Abstract
Backgound/Aim: Malignant mesothelioma (MM) is an aggressive and fatal pleural cancer. The cell secretome offers information allowing insight into the pathogenesis of MM while offering the possibility to identify potential therapeutic targets and biomarkers. In the present study the secretome protein profile of MM cell lines was compared to normal mesothelial cells. Materials and Methods: Six MM cell lines were compared against three primary mesothelial cell culture preparations using iTRAQ® mass spectrometry. Results: MM cell lines more abundantly secreted exosome-associated proteins than mesothelial cells. MM cell secretomes were enriched in proteins that are involved in response to stress, carbon metabolism, biosynthesis of amino acids, antigen processing and presentation and protein processing in the endoplasmic reticulum. Conclusion: The MM cell secretome is enriched in proteins that are likely to enhance its growth and response to stress and help it inhibit an adaptive immune response. These are potential targets for therapeutic and biomarker discovery.
Keywords: Malignant mesothelioma, proteomics, iTRAQ, secretome
Malignant mesothelioma (MM) is an incurable and aggressive tumour, predominantly of the pleura (1,2). MM has a median survival of less than 12 months and this poor survival is in part related to late diagnosis and resistance to radiotherapy and chemotherapy. Although many targeted investigations of MM have been designed in order to examine specific pathways that are significant to this disease, there are few investigations that have examined the overall protein expression landscape. Such an analysis has the potential to i) identify key structural and molecular characteristics of MM that can be further investigated, ii) identify molecules that might be useful biomarkers for diagnosis and monitoring and iii) may inform novel treatment strategies targeting the molecules/pathways identified. Proteins secreted from the cell, known as the cell protein “secretome” offer a valuable source of biological information. The secretome is a mixture of proteins that are secreted through both classical secretory pathways, and by non-classical mechanisms such as membrane-bound exosomes (3). We have comprehensively described the exosome “cargo” proteins that are secreted by MM cell lines (4), and have described a mesothelioma-enriched exosome protein cancer signature (mEXOS). We reported that MM exosome components are enriched in immunoregulatory components, tumour-derived antigens, components of cancer signalling networks and that they are able to regulate the tumour micro-environement (4).
What has not been determined is how the MM secretome compares to the secretome of non-malignant mesothelial cells and whether the secretome of MM cells is significantly enriched in proteins that are secreted via exosomes compared to non-malignant cells. Therefore, the aim of this study was to compare the secretome of MM cell lines with that of primary mesothelial cell cultures, in order to determine a protein secretion profile associated with MM. The information gained will increase our understanding of MM and may identify potential candidates for future biomarker investigation and potential therapeutic targets.
Materials and Methods
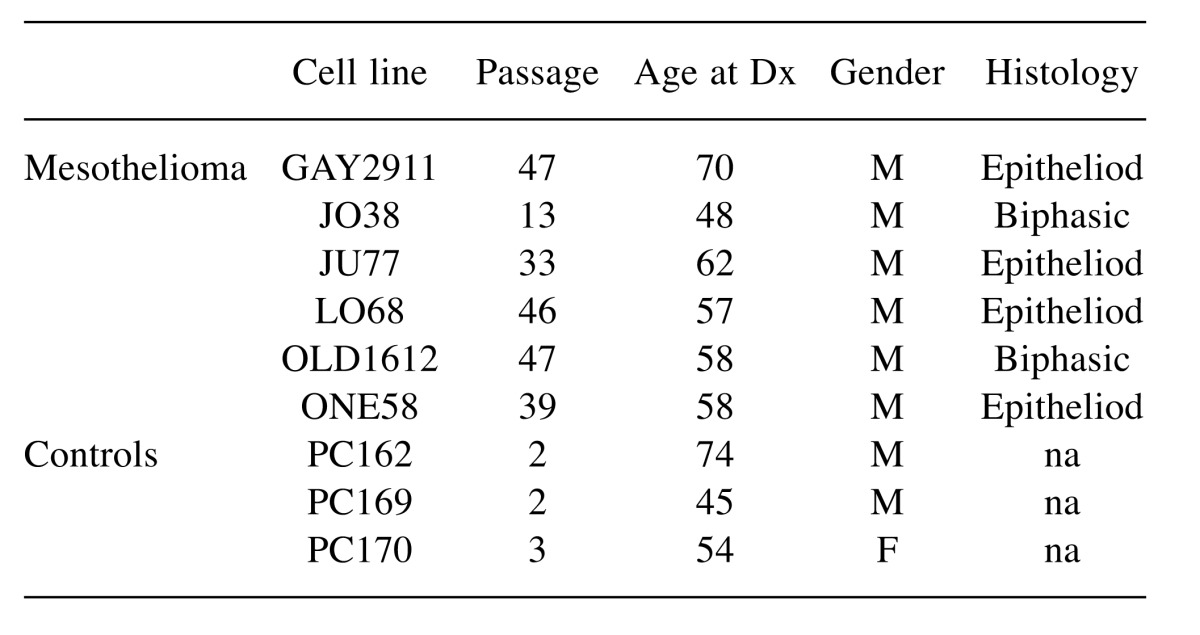
Cell culture. Six previously established MM cell lines, JU77, LO68, ONE58, OLD1612, JO38 and GAY2911, were maintained as previously described (5). The cell lines and the patient demographics from which the cell lines were derived are described in Table I. Cells were mycoplasma free and Short Tandem Repeat (STR) profiling was used to confirm that they were unique cell lines. Short term cultures of normal mesothelial cells were established from pericardial fluid of patients undergoing coronary artery bypass operations as described (6) and maintained in standard media for two to three passages for use as non-malignant normal mesothelial cell controls. Cultures contained >90% normal mesothelial cells as determined by immunohistochemical staining with calretinin and mesothelin antibodies (7). This study was approved by the Human Research Ethics Committee of Sir Charles Gairdner Hospital and all patients provided written informed consent.
Table I. Characteristics of cell lines and the MM patient they were derived from, that were used in the study.
Secretome samples for analysis were obtained from cells cultured in serum free RPMI-1640 (Life Technologies, Carlsbad CA, USA) for intervals between 0 and 48 hours. In order to determine the optimal cell culture conditions that were to be used for the iTRAQ experiments, cell viability was determined by trypan blue exclusion and by using the Guava via count DNA-binding dye method (Millipore, Hayward, CA, USA). The response of cells to the serum free conditions over time was assessed by measuring lactate deyhdrogenase (LDH) concentrations, an indicator of cell lysis, in order to determine a suitable time that the cells would remain viable in serum free conditions. LDH was measured using an NAD reduction, tetrazolium dye assay (Sigma Aldrich St Louis MO, USA). LDH measurements of corresponding freeze-thaw lysed cells were used as a positive control.
The proteins present in the cell culture secretomes were analysed by iTRAQ mass spectrometry. For each sample, MM or normal mesothelial control, cells (3×106) were seeded into two T175 flasks. After three days, the cells (~1×107 cells per flask) were washed with PBS and serum-free media was added. Flasks were cultured for a further 24 h and the serum-free conditioned media were collected and pooled from the two flasks. Cellular debris was removed by centrifugation (2000 × g for 10 min) and then the supernatant was stored at –80˚C until further processing. The total protein content of each pooled secretome available for iTRAQ processing was approximately 500 μg.
iTRAQ analysis. The iTRAQ® multiplex (4-plex) (Applied Biosystems, Foster City, CA, USA) isobaric labelling technique was used to compare the MM and normal mesothelial cell protein secretome and was performed by Proteomics International (Perth, Western Australia). Each secretome sample was concentrated using a 10-kDa mass cut-off Vivaspin filter. Samples were then acetone precipitated, re-solubilised, reduced, alkylated and finally digested with trypsin according to the iTRAQ® protocol. The resulting peptide solutions were desalted on a strata-X 33 μm polymeric reversed phase column (Phenomenex, Torrance, CA, USA) and dissolved in buffer (10 mM KH2PO4, pH3, in 10% acetonitrile) before separation by strong cation exchange liquid chromatography on an Agilent 1100 HPLC system using a PolySulfoethyl column (4.6×100 mm, 5 μm, 300 A). Peptides were eluted with a linear gradient of 0-400 mM KCl. SCX fractions were desalted and loaded onto an Ultimate 3000 nano HPLC system (Dionex, C18 PepMap100, 3 mm) and separated with a gradient of 10-40% acetonitrile (0.1% trifluoroacetic acid) with spotting using a ProBot (LC Packings) robotic spotter. The resultant spots were analysed on a 4800 MALDI TOF/TOF Analyzer (AB SCIEX, Framingham, MA, USA) and mass spectrometry data was generated. These data were generated according to ISO/IEC 17025 approved Standard Operating Procedures pertaining to: sample preparation for iTRAQ; solid phase extraction; operation of the 4800 TOF TOF Mass Spectrometer; operation of the Agilent HPLC; HPLC cation exchange; use of the Probot HPLC plate spotter.
Data analysis was performed using ProteinPilot™ 2.0.1 Software (Applied Biosytems). The results were searched against the SwissProt database (release 2015_11) with the taxonomy set to Homo sapien. A confident identification of a protein was defined as a ProtScore ( log (1-(percent confidence/100)) greater than 1.3 (95% confident).
Statistical analysis. For each identified protein, a ratio of the abundance of that protein present in each of the MM cell lines secretome with the abundance of that protein present in the normal mesothelial cell comparator secretome (the iTRAQ ratio for that protein) was calculated using the ProteinPilot™ software (8). A p-value for each iTRAQ ratio was also calculated by the ProteinPilot software. This was a measure of the level of significance that the iTRAQ ratio was different from 1 for the comparison of each protein’s expression in the MM cell line relative to the expression of that protein in the mesothelial comparator, with a calculated global and local false discovery rate (FDR) of <0.1%.
