Abstract
Cryptosporidiosis is a gastrointestinal illness caused by the protozoan parasite Cryptosporidium species, which is a leading cause of diarrhea in a variety of vertebrate hosts. The primary mode of transmission is through oral routes; infections spread with the ingestion of oocysts by susceptible animals or humans. In humans, Cryptosporidium infections are commonly found in children and immunocompromised individuals. The small intestine is the most common primary site of infection in humans while extraintestinal cryptosporidiosis occurs in immunocompromised individuals affecting the biliary tract, lungs, or pancreas. Both innate and adaptive immune responses play a critical role in parasite clearance as evident from studies with experimental infection in mice. However, the cellular immune responses induced during human infections are poorly understood. In this article, we review the currently available information with regard to epidemiology, diagnosis, therapeutic interventions, and strategies being used to control cryptosporidiosis infection. Since cryptosporidiosis may spread through zoonotic mode, we emphasis on more epidemiological surveillance-based studies in developing countries with poor sanitation and hygiene. These epidemiological surveys must incorporate fecal source tracking measures to identify animal and human populations contributing significantly to the fecal burden in the community, as mitigation measures differ by host type.
Keywords: Cryptosporidiosis, Cryptosporidium, diarrhea, microbial source tracking, zoonotic diseases
INTRODUCTION
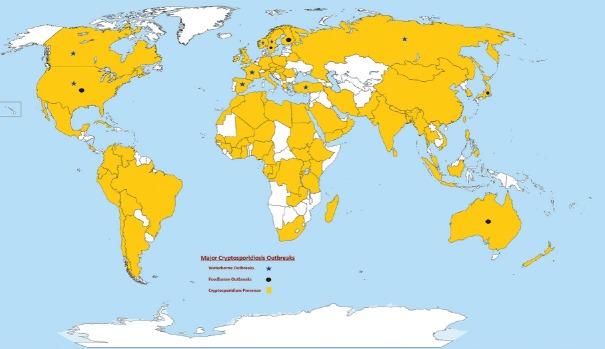
Cryptosporidiosis is a gastrointestinal illness caused by Cryptosporidium species, which is transmitted most often by direct fecal contamination or through waterborne routes and cause diarrhea in a variety of vertebrate hosts. In humans, Cryptosporidium infection is commonly found in children and immunocompromised individuals. In children, cryptosporidiosis has been associated with impairment in growth, physical fitness, and cognitive function and has been identified as the leading global cause of diarrheal mortality among infants aged between 12 and 23 months.[1,2] Cryptosporidiosis has emerged as the second major cause of diarrheal disease and death in infants as reported in the last few decades.[3] The first waterborne outbreak of Cryptosporidium was reported in Texas, USA in 1984.[4] A decade later, a major outbreak was documented in Milwaukee, Wisconsin, USA, affecting nearly 400,000 persons.[5] Recently, a major Cryptosporidium outbreak was reported in Sweden affecting 27,000 persons[6] and a foodborne outbreak of cryptosporidiosis was reported in the UK.[7] Since then, other outbreaks have been reported worldwide, highlighting the global significance of this parasite in Figure 1.[8]
Figure 1.
Major worldwide occurrence of human cryptosporidiosis outbreaks and sporadic cases: A color coded. distribution of major cases of cryptosporidiosis reported in different countries of the world between 1984-2013. Water-borne outbreaks represented with star symbol, round dot represents Food-borne outbreaks in the map, yellow color in the map represents the presence of cryptosporidiosis and white color represents no such reports are present
Cryptosporidiosis has great public health importance as it also poses occupational risks to the human population. For example, asymptomatic food workers infected with Cryptosporidium species can be the source of transmissions in the outbreaks.[9,10,11] Due to the public health concerns, the National Institutes of Health (NIH, USA) currently spends roughly US $4.3 million each year on carrying out basic research on Cryptosporidium, and apart from NIH, philanthropic organizations such as the Bill and Melinda Gates Foundation, are also focusing on interventional aspects for controlling Cryptosporidium infections.[3]
Cryptosporidiosis is usually a self-limiting illness in healthy individuals and lasts on average up to 9–15 days, while in immunocompromised individuals cryptosporidiosis can be life-threatening as there is no fully effective drug treatment. The small intestine is the primary site of Cryptosporidium infection in humans, and in immunocompromised individuals, the extraintestinal infectious may affect other organs such as the biliary tract, lungs, or pancreas.[12] Although cryptosporidiosis can be asymptomatic in some individuals, the most common clinical symptoms include profuse watery diarrhea, nausea, vomiting cramps such as abdominal pain and mild fever.[13,14] A recent study also suggested that infection with Cryptosporidium parvum could lead to digestive carcinogenesis in humans.[15] In recent years, the molecular biology of Cryptosporidium has advanced significantly, leading to the development of novel approaches for detecting Cryptosporidium. It is now important to revisit the current problem of cryptosporidiosis and address updates in the biology, epidemiology, transmission, detection, and treatment, where techniques and approaches emerging in developed countries can be applied to improve health and livelihoods in threshold and developing country settings as well.
EPIDEMIOLOGY
Cryptosporidium spp. has a wide host range and can infect all four classes of vertebrates.[16,17] To date, 27 species and more than 60 genotypes of Cryptosporidium have been identified worldwide.[18,19] In the genus, Cryptosporidium, Cryptosporidium hominis, and C. parvum are the major etiological agents of human clinical cryptosporidiosis. In addition, a novel spp. Cryptosporidium viatorum was recently identified among travelers returning to Great Britain from the Indian subcontinent.[20] Ng-Hublin et al. reported the occurrence of a mixed infection of Cryptosporidium species – Cryptosporidium meleagridis, the Cryptosporidium mink genotype, and an unknown Cryptosporidium species in immunocompetent individuals.[21]
Cryptosporidium accounts for almost 20% of diarrheal episodes in children in developing countries, and up to 9% of diarrheal episodes in developed nations.[22] A high prevalence of Cryptosporidium infections in children has been reported from various countries.[23,24,25] Cryptosporidium infection has also been observed in immunocompromised individuals such as HIV-infected persons.[16,17,18,19,20,21,22,23,24,25,26] Several Cryptosporidium seroprevalence studies in humans are available that showed immunoglobulin G (IgG) response specifically to 15/17-kDa Cryptosporidium sporozoite antigen complex and the 27-kDa antigen.[27] One study from Europe provides serological evidence of Cryptosporidium infection in blood donors (83% positive), perhaps explaining the infrequent occurrence of clinically detectable cryptosporidiosis in the study region.[28]
Epidemiological studies have shown that the geographical distributions of Cryptosporidium spp. vary around the world. C. hominis is more prevalent in North and South America, Australia, and Africa, while C. parvum is localized more in Europe, especially in the UK.[29] One study from the UK suggests C. parvum is more common in rural populations mainly in the spring season while C. hominis is more in urban residents and peaks in late summer and autumn.[27] Waterborne and foodborne transmission of Cryptosporidium are the major routes of infection, although person to person contact, especially in day care settings and between men who have sex with men have also been reported.[30] Cryptosporidium organisms have been reported in sputum and respiratory tract of humans,[31,32] thus suggesting a novel mode of respiratory transmission of cryptosporidiosis.
