Abstract
The delineation of signaling pathways to understand tumor biology combined with the rapid development of technologies that allow broad molecular profiling and data analysis, has led to a new era of personalized medicine in oncology. Many academic institutions now routinely profile patients and discuss them in personalized medicine tumor boards before making treatment recommendations. Clinical trials initiated by pharmaceutical companies often require specific markers for enrollment or at least explore multiple options for future markers. In addition to the still small number of targeted agents that are approved for the therapy of patients with histological and molecularly defined tumors, there is a broad range of novel targeted agents in development that are undergoing clinical studies with companion profiling to determine the best responding patient population. While the present focus of profiling are genetic analyses, additional testing of RNA, protein and immune parameters are being developed and incorporated in clinical research and are likely to contribute significantly to future patient selection and treatment approaches. As the advances in tumor biology and human genetics have identified promising tumor targets, the ongoing clinical evaluation of novel agents will now need to show if the promise can be translated into benefit for patients.
Background
Over the last decade, the delineation of signaling pathways to understand tumor biology has laid the foundation for the discovery of novel targets and development of multiple therapies for cancer. The identification of driver mutations and critical pathway dependencies, as well as genomic sequencing and other large-scale “-omics” approaches, have facilitated the discovery and development of novel targeted anti-cancer therapeutics, or at least provide the scientific rationale for their development. This increased molecular understanding of tumors has led to a new era of personalized medicine that has begun to influence common practice for oncologists beyond academic institutions.
The National Cancer Institute (NCI) defines Personalized Medicine, often also named “Precision Medicine”, as “A form of medicine that uses information about a person's genes, proteins, and environment to prevent, diagnose, and treat disease”. Personalized medicine uses specific markers in patients' tumors to diagnose particular cancers or make treatment decisions. A specific marker may have prognostic significance for biological behavior or predict therapeutic outcome with a particular anti-cancer agent. This review will focus on predictive markers associated with tumors and not discuss host genetic factors that also can determine treatment and prognosis.
Traditionally, the site of tumor origin, together with histology, was used to make treatment decisions. This approach has been changed to include molecular tumor parameters. Markers presently used to guide decisions for personalized medicine treatment with targeted agents are either protein-based (immunohistochemistry (IHC)) or detecting genetic aberrations and are summarized in Table 1. For genetic aberrations, a range of methodologies, including Fluorescent in situ Hybridization (FISH) -tests, polymerase-chain-reaction (PCR), sequencing – nowadays often Next-Generation-Sequencing (NGS) of either multiple selected genes in parallel or whole exome sequencing – are used, in particular in early clinical trials or for exploratory purposes. These large panels of genes offer the opportunity not only to detect aberrations, for which treatment options already exist, but also to generate data on additional driver mutations or resistance pathways (1, 2). Treatment decisions may be based on the data when tests have been performed in Clinical laboratory Improvement Amendment (CLIA)-certified laboratories.
Table 1. Molecular selection markers for approved anticancer agents.