For each experiment, normalized iTRAQ values for each protein within a MM cell line were calculated as the standard deviation difference between that proteins log(2) transformed iTRAQ value and the mean log(2) transformed iTRAQ value across the proteins observed for that MM cell line within that experiment. These normalized values were then scaled by dividing by the standard deviation for the iTRAQ results for each protein. Therefore, a transformed iTRAQ value was a measure of the relative magnitude of the iTRAQ ratio for a MM cell line - mesothelial cell control comparison for each identified protein, relative to the magnitude of all of the other iTRAQ ratios for that MM cell line - mesothelial cell comparison, which was then scaled for the variability of the normalized iTRAQ values measured across the 12 comparisons for that protein.
Principal components analysis (PCA) of the log(2) transformed, normalized and scaled data was performed using the R FactoMineR (9) program and the results were visualized using the factoextra program (10). Hierarchical cluster analysis was carried out using the ward agglomeration method of the R system hclust function to model the euclidian distance dissimilarity between samples and between proteins for the normalized and scaled iTRAQ values. For characterization of differentially expressed proteins a six cluster assignment was chosen which ensured euclidian height <20. For the purpose of hierarchical clustering and PCA analysis missing values from experiment 3A or 3B were imputed with a fully conditional specification using a linear regression analysis, with the number of imputations set to 100 (IBM® SPSS® statistics version 22 Armonk, NY, USA).
Secretion and pathway analysis. The proteins detected in the iTRAQ experiments were referenced to the Human Cancer Secretome Database (11) (www.cancersecretome.org) and the SignalP (12)and SecretomeP (13) scores were analysed for each protein in order to determine if the protein was secreted by classical or non-classical secretion pathways.
Proteins that were identified in the clusters produced by the hierarchical cluster analysis of the normalized data were analysed for Gene Ontology (GO) enrichment and KEGG pathways enrichment using the Search Tool for the Retrieval of Interacting Genes/Proteins version 10 (STRING) (14). A pathway was considered to be enriched if the FDR adjusted p-value was less than 0.05.
Comparison to MM exosomes. A comparison of the secretome data obtained in this study was performed with the MM exosome secreted proteins previously reported (4). There were four MM cell lines investigated common to both studies (JU77, LO68, OLD1612, JO38). The exosome data previously reported is available online (http://www.nature.com/articles/srep32643#supplementary-information). Of the 2,178 exosome proteins reported in this supplementary data set, 631 were found in all four exosome preparations studied. These were compared to the secretome proteins reported in this present study in order to determine the proportion of reported exosome proteins that can be detected in the MM secretome. The normalized spectral counts (SpC), a measure of peak intensity, were examined and difference between groups analysed. For statistical comparison of proportions (Pearson chi square) or continuous variables (Levene’s test), a p<0.05 was considered statistically significant.
Results
To establish culture conditions for the generation of secretome samples for analysis, cell death and cellular stress was monitored in cultures grown in serum-free media. Preliminary experiments demonstrated that no significant decrease in cell viability or evidence of cellular stress was observed in MM cell lines cultured in serum free media for 24 h, and therefore these conditions were used for subsequent experiments.
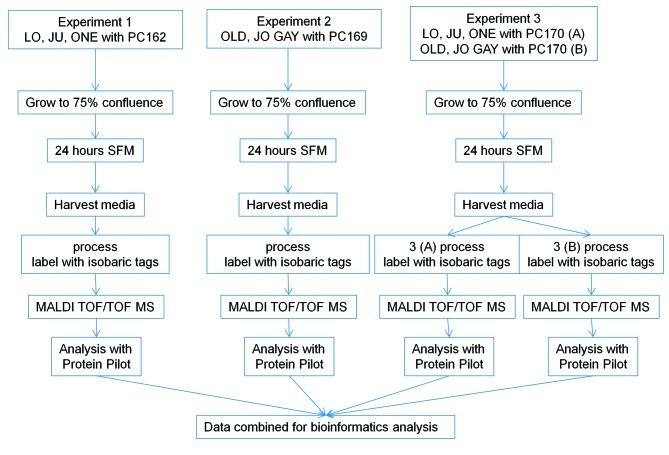
Three independent iTRAQ experiments were performed (Figure 1). In the first experiment the secretomes of three MM cell lines (LO68, JU77, ONE58) were compared to that of non-malignant mesothelial cells cultured from a 74-year-old male cardiac patient (PC162) and a total of 401 proteins were identified. In the second experiment three additional MM cell lines (OLD1612, JO38, GAY2911) were compared to normal mesothelial cells from a 45-year-old man (PC169) and a total of 292 proteins were identified. The third iTRAQ experiment repeated the analysis on the secretome of these six MM cell lines with a third independent normal mesothelial cell secretome as comparator from a 54-year-old woman (PC170). For experiment three, the four-label iTRAQ method meant that the six MM cell line secretomes and the non-malignant secretome had to be analyzed in two separate MS runs (A and B), which identified 241 and 281 proteins respectively. A total of 344 proteins were identified in the combined results from experiment 3, of which 178 were identified in both experiments 3A and 3B.
Figure 1. Experimental outline: LO68, JU77, ONE58, OLD1612, JO38 and GAY2911 are the human MM cell lines under investigation. PC162, PC169, PC170 are primary mesothelial cells derived from pericardial effusions; SFM,serum-free RPMI media.
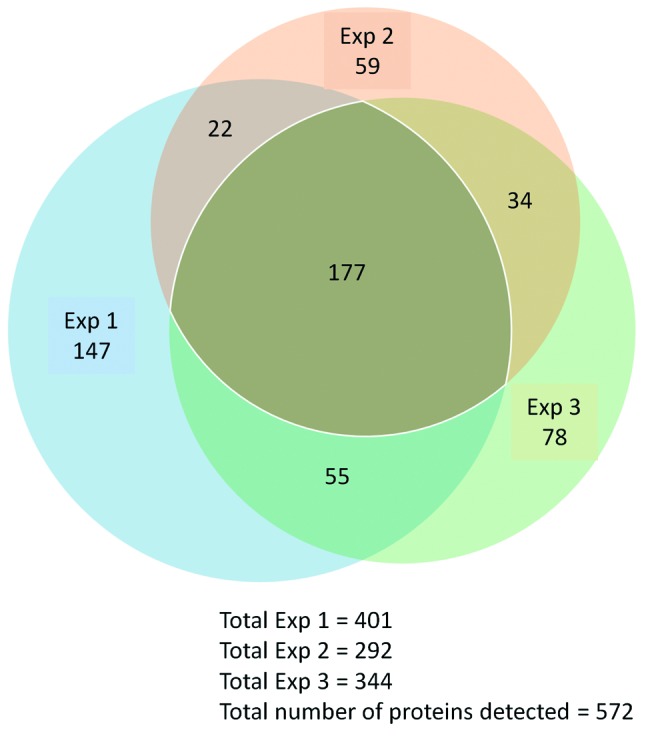
In total, 572 proteins were identified across the three experiments, with 147 proteins being identified only in experiment 1; 59 proteins being only identified in experiment 2; and 78 proteins only being identified in experiment 3. Of the total of 572 proteins that were identified across the three experiments, there were 177 proteins that were identified in both experiment 1, experiment 2 and in experiment 3A and/or experiment 3B. That is, these 177 proteins were identified in every MM cell line at least once, and were identified in all three experiments and were considered for further analysis (Figure 2).
Figure 2. Proportional Venn diagram displaying the overlap of proteins detected in the three iTRAQ experiments. Experiment 1 – JU77, LO68 and ONE58, Experiment 2 – OLD1612, JO38 and GAY2911, Experiment 3 – JU77, LO68, ONE58, OLD1612, JO38 and GAY2911 MM cell lines. Each experiment was compared to a different mesothelial cell culture.
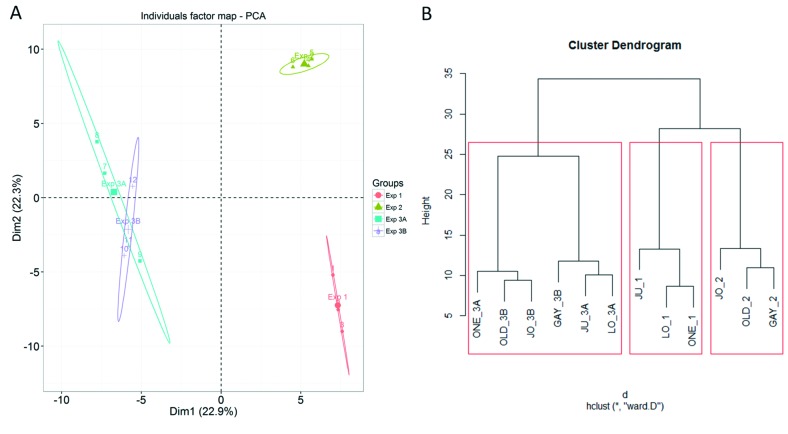
Analysis by PCA and hierarchical clustering (Figure 3) demonstrated the effect of experimental run on data variability. For the three independent MM - mesothelial cell comparisons, experiments one and two formed two distinct separate clusters, and the six MM – mesothelial cell comparisons in experiments 3A and 3B overlapped to form a third distinct cluster (Figure 3). This suggests that the differences in the experiments was due to a biological difference associated with comparison to primary normal mesothelial cells from different individuals, because experiment 3, where all six MM cell lines were compared to the same mesothelial cells, grouped together, even though they were analyzed in two separate iTRAQ procedures.
Figure 3. Between-experiment data evaluation. (A) Principal component analysis of 177 normalized and scaled iTRAQ ratios identified in three experiments. Plot displays component 1 against component 2 for the individual iTRAQ experiments. (B) Cluster analysis of the normalized and scaled iTRAQ ratios for 177 proteins that were detected in all three experiments. Experiment 3 is divided into 3A and 3B because they were separated into two iTRAQ labelling and MS procedures. Labels are the cell name and experiment number. Note: Values were imputed for 15 and 25 missing proteins in experiment 3A and 3B, respectively.
Of the 177 proteins commonly detected in all of the MM cell lines, 168 (95%) have been previously identified in cancer secretomes (www.cancersecretome.org). Secretory signal peptides were present in 77 (44%) of these proteins (SignalP D score >0.45) and 42 (24%) were identified as non-classically secreted proteins (SecretomeP NN score >0.5, with no secretory signal present). A further 58 (33%) were not classifiable by these criteria as either, but were identified as exosomal or as part of an extracellular region of known proteins by gene ontogeny.