Various reports have also highlighted the presence of Cryptosporidium infection in large categories of vertebrate animals.[33,34,35] Serological responses against Cryptosporidium in animals also indicate the continuous exposure of this parasite in animals.[36,37] One study from Uganda showed that Cryptosporidium has a prevalence rate of 11% in nonhuman primates and 2.2% in livestock,[38] while another study showed a prevalence of 6% in domestic and wild animals.[17] Few studies have also reported the existence of novel zoonotic Cryptosporidium species in fishes and giant panda.[39] Recently, Cryptosporidium ubiquitum has emerged as a zoonotic pathogen capable of causing cryptosporidiosis in humans.[40]
Cryptosporidium infection spreads with the ingestion of oocysts by susceptible animals or humans. Figure 2 shows detailed description of transmission route in humans and animals. The incubation period of this protozoan parasite ranges from 1 to 2 weeks. Most patients with symptomatic infection present with acute watery diarrhea that lasts for a few days to 2 weeks, but sometimes, it can be persistent and last for up to 5 weeks. Cryptosporidiosis can become endemic in developing countries, where the hygiene and sanitation conditions are not adequate. As Cryptosporidium spreads through contaminated food under un-hygienic conditions, it is expected that all individuals in developing countries may be exposed at an early age. In a birth cohort study from India, the serum IgG response and seroconversion pattern to Cryptosporidium gp-15 among children suggested a high rate of asymptomatic transmission of Cryptosporidium in the study region.[41]
Figure 2.
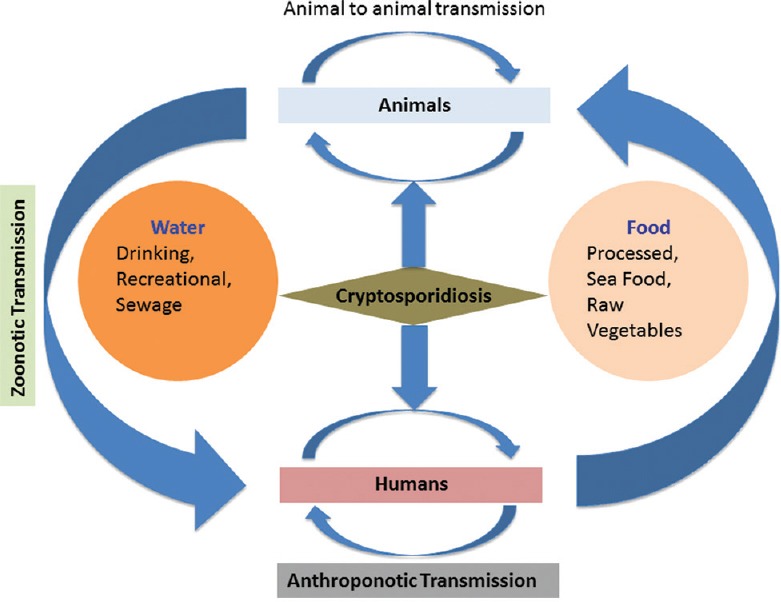
Transmission cycle of Cryptosporidium: Cryptosporidium transmission via zoonotic and anthroponotic routes
CRYPTOSPORIDIUM BIOLOGY
Cryptosporidium primarily infects the epithelium of small intestines but occasionally can infect lungs or bile ducts.[42,43,44] Complete genome sequences for C. parvum and C. hominis are now available.[45,46] The Cryptosporidium genome consists of eight chromosomes containing nearly 9.2 million base pairs. Genome alignment studies showed that both of these genomes consist of 3%–5% of sequence variations with no significant insertion deletion or rearrangement.[47] Several metabolic pathways, multiple organelles, and genes, which are common to eukaryotes or restricted to Apicomplexa, are either reduced or missing in Cryptosporidium. Cryptosporidium also lacks the enzymes for the synthesis of key biochemical building block such as sugars, amino acids, and nucleotides. Genome analysis of Cryptosporidium shows that it encodes more than 80 genes with strong similarities to known transporters and several hundred genes with transporter-like properties. Therefore, Cryptosporidium spp. relies heavily on scavenging nutrients from the host, salvage rather than de novo biosynthesis, and glycolysis or substrate-level phosphorylation for energy production.
Cryptosporidium has a complex life cycle, consisting of both sexual and asexual phases. It completes its life cycle within a single host, i.e., humans. The infective stage of Cryptosporidium is the oocyst, each containing four sporozoites.[48] Oocysts are released into the environment after the defecation of an infected host and are later ingested by a susceptible host. These oocysts can remain viable in the environment and can resist standard disinfectants such as chlorination of drinking water.[49] Cryptosporidium is highly infectious, with ingestion of as few as ten oocysts by a healthy human resulting in clinical infection.[50,51] After the ingestion of oocysts, spindle-shaped sporozoites are released into the gastrointestinal tract, which adhere to host intestinal epithelial cells (IECs). Encapsulation of sporozoites by parasitophorus vacuoles lead to the development of trophozoites. All stages of parasitic differentiation occur within the host cell plasma membrane. These trophozoites undergo merogony to produce meronts. Two consecutive generations of merogony occur, producing Type I and Type II meronts respectively. Type I merozoites, released from Type I meront, re-infect the nearby cells and thereby complete the asexual phase of reproduction.
The sexual phase of reproduction involves the Type II meronts, which differentiate into microgamonts (male gametes) and macrogamonts (female gametes). Macrogamonts, fertilized by the microgamonts, undergo successive division to form mature oocysts. As the result of sporogony two types of oocysts are formed, thick walled which are generally shed in the feces and thin walled which remain in the host intestine, and are responsible for autoinfection.
