Abstract
Objective
Overnutrition can alter gene expression patterns through epigenetic mechanisms that may persist through generations. However, it is less clear if overnutrition, for example a high fat diet, modifies epigenetic control of gene expression in adults, or by what molecular mechanisms, or if such mechanisms contribute to the pathology of the metabolic syndrome. Here we test the hypothesis that a high fat diet alters hepatic DNA methylation, transcription and gene expression patterns, and explore the contribution of such changes to the pathophysiology of obesity.
Methods
RNA-seq and targeted high-throughput bisulfite DNA sequencing were used to undertake a systematic analysis of the hepatic response to a high fat diet. RT-PCR, chromatin immunoprecipitation and in vivo knockdown of an identified driver gene, Phlda1, were used to validate the results.
Results
A high fat diet resulted in the hypermethylation and decreased transcription and expression of Phlda1 and several other genes. A subnetwork of genes associated with Phlda1 was identified from an existing Bayesian gene network that contained numerous hepatic regulatory genes involved in lipid and body weight homeostasis. Hepatic-specific depletion of Phlda1 in mice decreased expression of the genes in the subnetwork, and led to increased oil droplet size in standard chow-fed mice, an early indicator of steatosis, validating the contribution of this gene to the phenotype.
Conclusions
We conclude that a high fat diet alters the epigenetics and transcriptional activity of key hepatic genes controlling lipid homeostasis, contributing to the pathophysiology of obesity.
Keywords: DNA methylation, RNA-seq, Transcription, High fat diet, Liver, Phlda1
Highlights
-
•
Mice placed on a 14-week high fat diet displayed genes that were hypermethylated and underexpressed compared to controls.
-
•
Chromatin immunoprecipitation demonstrated decreased RNA pol II phosphorylation on Phlda1 and Onecut1 in the high fat fed mice, consistent with decreased transcriptional rates.
-
•
A subnetwork of genes downstream of Phlda1, involved in lipid homeostasis, were suppressed both by a high fat diet and by knockdown of Phlda1 in vivo using an adenovirus-delivered shRNA.
-
•
Knock-down of Phlda1 presented with increased lipid droplet sizes, consistent with the global Phlda1 knockout mouse model, which displays liver steatosis on a standard chow diet.
-
•
We conclude that a high fat diet is sufficient to alter DNA methylation in adults and contributes to the pathophysiology of obesity.
1. Introduction
Genetics and environment contribute to the development of obesity and its associated disorders, including type 2 diabetes, metabolic syndrome, and cardiovascular disease [1], [2]. There are multiple genetic risk factors for obesity that contribute to the pathological effects of energy oversupply and/or decreased energy expenditure [3]. However, the interactions between epigenetic changes and obesity in metabolically relevant tissues, such as liver, are not well understood. Many studies have focused on heritable epigenetic changes in response to environmental stress during gestation [4], [5]. For example, gestational over or undernutrition may lead to multiple generations of epigenetic modifications that promote increased caloric uptake and storage (the thrifty phenotype hypothesis [6]). It is not as clear, however, whether overnutrition in adult animals leads to epigenetic modifications in individuals that contribute to the pathophysiology of obesity. Understanding gene-environment interactions that underpin obesity and type 2 diabetes is an important goal for designing successful interventions for weight loss and management of metabolic disease.
Epigenetic regulatory influences are manifested via non-sequence based mechanisms including, for example, DNA methylation, post-translational histone modifications, histone variants, ATP-dependent chromatin remodeling complexes, polycomb/trithorax protein complexes, and small and other non-coding RNAs (lncRNAs and miRNAs). These epigenetic marks constitute multiple layers of control for modulating chromatin structure, to provide genomic homeostasis, and to regulate gene expression [7], [8]. Unlike the primary DNA sequence of the human genome, which is more or less identical in every somatic cell type, epigenetic information varies between different cell types and even between individual cells [7], [8], [9]. Furthermore, the epigenome undergoes dynamic change during development and across the adult age range and may be altered either as a requirement for, or as a consequence of, complex disease pathogenesis [10]. Given the emerging significance of epigenetics in gene regulation and the potential influence of environment on obesity and diabetes [11], there is substantial need to undertake a systematic dissection of the epigenome of metabolically relevant tissues as a consequence of obesogenic diets.
One particularly important epigenetic mark is DNA methylation. It has long been known that DNA can be chemically modified at CpG sequences by the addition of a methyl group on the 5 position of the cytosine pyrimidine ring and that these epigenetic marks can have a profound effect on gene expression [12]. For example, highly methylated genomic regions are associated with repressed chromatin states. In contrast, unmethylated regions, especially CpG islands located within gene promoters, are broadly associated with active transcription [13].
Of particular significance is the recent understanding that epigenetic changes in genome function can be triggered by environmental influences and that the resulting altered phenotypic states may be dynamic or sustainable and even heritable in a mitotic or meiotic fashion [4]. Consequently, environmental stress, whether it be dietary, chemical, or any one of a myriad of other exposures, can potentially alter the homeostatic regulation of the genome to an extent that is both chronic, thereby potentially driving pathogenesis, and trans-generational. This is significant because it suggests that epigenetic changes in chromatin function as a result of diet-induced obesity can be sustained within the gene pool as an environmentally dependent heritable trait.
Despite the public health significance of adult obesity and the emerging importance of epigenetics, there is little information relating to the epigenomic response to elevated caloric intake and its role in the regulation of gene expression. In the present study, we test the hypothesis that a high fat diet alters hepatic DNA methylation, transcription, and gene expression patterns, and we explore the contribution of such changes to the pathophysiology of a high fat diet. We performed targeted DNA Methyl-seq and RNA-seq analyses of livers from mice placed on a standard chow or high fat diet. A small set of genes were both hypermethylated and downregulated after a high fat diet and were evaluated by chromatin immunoprecipitation assays to confirm the relationship between transcriptional rate and DNA methylation. Phlda1 (Pleckstrin Homology Like Domain Family A Member 1, ascribed to multiple functions including regulation of apoptosis and adipogenesis) and Onecut1 (One Cut Homeobox 1, a transcription factor important for liver development and adult liver function) displayed decreased transcriptional elongation as determined by differential recruitment of phosphorylated RNA pol II. A subset of co-regulated genes was identified by investigating nearest neighbors of Phlda1 in a Bayesian network previously generated from F2 crossed mouse liver transcriptomic data [14], [15], [16]. Onecut1, and Foxa family members (Forkhead Box A 1–3) were among several genes in the subnetwork that were decreased by a high fat diet and are important for liver development, liver phenotype, and lipid homeostasis [17], [18]. Furthermore, knockdown of Phlda1 led to increased lipid droplet size. This observation was consistent with the whole animal knockout model of Phlda1, which presents with a phenotype including increased body mass and liver steatosis on a normal chow diet [19]. We conclude that a high fat diet alters specific genetic loci epigenetically, which contributes to the pathophysiology of obesity.
2. Materials and methods
2.1. Animals and diet
Six-week-old male C57Bl/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and housed at the University of Pittsburgh animal facility with ad libitum access to food and water. One week after arrival, the mice were exposed to a hypercaloric test high fat/high sucrose diet (HFD; 40% carbohydrate, 41% fat, 19% protein per calories; 4.4 kcal/g; #TD.96001 Harlan Teklad, Madison, WI) or a matched test standard chow (low fat/low sucrose) diet (SCD; 67% carbohydrate, 13% fat, 20% protein per calories; 3.6 kcal/g; #TD.110340 Harlan Teklad, Madison, WI) for 14 weeks, N = 7 for each diet. Animals were housed and cared for under the supervision of IACUC approved University of Pittsburgh. Weight and food intake was monitored 5 times weekly. A glucose tolerance test was performed as previously described [20].
2.2. Reverse transcription PCR
Reverse transcription PCR (RT-PCR) was performed as previously described [21]. The primer pairs used in this study are listed in Supplementary Table 1. For pre-mRNA RT-PCR, RNA was treated with DNase, and control experiments were performed to ensure intergenic regions were not amplified in PCR reactions.