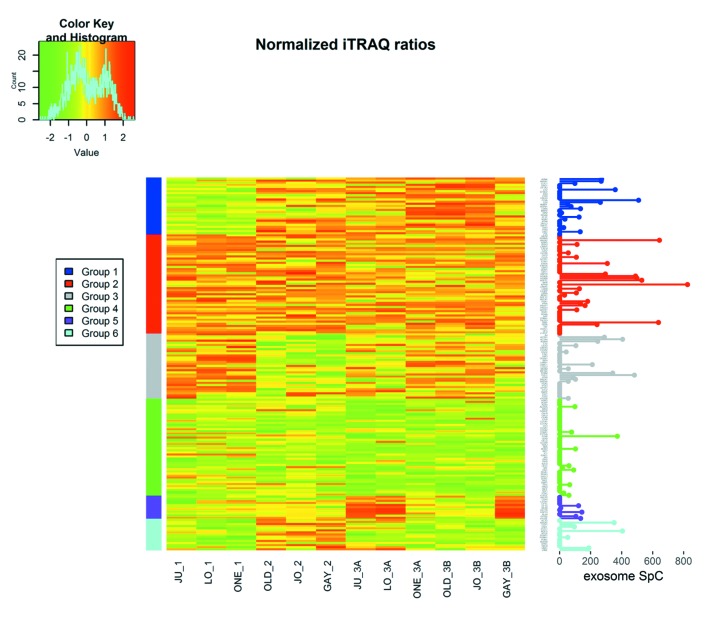
To characterise the proteins commonly differentially expressed between the MM and non-malignant mesothelial cell secretomes, cluster analysis was carried out. Proteins were clustered into 6 distinct groups (Figure 4). The resulting Group 2 cluster consisted of 47 proteins (26% of the total), which were in general over-expressed in the MM cell lines relative to the normal mesothelial cultures (Figure 4, Table II). The Group 4 cluster (46 proteins; 25% of the total) consisted predominantly of proteins less abundantly expressed across the MM cell lines (Figure 4, Table III). Biological component enrichment analysis confirmed that all of the cluster groups were enriched in proteins that were associated with exosomes and membrane bound vesicles (Table IV). Of the six protein clusters, only Group 2 and 4 (with the largest number of component proteins) were significantly enriched for pathways involved in biological processes. Group 2 was enriched with proteins that are associated with monosaccharide, glucose and nicotinamide metabolism and responses to topologically misfolded proteins. Group 4 was enriched in proteins that were associated with vesicle-mediated transport, platelet activation and degranulation, wound healing and cell adhesion (Table V). GO molecular function enrichment analysis indicated that the highly expressed Group 2 was enriched in proteins associated with protein binding, RNA binding, MHC Class II protein complex binding and extracellular matrix binding, whereas the under expressed Group 4 was enriched in proteins associated with peptidase and enzyme inhibitor activity, protein binding and platelet derived growth factor binding (Table VI). KEGG pathway enrichment was also variable across the groups, with the highly expressed Group 2 being enriched in carbon and amino acid metabolism, antigen processing and presenting, glycolysis/gluconeogenesis, and protein processing pathways (Table VII). Proteins in highly expressed Group 2 were more likely to be secreted only by non-classical mechanisms (i.e. no signal peptide present) compared to the under expressed Group 4 (62% vs. 36%, Pearson’s Chi-square 6.75, p=0.009).
Figure 4. Heatmap of iTRAQ results. Each iTRAQ ratio was log (2) transformed and normalized to the mean iTRAQ ratio for each experiment and scaled to the standard deviation across the experiments. Normalized iTRAQ results are indicated for each experiment for each of the 177 proteins selected for analysis. Exosome SpC values are exosome MS spectral values for each of the 177 proteins, as previously reported in MM cells (4).
Table II. Up-regulated proteins, identified in group 2. Gene and protein names are truncated from the UniProtKB IDs to the first highlighted entry for each. SigP, number of iTRAQ ratios significantly different from 1, out of a possible 12 samples. Median ITRAQ, median unadjusted iTRAQ value across all of the samples for that protein comparing MM cell lines to a non-malignant mesothelial cell secretome.
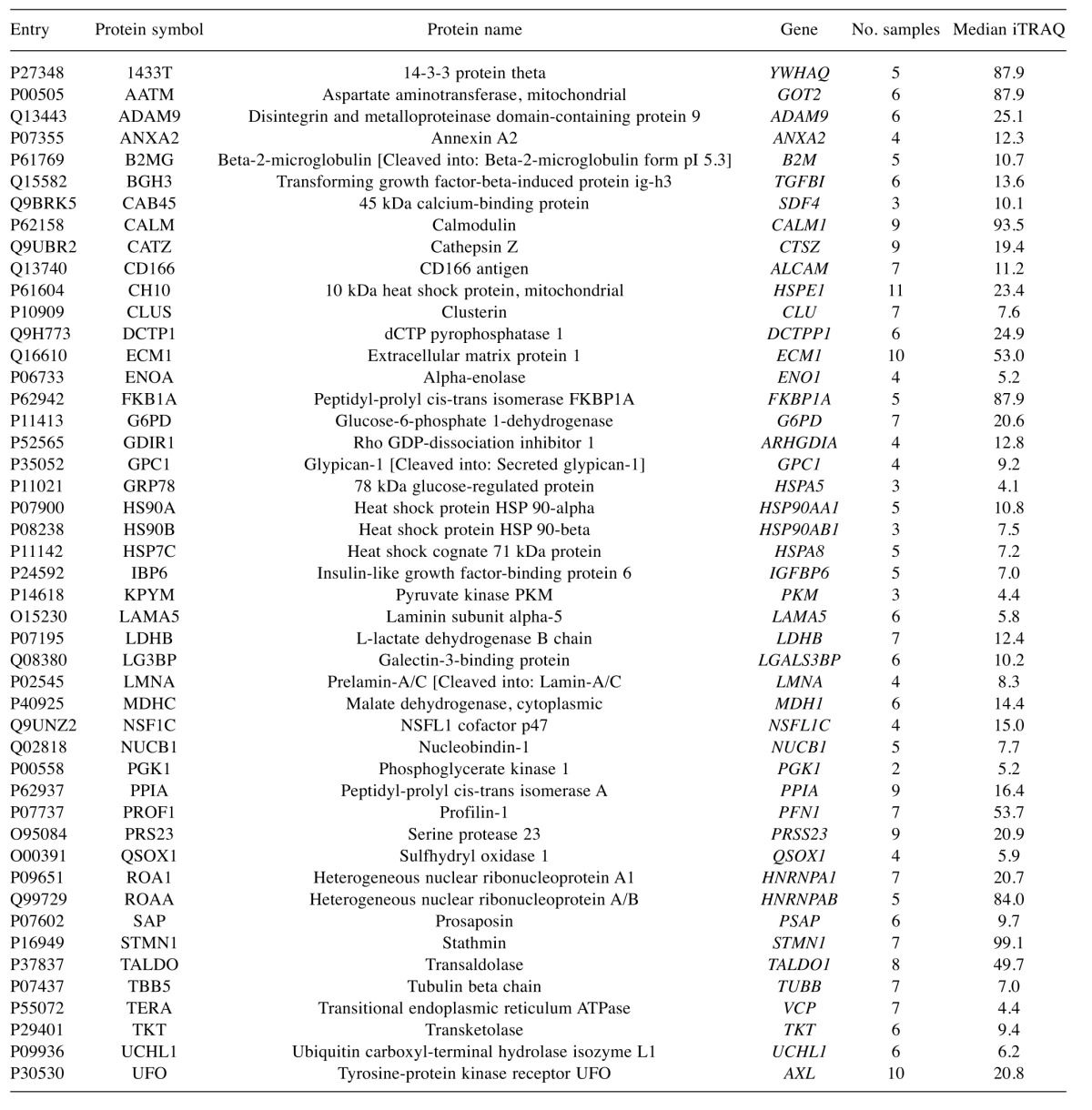
Table III. Down-regulated proteins, identified in group 4. Gene and protein names are truncated from the UniProtKB IDs to the first highlighted entry for each. SigP, the number of p-values for iTRAQ ratios significantly different from 1, out of a possible 12 samples. Median ITRAQ, the median unadjusted iTRAQ value across all of the samples for that protein comparing MM cell lines to a non-malignant mesothelial cell secretome.