| Target | Cancer | Variation type | Marker | Drug | test |
|---|---|---|---|---|---|
| EGFR | Lung cancer | Mutation | Predict benefit to EGFR TKIs | Erlotinib Gefitinib |
DNA |
| ALK | Lung Cancer | Rearrangement | Predict response to ALK-inhibitors | Crizotinib | FISH |
| ROS | Lung Cancer | Rearrangement | Predict response to TKIs | Crizotinib | FISH |
| RET | Lung Cancer | Rearrangement | Predict response to TKIs | Vandetanib | FISH |
| BRAF | Melanoma | Mutation | Predict response to BRAF-inhibitors | Vemurafenib Dabrafenib |
DNA |
| KRAS | CRC | Mutation | Predict lack of response to anti-EGFR antibodies | Panitumumab Cetuximab |
DNA |
| HER2 | Breast Cancer Gastric Cancer | Amplification Overexpression | Predict response to anti-HER2 antibodies | Trastuzumab Lapatinib Pertuzumab |
FISH, IHC |
| KIT | GIST | Mutation | Predict response to c-Kit inhibitors | Imatinib | IHC |
| Estrogen receptor | Breast Cancer | Overexpression | Predict response | Examestane Fulvestrant Letrozole Tamoxifen |
IHC |
| Progesterone receptor | Breast Cancer | Overexpression | Predict response | Examestane Letrozole |
IHC |
Anti-hormonal agents such as tamoxifen have been used for decades, initially without molecular profiling. Studies later correlated outcome with the expression of estrogen receptor (ER), progesterone receptor (PR) and androgen receptor (AR). The presence of ER, PR and AR not only categorizes breast and prostate cancers into different prognostic groups, but also determines treatment. Trastuzumab, a monoclonal antibody targeting human epithelial growth factor receptor-2 (HER-2) was approved for patient populations with a specific molecular profile, Her-2-overexpresing breast and gastric cancer (3-5). The first agent targeting a chromosomal translocation, imatinib, was originally approved for the treatment of Philadelphia chromosome positive (Ph+) chronic myelogenous leukemia, based on inhibiting the Bcr-Abl tyrosine kinase (6). In 2008, based on the additional activity of imatinib to inhibit c-Kit, approval by the FDA for the treatment of adult patients with c-Kit (CD117) positive unresectable and/or metastatic Gastrointestitinal Stromal Tumors (GIST) followed (7, 8) The development of vemurafenib for advanced BRAFV600- mutated melanoma (9) and crizotinib for patients with NSCLC with ALK translocations (10) are other recent successes of therapies targeting molecularly defined tumor subtypes. Molecular profiling can also lead to a negative selection, as the retrospective studies showing consistent lack of benefit from the anti-EGFR antibodies cetuximab or panitumumab in CRC patients with KRAS codon 12/13 mutations led to the recommendation to restrict the use of these antibodies to patients with wild-type Kras tumors (11-14).
Lung cancer has become the prototype for genetically tailored cancer therapy. The remarkable advances in understanding molecular drivers in NSCLC together with the development of targeted agents allows classification of NSCLC on a molecular basis and molecular profiling has become routine practice in thoracic oncology. EGFR mutations were discovered in 2004 in parallel to the development of gefitinib, an EGFR tyrosine kinase inhibitor (TKI), when retrospective studies revealed that patients with responses harbor exon 19 deletions or exon 21 point mutations (15, 16). This led to the use of the EGFR TKIs gefitinib and erlotinib as initial treatment for patients with these mutations. The discovery of EGFR mutations and the large benefit of targeted treatment has boosted profiling and the search for novel targets. The development of crizotinib to treat patients with ALK fusions, detected by a FISH-assay, highlights one of the outstanding successes in lung cancer with rapid development of a treatment (17). More recently the additional activity of crizotinib in patients with ROS-fusions (18, 19) was discovered and patients with RET-fusions are beginning to be treated with the RET inhibitors vandetanib and carbozantinib (20-24).
While these targeted therapies have brought significant improvements, all patients eventually develop therapeutic resistance. Multiple resistance mechanisms have been characterized, such as secondary mutations preventing inhibitor binding, EGFR or HER2 gene amplifications, HGF/Met pathway activations as well as PI3K and BRAF mutations (25, 26). Multiple resistance mechanisms have also been identified in patients with ALK fusions who progress on crizotinib. These include ALK mutations, copy gain number and mutations in alternative pathways, including EGFR mutations. Second and third generation inhibitors of EGFR and ALK, such as AZD9291 with activity against EGFRT790M and alectinib or ceritinib, especially active against the L11986M ALK mutation that confers resistance to crizotinib, are now in development with initial promising data (17, 27-32).
Multiple de-regulated pathways have been identified across a range of tumor types, which could potentially be targeted by novel agents (summarized in Table 2). However, often several genetic aberrations are found in the same tumor and it is not always clear which are driver mutations, which are secondary changes and which are the determinants of inherent or acquired resistance. Moreover, mutations that clearly act as driver mutations in one tumor type may not be of similar relevance in another tumor. For example, malignant melanoma patients with BRAF V600E mutations respond to treatment with vemurafenib, (33) whereas patients with colorectal cancer, that harbor the same mutation, seem to derive little benefit from BRAF inhibitors due to complex mechanisms that include a feedback loop that increases EGFR expression (34). There are multiple studies ongoing testing these hypotheses. Studies with preliminary results that are encouraging include PI3K/AKT/mTOR inhibitors in PIK3CAmt- or PTEN deficient cancers (35).