2.3. RNA and DNA analyses
After killing the mice, livers were harvested as previously described. Powdered liver was used to extract total RNA and DNA and prepared for RNAseq, RT-, and DNA methyl-seq analysis. RNA was extracted using Tri Reagent (Molecular Research Center, Cincinnati, OH), which was subsequently treated with DNase 1 (ThermoFisher, Waltham, MA).
2.4. RNAseq
RNA concentration and integrity were determined by Bioanalyzer using the RNA Nano Kit (Agilent Technologies, Santa Clara, CA). RNA-seq libraries were prepared using the TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA) per the manufacturer's protocol. Briefly, mRNA was purified from total RNA using oligo dT magnetic beads, fragmented, and then reverse transcribed via random hexamers and SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, CA). Second strand cDNA was synthesized using a proprietary master mix. The double stranded cDNA was then blunt ended, the 3′ ends were adenylated, and indexed Illumina adaptors were ligated. DNA fragments were enriched by PCR using the following conditions: initial denaturation of 98 °C for 30 s, 15 cycles of 98 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s, followed by a final extension of 72 °C for 5 min. RNA-seq libraries were visualized by Bioanalyzer (Agilent) and quantified by qPCR. Libraries were bar-coded and pooled by quantity and sequenced via 50 bp single reads on a HiSeq2000 (Illumina). Reads were trimmed to include only bases 9–57 for quality purposes. Alignment was done to mm9 using Tophat with the Bowtie 1 option [22], [23]. The number of reads intersecting with each gene was calculated, one was added, and counts were log2 transformed. DESeq was used for differential expression testing [24].
2.5. DNA Methyl-seq
Liver tissue was homogenized via Tissue Lyser II (Qiagen) and DNA was extracted via DNeasy Blood and Tissue Kit (Qiagen) per manufacturer's instructions. DNA libraries were prepared as per the SureSelectXT Methyl-Seq Target Enrichment System Protocol (Agilent) with a 9 cycle PCR post bisulfite conversion and a 6 cycle indexing PCR. SureSelect Mouse All Exon V1 52 Mbp was used for capture. Libraries were sequenced on a Hiseq2000 (Illumina) using 100 bp paired end reads. Quality and adaptor trimming were done using Trim Galore and alignment was performed using Bismark. Duplicates were removed. MethylSig was used for differential methylation analysis [25], [26]. The GEO accession number for this study is GSE85425.
2.6. Bioinformatics
We queried a mouse liver Bayesian network of ∼9400 nodes and ∼18,500 edges generated from integrating genotype and liver expression values generated in animals from multiple F2 crosses of mice [14], [15], [16]. Phlda1 was a node in this network, and we generated a subnetwork of its nearest neighbors by including nodes within two path lengths (undirected) from Phlda1. Genes in close proximity to Phlda1 in this subnetwork were predicted to be affected if expression of Phlda1 would be perturbed.
2.7. Chromatin immunoprecipitation assay
The liver was extracted and finely minced with a razorblade or flash frozen in liquid nitrogen. The ChIP assay protocol derived for this study was a modification of one published earlier [27], using components of the Millipore/Upstate ChIP kit (cat #17-295), with some additional modifications as previously described [21]. Standard curves were constructed using two-fold serial dilutions of the unbound DNA extracted from the fasted IgG treatment (2.5 μl) as a reference input. The abundance of the amplicons was expressed as a percentage of the total reference input after subtracting the signal from the control IgG group as previously described [21].
2.8. 5-Azacytidine experiments in isolated mouse hepatocytes
Mice on a standard chow diet were anesthetized, and the liver was perfused with 50 ml of preheated 37 °C perfusion buffer I (142 mM Sodium Chloride, 6.7 mM Potassium Chloride, 10 mM Hepes, 2.23 μM EGTA – pH7.4), then with 50–100 mL of preheated 37 °C perfusion buffer II (152 mM Albumin, 67 mM Sodium Chloride, 6.7 mM Potassium Chloride, 100 mM Hepes, 19 mM Calcium Chloride, 0.02% Type IV collagenase (Sigma) – pH7.6) at a flow rate of 8 ml/min. The liver was carefully excised and placed in 6 cm petri dish with ice-cold isolation media (RPMI, 10% Heat-inactivated FBS, Non-Essential Amino Acids, Sodium Pyruvate, Penicillin/Streptomycin, l-Glutamine). The liver was then manually disrupted and filtered through a gauze pad and the liquid was centrifuged at 4 °C at 400 rpm for 5 min. The pellet was re-suspended in isolation media, and 90% Percoll (GE Healthcare) was carefully added to cell suspension. Cells were centrifuged at 4 °C at 400 rpm for 10 min. The pellet was washed twice with isolation media, re-suspended in high glucose media (isolation media and 25 mM glucose), and the viability was calculated using trypan blue exclusion. Cells were plated on collagen-coated 6-well tissue culture plates at a density of 1.2 × 106 live cells per well. After incubating for 1 h at 37 °C and 5% CO2, media and any floating cells were removed and replaced with fresh high glucose treatment media.
Cells were treated for 18 h with or without 10 μM 5-azacytidine (Sigma, St. Louis, MO) prior to harvesting and isolating total RNA as described above.
2.9. Adenovirus-mediated knockdown of hepatic Phlda1
Eight week-old C57BL6 male mice were injected retro-orbitally with 5 × 109 pfu of control GFP-expressing adenovirus or adenovirus expressing an shRNA directed against Phlda1 (VectorBiolabs, Malvern, PA) according to the method of Yardeni et al. [28]. This method delivers adenovirus specifically to the liver with equal efficacy compared when compared to tail vein injection [29], [30]. After 2 weeks, the liver was collected.
2.10. Oil Red O stain and quantification of lipid droplet size by Image J
Liver samples were embedded in Tissue-Tek OCT Compound immediately after dissection, and the fresh frozen liver samples were cut in a cryostat, affixed to microscope slides, and air-dried at room temperature for 30 min. The liver sections were washed with 1 × PBS twice for 5 min, then stained in Oil Red O working solution for 1 h in the dark (Oil Red O stock solution: Oil Red O 500 mg, in 100 ml 60% triethyl-phosphate; and Oil Red O working solution: 15 ml deionized water added to ml 45 ml Oil Red O stock solution). The sections were rinsed with warm running tap water for 15 min and washed with PBS for 5 min. The air-dried sections were mounted with Fluroment G. The slides were then viewed under a Zeiss AxioImager light microscope. Photomicrographs of multiple fields of multiple fragments from five SC and 5 HFD male mice were imported into Image J and total red (representing neutral lipids) and lipid droplet size were quantified according to the method of Mehlem et al. [31]
2.11. Statistics
All the data shown are means ± standard errors (SE) for the numbers of experiments indicated in the figure legends. Significance was determined by unpaired two-tailed t tests. Differences were considered significant if the p value was <0.05.
3. Results
3.1. RNA-seq and DNA Methyl-seq analysis
Seven C57BL/6 mice were placed on a standard chow or a hypercaloric high fat diet for 14 weeks. These animals displayed the expected increase in body weight and decrease in glucose tolerance, which was accounted for by increased calories rather than hyperphagia, as the amount of food consumed was identical between the two groups (Figure S1). Total RNA and genomic DNA were isolated from the livers and RNAseq and Methylseq were performed. Summary statistics for RNA-seq and DNA Methyl-seq sequencing analyses are shown in Supplemental Tables 2 and 3, respectively.