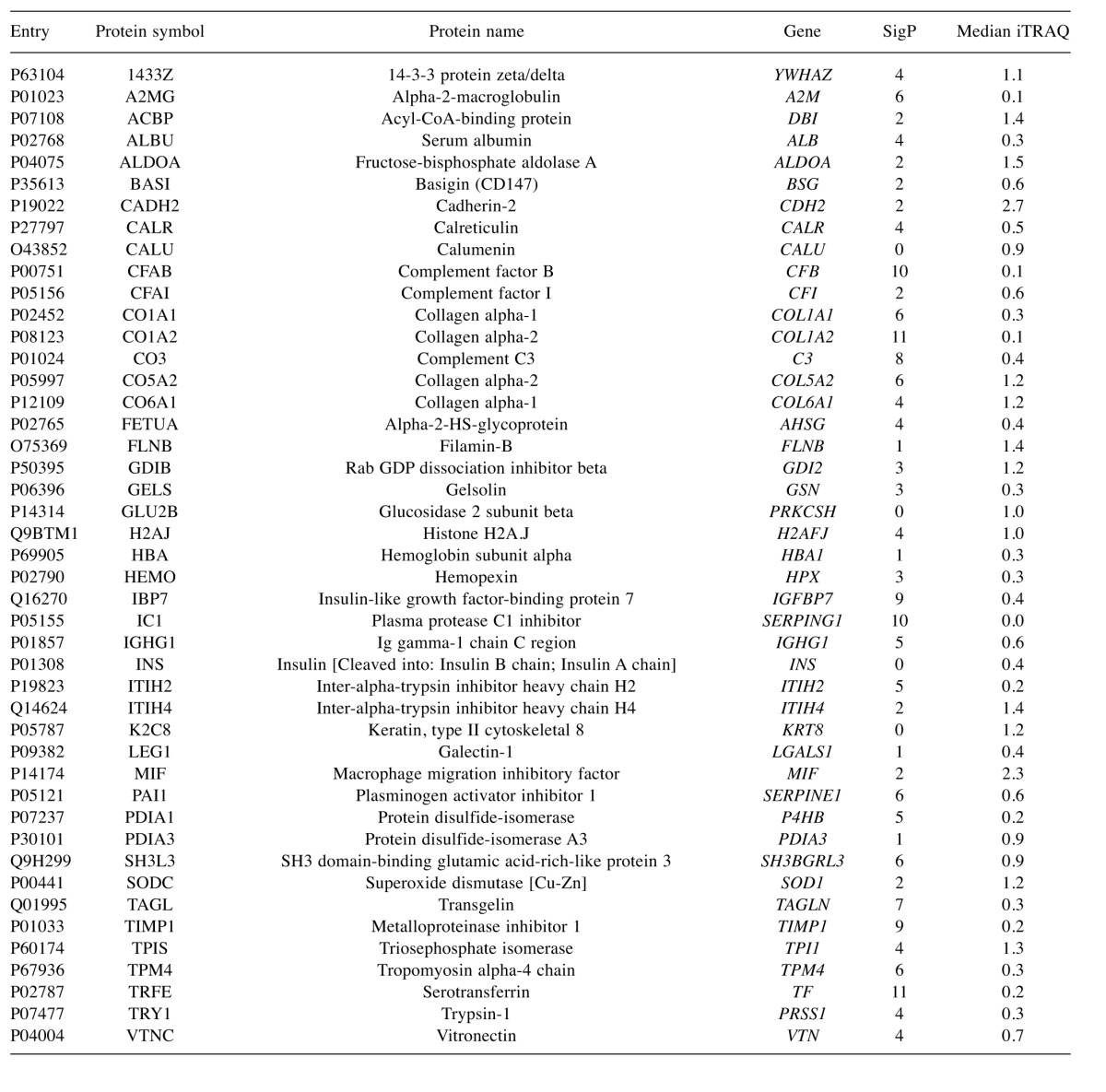
Table IV. The 5 most significant Gene Ontology biological component terms that were identified as being enriched in the six protein cluster groups.
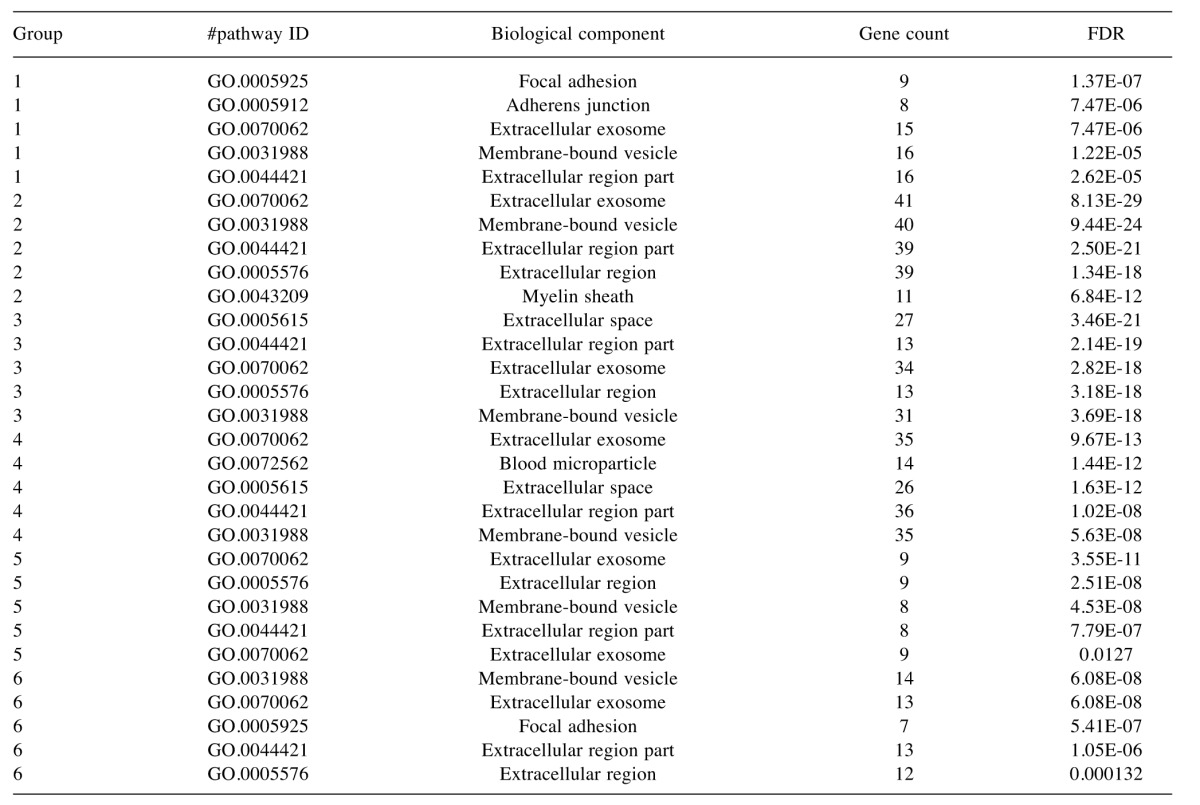
FDR, False discovery rate.
Table V. Top 5 most significant Gene Ontology process terms that were enriched in the various groups identified by cluster analysis.
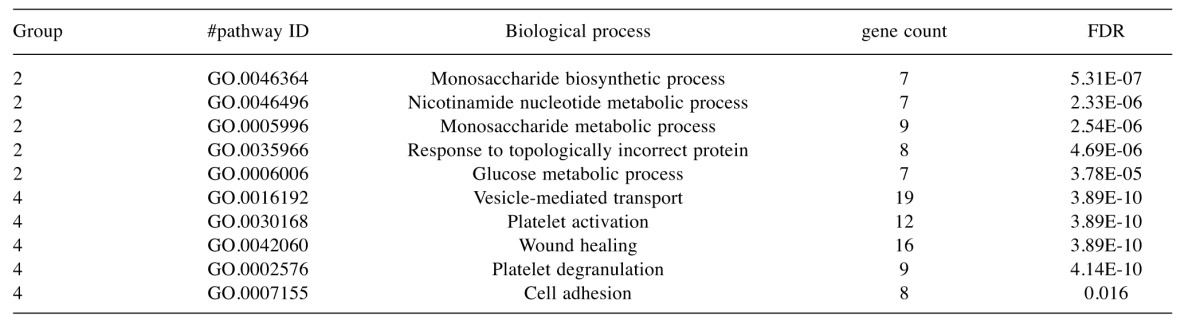
FDR, False discovery rate.
Table VI. Top 5 most significant Gene Ontology molecular function terms that were enriched in the various groups identified by cluster analysis.
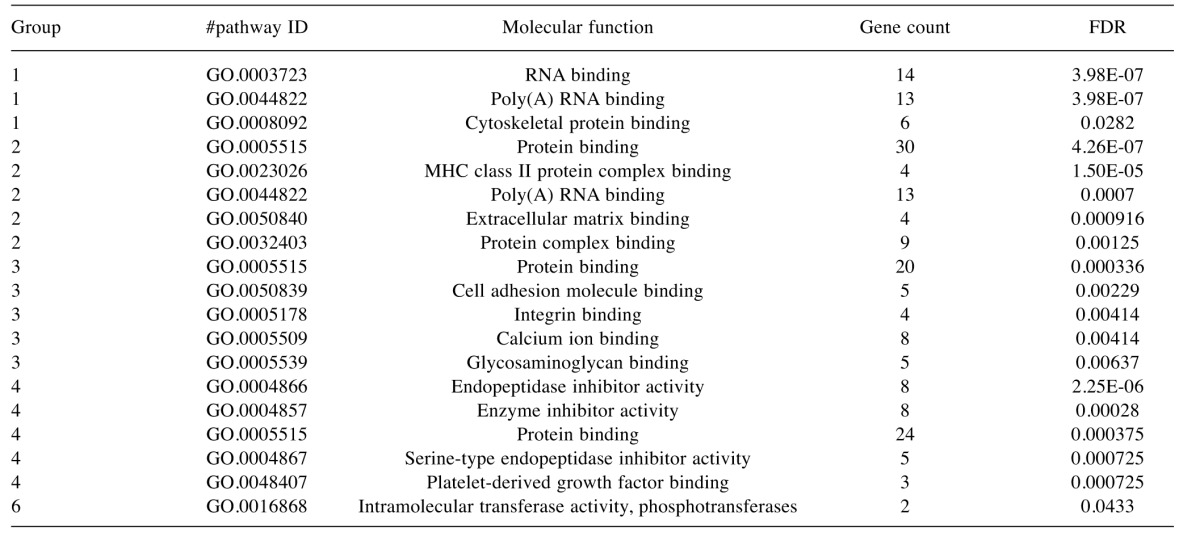
FDR, False discovery rate.
Table VII. KEGG pathway terms that were enriched in the various groups identified by cluster analysis.