Table 2. Genetic aberrations as putative predictive biomarkers for anticancer agents.
| Target/Pathway | Aberration type in solid tumors | Disease examples | Putative or proven drugs | Examples for drugs in clinical development | References |
|---|---|---|---|---|---|
| EGFR | Mutation Amplification | Lung cancer GBM |
EGFR inhibitors | Erlotinib Gefitinib Afatinib AZD9291 |
(15, 16, 25, 26, 30) |
| HER2 (ERB2) | Mutation Amplification | Breast cancer Gastric cancer Lung cancer |
ERB2/ ERB3 inhibitors | Lapatini Neratinib Trastuzumab Trastuzumab-emtansine |
(4, 5) |
| ALK | Rearrangement Mutation | Lung cancer Neuroblastoma CRC |
ALK inhibitors | Crizotinib Ceritinib Alectinib |
(10, 22, 28, 29, 65, 66) |
| RET | Rearrangement Mutation |
Thyroid cancer Lung cancer |
RET inhibitors | Vandetanib Carbozantinib |
(19, 20, 23, 67) |
| ROS1 | Rearrangement | Lung cancer | ROS1 inhibitors | Crizotinib | (18, 19) |
| DDR2 | Mutation | Lung cancer | DDR2 inhibitors | Dasatinib Nilotinib |
(68, 69) |
| FGFR1-4 | Amplification Mutation |
Lung cancer Gastric cancer Breast cancer Bladder cancer |
FGFR inhibitors | Ponatinib Dovitinib BGJ398 AZD4547 |
(70, 71) |
| MET/HGF | Mutation Amplification |
Lung cancer Gastric cancer CRC HCC RCC |
Met inhibitors | Onartuzumab Crizotinib Foretinib INC280 |
(72-74) |
| KIT | Mutation | GIST Mastocytosis Melanoma |
Kit inhibitors | Imatinib Nilotinib Sunitinib Dasatinib Ponatinib |
(7, 8) |
| PDGFRA and PDGFRB | Mutation Translocation |
GIST Sarcoma GBM Leukemia Dermatofibrosarcoma protuberans |
PDGFR inhibitors | Imatinib Sunitinib Ponatinib |
(8, 75) |
|
KRAS, NRAS, HRAS (RAS-RAF-MEK) |
Mutation | Most cancers, including CRC, Lung cancers | Mek inhibitors | Trametinib Selumetinib |
(76) |
| BRAF (RAS-RAF-MEK) | Mutation | Melanoma CRC HCC |
Braf inhibitors Mek inhibitors |
Vemurafenib Dabrafenib Trametinib Selumetinib |
(76, 77) |
| PI3KCA (PTEN/PI3K AKT/mTOR) | Mutation Amplifcation |
Multiple, including: Breast Cancer CRC, GBM Lung Cancer Endometrial |
PI3K inhibitors AKT inhibitors |
BKM-120 BEZ235 BYL719 GDC0941 GDC0032 MLN1117 AKT inhibitors- see below |
(78, 79) |
| PIK3R1 (PTEN/PI3K AKT/mTOR) | Mutation | Endometrial cancer CRC |
PI3K inhibitors | PI3K inhibitors- see above | (79) |
| AKT1-3(PTEN/PI3K/AKT/mTOR) | Mutation Amplification |
Breast cancer CRC Meningeal cancer Urinary tract cancers Endometrial cancer |
PI3K inhibitors AKT inhibitors |
GDC0086 MK2206 AZD5363 PI3K inhibitors- see above |
(78-80) |
| PTEN (PTEN/PI3K/AKT/mTOR) | Deletion Mutation |
Most cancers, including breast cancer Lung cancer CRC |
PI3K inhibitors AKT inhibitors |
PI3K and AKT inhibitors- see above | (78-81) |
| mTOR (PTEN/PI3K/AKT/mTOR) | Mutation | Endometrial cancer RCC CRC Lung cancer |
mTOR inhibitors | Everolimus Temsirolimus MLN128 AZD2014 GDC00980 BEZ235 |
(79, 82, 83) |
| TSC1/2 (PTEN/PI3K/AKT/mTOR) | Mutation | Tuberous sclerosis Urinary tract cancers Endometrial cancer Cervical cancer HCC, CRC |
mTOR inhibitors | mTOR inhibitors-see above | (79, 84) |
| LKB1 (PTEN/PI3K/AKT/mTOR) | Mutation | Cervical cancer Small intestine cancer Lung cancer Skin cancer |