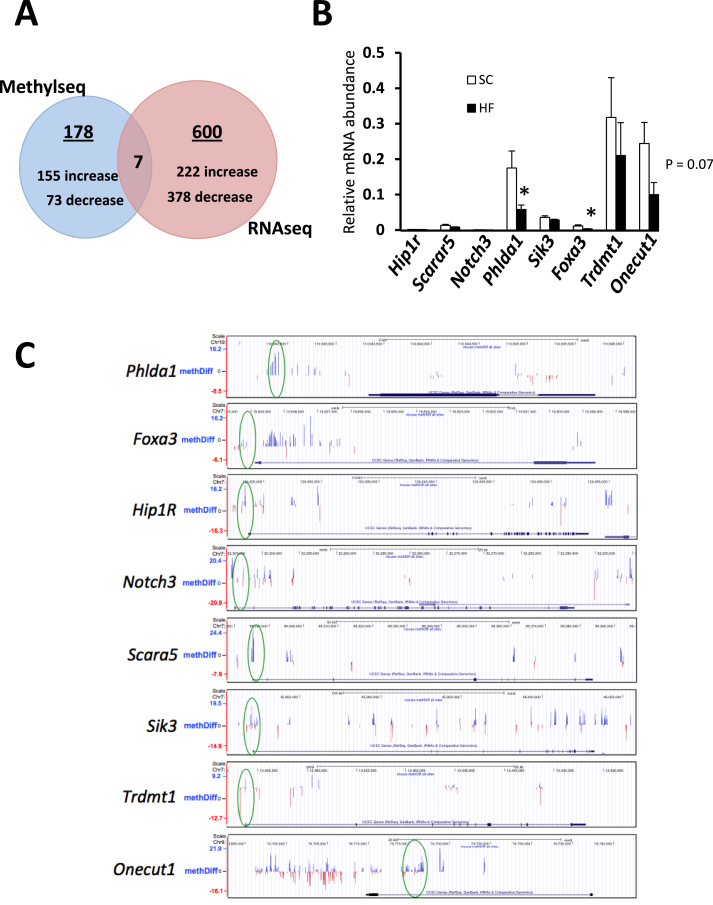
Methyl-seq results were first ranked by the lowest q value of the differential methylation scores. Data were filtered to identify differentially methylated CpG sites that were within 1,000 bp of the transcriptional start site (TSS) of known genes, with a q value less than 10−4, without regard to direction (hypermethylated and hypomethylated genes were treated equally). In addition, we found one gene (Onecut1) that stood out because of highly significantly suppressed expression in the RNAseq data and significantly differentially methylated CpG sites at multiple sites within 10,000 bp of the TSS and because of its importance to liver development [18]. We note that the very stringent parameters used to identify differential Methyl-seq loci were often associated with CpG shores (defined as near CpG islands, and more likely than islands to contains regulatory alterations in DNA methylation [32]) and were often imbedded in a larger grouping of less significantly differentially methylated regions (see Supplemental Table 4). RNA-seq data were filtered by listing the top 600 genes with significantly differential expression by p value, without regard to direction. Seven genes were found that were both differentially methylated and differentially expressed (Figure 1A). The seven genes had differentially methylated sites that ranged from 5 to 22% different (Supplemental Table 4). We were surprised by the relatively small number of overlapping genes and found that we could dramatically increase the number by decreasing the stringency of inclusion of genes associated with differentially methylated CpGs. If we included differentially methylated CpGs within 10,000 bp of transcription start sites, we found many more overlapping genes. A summary of this analysis is presented in Supplementary Figure 2 and Tables 5–8. In short, 147 genes were found to be differentially expressed and associated with differentially methylated CpGs. Importantly, a ToppGene [33] gene enrichment analysis of these 147 genes found many differentially methylated and expressed genes that also associated with diseases relevant to the metabolic syndrome [34]. These results were reassuring and we proceeded with interrogation of the 7 genes in Figure 1.
Figure 1.
RNA-seq and DNA Methyl-seq of standard chow- and high fat–fed mouse livers. A. Venn diagram of genes found to be significantly differentially methylated within 1,000 bp of their TSS, and those with significantly different RNA expression in response to a high fat diet. B. Quantitative RT-PCR of the 7 genes plus Onecut1 added due to remarkable number of hypermethylated CpGs and important role in liver development. N = 7, *, p < 0.05. C. Screenshots of UCSC genome browser showing significantly hypermethylated sites (blue bars, circled in green) near the TSSs of the indicated genes (oriented with upstream regions to the left). MethDiff, average percentage differentially methylated CpGs; blue bars, hypermethylated, red bars hypomethylated. http://genome.ucsc.edu[35].
Two of these genes were confirmed to have significant decreases in RNA expression after a high fat diet as determined by RT-PCR, and Onecut1 was not significantly different but trended down with a p value of 0.07 (Figure 1B). The other genes trended by RT-PCR in a qualitatively similar manner to the RNA-seq data with the exception of Scara5, which was increased in RNA-seq (with corresponding hypomethylation) but decreased by RT-PCR. The differential methylation near the TSS of these genes can be seen in screenshots from the UCSC genome browser [http://genome.ucsc.edu [35]] in Figure 1C. Summaries of these genes, along with a brief description of their functions, are listed in Supplemental Table 5. Of note is that Onecut1 and Foxa3 are both important for liver development [17], [18], and the Phlda1 global knockout model presents with a phenotype of increased adult body mass and liver steatosis, even on a standard chow diet [19].
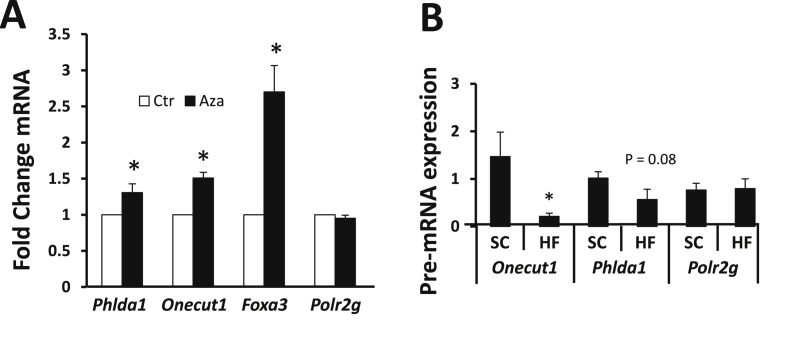
If genes were regulated by hypermethylation, such that increased promoter methylation decreases gene expression, then hypomethylation might reasonably be expected to increase the expression of that gene [36]. To test this possibility for our target genes, we treated mouse primary hepatocytes overnight with a vehicle control or with 5-azacytodine, an agent that removes DNA methylation [37]. We found that Phlda1, Foxa3, and Onecut1 all displayed increased expression after 5-azacytidine treatment (Figure 2A).
Figure 2.
Genes potentially regulated by methylation status, transcriptional and post-transcriptional mechanisms. A. Primary mouse hepatocytes were treated with 10 μM 5-azacytodine (Aza) for 18 h. Total RNA was collected and mRNA was measured using RT-PCR with primers for the indicated genes relative to β-actin using the ΔΔCT method. B. Measurement of pre-mRNA of Phlda1, Onecut1, and Foxa3 using DNase-treated RNA and primers specifically spanning intron/exon boundaries. Error bars represent SEM, *, p < 0.05, N = 7.
To test if the differential RNA expression was due to transcriptional regulation we first measured the relative abundance of pre-mRNA of several genes, including Phlda1, Onecut1, and Foxa3 (Figure 2B). Pre-mRNA levels were determined by RT-PCR using primers that spanned intron-exon boundaries after insuring that no DNA was present. This readout represents newly transcribed mRNA that has not yet been spliced. We found that Onecut1 pre-mRNA significantly decreased after a high fat diet, Phlda1 trended downward, and Foxa3 pre-mRNA expression did not change. These results suggested that the decreased Foxa3 mRNA was due to a post-transcriptional mechanism, while the decreases in Onecut1 and Phlda1 mRNA were more likely due to transcriptional mechanisms.
