.1038/sj.ijo.0803110. [DOI] [PubMed] [Google Scholar]
- 26.Andrikopoulos S., Massa C.M., Aston-Mourney K., Funkat A., Fam B.C., Hull R.L. Differential effect of inbred mouse strain (C57BL/6, DBA/2, 129T2) on insulin secretory function in response to a high fat diet. The Journal of Endocrinology. 2005;187(1):45–53. doi: 10.1677/joe.1.06333. [DOI] [PubMed] [Google Scholar]
- 27.Gallou-Kabani C., Vige A., Gross M.S., Rabes J.P., Boileau C., Larue-Achagiotis C. C57BL/6J and A/J mice fed a high-fat diet delineate components of metabolic syndrome. Obesity (Silver Spring) 2007;15(8):1996–2005. doi: 10.1038/oby.2007.238. [DOI] [PubMed] [Google Scholar]
- 28.Goren H.J., Kulkarni R.N., Kahn C.R. Glucose homeostasis and tissue transcript content of insulin signaling intermediates in four inbred strains of mice: C57BL/6, C57BLKS/6, DBA/2, and 129X1. Endocrinology. 2004;145(7):3307–3323. doi: 10.1210/en.2003-1400. [DOI] [PubMed] [Google Scholar]
- 29.Kooptiwut S., Zraika S., Thorburn A.W., Dunlop M.E., Darwiche R., Kay T.W. Comparison of insulin secretory function in two mouse models with different susceptibility to beta-cell failure. Endocrinology. 2002;143(6):2085–2092. doi: 10.1210/endo.143.6.8859. [DOI] [PubMed] [Google Scholar]
- 30.Berglund E.D., Li C.Y., Poffenberger G., Ayala J.E., Fueger P.T., Willis S.E. Glucose metabolism in vivo in four commonly used inbred mouse strains. Diabetes. 2008;57(7):1790–1799. doi: 10.2337/db07-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warden C.H., Fisler J.S. Comparisons of diets used in animal models of high-fat feeding. Cell Metabolism. 2008;7(4):277. doi: 10.1016/j.cmet.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravassard P., Hazhouz Y., Pechberty S., Bricout-Neveu E., Armanet M., Czernichow P. A genetically engineered human pancreatic beta cell line exhibiting glucose-inducible insulin secretion. The Journal of Clinical Investigation. 2011;121(9):3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 34.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang B., Mezlini A.M., Demir F., Fiume M., Tu Z., Brudno M. Similarity network fusion for aggregating data types on a genomic scale. Nature Methods. 2014;11(3):333–337. doi: 10.1038/nmeth.2810. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Statistical Applications in Genetics and Molecular Biology. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 38.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahdi T., Hanzelmann S., Salehi A., Muhammed S.J., Reinbothe T.M., Tang Y. Secreted frizzled-related protein 4 reduces insulin secretion and is overexpressed in type 2 diabetes. Cell Metabolism. 2012;16(5):625–633. doi: 10.1016/j.cmet.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 40.Scharfmann R., Pechberty S., Hazhouz Y., von Bulow M., Bricout-Neveu E., Grenier-Godard M. Development of a conditionally immortalized human pancreatic beta cell line. The Journal of Clinical Investigation. 2014;124(5):2087–2098. doi: 10.1172/JCI72674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weir G.C., Bonner-Weir S. Finally! A human pancreatic beta cell line. The Journal of Clinical Investigation. 2011;121(9):3395–3397. doi: 10.1172/JCI58899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guillou H., Zadravec D., Martin P.G., Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: insights from transgenic mice. Progress in Lipid Research. 2010;49(2):186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 43.Ohno Y., Suto S., Yamanaka M., Mizutani Y., Mitsutake S., Igarashi Y. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(43):18439–18444. doi: 10.1073/pnas.1005572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tvrdik P., Westerberg R., Silve S., Asadi A., Jakobsson A., Cannon B. Role of a new mammalian gene family in the biosynthesis of very long chain fatty acids and sphingolipids. The Journal of Cell Biology. 2000;149(3):707–718. doi: 10.1083/jcb.149.