Several studies have elucidated various virulence mechanisms involved during sporozoite excystation, host cell adherence, invasion, intracellular maintenance, and host cell destruction occurring during Cryptosporidium infection.[52] A recent study suggests that C. parvum elongation factor 1α and novel Ca-activated apyrase plays an important role in host cell invasion.[53] Sporozoite apical protein also plays an important role in target cell attachment and parasitophorous vacuole formation.[54] Various host factors have also been implicated as critical determinants of the outcomes of host-pathogen interactions during Cryptosporidium infection.[55] The host immune response plays an important role in host-pathogen interactions by affecting both the probability of an infection and the severity of a subsequent disease.[56]
HOST RESPONSE TO CRYPTOSPORIDIUM
During Cryptosporidium infection, both innate and adaptive immune responses play critical roles in parasite clearance. Most of the information regarding host-pathogen interactions and immune responses to this protozoan parasite come from studies with Cryptosporidium-infected mice models. Many studies have reported the involvement of IECs in inflammatory response and parasite killing.[57,58] IECs infected with C. parvum showed an elevated inflammatory response by upregulating numerous C, CC, and C-X-C classes of chemokine genes.[59] A recent study demonstrated that C. parvum infection suppresses transcription of the mir-424-503 gene through a nuclear factor-kappa B (NF-kB)-and histone deacetylases-dependent manner.[60] Another study suggests that C. parvum infection induces ileocecal adenocarcinoma and Wnt signaling in mice.[15] One member of the sirtuin family of proteins nicotine adenine dinucleotide (NAD)-dependent deacetylase sirtuin-1 (SIRT1), and an NAD-dependent deacetylase have been concerned in the regulation of multiple cellular processes, including inflammation, longevity, and metabolism. Recent gain and loss-of-function studies revealed that let-7i could modulate NF-κB activation through modification of SIRT1 protein expression these findings suggested a new role of let-7i in the regulation of NF-κB-mediated epithelial innate immune response.[61]
Cellular immune responses induced during Cryptosporidium infections are poorly understood. Various studies have reported the activation of toll-like receptors (TLRs) during Cryptosporidium infection leading to the release of various pro-inflammatory cytokines. The TLRs activate innate immune responses via NK-κβ pathway or by activation of caspase 1 inflammasome that induces proinflamatory cytokines, chemokines, and antimicrobial peptides.[62] TLR4 promotes C. parvum clearance in C. parvum infected mouse model.[63] In one such study involving C. parvum infected mouse, TLR4 was observed to be involved in promoting clearance of C. parvum.[64] In another study, Barakat et al. showed that mice deficient in B, T, and NK cells excrete higher oocysts concentration in comparison to mice that are deficient in B and T cells.[65] Higher levels of neutrophils, eosinophils, NK cells, and CD4 (+) CD25 T cells were found in C. parvum-infected BALB/C mice.[66]
Dendritic cells (DCs) play an important role in host immune responses during Cryptosporidium infection.[67] A few animal studies have reported the interaction between mouse DCs and C. parvum.[68] In one such study, it was observed that DCs expressed interferon-alpha (IFN-α) and IFN-β when exposed to live C. parvum sporozoites.[65] Bedi et al. studied the role of DCs using CD11c+ depleted mice model and observed a considerable increase in susceptibility to C. parvum infection.[67] Another study showed that CD103+ DCs are responsible for C. parvum clearance during infection.[69] In a study using human DCs stimulated with Cryptosporidium soluble and recombinant antigens, elevated secretion of Th-1 cytokines, specifically interleukin-12 (IL-12) p70, IL-2, IL-beta, and IL-6, was reported.[70] A few studies have also evaluated the role of CD+ T cells during Cryptosporidium infection; it was reported that CD4+ T cells significantly contribute toward the clearance of parasite.[71,72] Human and mouse CD8+ T cells contribute significantly in the clearance of Cryptosporidium infection.[71,72] The CD4+ T cells also provide a protective barrier against Cryptosporidium infection. In recent reports of immunodeficient mice infected with the parasite mounted parasite-specific serum IgG response as well as a systemic and mucosal IgA response; moreover, challenge infection led to a booster effect in immunoglobulin response despite the absence of oocyst shedding.[73] Non coding RNAs (ncRNAs) may modulate epithelial immune responses such as production of antimicrobial molecules and expression of cytokines during Cryptosporidium infection.[74]
Macrophages and neutrophils are important immune effector cells and contribute significantly to the intestinal inflammatory response during C. parvum infection.[75] These cells produce an important free radical called nitric oxide (NO) and NO synthesis is significantly increased in IECs that stimulate prostaglandin synthesis in the C. parvum-infected ileum.[76] A study on undernourished mice revealed that L-arginine plays a protective role in C. parvum infection, with involvement of arginase-I and NO synthase enzymatic actions.[77] One recent study also suggested that macrophages and IL-18 play prominent parts in NK cell–independent, IFN-γ–mediated innate immune pathway against C. parvum.[78]
LABORATORY DIAGNOSIS OF CRYPTOSPORIDIOSIS
Various methods are available for the detection and diagnosis of Cryptosporidium as shown in Table 1. Most commonly used diagnostic methods involve detection of Cryptosporidium oocysts, antigens, or nucleic acid in stool samples. For routine examination, bright field microscopy is used along with Romanowsky stain, modified acid-fast staining, negative staining, or dimethyl sulfoxide modified acid-fast staining procedures.[79] Although these methods are low cost, they have low analytic sensitivity for detecting oocysts. On the other hand, direct and indirect immunofluorescent microscopy, although expensive, are more sensitive and oocysts are readily identified by this method. Various reference laboratories in the US and Europe use immunofluorescent microscopy as a gold standard for detection of Cryptosporidium.[80,81] In the past decade, diagnostic interest has been greatly developed toward using Enzyme Immunoassays for detecting oocyst antigens, but still we are lacking clinic tools that can differentiate between oocysts of different species.[81] Rapid detection techniques such as immunochromatographic strip tests or immunochromatographic lateral flow tests are also available that show higher throughput with variable sensitivities (70%–100%).[82] A recent study suggested that the points-of-care tests for Cryptosporidium can be used as an alternative to conventional microscopy where diagnosis of this parasite is limited due to resource-poor settings.[83]
Table 1.
Advantages and disadvantages of various techniques used for Cryptosporidium detection
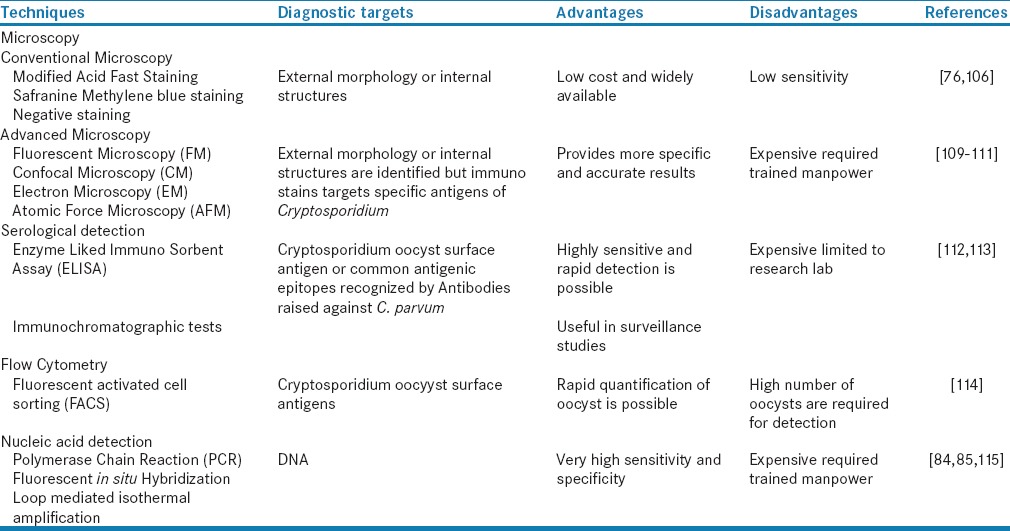
Polymerase chain reaction (PCR) is widely used for genotyping Cryptosporidium, and a variety of PCR assays are available for the sensitive and accurate detection of Cryptosporidium species in clinical samples.[79] Amplification of Cryptosporidium genes encoding 18S rRNA is commonly used for this purpose. With the help of these assays, Cryptosporidium species identification and genotype differentiation can be done easily. Post-PCR analyses are usually based on the direct sequencing of the amplified products, or on the digestion with endonucleases followed by gel electrophoresis of the restriction fragments.[84] The conventional PCR approach does not provide information on pathogen viability and infectivity of pathogens; therefore, with the help of real-time PCR and inclusion/exclusion assays using vital dyes, we can get additional biologic information.[85] When needed, low amounts of Cryptosporidium oocyst DNA can be detected with the help of Loop mediated isothermal amplification (LAMP) PCR, and a recent study suggested that LAMP can be an effective and efficient way for conducting epidemiological studies of Cryptosporidium.[85]
Few studies have highlighted the importance of serological assays to detect IgG antibodies against cryptosporidium as an important tool for epidemiological studies.[16,86] Antibody against Cp23 seems to correlate with distant infection, whereas responses to Cp17 suggest recent infections and antibody to P2 are associated with repeated infection.[87] There are numerous reports of in vitro cultivation techniques to grow Cryptosporidium in cell culture systems.[88] Different cell lines like MDCK, MA-104, HEP-2, Caco-2, AGS, HCT-8 can be used for this parasite culture and HCT-8 cell lines are the most compatible cell lines for Cryptosporidium growth.[89,90] In spite of all these developments, it is still difficult to maintain Cryptosporidium in cell culture system that limits an in-depth understanding on various aspects of Cryptosporidium life cycles, host pathogen responses, and signaling pathways.