3.2. Transcriptional validation
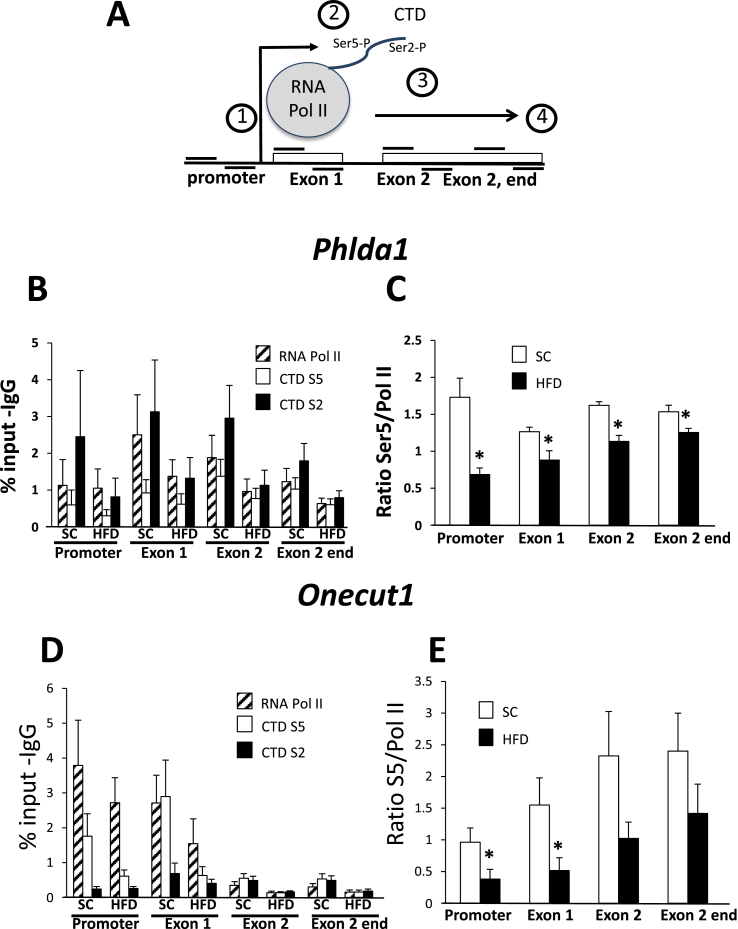
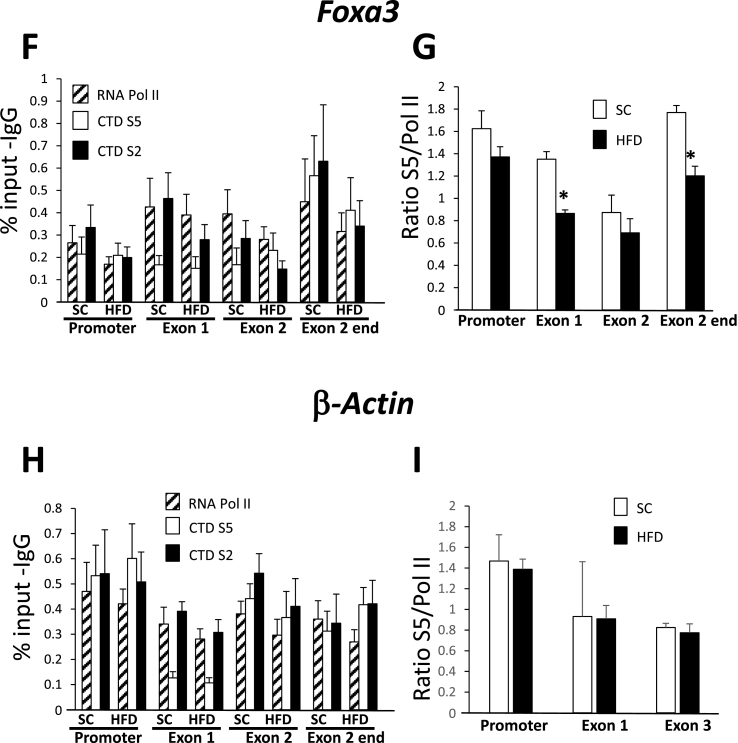
To confirm that hypermethylation from a high fat diet is associated with changes in transcription, we performed chromatin immunoprecipitation (ChIP) assays using antibodies against RNA pol II and the phosphorylated C-terminal domain of RNA pol II (Figure 3). Transcription proceeds after recruitment of the RNA pol II holocomplexes and often results in pausing approximately 20–60 bp downstream of the TSS [38]. Here, the C-terminal domain, comprising approximately 50 heptad repeats of a conserved sequence (Y1S2P3T4S5P6S7), and with serine phospho-acceptor sites at positions 2 and 5, is critical for regulation of the polymerase. Phosphorylation of Ser5 of the CTD by TFIIH occurs after binding and before elongation of the RNA pol II complex while a second phosphorylation event mediated by CTDK1 of PTEFb on Ser2 occurs after release from pausing and during transcriptional elongation [39]. Thus, by examining the relative recovery of RNA pol II, and CTD Ser5 and Ser2 from chromatin immunoprecipitates along a gene, from promoter to polyA signal, one is able to reconstruct the procession of RNA pol II through the gene (see Figure 3A) [40]. In Figure 3B,C we found that Phlda1 is transcribed in a typical fashion after a standard chow diet with abundant RNA pol II in the promoter region upstream and just downstream of the TSS in the 5′ region of exon 1 near the TSS, and less RNA pol II apparent in the second (last) exon of the gene. Likewise, the CTD Ser5 phosphorylation is most abundant at the 5′ regions of the gene, while CTD Ser2 phosphorylation is most abundant in the coding region and towards the 3′ end of the gene. There was a trend of decreased recovery of immunoprecipitated RNA pol II from chromatin isolated from mice fed a high fat diet compared to the standard chow diet at all of the sites interrogated, though this did not reach the level of statistical significance. However, the ratio of recovery of CTD Ser5 to total RNA pol II showed a very significant difference between the two diets (Figure 3C). Since the CTD of RNA pol II must be phosphorylated on Ser5 for release from pausing prior to elongation [41], these data reflect a decrease in the transcriptional rate of this gene after a high fat diet. The pattern for Onecut1 was similar, with RNA pol II most prominent near the 5′ end of the gene (Figure 3D), and with a pronounced decreased ratio (either significant or trending) of Ser5 phosphorylation to total RNA pol II after a high fat diet (Figure 3E). These data also reflect decreased transcription through the Onecut1 gene. By contrast, the Foxa3 gene displayed a more uniform distribution of RNA pol II (Figure 3F) and showed inconsistently decreased ratios of Ser5 phosphorylation to total RNA pol II only in exon 1 and at the end of exon 2 (Figure 3G). While there may be some transcriptional control, when combined with the pre-mRNA data, the main form of regulation of the Foxa3 gene appears to be post-transcriptional. As a control, we measured RNA pol II occupancy of the β-actin housekeeping gene and found no change of recruitment near the promoter or in exons 1 or 3 with a HFD, consistent with a lack of transcriptional regulation (Figure 3H,I). We note that many genes increase in response to a HFD (165 genes increased greater than 2-fold with a p value less than 0.05), so there is not a general decrease in transcription in response to this diet.
Figure 3.
Molecular validation of transcriptional regulation. A. Schematic of chromatin immunoprecipitation experiment. A time line is included: (1) RNA polymerase in recruited to the gene and pauses approximately 40 bp from the transcription start site; (2) the heptad repeat of the C-terminal domain (CTD) is “primed” by phosphorylation on Ser5; (3) Ser2 is phosphorylated and Ser5 is gradually dephosphorylated, allowing efficient transcriptional elongation; (4). Notably, Phlda1, Onecut1, and Foxa3 all have only 2 exons. B–I. Chromatin immunoprecipitation was performed using antibodies against total RNA pol II, C-terminal domain (CTD) phospho-Ser5, CTD phospho-Ser2, and control IgG. B,D,F,H. Data shown were generated from quantitative PCR using primers specific for the indicated regions of the indicated genes. Shown is the signal as a percentage of input after subtraction of the IgG control. C, E, G, I. Data shown are the mean of the ratio between the signal from the CTD phospho-Ser5 and total RNA pol II signals at the indicated region of the indicated gene. Error bars represent SEM, *, p < 0.05, N = 6–7.