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gharami K., Das M., Das S. Essential role of docosahexaenoic acid towards development of a smarter brain. Neurochemistry International. 2015;89:51–62. doi: 10.1016/j.neuint.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 46.Sato A., Kawano H., Notsu T., Ohta M., Nakakuki M., Mizuguchi K. Antiobesity effect of eicosapentaenoic acid in high-fat/high-sucrose diet-induced obesity: importance of hepatic lipogenesis. Diabetes. 2010;59(10):2495–2504. doi: 10.2337/db09-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X., Xue Y., Liu C., Lou Q., Wang J., Yanagita T. Eicosapentaenoic acid-enriched phospholipid ameliorates insulin resistance and lipid metabolism in diet-induced-obese mice. Lipids in Health and Disease. 2013;12:109. doi: 10.1186/1476-511X-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhaswant M., Poudyal H., Brown L. Mechanisms of enhanced insulin secretion and sensitivity with n-3 unsaturated fatty acids. The Journal of Nutritional Biochemistry. 2015;26(6):571–584. doi: 10.1016/j.jnutbio.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Zadravec D., Tvrdik P., Guillou H., Haslam R., Kobayashi T., Napier J.A. ELOVL2 controls the level of n-6 28:5 and 30:5 fatty acids in testis, a prerequisite for male fertility and sperm maturation in mice. Journal of Lipid Research. 2011;52(2):245–255. doi: 10.1194/jlr.M011346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pauter A.M., Olsson P., Asadi A., Herslof B., Csikasz R.I., Zadravec D. Elovl2 ablation demonstrates that systemic DHA is endogenously produced and is essential for lipid homeostasis in mice. Journal of Lipid Research. 2014;55(4):718–728. doi: 10.1194/jlr.M046151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scharfmann R., Didiesheim M., Richards P., Chandra V., Oshima M., Albagli O. Mass production of functional human pancreatic beta-cells: why and how? Diabetes, Obesity & Metabolism. 2016;18(Suppl. 1):128–136. doi: 10.1111/dom.12728. [DOI] [PubMed] [Google Scholar]
- 52.Blodgett D.M., Nowosielska A., Afik S., Pechhold S., Cura A.J., Kennedy N.J. Novel observations from next-generation RNA sequencing of highly purified human adult and fetal islet cell subsets. Diabetes. 2015;64(9):3172–3181. doi: 10.2337/db15-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bramswig N.C., Everett L.J., Schug J., Dorrell C., Liu C., Luo Y. Epigenomic plasticity enables human pancreatic alpha to beta cell reprogramming. The Journal of Clinical Investigation. 2013;123(3):1275–1284. doi: 10.1172/JCI66514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kone M., Pullen T.J., Sun G., Ibberson M., Martinez-Sanchez A., Sayers S. LKB1 and AMPK differentially regulate pancreatic beta-cell identity. FASEB Journal. 2014;28(11):4972–4985. doi: 10.1096/fj.14-257667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swisa A., Granot Z., Tamarina N., Sayers S., Bardeesy N., Philipson L. Loss of liver kinase B1 (LKB1) in beta cells enhances glucose-stimulated insulin secretion despite profound mitochondrial defects. The Journal of Biological Chemistry. 2015;290(34):20934–20946. doi: 10.1074/jbc.M115.639237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinel A., Rigaudiere J.P., Laillet B., Pouyet C., Malpuech-Brugere C., Prip-Buus C. N-3PUFA differentially modulate palmitate-induced lipotoxicity through alterations of its metabolism in C2C12 muscle cells. Biochimica et Biophysica Acta. 2016;1861(1):12–20. doi: 10.1016/j.bbalip.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 57.Bjursell M., Xu X., Admyre T., Bottcher G., Lundin S., Nilsson R. The beneficial effects of n-3 polyunsaturated fatty acids on diet induced obesity and impaired glucose control do not require Gpr120. PLoS One. 2014;9(12):e114942. doi: 10.1371/journal.pone.0114942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A. The FAIR guiding principles for scientific data management and stewardship. Scientific Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.