TREATMENT AND CONTROL STRATEGIES
Present cryptosporidiosis treatments strategies are extremely limited. One promising treatment is nitazoxanide (NTZ), where a prospective randomized, placebo-controlled study showed that NTZ treatment reduced the duration of both diarrhea and oocyst shedding in patients.[91] Another randomized controlled trial of NTZ showed this drug is more effective in HIV-seronegative children in comparison to HIV-seropositive children.[92] Currently, NTZ is the only Food and Drug administration-approved drug for cryptosporidiosis that shows a moderate efficacy in immunocompromised individuals and children.[93] Other drugs including paromomycin, clarithomycin, azithromycin, rifaximin, rifabutin, and roxithromycinmay also be effective in inhibiting Cryptosporidium growth.[94,95] Gargala et al. reported that Halogeno-Thiazolides (RM-5038) was more effective in comparison to NTZ in inhibiting the growth of Cryptosporidium in Mongolian gerbils.[96] Recent reports have also highlighted the effectiveness of pomegranate peel against C. parvum infection in a murine model.[97] More such studies are required to develop cheap and effective drugs to control Cryptosporidium. Nanotherapy-based approaches might be an effective drug delivery strategy, as a recent study showed Cryptosporidium-specific CP2 protein labeled NP-906 nanoparticles reduced the parasite level 200 folds in a cell culture model.[98]
Lack of long-term maintenance in cell culture and inability to carry out in vitro genetic modification in Cryptosporidium genome are hampering the development of new therapeutic strategies for Cryptosporidium. Various developmental stages of Cryptosporidium and its unique intracellular location, further complicates the drug development procedure. Recent emerging reports highlight the importance of protozoan kinases as a possible drug target for Cryptosporidium.[99]
Availability of full genome sequences of C. parvum and C. hominis has opened new avenues for developing effective drugs and vaccines for Cryptosporidium. In comparison to other apicomplexan parasites, the Cryptosporidium genome has undergone substantial gene lose and horizontal transfer; therefore, it contains highly streamlined metabolic pathways.[100,101] Recently, using a novel in silico approach of reverse vaccinology, three new potential vaccine candidates Cp-15, Prolofin, and Cryptosporidium apyrase have been identified.[102] A DNA vaccine encoding Cp-12 and Cp-21 has shown protective immunity in BALB/c challenged with C. parvum.[103] Although no vaccines are commercially available for cryptosporidiosis yet, various experimental studies to develop attenuated vaccines and DNA vaccines are in progress.[104] At the present time, disease prevention efforts must focus on proper hygiene and clean sanitary conditions to minimize Cryptosporidium outbreaks, especially in developing countries where the prevalence is relatively high in human and animal populations.
RESEARCH NEEDS AND RECOMMENDATIONS
As cryptosporidiosis is a zoonotic disease that spreads through contaminated food and water, epidemiological surveillance is warranted in regions with poor sanitation practice. More attention should be given toward the pathogen source tracking as a complementary tool using molecular studies to identify the actual human and animal contributor sources for infection transmission in a given geographic region. Microbial source tracking (MST) methods are helpful to discriminate between human and various nonhuman sources of fecal contamination.[105,106] Recently, in one such MST study from India, it was observed that both human and animal sources are often present in the environment,[107] and contributing proportions of fecal loading can be estimated using quantitative PCR in combination with analytic modeling. In our recent study form rural India, we showed a high prevalence of Cryptosporidium in community water sources, humans, and domestic animals in the same geographical region.[108] Thus, MST is an active area of research and can be an effective method for source identification and characterization to address pathogen pollution and associated health risks.
The currently available diagnostic techniques for detection of Cryptosporidium have variable degrees of specificity and sensitivity; thereby none of these are unanimously recommended for routine laboratory diagnosis or in epidemiological screening studies. Therefore, it is now essential to develop more affordable diagnostic assays with high specificity and sensitivity for both medical as well as veterinary needs. Availability of such an assay would be boon to many developing countries with poor sanitation practice, where neonatal death due to Cryptosporidium-induced diarrhea is on the rise.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Desai NT, Sarkar R, Kang G. Cryptosporidiosis: An under-recognized public health problem. Trop Parasitol. 2012;2:91–8. doi: 10.4103/2229-5070.105173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–22. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 3.Striepen B. Parasitic infections: Time to tackle cryptosporidiosis. Nature. 2013;503:189–91. doi: 10.1038/503189a. [DOI] [PubMed] [Google Scholar]
- 4.D’Antonio RG, Winn RE, Taylor JP, Gustafson TL, Current WL, Rhodes MM, et al. A waterborne outbreak of cryptosporidiosis in normal hosts. Ann Intern Med. 1985;103(Pt 1):886–8. doi: 10.7326/0003-4819-103-6-886. [DOI] [PubMed] [Google Scholar]
- 5.Mac Kenzie WR, Hoxie NJ, Proctor ME, Gradus MS, Blair KA, Peterson DE, et al. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N Engl J Med. 1994;331:161–7. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 6.Widerström M, Schönning C, Lilja M, Lebbad M, Ljung T, Allestam G, et al. Large outbreak of Cryptosporidium hominis infection transmitted through the public water supply, Sweden. Emerg Infect Dis. 2014;20:581–9. doi: 10.3201/eid2004.121415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puleston RL, Mallaghan CM, Modha DE, Hunter PR, Nguyen-Van-Tam JS, Regan CM, et al. The first recorded outbreak of cryptosporidiosis due to Cryptosporidium cuniculus (formerly rabbit genotype), following a water quality incident. J Water Health. 2014;12:41–50. doi: 10.2166/wh.2013.097. [DOI] [PubMed] [Google Scholar]
- 8.Karanis P, Kourenti C, Smith H. Waterborne transmission of protozoan parasites: a worldwide review of outbreaks and lessons learnt. J Water Health. 2007;5:1–38. doi: 10.2166/wh.2006.002. [DOI] [PubMed] [Google Scholar]
- 9.Lassen B, Ståhl M, Enemark HL. Cryptosporidiosis – An occupational risk and a disregarded disease in Estonia. Acta Vet Scand. 2014;56:36. doi: 10.1186/1751-0147-56-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciçek M, Körkoca H, Gül A. Investigation of Cryptosporidium sp. in workers of the Van municipality slaughterhouse and in slaughtered animals. Turkiye Parazitol Derg. 2008;32:8–11. [PubMed] [Google Scholar]