3.3. Prioritization of candidate Phlda1 gene targets via interrogation of an existing mouse Bayesian network
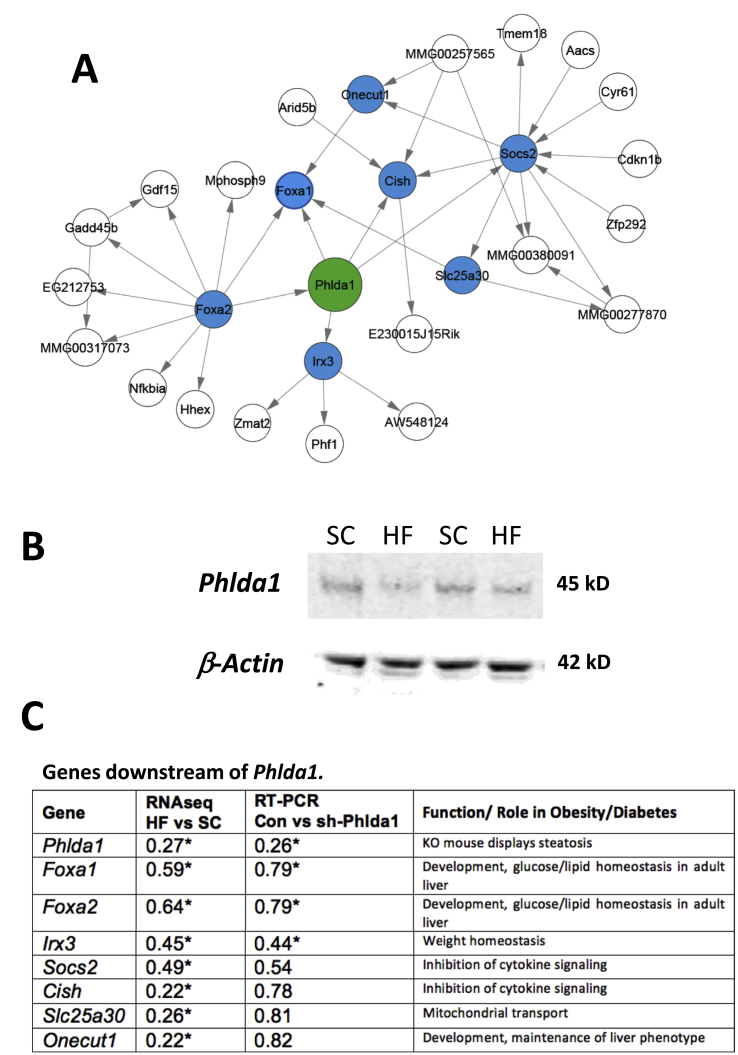
One of the most vexing problems for modern biologists is to decide which genes to interrogate after a whole genome analysis of differential gene regulation. We used a data-driven approach to identify a subnetwork of genes that may be influenced by perturbations in Phlda1. Phlda1 is a node in an existing causal (Bayesian) network model of metabolic diseases (including obesity, diabetes and cardiovascular disease) that was assembled using liver transcriptome and genotype information scored in individuals of F2 mouse crosses of various strains and genotypes [14], [15], [16]. As these Bayesian networks have proven effective in predicting causal gene–gene relationships [42], [43], [44], we identified the subnetwork of genes comprising the nearest neighbors of Phlda1 as in Figure 4A. Hepatic PHLDA1 was decreased after a high fat diet as determined by immunoblots (Figure 4B). We then evaluated each of the genes in the neighborhood in our RNA-seq dataset and noted that the highlighted genes in Figure 4A were found to be decreased by a high fat diet (Figure 4C). This strongly suggests that diet-induced changes in Phlda1 transcription may in turn control expression of these genes. Notably, several of these genes are involved in liver phenotype, lipid homeostasis, and inflammation. With these molecular observations in hand, we proceeded to test if perturbing Phlda1 in vivo could account for at least part of the pathophysiological consequences of a high fat diet in liver.
Figure 4.
Generation of a subnetwork from an existing Bayesian network of obese mouse liver. A. A diagram of the small subnetwork immediately surrounding Phlda1 (green) generated from an existing Bayesian gene expression network that was assembled using liver transcriptome and genotype information scored in individuals of F2 mouse crosses [14], [15], [16]. Nearest neighbors that were interrogated further by RT-PCR are highlighted in blue. B. Representative immunoblots of liver extracts from mice on a standard chow or high fat diet using antibodies against PHLDA1 or β-ACTIN. C. Mean of fold change comparing high fat diet (HF) to standard chow diet (SC), or intraorbital injection of control (GFP) to sh-Phlda1-expressing adenovirus, using data from RNA-seq or RT-PCR, respectively. N = 5–7, *p < 0.05.
3.4. In vivo validation
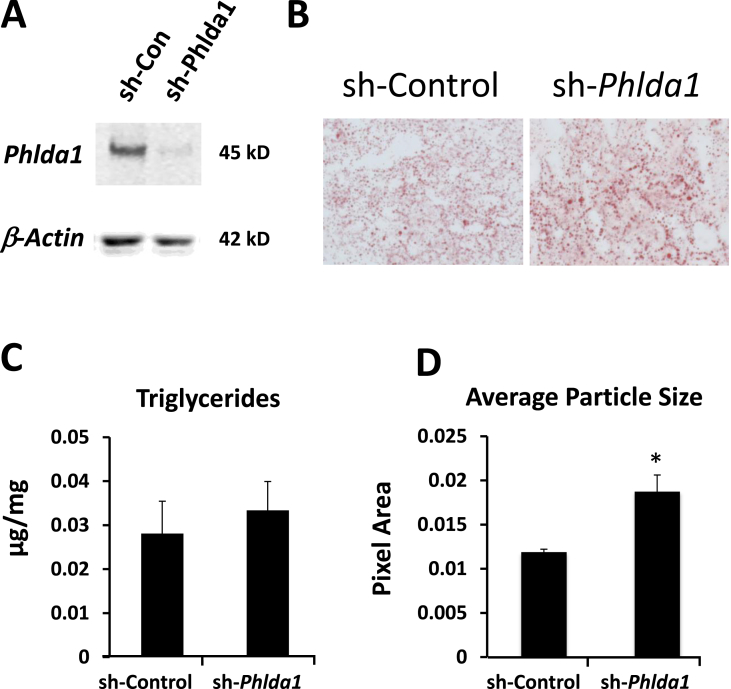
Global knock out of Phlda1 presents with a phenotype that includes liver steatosis on a standard chow diet [19]. We tested if liver-specific knock down of Phlda1 was sufficient to alter liver phenotype. C57Bl/6 mice on a standard chow diet were injected with an adenovirus expressing shRNA directed against Phlda1. We found that 5 × 109 PFU was sufficient to knock down hepatic Phlda1 mRNA by approximately 80% and substantially decreased PHLDA1 at the protein level as well (Figure 4, Figure 5A). Furthermore, knockdown of Phlda1 resulted in a general decrease in genes that were in the same local subnetwork as Phlda1 (Figure 4C) including significant decreases in the expression of Foxa1, Foxa2 and Irx3 (Iroquois Homeobox 3, involved in neural development and also has been implicated in control of weight homeostasis, [45]). These data support the idea that Phlda1 is a key driver of the expression of the neighboring genes in the subnetwork. In addition, we tested if lipid content was altered after depletion of Phlda1. As shown in Figure 5B–D, whereas triglyceride levels and total Oil-red O staining were unchanged, livers from animals with the Phlda1 knockdown displayed significantly increased lipid droplet size (Figure 5D). Increased lipid droplets in the liver can be an early indicator of liver steatosis [46]. Together these data support the following interpretation presented as a model in Figure 6: a high-fat diet increases DNA methylation on select genes including the key driver gene, Phlda1, leading to decreased RNA polymerase activity, decreased gene transcription of Phlda1, and decreased expression of genes downstream in the Bayesian network, which led to increased lipid droplet size and contributed to the hepatic steatosis of a high fat diet.