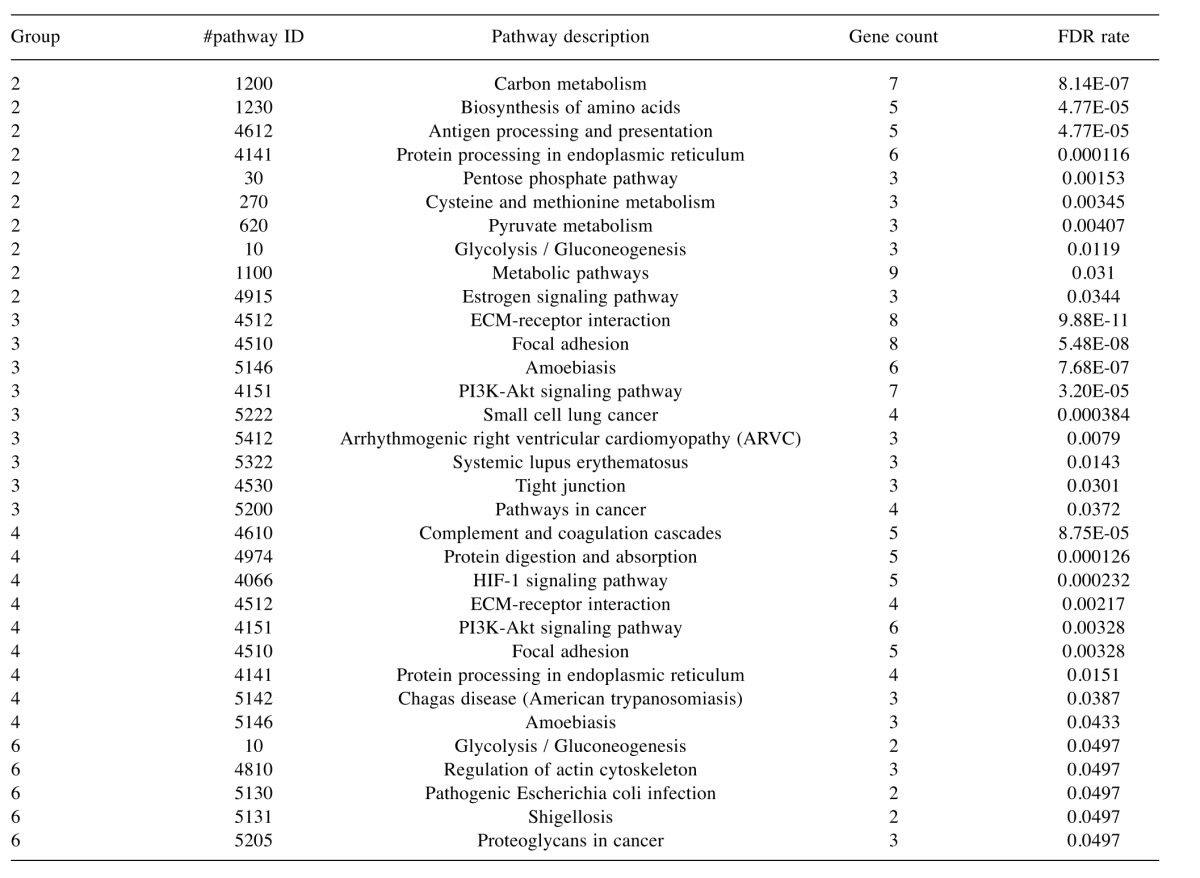
FDR, False discovery rate.
The list of 177 proteins commonly detected in the secretomes of MM cells were compared to the list of proteins previously reported to be present in the exosome cargo of MM cells (4) (Figure 4). Of the 177 secretome proteins identified in the present study, 71 (40%) were also reported to be present in MM exosome preparations (4). Of the Group 2 highly expressed secretome proteins 19 of 47 (40%) were found in the exosomes of the four MM cell lines and of the Group 4 proteins with relatively low expression, 10 of 46 (22%) were found in the exosomes of all four MM cell lines. The Pearson chi square p-value for the difference in these proportions was 0.051.
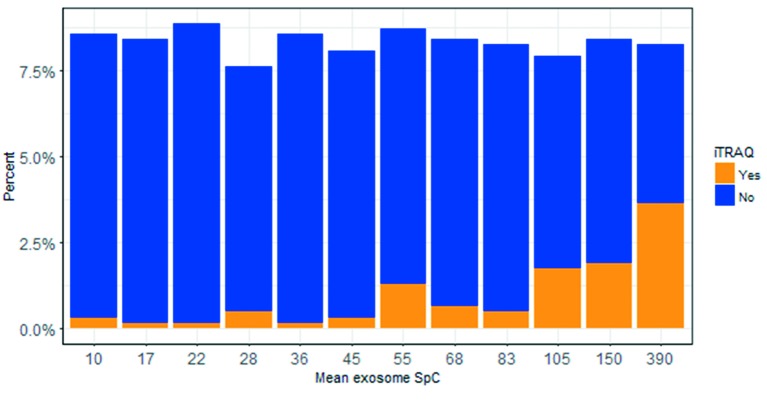
A comparison of the exosome normalized spectral count (SpC) between those exosome proteins that were and were not identified in the secretome by iTRAQ in the present study, demonstrated that the 71 proteins that were also identified in the MM secretome by iTRAQ were more abundant in exosomes than those that were not identified in the MM secretome (n=560) (SpC values 188±176 vs. 70±87, Levene’s test p<0.001, Figure 5). Thus the MM secretome contains large numbers of both exosome and non-exosome proteins.
Figure 5. Stacked histogram (approximately 50 spectral count values (SpC) per bar) from exosomal proteins, as previously reported in MM cells (4). iTRAQ “Yes” are the exosme SpC values for the 71 (of 177) proteins that were identified in the iTRAQ experiments and iTRAQ “No” are the exosme SpC values for the other 560 proteins in the exosome study that were not detected in the iTRAQ experiments. “Mean exosome SpC” is the mean SpC value for each group of approximately 50 SpC values.
Discussion
This study was designed to identify differential protein expression in MM cell lines compared to normal mesothelial cells, with the aim of gaining insight into the pathogenesis of MM and also with the intention of identifying potential new biomarkers for MM diagnosis that can be investigated in future studies. Due to the replication that we undertook in this study of multiple MM cell lines, compared to multiple non-malignant mesothelial cells, the proteins that we identified in the overlap between the three experiments and the estimate of their abundance relative to the comparator mesothelial cells is likely to be robust. Therefore this experiment has identified some fundamental shared characteristics of the six MM cell lines examined, relative to normal mesothelial cells.
Overall, proteins that were expressed in the secretome of MM cells were associated with enrichment in exosome-associated proteins, regardless of their relative expression compared to the normal mesothelial secretome. Proteins that were more highly expressed in the MM secretome were more likely to be exosome proteins, but some proteins that had a lower protein expression in MM secretomes compared to mesothelial cell secretomes were also exosome proteins. Exosomes are extracellular, lipid bilayer, vesicles released by cells that contain cell derived protein, DNA and RNA (15) and there is now convincing evidence that they are important in many aspects associated with malignancy, including epithelial– mesenchymal transition (16), angiogensis (17) and tumour immunity (17) and MM-isolated exosomes enhance endothelial cell migration (4). A comparison of the highly expressed proteins compared to proteins that had a low expression suggests that MM cells rely on non-classical mechanisms such as exosomes to excrete many of those proteins that are relatively more abundantly expressed compared to normal mesothelial cells. However some proteins that are found in exosomes of MM cells were also found to have reduced expression in the secretome of MM cells compared to normal mesothelial cells, suggesting that these proteins are secreted through mechanisms other than exosomes by normal mesothelial cells, or alternatively that normal mesothelial cells also secrete these proteins within exosomes.
A number of proteins were identified that were reproducibly more highly expressed in the MM cells lines, compared to normal mesothelial cells. Glypican-1 (GPC1) is a cell surface proteoglycan that has been identified as a cancer-specific exosome marker. The presence of GPC1 positive exosomes was reported to be a highly sensitive and specific serum biomarker for pancreatic cancer (18) and has been reported as a MM exosome protein (4), suggesting that this protein should be further studied for its role in MM.
One of the most highly up-regulated proteins in the MM secretome was calmodulin (CALM). CALM is a calcium receptor protein that regulates a multitude of cellular functions including proliferation, apoptosis, autophagy, proliferation, differentiation and cell migration, cell adhesion and angiogenesis (19). It operates by sequestering calcium in the cell, thus allowing for transient changes in ionized calcium across the cell cytosol to the nucleus as part of many cell signalling pathways (19). Its marked increase in the MM secretome is indicative of the many cell signalling pathways that are activated in MM and associated with many of the pathways discussed below.
Metabolic reprogramming of cancer cells is recognized as being an important “hallmark of cancer” (20). A number of up-regulated proteins identified in this study are associated with important metabolic functions that have been identified as being altered in cancer. These include enzymes catalysing critical metabolic functions, such as the tricarboxylic (TCA) cycle and pentose phosphate pathways (PPP). Up-regulated PPP enzymes identified in this study, glucose-6-phosphate dehydrogenase (G6PD), transaldolase (TALDO) and transketolase (TKT), have been previously identified as important exosome cargo proteins in ovarian cancer cells (21). Activation of this pathway diverts glucose metabolism from energy metabolism to NADPH production, an essential cofactor in lipid and DNA synthesis, as well as protection from oxidative damage via reduction of glutathione. Another product of the PPP is ribose-5-P production for nucleotide synthesis. Cytoplasmic malate dehydrogenase (MDHC) and mitochondrial aspartate amino-transferase (AATM) catalyze the NAD dependant reversible conversion of malate to oxaloacetate. This is an essential component of the malate aspartate shuttle, which is necessary pathway for the net transfer of cytosolic NADH into mitochondria in order to maintain a high rate of glycolysis and has been shown to be up-regulated in pancreatic cancer, with a consequent increase in oxidative protection (17). Another metabolic enzyme, pyruvate kinase M (KPYM), which is a key enzyme that regulates aerobic glycolysis, the “Warburg effect”, in cancer (22), was also part of this complex of up-regulated metabolic proteins. KPYM is pivotal in directing glucose from catabolic to anabolic pathways by inhibiting the entry of pyruvate into the TCA cycle, with the result of glycolytic intermediates accumulating, which are then available for nucleotide and lipid synthesis via the PPP. The subsequent accumulation of cytosolic pyruvate resulting from this is then oxidized to lactate by lactate dehydrogenase, (LDH). LDHB is the B subunit of LDH and is also a component of the up-regulated suite of metabolic proteins identified in this study. Other metabolic enzymes included in this up-regulated group of metabolic enzymes are the Warburg effect-associated enzymes alpha enolase (ENOA) (23) and phosphoglycerate kinase-1 (PGK1), which have a role in coordinating glycolysis and the TCA cycle (24). Extracellular matrix protein 1 (ECM1), one of the most highly up-regulated proteins observed in this study, is a secreted glycoprotein that promotes cell proliferation by activating epidermal growth factor receptor (EGFR) signalling (25). ECM1 has been shown to induce Warburg effect associated genes in breast cancer cells via the EGFR/ERK pathway (26). Despite the recent renewed interest in metabolism in cancer, there are very few studies that have reported metabolic regulation in MM and the results from this study strongly suggest that this should be further considered from a therapeutic perspective, particularly as the glycolysis inhibitor 3-bromopyruvate has been shown to increase the survival of nude mice injected interperitonealy with human MM cells (27,28).