- 11.Körkoca H, Göz Y, Atas AD, Kurtoglu MG, Ekici K, Berktas M. Prevalence of Cryptosporidium spp.in asymptomatic food workers. Turkiye Parazitol Derg. 2013;37:241–4. doi: 10.5152/tpd.2013.2981. [DOI] [PubMed] [Google Scholar]
- 12.Chen XM, Keithly JS, Paya CV, LaRusso NF. Cryptosporidiosis. N Engl J Med. 2002;346:1723–31. doi: 10.1056/NEJMra013170. [DOI] [PubMed] [Google Scholar]
- 13.Cegielski JP, Ortega YR, McKee S, Madden JF, Gaido L, Schwartz DA, et al. Cryptosporidium, enterocytozoon, and cyclospora infections in pediatric and adult patients with diarrhea in Tanzania. Clin Infect Dis. 1999;28:314–21. doi: 10.1086/515131. [DOI] [PubMed] [Google Scholar]
- 14.Fayer R. Cryptosporidium: a water-borne zoonotic parasite. Vet Parasitol. 2004;126:37–56. doi: 10.1016/j.vetpar.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Benamrouz S, Conseil V, Chabé M, Praet M, Audebert C, Blervaque R, et al. Cryptosporidium parvum-induced ileo-caecal adenocarcinoma and Wnt signaling in a mouse model. Dis Model Mech. 2014;7:693–700. doi: 10.1242/dmm.013292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wanyiri JW, Kanyi H, Maina S, Wang DE, Steen A, Ngugi P, et al. Cryptosporidiosis in HIV/AIDS patients in Kenya: clinical features, epidemiology, molecular characterization and antibody responses. Am J Trop Med Hyg. 2014;91:319–28. doi: 10.4269/ajtmh.13-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oates SC, Miller MA, Hardin D, Conrad PA, Melli A, Jessup DA, et al. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California Coast. Appl Environ Microbiol. 2012;78:8762–72. doi: 10.1128/AEM.02422-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouzid M, Hunter PR, McDonald V, Elwin K, Chalmers RM, Tyler KM. A new heterogeneous family of telomerically encoded Cryptosporidium proteins. Evol Appl. 2013;6:207–17. doi: 10.1111/j.1752-4571.2012.00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan U, Paparini A, Tong K, Yang R, Gibson-Kueh S, O’Hara A, et al. Cryptosporidium huwi n. sp. (Apicomplexa: Eimeriidae) from the guppy (Poecilia reticulata) Exp Parasitol. 2015;150:31–5. doi: 10.1016/j.exppara.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Elwin K, Hadfield SJ, Robinson G, Crouch ND, Chalmers RM. Cryptosporidium viatorum n. sp. (Apicomplexa: Cryptosporidiidae) among travellers returning to Great Britain from the Indian subcontinent, 2007-2011. Int J Parasitol. 2012;42:675–82. doi: 10.1016/j.ijpara.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 21.Ng-Hublin JS, Combs B, Mackenzie B, Ryan U. Human cryptosporidiosis diagnosed in Western Australia: a mixed infection with Cryptosporidium meleagridis, the Cryptosporidium mink genotype, and an unknown Cryptosporidium species. J Clin Microbiol. 2013;51:2463–5. doi: 10.1128/JCM.00424-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rimšeliene G, Vold L, Robertson L, Nelke C, Søli K, Johansen ØH, et al. An outbreak of gastroenteritis among schoolchildren staying in a wildlife reserve: thorough investigation reveals Norway's largest cryptosporidiosis outbreak. Scand J Public Health. 2011;39:287–95. doi: 10.1177/1403494810396557. [DOI] [PubMed] [Google Scholar]
- 23.Sarkar R, Tate JE, Ajjampur SS, Kattula D, John J, Ward HD, et al. Burden of diarrhea, hospitalization and mortality due to cryptosporidial infections in Indian children. PLoS Negl Trop Dis. 2014;8:e3042. doi: 10.1371/journal.pntd.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansour AM, Mohammady HE, Shabrawi ME, Shabaan SY, Zekri MA, Nassar M, et al. Modifiable diarrhoea risk factors in Egyptian children aged Cryptosporidium hominis subtypes and Enterocytozoon bieneusi genotypes in HIV-infected persons in Ibadan, Nigeria. Zoonoses Public Health. 2014;61:297–303. doi: 10.1111/zph.12072. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Zhu H, Chen Y, Zhang Y. Epidemiological analysis on 589 children with hand-foot-mouth disease from Xianju county of Zhejiang province. Zhonghua Liu Xing Bing Xue Za Zhi. 2014;35:708–9. [PubMed] [Google Scholar]
- 26.Ayinmode AB, Zhang H, Dada-Adegbola HO, Xiao L. Cryptosporidium hominis subtypes and Enterocytozoon bieneusi genotypes in HIV-infected persons in Ibadan, Nigeria. Zoonoses Public Health. 2014;61:297–303. doi: 10.1111/zph.12072. [DOI] [PubMed] [Google Scholar]
- 27.Elwin K, Hadfield SJ, Robinson G, Chalmers RM. The epidemiology of sporadic human infections with unusual cryptosporidia detected during routine typing in England and Wales, 2000-2008. Epidemiol Infect. 2012;140:673–83. doi: 10.1017/S0950268811000860. [DOI] [PubMed] [Google Scholar]
- 28.Frost FJ, Muller T, Craun GF, Fraser D, Thompson D, Notenboom R, et al. Serological analysis of a cryptosporidiosis epidemic. Int J Epidemiol. 2000;29:376–9. doi: 10.1093/ije/29.2.376. [DOI] [PubMed] [Google Scholar]
- 29.Cacciò SM. Molecular epidemiology of human cryptosporidiosis. Parassitologia. 2005;47:185–92. [PubMed] [Google Scholar]
- 30.Soonawala D, van Lieshout L, den Boer MA, Claas EC, Verweij JJ, Godkewitsch A, et al. Post-travel screening of asymptomatic long-term travelers to the tropics for intestinal parasites using molecular diagnostics. Am J Trop Med Hyg. 2014;90:835–9. doi: 10.4269/ajtmh.13-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mor SM, Tumwine JK, Ndeezi G, Srinivasan MG, Kaddu-Mulindwa DH, Tzipori S, et al. Respiratory cryptosporidiosis in HIV-seronegative children in Uganda: potential for respiratory transmission. Clin Infect Dis. 2010;50:1366–72. doi: 10.1086/652140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercado R, Buck GA, Manque PA, Ozaki LS. Cryptosporidium hominis infection of the human respiratory tract. Emerg Infect Dis. 2007;13:462–4. doi: 10.3201/eid1303.060394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzanidakis N, Sotiraki S, Claerebout E, Ehsan A, Voutzourakis N, Kostopoulou D, et al. Occurrence and molecular characterization of Giardia duodenalis and Cryptosporidium spp. in sheep and goats reared under dairy husbandry systems in Greece. Parasite. 2014;21:45. doi: 10.1051/parasite/2014048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al Mawly J, Grinberg A, Velathanthiri N, French N. Cross sectional study of prevalence, genetic diversity and zoonotic potential of Cryptosporidium parvum cycling in New Zealand dairy farms. Parasit Vectors. 2015;8:240. doi: 10.1186/s13071-015-0855-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abe N, Matsubara K. Molecular identification of Cryptosporidium isolates from exotic pet animals in Japan. Vet Parasitol. 2015;209:254–7. doi: 10.1016/j.vetpar.2015.02.035. [DOI] [PubMed] [Google Scholar]
- 36.Helmy YA, Krücken J, Nöckler K, von Samson-Himmelstjerna G, Zessin KH. Comparison between two commercially available serological tests and polymerase chain reaction in the diagnosis of Cryptosporidium in animals and diarrhoeic children. Parasitol Res. 2014;113:211–6. doi: 10.1007/s00436-013-3645-3. [DOI] [PubMed] [Google Scholar]