mTOR inhibitors | mTOR inhibitors- see above | (79) |
| SMO, PTCH1 (Hedgehog) | Mutation | Basal cell carcinoma Medulloblastoma Meningioma Breast cancer |
Hedgehog inhibitors | Vismodegib | (85, 86) |
| MDM2 | Amplification | GBM Sarcoma |
MDM2 inhibitors/antagonists disrupting p53-MDM2 interaction | RG7388 | (53, 54) |
| P53 | Mutation | Most tumors | P53 activators | (53, 54) | |
| NOTCH | Mutation Rearrangement |
Breast cancer Lung cancer Ovarian cancer GBM H&N cancer |
Gamma-secretase inhibitors ABs to Notch receptors or ligands |
MK0752 PF03084014 Demcizumab OMP59R5 OMP52M51 Enoticumab |
(87, 88) |
| CDKS | Amplification Mutation Rearrangement |
Sarcoma Melanoma GBM |
CDK inhibitors | Flavopiridol Palbociclib |
(89) |
| CHK1/2 | Mutation | Multiple tumors, including: CRC Gastric cancer Endometrial cancer Breast cancer |
CHK inhibitors | RG7741 LY2606368 |
(89) |
| AURKA (Aurora Kinases) | Amplification | Multiple tumors | Aurora kinase inhibitors | Alisertib | (89, 90) |
| ATR | Mutation Deletion |
Gastric cancer Breast cancer Endometrial cancer |
ATM inhibitors PARP inhibitors |
No ATM inhibitors in clinical development PARP inhibitors- see below | (89, 91) |
| ATM | Mutation Deletion |
Multiple tumors, including Breast cancer | ATR inhibitors PARP inhibitors |
VX970 PARP inhibitors- see below |
(89, 91) |
| BRCA1/2 | Mutation | Breast cancer Ovarian cancer |
PARP inhibitors | Olaparib Veliparib Rucaparib BMN673 |
(92, 93) |
On The Horizon
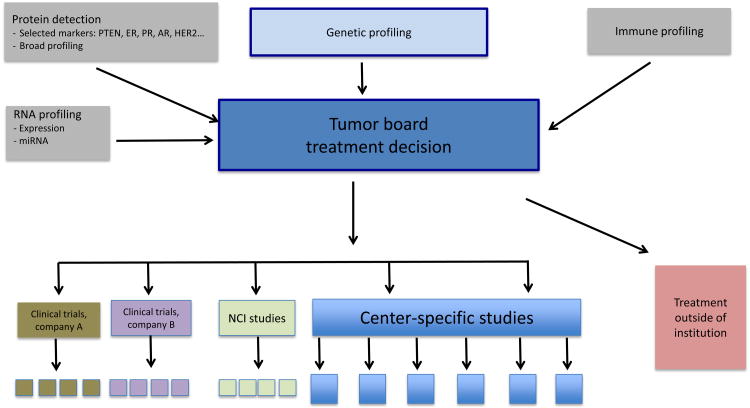
Many cancer centers now profile patients by genetic testing, RNA expression profiling, or protein analyses and use the obtained data to direct patients to clinical trials at their centers or elsewhere (36, 37). Genetic profiling is booming and molecular profiling is discussed in multi-disciplinary tumor boards and contributes to treatment recommendations. Some academic cancer centers now hold a weekly Precision Tumor Board in which oncologists together with radiologists and molecular pathologists present cases. The multi-disciplinary team, that also includes basic and translational scientists, surgeons and nurses, discusses the cases and potential treatment options. Presently, molecular profiling is mostly limited to genetic aberrations (mutations, translocations, fusions, copy number variation (CNV)), but it is planned to extend the profiling to include RNA-, and immune-profiling (Figure 1). While currently genetic profiling is only performed for patients for whom additional data are critical to guide treatment, it is envisioned that in the near future all patients treated at academic cancer centers will have a diagnostic tumor sample analyzed for genetic aberrations, and with repeat-biopsy programs in place, ideally also at progression. The ultimate goal of the profiling is to find the best available treatment for the patient, mostly by enrollment in a clinical trial, if such a trial exists. There is an urgent need for more studies with molecularly targeted agents that are open for patients across tumor types to investigate if these treatments can benefit other patient populations.