Figure 5.
Knockdown of Phlda1 inhibits expression of candidate target genes and increases lipid droplet size in mouse liver. Adenoviruses expressing shRNA against Phlda1, or a control adenovirus, were injected retro-orbitally into C57/Bl6 mice on a standard chow diet. Two weeks later, liver sections were prepared for immunoblots using antibodies against PHLDA1 or β-ACTIN (A) and stained with Oil Red O (B). The concentration of triglycerides in liver tissue was measured using a kit (C). The average size of the Oil-red O stained particles was quantified using Image J software (D). Data are the mean ± SEM. *, p < 0.05; N = 5.
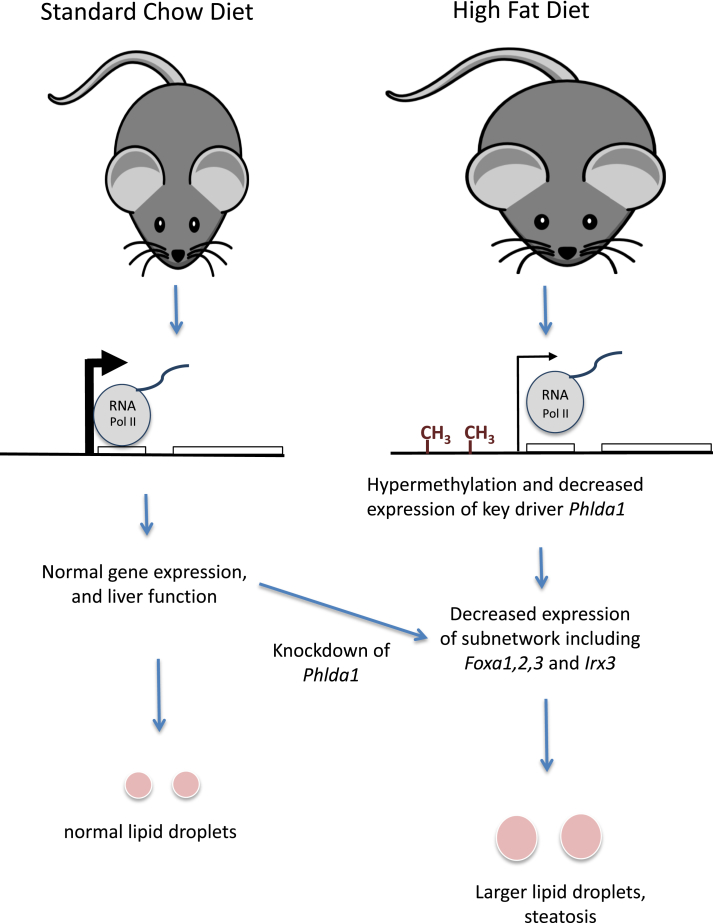
Figure 6.
Model depicting how high fat diet affects DNA methylation, gene expression, and steatosis. A high fat diet results in hypermethylation and decreased mRNA and pre-mRNA levels of specific genes, including Phlda1. The hypermethylation results in decreased transcription, providing a mechanistic explanation for decreased RNA expression. Phlda1 was found to be a key driver controlling a small set of genes including Foxa1, Foxa2, and Irx3. These genes are known to be involved in liver function, lipid homeostasis, and inflammation. Knockdown of Phlda1 resulted in larger lipid droplets in standard chow fed mice, an early manifestation of liver steatosis.
4. Discussion
Obesity is often associated with hepatic steatosis or fatty liver, which is on a spectrum of non-alcoholic fatty liver disease (NAFLD). NAFLD, if left unchecked, may progress to more inflammation and some fibrosis, identified pathologically as non-alcoholic steatohepatitis (NASH), and then to the more deadly cirrhosis or hepatocellular carcinoma [47]. With the worldwide epidemic in obesity and diabetes, it is estimated that hundreds of millions of people have steatosis and are therefore at risk for advanced NAFLD, including cirrhosis and cancer [48]. The molecular mechanisms underlying the pathophysiology of the disease are not completely understood. In the current study, we hypothesized that DNA methylation is altered in response to a high fat diet and that this epigenetic mechanism contributes to the pathophysiology of steatosis.
We found a set of genes that were hypermethylated and differentially underexpressed after 14 weeks on a high fat diet, including Phlda1, Onecut1, and Foxa3. PHLDA1 has 2 plextrin homology domains and polyglutamine domains suggestive of being a transcription factor [49]. Phlda1 is expressed in a wide variety of tissues and is found in different cellular compartments, including the nucleus, cytoplasm and endoplasmic reticulum. However, the exact biochemical role and mechanism of action of PHLDA1 is poorly elucidated. Phlda1 appears to influence apoptosis and proliferation in cancer, but in a complex and tissue-specific manner [49], [50], [51]. For example, Phlda1 expression is reduced in melanoma and breast cancer but expressed at high levels in colon cancer and osteosarcomas [49]. Notably, global Phlda1 knockout mice have a phenotype with increased body mass and hepatic steatosis on a standard chow diet [19]. Onecut1 plays an important role in liver development and is conspicuous by often being associated with hepatic transcription factor networks that include the FOXA proteins, including FOXA3 [18], [52]. Foxa3 has functional redundancy with Foxa1 and Foxa2 in the liver, and liver-specific knock down of Foxa3 results in a relative hypoglycemia during a prolonged fast [53].
Increased DNA methylation, particularly in promoter and enhancer regions enriched with CpG islands, is often accompanied with decreased expression of the associated gene, with the assumption that transcription factors have reduced binding at elements containing methylated CpGs and that transcription is reduced on these genes [5]. However, the exact molecular mechanisms of altered gene expression via increased DNA methylation are rarely explained or validated. Here we found that a high fat diet in mice led to hypermethylation of DNA near the TSSs of several genes that also underwent significant decreases in RNA expression. Additionally, in genes that were regulated at the transcriptional level via the analysis of pre-mRNA expression (Phlda1 and Onecut1), increased DNA methylation was accompanied by decreased activation and transcriptional procession of RNA polymerase in response to a high fat diet. Numerous studies have shown differential expression of genes related to lipid homeostasis and inflammation as a signature of the consequences of a high fat diet [54], [55], [56]. However, relatively few studies have correlated changes in DNA methylation with changes in gene expression and, of those, most involve the inheritance of epigenetic marks and altered gene expression from high fat diets consumed by the mother [57] or father [58], or involved the restoration of DNA methylation with nutritional supplements that augment one carbon metabolism, such as folate or betaine [59], [60]. Recently, Murphy et al. evaluated the changes in DNA methylation and corresponding gene expression in humans with varying degrees of NAFLD, defined and measured from liver biopsies [61]. This study focused on hypomethylated genes and the correspondingly increased expression of genes involved in the pathogenesis of NAFLD. (Note, while some genes are similar in function, tissue repair for example, the actual genes were different.) By comparison, our study focused on genes that were hypermethylated and decreased with the high fat diet.
We found that knockdown of Phlda1 in mouse liver for 2 weeks, using an adenovirus expressing a specific shRNA, resulted in downregulation of all of the nearest neighbor candidate genes predicted by a mouse population based liver molecular network. Importantly, all of the genes in the subnetwork were significantly decreased after a high fat diet as determined by RNAseq (see comparison, Supplemental Table 5). Included in this gene list were Foxa1 and Foxa2, paralogs of Foxa3, which are required for liver development and are important for the expression of numerous metabolic enzymes involved in glucose and lipid homeostasis [17]. Also included was Irx3, which is involved in weight homeostasis in the hypothalamus, though its role in the liver is not fully understood [45]. Finally, the Socs2 (Suppressor Of Cytokine Signaling 2) and Cish (Cytokine Inducible SH2 Containing Protein) genes express anti-inflammatory proteins. Decreased expression of these genes should promote inflammation, a hallmark of obesity and liver steatosis [62].