- 37.Vergara-Castiblanco CA, Quílez-Cinca J, Freire-Santos F, Castro-Hermida JA, Ares-Mazás ME. Serological response to Cryptosporidium parvum in adult cattle from the Andean region of Colombia. Parasitol Res. 2001;87:500–4. doi: 10.1007/s004360100375. [DOI] [PubMed] [Google Scholar]
- 38.Salyer SJ, Gillespie TR, Rwego IB, Chapman CA, Goldberg TL. Epidemiology and molecular relationships of Cryptosporidium spp.in people, primates, and livestock from Western Uganda. PLoS Negl Trop Dis. 2012;6:e1597. doi: 10.1371/journal.pntd.0001597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koinari M, Karl S, Ng-Hublin J, Lymbery AJ, Ryan UM. Identification of novel and zoonotic Cryptosporidium species in fish from Papua New Guinea. Vet Parasitol. 2013;198:1–9. doi: 10.1016/j.vetpar.2013.08.031. [DOI] [PubMed] [Google Scholar]
- 40.Li N, Xiao L, Alderisio K, Elwin K, Cebelinski E, Chalmers R, et al. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg Infect Dis. 2014;20:217–24. doi: 10.3201/eid2002.121797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarkar R, Ajjampur SS, Muliyil J, Ward H, Naumova EN, Kang G. Serum IgG responses and seroconversion patterns to Cryptosporidium gp15 among children in a birth cohort in south India. Clin Vaccine Immunol. 2012;19:849–54. doi: 10.1128/CVI.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varughese EA, Bennett-Stamper CL, Wymer LJ, Yadav JS. A new in vitro model using small intestinal epithelial cells to enhance infection of Cryptosporidium parvum. J Microbiol Methods. 2014;106:47–54. doi: 10.1016/j.mimet.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 43.Albuquerque YM, Silva MC, Lima AL, Magalhães V. Pulmonary cryptosporidiosis in AIDS patients, an underdiagnosed disease. J Bras Pneumol. 2012;38:530–2. doi: 10.1590/s1806-37132012000400017. [DOI] [PubMed] [Google Scholar]
- 44.Velásquez JN, di Risio C, Marta E, Astudillo OG, Etchart C, Cucher MA, et al. Occurrence of bile-duct/duodenal abnormalities in nine AIDS patients co-infected with Cryptosporidium hominis and/or C. parvum. Ann Trop Med Parasitol. 2010;104:257–63. doi: 10.1179/136485910X12647085215732. [DOI] [PubMed] [Google Scholar]
- 45.Deng M, Rutherford MS, Abrahamsen MS. Host intestinal epithelial response to Cryptosporidium parvum. Adv Drug Deliv Rev. 2004;56:869–84. doi: 10.1016/j.addr.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 46.Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, et al. The genome of Cryptosporidium hominis. Nature. 2004;431:1107–12. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- 47.Mazurie AJ, Alves JM, Ozaki LS, Zhou S, Schwartz DC, Buck GA. Comparative genomics of Cryptosporidium. Int J Genomics. 2013;2013:832756. doi: 10.1155/2013/832756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao L, Fayer R, Ryan U, Upton SJ. Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev. 2004;17:72–97. doi: 10.1128/CMR.17.1.72-97.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graczyk TK, Fried B. Human waterborne trematode and protozoan infections. Adv Parasitol. 2007;64:111–60. doi: 10.1016/S0065-308X(06)64002-5. [DOI] [PubMed] [Google Scholar]
- 50.Chappell CL, Okhuysen PC, Langer-Curry R, Widmer G, Akiyoshi DE, Tanriverdi S, et al. Cryptosporidium hominis: experimental challenge of healthy adults. Am J Trop Med Hyg. 2006;75:851–7. [PubMed] [Google Scholar]
- 51.Okhuysen PC, Chappell CL, Crabb JH, Sterling CR, DuPont HL. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J Infect Dis. 1999;180:1275–81. doi: 10.1086/315033. [DOI] [PubMed] [Google Scholar]
- 52.Fayer R, Orlandi P, Perdue ML. Virulence factor activity relationships for hepatitis E and Cryptosporidium. J Water Health. 2009;7(Suppl 1):S55–63. doi: 10.2166/wh.2009.044. [DOI] [PubMed] [Google Scholar]
- 53.Manque PA, Woehlbier U, Lara AM, Tenjo F, Alves JM, Buck GA. Identification and characterization of a novel calcium-activated apyrase from Cryptosporidium parasites and its potential role in pathogenesis. PLoS One. 2012;7:e31030. doi: 10.1371/journal.pone.0031030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borowski H, Clode PL, Thompson RC. Active invasion and/or encapsulation? A reappraisal of host-cell parasitism by Cryptosporidium. Trends Parasitol. 2008;24:509–16. doi: 10.1016/j.pt.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Bouzid M, Hunter PR, Chalmers RM, Tyler KM. Cryptosporidium pathogenicity and virulence. Clin Microbiol Rev. 2013;26:115–34. doi: 10.1128/CMR.00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petry F, Jakobi V, Tessema TS. Host immune response to Cryptosporidium parvum infection. Exp Parasitol. 2010;126:304–9. doi: 10.1016/j.exppara.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 57.Chen XM, LaRusso NF. Mechanisms of attachment and internalization of Cryptosporidium parvum to biliary and intestinal epithelial cells. Gastroenterology. 2000;118:368–79. doi: 10.1016/s0016-5085(00)70219-8. [DOI] [PubMed] [Google Scholar]
- 58.Guk SM, Yong TS, Chai JY. Role of murine intestinal intraepithelial lymphocytes and lamina propria lymphocytes against primary and challenge infections with Cryptosporidium parvum. J Parasitol. 2003;89:270–5. doi: 10.1645/0022-3395(2003)089[0270:ROMIIL]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 59.Wang HC, Dann SM, Okhuysen PC, Lewis DE, Chappell CL, Adler DG, et al. High levels of CXCL10 are produced by intestinal epithelial cells in AIDS patients with active cryptosporidiosis but not after reconstitution of immunity. Infect Immun. 2007;75:481–7. doi: 10.1128/IAI.01237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou R, Gong AY, Chen D, Miller RE, Eischeid AN, Chen XM. Histone deacetylases and NF-kB signaling coordinate expression of C×3CL1 in epithelial cells in response to microbial challenge by suppressing miR-424 and miR-503. PLoS One. 2013;8:e65153. doi: 10.1371/journal.pone.0065153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie H, Lei N, Gong AY, Chen XM, Hu G. Cryptosporidium parvum induces SIRT1 expression in host epithelial cells through downregulating let-7i. Hum Immunol. 2014;75:760–5. doi: 10.1016/j.humimm.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen XM, O’Hara SP, Nelson JB, Splinter PL, Small AJ, Tietz PS, et al. Multiple TLRs are expressed in human cholangiocytes and mediate host epithelial defense responses to Cryptosporidium parvum via activation of NF-kappaB. J Immunol. 2005;175:7447–56. doi: 10.4049/jimmunol.175.11.7447. [DOI] [PubMed] [Google Scholar]
- 63.O’Hara SP, Bogert PS, Trussoni CE, Chen X, LaRusso NF. TLR4 promotes Cryptosporidium parvum clearance in a mouse model of biliary cryptosporidiosis. J Parasitol. 2011;97:813–21. doi: 10.1645/GE-2703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen XM, Splinter PL, O’Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–38. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barakat FM, McDonald V, Foster GR, Tovey MG, Korbel DS. Cryptosporidium parvum infection rapidly induces a protective innate immune response involving type I interferon. J Infect Dis. 2009;200:1548–55. doi: 10.1086/644601. [DOI] [PubMed] [Google Scholar]