In addition to these metabolic proteins a number of proteins associated with other cellular functions were up-regulated in MM cells. A number of heat shock and chaperone proteins were found to be up-regulated, including CH10, HSP90A, HSP90B, HSP7C, GRP78 and CLUS. One of these, HSPA8 (4,29) has been identified in two separate studies to be present in MM cell exosomes and the others were all reported in our MM exosome study (4). HSP90A, HSP90B and HSPA8 have all been shown to be up-regulated in another proteomic analysis of a MM cell secretome (30). In MM cells, inhibition of HSP90 by 17-AAG results in cell cycle arrest and apoptosis (31). Quiescin sulfhydryl oxidase 1 (QSOX1) is involved in protecting cells from ROS induced apoptosis by preserving mitochondrial polarity (32) and in the activation of matrix metalloproteinases (MMP) 2 and 9 (33).
A number of up-regulated proteins identified in this study have been associated with cancer invasion and metastasis. EFHD2 (swiprosin-1) is an EGF induced actin binding protein that stimulates cancer invasion and metastasis(34). The receptor tyrosine kinase UFO (AXL) is important in EMT and tumour progression in MM and is over-expressed in most MM tumours. Its expression is an independent indicator of overall survival (35) and the inhibition of AXL in MM tumours with strong AXL expression is associated with inhibition of proliferation and invasiveness(36). PPIA (CypA) is a peptidyl-prolyl cis–trans isomerase, which is up-regulated in a range of solid cancers (37). CypA has been shown to stimulate cell motility and invasion by binding to the adaptor protein CrkII and preventing it from switching to its inactive form (38). Another potentially important up-regulated protein identified in this study is Ubiquitin C-terminal hydrolase-L1 (UCHL1), which promotes metastasis by deubiquitinating hypoxia inducible factor (HIF-1α) (39). The expression levels of UCHL1 in lung and breast cancer patients are a strong negative prognostic factor (39).
Lectin galactoside-binding soluble 3 binding protein (LGALS3BP) has been previously shown to be elevated in pleural effusions from MM patients (40) and, paradoxically, its effusion level is positively associated with survival (40). This positive association with survival may be due to inhibition of WNT signalling as has been demonstrated in colorectal cancer (41,42). LGALS3BP may inhibit immune responses to cancer (43).
Stathmin is a microtubule-destabilizing phosphoprotein that is considered a candidate for cancer therapy (44). It has been shown to be over-expressed in MM cell lines and tumour samples (45) and it is over expression is consistently associated with increased metastasis and malignant growth (46). Over-expression of miR-223 and siRNA knock-down in MM cell lines reduces stathmin expression, resulting in inhibition of motility and tubulin acetylation via a JNK signalling pathway-dependant mechanism (47).
Two heterogeneous nuclear ribonucleoproteins (HNRNPs) were observed to be consistently up-regulated in the MM cell lines. They are associated with pre-mRNAs in the nucleus and are associated with various aspects of pre-mRNA processing and mRNA metabolism and transport. HNRNPA1 has been shown to be up-regulated in lung adenocarcinoma and knockdown of the HNRNPA1 gene induces cell cycle arrest in G0/G1 phase (48). In hepatocellular carcinoma (HCC), HNRNPAB induces EMT and metastasis by transcriptionally activating SNAIL and down-regulating E cadherin (49). High HNRNPAB expression was associated with a poor prognosis in HCC patients (49). Transitional endoplasmic reticulum ATPase (p97/VCP) interacts with NSFL1 cofactor p47 resulting in the post-mitotic regulation of membrane fusion involved in the morphogenesis of the endoplasmic reticulum and golgi (50). High p97 expression has been associated with tumour aggressiveness (51) and is another potential therapeutic target (52,53) that could be further investigated in MM. Beta 2 microglobulin (B2MG) is a component of the major histocompatibility complex class I molecule. It is secreted by solid tumours and has been shown to stimulate cancer growth and metastasis in a number of solid and hematological cancers due to its action as a signal transducing molecule (54). Elevated B2MG serum levels are associated with increased risk of colorectal cancer (55). B2MG has been identified as a potential therapeutic target for cancer as siRNA or antibodies targeted against B2MG result in caspase induced apoptosis in prostate, renal and hematological malignancies (54).
A complex of relatively down-regulated proteins was also observed in this study, which may be relevant to the pathogenesis of MM. In particular a decrease in the expression of a number of collagen proteins that are associated with cell adhesion was observed that could be associated with the observed up-regulation of annexin A2, which is a regulator of the COL1A2 promoter region (56). A down-regulation of TIMP1, which is an MMP inhibitor, was observed. A change in the balance between MMP and TIMPs are associated with tumour progression and metastasis (57). MMP2 was identified in the secretomes of all cell lines in this study, confirming an earlier identification of MMP2 in a MM cell line (58). MMP2 is the only MMP identified in MM exosomes (4). MMP2 levels, measured in tissue homogenates, are a negative prognostic indicator in MM (59). Thus, this study suggests that MM is characterized by an increase in the activity of MMPs, particularly exosome secreted MMP2, which may be due to decreased TIMP1 expression and activation by QSOX1 as discussed previously.
Two serpin gene family proteins plasma protease C1 inhibitor (ICI), and plasminogen activator inhibitor (PAI-1), were down regulated across the MM cell lines. These proteins are complement cascade regulators. PAI-1 is a marker of cell senescence, acting downstream of p53 and upstream of insulin-like growth factor binding protein-3 in inducing cell senescence. In vitro, PAI-1 deficiency retards the development of senescence. Other senescence markers, IGFBP7 (60) and transgelin (61) were also relatively under-expressed across the MM cell lines. Cellular senescence occurs both in culture and in vivo as a response to excessive extracellular or intracellular stress (62). This suggests that the primary cell culture mesothelial cells were relatively senescent compared to the MM cell lines, which were protected by the activated stress response mechanisms discussed above.
The inter-alpha-trypsin inhibitor (ITI) proteins are serine protease inhibitors and contribute to extracellular matrix stability by binding covalently to hyaluronan. Two of these ICIs, ITIH2 and ITIH4, were down-regulated in the MM cell lines investigated in this study. Low expression of ITIH2 and ITH4 has been previously shown to exist in a range of solid cancers, including breast, colon and lung cancer (63), indicating that they are potential tumour suppressor proteins.
Another “hallmark of cancer” is the “evasion of immune destruction” (20). MM is potentially amenable to immunotherapy (2) and the differential regulation of proteins associated with immune response mechanisms were observed in this study. Calreticulin (CALR) is a calcium binding protein that is part of the folding process of MHC class 1 molecules and is a cell surface “eat me” signal for phagacytosis (64). Even though it has been identified as a MM exosome protein (4) CALR was down regulated in the MM compared to the mesothelial cell secretome. CALR has been associated with a poor response to anti-cancer vaccination (65). Subsets of patients with various cancers with low CALR expression responded less well to inducers of immunogenic cell death than patients with high CALR expressing tumours (65). Cells from a variety of cancers that have survived exposure to radiation express more cell surface CALR and this was associated with an enhanced sensitivity to cytotoxic T-cell killing (66). CD166 antigen (ALCAM), that was consistently up-regulated in this study, is an immunoglobulin receptor that binds to the CD6 T-cell differentiation antigen. This ALCAM - CD6 interaction is essential for dendritic cell (DC) induced proliferation of T cells, both for the initial DC – T-cell interaction and for sustaining DC induced T-cell proliferation (67). Crucially, soluble CD166 and CD6 have been shown to inhibit the CD6-CD166 interaction with a consequent inhibition of antigen-specific human T cell responses (68), suggesting that the extracellular secretion of ALCAM by MM cells observed in this study could inhibit T-cell responses to MM neo-antigens. In melanoma (69) and endometrial cancer (70) high ALCAM protein expression is associated with a poor prognosis, indicating an association of this protein with cancer. These findings have potentially important implications for immunotherapy in MM.
A limitation of this study, which was carried out using a conventional data-dependent acquisition (DDA) discovery approach, is that a number of secreted proteins that could have been of relevance may have been missed. Due to the inherent limitations of MS operating in a DDA mode to acquire fragmentation spectra for all of the peptides entering the instrument at a given time, peptide selection for fragmentation is based on their signal intensity resulting in low-abundance peptides being only randomly detected with a consequent lack of reproducibility (71).
In conclusion, this study indicates that the MM cell secretome is highly enriched in proteins that are secreted in exosomes. The MM cell secretome is characterized by up-regulated secretion of proteins that are associated with an enhanced stress response and with metabolic pathway enzymes associated with growth, as well as proteins associated with metastasis. Conversely the MM secretome has a decrease in proteins associated with cell senescence and intercellular communication. Finally, MM cells differentially secrete proteins associated with MHC class 1 expression and DC- T-cell interaction, suggesting that the MM secretome assists in its evasion of an adaptive immune response.
Acknowledgements
The Authors thank Proteomics International for performing the iTRAQ analysis and Dr Katie Meehan of the School of Pathology and Laboratory Medicine, University of Western Australia for reviewing the manuscript. This study was supported by funding from the National Health and Medical Research Council of Australia Project Grant 1063067.