Knockdown of Phlda1 conferred a phenotype that included increased lipid droplet size. Altered lipid droplets in the liver suggest a deficiency in lipid droplet turnover and may be an early manifestation of liver steatosis [46]. Mice treated with adenovirus expressing an shRNA directed against Phlda1 did not display frank steatosis as in the global knockout animal [19]. We suspect that the loss of Phlda1 in additional tissues, such as fat, and the lifelong absence of Phlda1 in the global knockout animal contributed to the stronger phenotype seen in the knockout animals. Our model proposes that decreased expression of Phlda1 decreases other genes involved in liver function and lipid homeostasis, and, indeed, knocking down Phlda1 with adenovirus led to a decrease in Foxa1. In support of this idea, Moya et al. demonstrated that Foxa1 controls lipid accumulation in rat and human hepatocytes, with increased Foxa1 depleting hepatocytes via several mechanisms including synthesis and storage of triglycerides [63]. Conversely, samples of liver with nonalcoholic fatty liver displayed reduced expression of Foxa1.
Several key questions arise from our study. How much time on a high fat diet is required to initiate gene-specific DNA methylation, alterations in downstream network genes, and liver phenotype? Another important question is the plasticity of the response. Once methylated by a high fat diet, can the DNA be demethylated by reversion to a standard chow or a calorically restricted diet? If so, how long does it take? Careful time course experiments and experiments switching from a high fat diet to a standard chow or low caloric diet will be needed to answer these important questions.
In summary, we found that 14 weeks on a high fat diet leads to hypermethylation and decreased expression of a relatively small set of genes. Among these were Onecut1 and Phlda1, which we confirmed displayed decreased RNA pol II activation in the coding regions of their genes, consistent with decreased transcription. A subnetwork was generated using Phlda1 and its nearest neighbors from an existing Bayesian gene expression network [15]. Strikingly, knockdown of Phlda1 decreased expression of the regulatory genes in the subnetwork and increased lipid droplet size, consistent with the whole body knockout phenotype, which includes hepatic steatosis. We conclude that a high fat diet alters the epigenetics of hepatocytes, contributing to the pathophysiology of obesity.
Author contributions
Conceptualization, DKS, DGP, and RMO; Investigation, PZ, TC, ND, LK, BSM, IS, LL, KB, and PAS; Writing – Original Draft, DKS and DGP; Writing – Review & Editing, DKS, DGP, RMO, CA, and JZ; Funding Acquisition, DKS and DGP; Resources, DKS, DGP, RMO, CA, and JZ; Supervision, DKS, DGP, RMO, CA, and JZ. DKS and DGP are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Acknowledgements
R21DK091673 to DKS and DGP. The authors would like to thank Dr. Susan Ozannne (Cambridge University), Dr. Clark Glymour (Carnegie Mellon University) and Jen-Chywan Wang (Berkeley University) for helpful discussions.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2017.02.001.
Contributor Information
David G. Peters, Email: dgp6@pitt.edu.
Donald K. Scott, Email: donald.scott@mssm.edu.
Conflict of interest
The authors declare no competing financial interests.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Permutt M.A., Wasson J., Cox N. Genetic epidemiology of diabetes. The Journal of Clinical Investigation. 2005;115:1431–1439. doi: 10.1172/JCI24758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qi L., Cho Y.A. Gene-environment interaction and obesity. Nutrition Reviews. 2008;66:684–694. doi: 10.1111/j.1753-4887.2008.00128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grarup N., Sandholt C.H., Hansen T., Pedersen O. Genetic susceptibility to type 2 diabetes and obesity: from genome-wide association studies to rare variants and beyond. Diabetologia. 2014;57:1528–1541. doi: 10.1007/s00125-014-3270-4. [DOI] [PubMed] [Google Scholar]
- 4.Heard E., Martienssen R.A. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozanne S.E., Constancia M. Mechanisms of disease: the developmental origins of disease and the role of the epigenotype. Nature Clinical Practice, Endocrinology & Metabolism. 2007;3:539–546. doi: 10.1038/ncpendmet0531. [DOI] [PubMed] [Google Scholar]
- 6.McMillen I.C., Robinson J.S. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiological Reviews. 2005;85:571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- 7.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature Reviews Genetics. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 8.Torres I.O., Fujimori D.G. Functional coupling between writers, erasers and readers of histone and DNA methylation. Current Opinion in Structural Biology. 2015;35:68–75. doi: 10.1016/j.sbi.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farlik M., Sheffield N.C., Nuzzo A., Datlinger P., Schonegger A., Klughammer J. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Reports. 2015;10:1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaelin W.G., Jr., McKnight S.L. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling C., Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monk M. Genomic imprinting. Genes Development. 1988;2:921–925. doi: 10.1101/gad.2.8.921. [DOI] [PubMed] [Google Scholar]
- 13.Ziller M.J., Gu H., Muller F., Donaghey J., Tsai L.T., Kohlbacher O. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derry J.M., Zhong H., Molony C., MacNeil D., Guhathakurta D., Zhang B. Identification of genes and networks driving cardiovascular and metabolic phenotypes in a mouse F2 intercross. PLoS One. 2010;5:e14319. doi: 10.1371/journal.pone.0014319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y. Mapping the genetic architecture of gene expression in human liver. PLoS Biology. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu Z., Keller M.P., Zhang C., Rabaglia M.E., Greenawalt D.M., Yang X. Integrative analysis of a cross-loci regulation network identifies App as a gene regulating insulin secretion from pancreatic islets. PLoS Genetics. 2012;8:e1003107. doi: 10.1371/journal.pgen.1003107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Lay J., Kaestner K.H. The Fox genes in the liver: from organogenesis to functional integration. Physiological Reviews. 2010;90:1–22. doi: 10.1152/physrev.00018.2009. [DOI] [PubMed] [Google Scholar]
- 18.Vanderpool C., Sparks E.E., Huppert K.A., Gannon M., Means A.L., Huppert S.S. Genetic interactions between hepatocyte nuclear factor-6 and Notch signaling regulate mouse intrahepatic bile duct development in vivo. Hepatology. 2012;55:233–243. doi: 10.1002/hep.24631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basseri S., Lhotak S., Fullerton M.D., Palanivel R., Jiang H., Lynn E.G. Loss of TDAG51 results in mature-onset obesity, hepatic steatosis, and insulin resistance by regulating lipogenesis. Diabetes. 2013;62:158–169. doi: 10.2337/db12-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mantell B.S., Stefanovic-Racic M., Yang X., Dedousis N., Sipula I.J., O'Doherty R.M. Mice lacking NKT cells but with a complete complement of CD8+ T-cells are not protected against the metabolic abnormalities of diet-induced obesity. PLoS One. 2011;6:e19831. doi: 10.1371/journal.pone.0019831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang P., Metukuri M.R., Bindom S.M., Prochownik E.V., O'Doherty R.M., Scott D.K. c-Myc is required for the CHREBP-dependent activation of glucose-responsive genes. Molecular Endocrinology. 2010;24:1274–1286. doi: 10.1210/me.2009-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park Y., Figueroa M.E., Rozek L.S., Sartor M.A. MethylSig: a whole genome DNA methylation analysis pipeline. Bioinformatics. 2014;30:2414–2422. doi: 10.1093/bioinformatics/btu339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cha J.Y., Repa J.J. The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. Journal of Biological Chemistry. 2007;282:743–751. doi: 10.1074/jbc.M605023200. [DOI] [PubMed] [Google Scholar]