- 66.Codices V, Martins C, Novo C, Pinho M, de Sousa B, Lopes A, et al. Cell phenotypic change due to Cryptosporidium parvum infection in immunocompetent mice. Acta Parasitol. 2013;58:70–9. doi: 10.2478/s11686-013-0113-2. [DOI] [PubMed] [Google Scholar]
- 67.Bedi B, McNair NN, Mead JR. Dendritic cells play a role in host susceptibility to Cryptosporidium parvum infection. Immunol Lett. 2014;158:42–51. doi: 10.1016/j.imlet.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 68.Perez-Cordon G, Yang G, Zhou B, Nie W, Li S, Shi L, et al. Interaction of Cryptosporidium parvum with mouse dendritic cells leads to their activation and parasite transportation to mesenteric lymph nodes. Pathog Dis. 2014;70:17–27. doi: 10.1111/2049-632X.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lantier L, Lacroix-Lamandé S, Potiron L, Metton C, Drouet F, Guesdon W, et al. Intestinal CD103+dendritic cells are key players in the innate immune control of Cryptosporidium parvum infection in neonatal mice. PLoS Pathog. 2013;9:e1003801. doi: 10.1371/journal.ppat.1003801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bedi B, Mead JR. Cryptosporidium parvum antigens induce mouse and human dendritic cells to generate Th1-enhancing cytokines. Parasite Immunol. 2012;34:473–85. doi: 10.1111/j.1365-3024.2012.01382.x. [DOI] [PubMed] [Google Scholar]
- 71.Pantenburg B, Castellanos-Gonzalez A, Dann SM, Connelly RL, Lewis DE, Ward HD, et al. Human CD8(+) T cells clear Cryptosporidium parvum from infected intestinal epithelial cells. Am J Trop Med Hyg. 2010;82:600–7. doi: 10.4269/ajtmh.2010.09-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kvác M, Kodádková A, Sak B, Kvetonová D, Jalovecká M, Rost M, et al. Activated CD8+T cells contribute to clearance of gastric Cryptosporidium muris infections. Parasite Immunol. 2011;33:210–6. doi: 10.1111/j.1365-3024.2010.01271.x. [DOI] [PubMed] [Google Scholar]
- 73.Jakobi V, Petry F. Humoral immune response in IL-12 and IFN-gamma deficient mice after infection with Cryptosporidium parvum. Parasite Immunol. 2008;30:151–61. doi: 10.1111/j.1365-3024.2007.01013.x. [DOI] [PubMed] [Google Scholar]
- 74.Zhou R, Feng Y, Chen XM. Non-coding RNAs in epithelial immunity to Cryptosporidium infection. Parasitology. 2014;141:1233–43. doi: 10.1017/S0031182014000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takeuchi D, Jones VC, Kobayashi M, Suzuki F. Cooperative role of macrophages and neutrophils in host Antiprotozoan resistance in mice acutely infected with Cryptosporidium parvum. Infect Immun. 2008;76:3657–63. doi: 10.1128/IAI.00112-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gookin JL, Duckett LL, Armstrong MU, Stauffer SH, Finnegan CP, Murtaugh MP, et al. Nitric oxide synthase stimulates prostaglandin synthesis and barrier function in C. parvum-infected porcine ileum. Am J Physiol Gastrointest Liver Physiol. 2004;287:G571–81. doi: 10.1152/ajpgi.00413.2003. [DOI] [PubMed] [Google Scholar]
- 77.Castro IC, Oliveira BB, Slowikowski JJ, Coutinho BP, Siqueira FJ, Costa LB, et al. Arginine decreases Cryptosporidium parvum infection in undernourished suckling mice involving nitric oxide synthase and arginase. Nutrition. 2012;28:678–85. doi: 10.1016/j.nut.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Choudhry N, Petry F, van Rooijen N, McDonald V. A protective role for interleukin 18 in interferon γ-mediated innate immunity to Cryptosporidium parvum that is independent of natural killer cells. J Infect Dis. 2012;206:117–24. doi: 10.1093/infdis/jis300. [DOI] [PubMed] [Google Scholar]
- 79.Vohra P, Sharma M, Chaudary U. A comprehensive review of diagnostic techniques for detection of Cryptosporidium parvum in stool samples. J Pharm. 2012;2:15–26. [Google Scholar]
- 80.Chalmers RM, Campbell B, Crouch N, Davies AP. Clinical laboratory practices for detection and reporting of Cryptosporidium in community cases of diarrhoea in the United Kingdom, 2008. Euro Surveill. 2010;15 doi: 10.2807/ese.15.48.19731-en. pii: 19731. [DOI] [PubMed] [Google Scholar]
- 81.Mittal S, Sharma M, Chaudhary U, Yadav A. Comparison of ELISA and microscopy for detection of Cryptosporidium in stool. J Clin Diagn Res. 2014;8:DC07–8. doi: 10.7860/JCDR/2014/9713.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goñi P, Martín B, Villacampa M, García A, Seral C, Castillo FJ, et al. Evaluation of an immunochromatographic dip strip test for simultaneous detection of Cryptosporidium spp, Giardia duodenalis, and Entamoeba histolytica antigens in human faecal samples. Eur J Clin Microbiol Infect Dis. 2012;31:2077–82. doi: 10.1007/s10096-012-1544-7. [DOI] [PubMed] [Google Scholar]
- 83.Shimelis T, Tadesse E. Performance evaluation of point-of-care test for detection of Cryptosporidium stool antigen in children and HIV infected adults. Parasit Vectors. 2014;7:227. doi: 10.1186/1756-3305-7-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Amar CF, Dear PH, McLauchlin J. Detection and identification by real time PCR/RFLP analyses of Cryptosporidium species from human faeces. Lett Appl Microbiol. 2004;38:217–22. doi: 10.1111/j.1472-765x.2004.01473.x. [DOI] [PubMed] [Google Scholar]
- 85.Bakheit MA, Torra D, Palomino LA, Thekisoe OM, Mbati PA, Ongerth J, et al. Sensitive and specific detection of Cryptosporidium species in PCR-negative samples by loop-mediated isothermal DNA amplification and confirmation of generated LAMP products by sequencing. Vet Parasitol. 2008;158:11–22. doi: 10.1016/j.vetpar.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 86.Farkas K, Plutzer J, Moltchanova E, Török A, Varró MJ, Domokos K, et al. Serological responses to Cryptosporidium antigens in inhabitants of Hungary using conventionally filtered surface water and riverbank filtered drinking water. Epidemiol Infect. 2015;143:2743–7. doi: 10.1017/S0950268814003859. [DOI] [PubMed] [Google Scholar]
- 87.Priest JW, Bern C, Xiao L, Roberts JM, Kwon JP, Lescano AG, et al. Longitudinal analysis of Cryptosporidium species-specific immunoglobulin G antibody responses in Peruvian children. Clin Vaccine Immunol. 2006;13:123–31. doi: 10.1128/CVI.13.1.123-131.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Müller J, Hemphill A. In vitro culture systems for the study of apicomplexan parasites in farm animals. Int J Parasitol. 2013;43:115–24. doi: 10.1016/j.ijpara.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 89.Siripanth C, Punpoowong B, Amarapal P, Thima N, Eampokalap B, Kaewkungwal J. Comparison of Cryptosporidium parvum development in various cell lines for screening in vitro drug testing. Southeast Asian J Trop Med Public Health. 2004;35:540–6. [PubMed] [Google Scholar]