References
- 1.Robinson BWS, Lake RA. Advances in malignant mesothelioma. N Engl J Med. 2005;353(15):1591–1603. doi: 10.1056/NEJMra050152. [DOI] [PubMed] [Google Scholar]
- 2.Robinson BW, Musk AW, Lake RA. Malignant mesothelioma. Lancet. 2005;366(9483):397–408. doi: 10.1016/S0140-6736(05)67025-0. [DOI] [PubMed] [Google Scholar]
- 3.Schaaij-Visser TB, de Wit M, Lam SW, Jimenez CR. The cancer secretome, current status and opportunities in the lung, breast and colorectal cancer context. Biochim Biophys Acta. 2013;1834(11):2242–2258. doi: 10.1016/j.bbapap.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 4.Greening DW, Ji H, Chen M, Robinson BW, Dick IM, Creaney J, Simpson RJ. Secreted primary human malignant mesothelioma exosome signature reflects oncogenic cargo. Sci Rep. 2016;6:32643. doi: 10.1038/srep32643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manning LS, Whitaker D, Murch AR, Garlepp MJ, Davis MR, Musk AW, Robinson BW. Establishment and characterization of five human malignant mesothelioma cell lines derived from pleural effusions. Int J Cancer. 1991;47(2):285–290. doi: 10.1002/ijc.2910470219. [DOI] [PubMed] [Google Scholar]
- 6.Holloway AJ, Diyagama DS, Opeskin K, Creaney J, Robinson BW, Lake RA, Bowtell DD. A molecular diagnostic test for distinguishing lung adenocarcinoma from malignant mesothelioma using cells collected from pleural effusions. Clin Cancer Res. 2006;12(17):5129–5135. doi: 10.1158/1078-0432.CCR-06-1027. [DOI] [PubMed] [Google Scholar]
- 7.Hjerpe A, Ascoli V, Bedrossian CWM, Boon ME, Creaney J, Davidson B, Dejmek A, Dobra K, Fassina A, Field A, Firat P, Kamei T, Kobayashi T, Michael CW, Önder S, Segal A, Vielh P. Guidelines for the cytopathologic diagnosis of epithelioid and mixed-type malignant mesothelioma. Acta Cytol. 2015;59(1):2–16. doi: 10.1159/000377697. [DOI] [PubMed] [Google Scholar]
- 8.Song X, Bandow J, Sherman J, Baker JD, Brown PW, McDowell MT, Molloy MP. iTRAQ experimental design for plasma biomarker discovery. J Proteome Res. 2008;7(7):2952–2958. doi: 10.1021/pr800072x. [DOI] [PubMed] [Google Scholar]
- 9.Lê S, Josse J, Husson F. FactoMineR: An R Package for Multivariate Analysis. 2008. 2008;25(1):18. [Google Scholar]
- 10.Kassambara AMF: Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R package version 1.0.3.9000. 2015. Available at: http://www.sthda.com/english/ wiki/factoextra.
- 11.Feizi A, Banaei-Esfahani A, Nielsen J. HCSD: the human cancer secretome database. Database (Oxford) 2015;2015:bav051. doi: 10.1093/database/bav051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 13.Bendtsen JD, Jensen LJ, Blom N, von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Engineering Design and Selection. 2004;17(4):349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 14.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43(Database issue) 2015:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greening DW, Gopal SK, Mathias RA, Liu L, Sheng J, Zhu HJ, Simpson RJ. Emerging roles of exosomes during epithelial-mesenchymal transition and cancer progression. Semin Cell Dev Biol. 2015;40:60–71. doi: 10.1016/j.semcdb.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Yi H, Ye J, Yang XM, Zhang LW, Zhang ZG, Chen YP. High-grade ovarian cancer secreting effective exosomes in tumor angiogenesis. Int J Clin Exp Pathol. 2015;8(5):5062–5070. [PMC free article] [PubMed] [Google Scholar]
- 18.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berchtold MW, Villalobo A. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2014;1843(2):398–435. doi: 10.1016/j.bbamcr.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Yi H, Zheng X, Song J, Shen R, Su Y, Lin D. Exosomes mediated pentose phosphate pathway in ovarian cancer metastasis: a proteomics analysis. Int J Clin Exp Pathol. 2015;8(12):15719–15728. [PMC free article] [PubMed] [Google Scholar]
- 22.Tamada M, Suematsu M, Saya H. Pyruvate kinase M2: multiple faces for conferring benefits on cancer cells. Clin Cancer Res. 2012;18(20):5554–5561. doi: 10.1158/1078-0432.CCR-12-0859. [DOI] [PubMed] [Google Scholar]
- 23.Capello M, Ferri-Borgogno S, Riganti C, Chattaragada MS, Principe M, Roux C, Zhou W, Petricoin EF, Cappello P, Novelli F. Targeting the Warburg effect in cancer cells through ENO1 knockdown rescues oxidative phosphorylation and induces growth arrest. Oncotarget. 2016;7(5):5598–5612. doi: 10.18632/oncotarget.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y, Xia Y, Aldape K, He J, Hunter T, Wang L, Lu Z. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol Cell. 2016;61(5):705–719. doi: 10.1016/j.molcel.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K-m, Nam K, Oh S, Lim J, Kim Y-P, Lee JW, Yu J-H, Ahn S-H, Kim S-B, Noh D-Y, Lee T, Shin I. Extracellular matrix protein 1 regulates cell proliferation and trastuzumab resistance through activation of epidermal growth factor signaling. Breast Cancer Res. 2014;16(6):1–17. doi: 10.1186/s13058-014-0479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee K-m, Nam K, Oh S, Lim J, Lee T, Shin I. ECM1 promotes the Warburg effect through EGF-mediated activation of PKM2. Cell Signal. 2015;27(2):228–235. doi: 10.1016/j.cellsig.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Varin E, Briand M, Allouche S, Heutte N, Schwartz L, Poulain L, Icard P. Novel therapy for malignant pleural mesothelioma based on anti-energetic effect: an experimental study using 3-Bromopyruvate on nude mice. Anticancer Res. 2009;29(4):1443–1448. [PubMed] [Google Scholar]
- 28.Icard P, Zhang XD, Lemoisson E, Louis MH, Allouche S, Lincet H, Poulain L. Experimental results using 3-bromopyruvate in mesothelioma: in vitro and in vivo studies. J Bioenerg Biomembr. 2012;44(1):81–90. doi: 10.1007/s10863-012-9415-6. [DOI] [PubMed] [Google Scholar]
- 29.Hegmans JP, Bard MP, Hemmes A, Luider TM, Kleijmeer MJ, Prins JB, Zitvogel L, Burgers SA, Hoogsteden HC, Lambrecht BN. Proteomic analysis of exosomes secreted by human mesothelioma cells. Am J Pathol. 2004;164(5):1807–1815. doi: 10.1016/S0002-9440(10)63739-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manfredi M, Martinotti S, Gosetti F, Ranzato E, Marengo E. The secretome signature of malignant mesothelioma cell lines. J Proteomics. 2016;145:3–10. doi: 10.1016/j.jprot.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 31.Okamoto J, Mikami I, Tominaga Y, Kuchenbecker KM, Lin Y-C, Bravo DT, Clement G, Yagui-Beltran A, Ray MR, Koizumi K, He B, Jablons DM. Inhibition of Hsp90 leads to cell cycle arrest and apoptosis in human malignant pleural mesothelioma. J Thorac Oncol. 2008;3(10):1089–1095. doi: 10.1097/JTO.0b013e3181839693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morel C, Adami P, Musard JF, Duval D, Radom J, Jouvenot M. Involvement of sulfhydryl oxidase QSOX1 in the protection of cells against oxidative stress-induced apoptosis. Exp Cell Res. 2007;313(19):3971–3982. doi: 10.1016/j.yexcr.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Katchman BA, Antwi K, Hostetter G, Demeure MJ, Watanabe A, Decker GA, Miller LJ, Von Hoff DD, Lake DF. Quiescin sulfhydryl oxidase 1 promotes invasion of pancreatic tumor cells mediated by matrix metalloproteinases. Mol Cancer Res. 2011;9(12):1621–1631. doi: 10.1158/1541-7786.MCR-11-0018. [DOI] [PubMed] [Google Scholar]
- 34.Huh YH, Oh S, Yeo YR, Chae IH, Kim SH, Lee JS, Yun SJ, Choi KY, Ryu JH, Jun CD, Song WK. Swiprosin-1 stimulates cancer invasion and metastasis by increasing the Rho family of GTPase signaling. Oncotarget. 2015;6(15):13060–13071. doi: 10.18632/oncotarget.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinato DJ, Mauri FA, Lloyd T, Vaira V, Casadio C, Boldorini RL, Sharma R. The expression of Axl receptor tyrosine kinase influences the tumour phenotype and clinical outcome of patients with malignant pleural mesothelioma. Br J Cancer. 2013;108(3):621–628. doi: 10.1038/bjc.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ou WB, Corson JM, Flynn DL, Lu WP, Wise SC, Bueno R, Sugarbaker DJ, Fletcher JA. AXL regulates mesothelioma proliferation and invasiveness. Oncogene. 2011;30(14):1643–1652. doi: 10.1038/onc.2010.555. [DOI] [PubMed] [Google Scholar]
- 37.Lee J, Kim SS. An overview of cyclophilins in human cancers. J Int Med Res. 2010;38(5):1561–1574. doi: 10.1177/147323001003800501. [DOI] [PubMed] [Google Scholar]