- 28.Yardeni T., Eckhaus M., Morris H.D., Huizing M., Hoogstraten-Miller S. Retro-orbital injections in mice. Lab Animal. 2011;40:155–160. doi: 10.1038/laban0511-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becard D., Hainault I., Azzout-Marniche D., Bertry-Coussot L., Ferre P., Foufelle F. Adenovirus-mediated overexpression of sterol regulatory element binding protein-1c mimics insulin effects on hepatic gene expression and glucose homeostasis in diabetic mice. Diabetes. 2001;50:2425–2430. doi: 10.2337/diabetes.50.11.2425. [DOI] [PubMed] [Google Scholar]
- 30.Steel C.D., Stephens A.L., Hahto S.M., Singletary S.J., Ciavarra R.P. Comparison of the lateral tail vein and the retro-orbital venous sinus as routes of intravenous drug delivery in a transgenic mouse model. Lab Animal. 2008;37:26–32. doi: 10.1038/laban0108-26. [DOI] [PubMed] [Google Scholar]
- 31.Mehlem A., Hagberg C.E., Muhl L., Eriksson U., Falkevall A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nature Protocols. 2013;8:1149–1154. doi: 10.1038/nprot.2013.055. [DOI] [PubMed] [Google Scholar]
- 32.Irizarry R.A., Ladd-Acosta C., Wen B., Wu Z., Montano C., Onyango P. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature Genetics. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Research. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaur J. A comprehensive review on metabolic syndrome. Cardiology Research and Practice. 2014;2014:943162. doi: 10.1155/2014/943162. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M. The UCSC Genome Browser database: 2015 update. Nucleic Acids Research. 2015;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 37.Christman J.K. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 38.Liu X., Kraus W.L., Bai X. Ready, pause, go: regulation of RNA polymerase II pausing and release by cellular signaling pathways. Trends in Biochemical Sciences. 2015;40:516–525. doi: 10.1016/j.tibs.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heidemann M., Hintermair C., Voss K., Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochimica et Biophysica Acta. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 40.Kim H., Erickson B., Luo W., Seward D., Graber J.H., Pollock D.D. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nature Structural & Molecular Biology. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang D.W., Rodriguez-Molina J.B., Tietjen J.R., Nemec C.M., Ansari A.Z. Emerging views on the CTD code. Genetics Research International. 2012;2012:347214. doi: 10.1155/2012/347214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y., Zhu J., Lum P.Y., Yang X., Pinto S., MacNeil D.J. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schadt E.E., Lamb J., Yang X., Zhu J., Edwards S., Guhathakurta D. An integrative genomics approach to infer causal associations between gene expression and disease. Nature Genetics. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu J., Zhang B., Smith E.N., Drees B., Brem R.B., Kruglyak L. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nature Genetics. 2008;40:854–861. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smemo S., Tena J.J., Kim K.-H., Gamazon E.R., Sakabe N.J., Gomez-Marin C. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mashek D.G., Khan S.A., Sathyanarayan A., Ploeger J.M., Franklin M.P. Hepatic lipid droplet biology: getting to the root of fatty liver. Hepatology. 2015;62:964–967. doi: 10.1002/hep.27839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeh M.M., Brunt E.M. Pathological features of fatty liver disease. Gastroenterology. 2014;147:754–764. doi: 10.1053/j.gastro.2014.07.056. [DOI] [PubMed] [Google Scholar]
- 48.Ahmed M.H., Husain N.E., Almobarak A.O. Nonalcoholic fatty liver disease and risk of diabetes and cardiovascular disease: what is important for primary care physicians? Journal of Family Medicine and Primary Care. 2015;4:45–52. doi: 10.4103/2249-4863.152252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagai M.A. Pleckstrin homology-like domain, family A, member 1 (PHLDA1) and cancer. Biomedical Reports. 2016;4:275–281. doi: 10.3892/br.2016.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kastrati I., Canestrari E., Frasor J. PHLDA1 expression is controlled by an estrogen receptor-NFkappaB-miR-181 regulatory loop and is essential for formation of ER+ mammospheres. Oncogene. 2015;34:2309–2316. doi: 10.1038/onc.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toyoshima Y., Karas M., Yakar S., Dupont J., Lee H., LeRoith D. TDAG51 mediates the effects of insulin-like growth factor I (IGF-I) on cell survival. Journal of Biological Chemistry. 2004;279:25898–25904. doi: 10.1074/jbc.M400661200. [DOI] [PubMed] [Google Scholar]
- 52.Tomaru Y., Nakanishi M., Miura H., Kimura Y., Ohkawa H., Ohta Y. Identification of an inter-transcription factor regulatory network in human hepatoma cells by Matrix RNAi. Nucleic Acids Research. 2009;37:1049–1060. doi: 10.1093/nar/gkn1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen W., Scearce L.M., Brestelli J.E., Sund N.J., Kaestner K.H. Foxa3 (hepatocyte nuclear factor 3gamma ) is required for the regulation of hepatic GLUT2 expression and the maintenance of glucose homeostasis during a prolonged fast. Journal of Biological Chemistry. 2001;276:42812–42817. doi: 10.1074/jbc.M106344200. [DOI] [PubMed] [Google Scholar]
- 54.de Fourmestraux V., Neubauer H., Poussin C., Farmer P., Falquet L., Burcelin R. Transcript profiling suggests that differential metabolic adaptation of mice to a high fat diet is associated with changes in liver to muscle lipid fluxes. Journal of Biological Chemistry. 2004;279:50743–50753. doi: 10.1074/jbc.M408014200. [DOI] [PubMed] [Google Scholar]
- 55.Kirpich I.A., Gobejishvili L.N., Homme M.B., Waigel S., Cave M., Arteel G. Integrated hepatic transcriptome and proteome analysis of mice with high-fat diet-induced nonalcoholic fatty liver disease. The Journal of Nutritional Biochemistry. 2011;22:38–45. doi: 10.1016/j.jnutbio.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waller-Evans H., Hue C., Fearnside J., Rothwell A.R., Lockstone H.E., Calderari S. Nutrigenomics of high fat diet induced obesity in mice suggests relationships between susceptibility to fatty liver disease and the proteasome. PLoS One. 2013;8:e82825. doi: 10.1371/journal.pone.0082825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borengasser S.J., Kang P., Faske J., Gomez-Acevedo H., Blackburn M.L., Badger T.M. High fat diet and in utero exposure to maternal obesity disrupts circadian rhythm and leads to metabolic programming of liver in rat offspring. PLoS One. 2014;9:e84209. doi: 10.1371/journal.pone.0084209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ng S.-F., Lin R.C.Y., Laybutt D.R., Barres R., Owens J.A., Morris M.J. Chronic high-fat diet in fathers programs [bgr]-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 59.Cordero P., Gomez-Uriz A.M., Campion J., Milagro F.I., Martinez J.A. Dietary supplementation with methyl donors reduces fatty liver and modifies the fatty acid synthase DNA methylation profile in rats fed an obesogenic diet. Genes & Nutrition. 2013;8:105–113. doi: 10.1007/s12263-012-0300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L.J., Zhang H.W., Zhou J.Y., Liu Y., Yang Y., Chen X.L. Betaine attenuates hepatic steatosis by reducing methylation of the MTTP promoter and elevating genomic methylation in mice fed a high-fat diet. Journal of Nutritional Biochemistry. 2014;25:329–336. doi: 10.1016/j.jnutbio.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 61.Murphy S.K., Yang H., Moylan C.A., Pang H., Dellinger A., Abdelmalek M.F. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145:1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trengove M.C., Ward A.C. SOCS proteins in development and disease. American Journal of Clinical and Experimental Immunology. 2013;2:1–29. [PMC free article] [PubMed] [Google Scholar]
- 63.Moya M., Benet M., Guzman C., Tolosa L., Garcia-Monzon C., Pareja E. Foxa1 reduces lipid accumulation in human hepatocytes and is down-regulated in nonalcoholic fatty liver. PLoS One. 2012;7:e30014. doi: 10.1371/journal.pone.0030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.