- 90.Sifuentes LY, Di Giovanni GD. Aged HCT-8 cell monolayers support Cryptosporidium parvum infection. Appl Environ Microbiol. 2007;73:7548–51. doi: 10.1128/AEM.01579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rossignol JF, Ayoub A, Ayers MS. Treatment of diarrhea caused by Cryptosporidium parvum: a prospective randomized, double-blind, placebo-controlled study of Nitazoxanide. J Infect Dis. 2001;184:103–6. doi: 10.1086/321008. [DOI] [PubMed] [Google Scholar]
- 92.Amadi B, Mwiya M, Musuku J, Watuka A, Sianongo S, Ayoub A, et al. Effect of nitazoxanide on morbidity and mortality in Zambian children with cryptosporidiosis: a randomised controlled trial. Lancet. 2002;360:1375–80. doi: 10.1016/S0140-6736(02)11401-2. [DOI] [PubMed] [Google Scholar]
- 93.Abubakar I, Aliyu SH, Arumugam C, Usman NK, Hunter PR. Treatment of cryptosporidiosis in immunocompromised individuals: systematic review and meta-analysis. Br J Clin Pharmacol. 2007;63:387–93. doi: 10.1111/j.1365-2125.2007.02873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hewitt RG, Yiannoutsos CT, Higgs ES, Carey JT, Geiseler PJ, Soave R, et al. Paromomycin: No more effective than placebo for treatment of cryptosporidiosis in patients with advanced human immunodeficiency virus infection. AIDS Clinical Trial Group. Clin Infect Dis. 2000;31:1084–92. doi: 10.1086/318155. [DOI] [PubMed] [Google Scholar]
- 95.Cabada MM, White AC., Jr Treatment of cryptosporidiosis: do we know what we think we know? Curr Opin Infect Dis. 2010;23:494–9. doi: 10.1097/QCO.0b013e32833de052. [DOI] [PubMed] [Google Scholar]
- 96.Gargala G, François A, Favennec L, Rossignol JF. Activity of halogeno-thiazolides against Cryptosporidium parvum in experimentally infected immunosuppressed gerbils (Meriones unguiculatus) Antimicrob Agents Chemother. 2013;57:2821–3. doi: 10.1128/AAC.01538-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Al-Mathal EM, Alsalem AA. Pomegranate (Punica granatum) peel is effective in a murine model of experimental Cryptosporidium parvum ultrastructural studies of the ileum. Exp Parasitol. 2013;134:482–94. doi: 10.1016/j.exppara.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 98.Mukerjee A, Iyidogan P, Castellanos-Gonzalez A, Cisneros JA, Czyzyk D, Ranjan AP, et al. A nanotherapy strategy significantly enhances anticryptosporidial activity of an inhibitor of bifunctional thymidylate synthase-dihydrofolate reductase from Cryptosporidium. Bioorg Med Chem Lett. 2015;25:2065–7. doi: 10.1016/j.bmcl.2015.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Artz JD, Wernimont AK, Allali-Hassani A, Zhao Y, Amani M, Lin YH, et al. The Cryptosporidium parvum kinome. BMC Genomics. 2011;12:478. doi: 10.1186/1471-2164-12-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–5. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 101.Striepen B, Kissinger JC. Genomics meets transgenics in search of the elusive Cryptosporidium drug target. Trends Parasitol. 2004;20:355–8. doi: 10.1016/j.pt.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 102.Manque PA, Tenjo F, Woehlbier U, Lara AM, Serrano MG, Xu P, et al. Identification and immunological characterization of three potential vaccinogens against Cryptosporidium species. Clin Vaccine Immunol. 2011;18:1796–802. doi: 10.1128/CVI.05197-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yu Q, Li J, Zhang X, Gong P, Zhang G, Li S, et al. Induction of immune responses in mice by a DNA vaccine encoding Cryptosporidium parvum Cp12 and Cp21 and its effect against homologous oocyst challenge. Vet Parasitol. 2010;172:1–7. doi: 10.1016/j.vetpar.2010.04.036. [DOI] [PubMed] [Google Scholar]
- 104.Mead JR. Prospects for immunotherapy and vaccines against Cryptosporidium. Hum Vaccin Immunother. 2014;10:1505–13. doi: 10.4161/hv.28485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Griffith JF, Weisberg SB, McGee CD. Evaluation of microbial source tracking methods using mixed fecal sources in aqueous test samples. J Water Health. 2003;1:141–51. [PubMed] [Google Scholar]
- 106.Chalmers RM, Campbell BM, Crouch N, Charlett A, Davies AP. Comparison of diagnostic sensitivity and specificity of seven Cryptosporidium assays used in the UK. J Med Microbiol. 2011;60(Pt 11):1598–604. doi: 10.1099/jmm.0.034181-0. [DOI] [PubMed] [Google Scholar]
- 107.Schriewer A, Odagiri M, Wuertz S, Misra PR, Panigrahi P, Clasen T, et al. Human and animal fecal contamination of community water sources, stored drinking water and hands in rural India measured with validated microbial source tracking assays. Am J Trop Med Hyg. 2015;93:509–16. doi: 10.4269/ajtmh.14-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Daniels ME, Shrivastava A, Smith WA, Sahu P, Odagiri M, Misra PR, et al. Cryptosporidium and giardia in humans, domestic animals, and village water sources in rural India. Am J Trop Med Hyg. 2015;93:596–600. doi: 10.4269/ajtmh.15-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Considine RF, Dixon DR, Drummond CJ. Oocysts of Cryptosporidium parvum and model sand surfaces in aqueous solutions: an atomic force microscope (AFM) study. Water Res. 2002;36:3421–8. doi: 10.1016/s0043-1354(02)00082-9. [DOI] [PubMed] [Google Scholar]
- 110.de Roubin MR, Pharamond JS, Zanelli F, Poty F, Houdart S, Laurent F, et al. Application of laser scanning cytometry followed by epifluorescent and differential interference contrast microscopy for the detection and enumeration of Cryptosporidium and Giardia in raw and potable waters. J Appl Microbiol. 2002;93:599–607. doi: 10.1046/j.1365-2672.2002.01736.x. [DOI] [PubMed] [Google Scholar]
- 111.Pohlenz J, Bemrick WJ, Moon HW, Cheville NF. Bovine cryptosporidiosis: a transmission and scanning electron microscopic study of some stages in the life cycle and of the host-parasite relationship. Vet Pathol. 1978;15:417–27. doi: 10.1177/030098587801500318. [DOI] [PubMed] [Google Scholar]
- 112.Van den Bossche D, Cnops L, Verschueren J, Van Esbroeck M. Comparison of four rapid diagnostic tests, ELISA, microscopy and PCR for the detection of Giardia lamblia, Cryptosporidium spp. and Entamoeba histolytica in feces. J Microbiol Methods. 2015;110:78–84. doi: 10.1016/j.mimet.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 113.Abdel Hameed DM, Elwakil HS, Ahmed MA. A single-step immunochromatographic lateral-flow assay for detection of Giardia lamblia and Cryptosporidium parvum antigens in human fecal samples. J Egypt Soc Parasitol. 2008;38:797–804. [PubMed] [Google Scholar]
- 114.Barbosa JM, Costa-de-Oliveira S, Rodrigues AG, Hanscheid T, Shapiro H, Pina-Vaz C. A flow cytometric protocol for detection of Cryptosporidium spp. Cytometry A. 2008;73:44–7. doi: 10.1002/cyto.a.20502. [DOI] [PubMed] [Google Scholar]
- 115.Verweij JJ, Blangé RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, et al. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol. 2004;42:1220–3. doi: 10.1128/JCM.42.3.1220-1223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]