- 38.Saleh T, Jankowski W, Sriram G, Rossi P, Shah S, Lee K-B, Cruz LA, Rodriguez AJ, Birge RB, Kalodimos CG. Cyclophilin A promotes cell migration via the Abl-Crk signaling pathway. Nat Chem Biol. 2016;12(2):117–123. doi: 10.1038/nchembio.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goto Y, Zeng L, Yeom CJ, Zhu Y, Morinibu A, Shinomiya K, Kobayashi M, Hirota K, Itasaka S, Yoshimura M, Tanimoto K, Torii M, Sowa T, Menju T, Sonobe M, Kakeya H, Toi M, Date H, Hammond EM, Hiraoka M, Harada H. UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF-1alpha. Nat Commun. 2015;6:6153. doi: 10.1038/ncomms7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strizzi L, Muraro R, Vianale G, Natoli C, Talone L, Catalano A, Mutti L, Tassi G, Procopio A. Expression of glycoprotein 90K in human malignant pleural mesothelioma: correlation with patient survival. J Pathol. 2002;197(2):218–223. doi: 10.1002/path.1125. [DOI] [PubMed] [Google Scholar]
- 41.Lee JH, Bae JA, Lee JH, Seo YW, Kho DH, Sun EG, Lee SE, Cho SH, Joo YE, Ahn KY, Chung IJ, Kim KK. Glycoprotein 90K, downregulated in advanced colorectal cancer tissues, interacts with CD9/CD82 and suppresses the Wnt/beta-catenin signal via ISGylation of beta-catenin. Gut. 2010;59(7):907–917. doi: 10.1136/gut.2009.194068. [DOI] [PubMed] [Google Scholar]
- 42.Piccolo E, Tinari N, D'Addario D, Rossi C, Iacobelli V, La Sorda R, Lattanzio R, D'Egidio M, Di Risio A, Piantelli M, Natali PG, Iacobelli S. Prognostic relevance of LGALS3BP in human colorectal carcinoma. J Transl Med. 2015;13:248. doi: 10.1186/s12967-015-0606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laubli H, Alisson-Silva F, Stanczak MA, Siddiqui SS, Deng L, Verhagen A, Varki N, Varki A. Lectin galactoside-binding soluble 3 binding protein (LGALS3BP) is a tumor-associated immunomodulatory ligand for CD33-related Siglecs. J Biol Chem. 2014;289(48):33481–33491. doi: 10.1074/jbc.M114.593129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rana S, Maples PB, Senzer N, Nemunaitis J. Stathmin 1: a novel therapeutic target for anticancer activity. Expert Rev Anticancer Ther. 2008;8(9):1461–1470. doi: 10.1586/14737140.8.9.1461. [DOI] [PubMed] [Google Scholar]
- 45.Kim JY, Harvard C, You L, Xu Z, Kuchenbecker K, Baehner R, Jablons D. Stathmin is overexpressed in malignant mesothelioma. Anticancer Res. 2007;27(1a):39–44. [PubMed] [Google Scholar]
- 46.Biaoxue R, Hua L, Wenlong G, Shuanying Y. Overexpression of stathmin promotes metastasis and growth of malignant solid tumors: a systemic review and meta-analysis. Oncotarget. 2016;7(48):78994–79007. doi: 10.18632/oncotarget.12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Birnie KA, Yip YY, Ng DC, Kirschner MB, Reid G, Prele CM, Musk AW, Lee YC, Thompson PJ, Mutsaers SE, Badrian B. Loss of miR-223 and JNK Signaling Contribute to Elevated Stathmin in Malignant Pleural Mesothelioma. Mol Cancer Res. 2015;13(7):1106–1118. doi: 10.1158/1541-7786.MCR-14-0442. [DOI] [PubMed] [Google Scholar]
- 48.Liu X, Zhou Y, Lou Y, Zhong H. Knockdown of HNRNPA1 inhibits lung adenocarcinoma cell proliferation through cell cycle arrest at G0/G1 phase. Gene. 2016;576(2, Part 2):791–797. doi: 10.1016/j.gene.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 49.Zhou ZJ, Dai Z, Zhou SL, Hu ZQ, Chen Q, Zhao YM, Shi YH, Gao Q, Wu WZ, Qiu SJ, Zhou J, Fan J. HNRNPAB induces epithelial-mesenchymal transition and promotes metastasis of hepatocellular carcinoma by transcriptionally activating SNAIL. Cancer Res. 2014;74(10):2750–2762. doi: 10.1158/0008-5472.CAN-13-2509. [DOI] [PubMed] [Google Scholar]
- 50.Uchiyama K, Kondo H. p97/p47-Mediated biogenesis of Golgi and ER. J Biochem. 2005;137(2):115–119. doi: 10.1093/jb/mvi028. [DOI] [PubMed] [Google Scholar]
- 51.Fessart D, Marza E, Taouji S, Delom F, Chevet E. P97/CDC-48: Proteostasis control in tumor cell biology. Cancer Lett. 2013;337(1):26–34. doi: 10.1016/j.canlet.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 52.Deshaies RJ. Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biol. 2014;12(1):1–14. doi: 10.1186/s12915-014-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, Madriaga A, Soriano F, Menon MK, Wu ZY, Kampmann M, Chen Y, Weissman JS, Aftab BT, Yakes FM, Shawver L, Zhou HJ, Wustrow D, Rolfe M. Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell. 2015;28(5):653–665. doi: 10.1016/j.ccell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi C, Zhu Y, Su Y, Chung LWK, Cheng T. β2-Microglobulin: emerging as a promising cancer therapeutic target. Drug Discovery Today. 2009;14(1-2):25–30. doi: 10.1016/j.drudis.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 55.Prizment AE, Linabery AM, Lutsey PL, Selvin E, Nelson HH, Folsom AR, Church TR, Drake CG, Platz EA, Joshu C. Circulating Beta-2 Microglobulin and Risk of Cancer: The Atherosclerosis Risk in Communities Study (ARIC) Cancer Epidemiology Biomarkers & Prevention. 2016;25(4):657–664. doi: 10.1158/1055-9965.EPI-15-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schafer G, Hitchcock JK, Shaw TM, Katz AA, Parker MI. A novel role of annexin A2 in human type I collagen gene expression. J Cell Biochem. 2015;116(3):408–417. doi: 10.1002/jcb.24989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polette M, Nawrocki-Raby B, Gilles C, Clavel C, Birembaut P. Tumour invasion and matrix metalloproteinases. Crit Rev Oncol Hematol. 2004;49(3):179–186. doi: 10.1016/j.critrevonc.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 58.Roomi MW, Kalinovsky T, Niedzwiecki A, Rath M. Modulation of u-PA, MMPs and their inhibitors by a novel nutrient mixture in human lung cancer and mesothelioma cell lines. Int J Oncol. 2013;42(6):1883–1889. doi: 10.3892/ijo.2013.1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edwards JG, McLaren J, Jones JL, Waller DA, O'Byrne KJ. Matrix metalloproteinases 2 and 9 (gelatinases A and B) expression in malignant mesothelioma and benign pleura. Br J Cancer. 2003;88(10):1553–1559. doi: 10.1038/sj.bjc.6600920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Acosta JC, O'Loghlen A, Banito A, Raguz S, Gil J. Control of senescence by CXCR2 and its ligands. Cell Cycle. 2008;7(19):2956–2959. doi: 10.4161/cc.7.19.6780. [DOI] [PubMed] [Google Scholar]
- 61.Herzog R, Tarantino S, Rudolf A, Aufricht C, Kratochwill K, Witowski J. Senescence-Associated Changes in Proteome and O-GlcNAcylation Pattern in Human Peritoneal Mesothelial Cells. Biomed Res Int. 2015;2015:382652. doi: 10.1155/2015/382652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamm A, Veeck J, Bektas N, Wild PJ, Hartmann A, Heindrichs U, Kristiansen G, Werbowetski-Ogilvie T, Del Maestro R, Knuechel R, Dahl E. Frequent expression loss of Inter-alpha-trypsin inhibitor heavy chain (ITIH) genes in multiple human solid tumors: a systematic expression analysis. BMC Cancer. 2008;8:25. doi: 10.1186/1471-2407-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raghavan M, Wijeyesakere SJ, Peters LR, Del Cid N. Calreticulin in the immune system: ins and outs. Trends Immunol. 2013;34(1):13–21. doi: 10.1016/j.it.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garg AD, Elsen S, Krysko DV, Vandenabeele P, de Witte P, Agostinis P. Resistance to anticancer vaccination effect is controlled by a cancer cell-autonomous phenotype that disrupts immunogenic phagocytic removal. Oncotarget. 2015;6(29):26841–26860. doi: 10.18632/oncotarget.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gameiro SR, Malamas AS, Bernstein MB, Tsang KY, Vassantachart A, Sahoo N, Tailor R, Pidikiti R, Guha CP, Hahn SM, Krishnan S, Hodge JW. Tumor Cells Surviving Exposure to Proton or Photon Radiation Share a Common Immunogenic Modulation Signature, Rendering Them More Sensitive to T Cell–Mediated Killing. International Journal of Radiation Oncology*Biology*Physics. 2016;95(1):120–130. doi: 10.1016/j.ijrobp.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zimmerman AW, Joosten B, Torensma R, Parnes JR, van Leeuwen FN, Figdor CG. Long-term engagement of CD6 and ALCAM is essential for T-cell proliferation induced by dendritic cells. Blood. 2006;107(8):3212–3220. doi: 10.1182/blood-2005-09-3881. [DOI] [PubMed] [Google Scholar]
- 68.Hassan NJ, Barclay AN, Brown MH. Frontline: Optimal T cell activation requires the engagement of CD6 and CD166. Eur J Immunol. 2004;34(4):930–940. doi: 10.1002/eji.200424856. [DOI] [PubMed] [Google Scholar]
- 69.Weidle UH, Eggle D, Klostermann S, Swart GWM. ALCAM/CD166: Cancer-related Issues. Cancer Genomics - Proteomics. 2010;7(5):231–243. [PubMed] [Google Scholar]
- 70.Devis L, Moiola CP, Masia N, Martinez-Garcia E, Santacana M, Stirbat TV, Brochard-Wyart F, Garcia A, Alameda F, Cabrera S, Palacios J, Moreno-Bueno G, Abal M, Thomas W, Dufour S, Matias-Guiu X, Santamaria A, Reventos J, Gil-Moreno A, Colas E. Activated leukocyte cell adhesion molecule (ALCAM) is a marker of recurrence and promotes cell migration, invasion and metastasis in early stage endometrioid endometrial cancer. J Pathol. 2016;241(4):475–487. doi: 10.1002/path.4851. [DOI] [PubMed] [Google Scholar]
- 71.Michalski A, Cox J, Mann M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J Proteome Res. 2011;10(4):1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]