Figure 1. Academic Medical Center Precision Medicine Tumor Board Model.
Several institutions and companies have launched trials, that assign patients based on molecular profiling of tumors in specific cancer types (BATTLE I and II) (38), but also independent of cancer type. These studies include observational studies, as well as non-randomized and randomized studies (2, 39-41). Non-randomized studies building on the ability of academic cancer centers to perform molecular profiling in CLIA certified laboratories have been initiated in 2013 by pharmaceutical companies making a number of agents available for use in molecular defined patient populations. These include agents for which safety data already exist and a Phase II dose is defined, and exclude patient populations for which the drugs are either already registered or for which dedicated randomized studies exist or populations where a lack of benefit for the agent was already observed. For example, the Novartis SIGNATURE studies offer independent trials with investigational agents from a pharmaceutical company for patients with a specific molecular profile (42). These agents would otherwise not be available for patients with many tumor types, as specific studies in all tumor types do not exist. The NCI is planning to open a study of a similar type in 2014, NCI-MATCH, which is expected to include agents from multiple companies (43). For the NCI-MATCH studies, genomic profiling will be performed by a consortium of NCI-selected CLIA-certified laboratories using NGS of a defined number of genes as the basis for enrollment. The arms of the NCI-MATCH study are still under discussion. It is expected to include agents from multiple companies under NCI sponsorship and will validate the proposed broad sequencing platform. The SHIVA study randomizes patients with a particular molecular abnormality with any type of cancer between the matched agent and conventional cytotoxic therapy, with crossover on progression (39). The SHIVA study is different to the other two mentioned studies in that it has SOC comparator arms for each newly tested agent and will therefore collect data on the outcome of SOC therapies in molecular segments. More studies of this type with agents in development would be of high interest, but are difficult to initiate and coordinate if they involve multiple companies. The results of these ongoing studies are likely to identify additional patient populations for targeted therapy and hopefully will aid registration for additional indications. Data collection is critical for the future direction of personalized medicine and may trigger additional basic research to fully understand the contributions of newly discovered mutations to tumor development and progression.
Is testing for certain mutations sufficient or should broad testing be applied to all samples? Single mutation or small panel testing has the clear advantage of requiring smaller amounts of tissue, is less costly and interpretation of data is simpler and hence often quicker. While there are only a few tests that clearly direct patient treatment at this time (see Table 1), only broader testing will facilitate better understanding of tumor drivers and mechanisms of resistance critical for future direction. Multiplexing also requires less tissue than multiple single tests. The latter is becoming an increasing problem for Phase I trials, where companies require large amounts of tissue for testing to evaluate eligibility that exhaust archival diagnostic tissue resulting in patients requiring new biopsies or even deprives them of enrollment if a new sample cannot be obtained.
Advanced technology has revealed intra- and inter-tumor heterogeneity at protein, genetic and epigenetic level (44, 45). Genetic analyses, RNA- and protein - profiling of primary carcinomas and metastases from the same patients with renal carcinomas showed a large heterogeneity with most detected genetic aberrations not consistent between lesions and tumor regions that confirm that multiple clonal subpopulations exist within a single lesion (46, 47). In contrast, genetic analyses of colorectal and lung cancer lesions showed more concordant data for a limited gene set (48, 49). Indeed, the effectiveness of available targeted treatments for advanced cancer patients such as gefitinib in EGFRmt lung cancer and cetuximab or panitumumab in KRASwt CRC has largely been demonstrated in clinical trials that identified the mutations from archival diagnostic biopsies. In light of the heterogeneity, it is questionable, if a single archival biopsy taken for diagnostic purposes in early stages of the disease is sufficient as a sample for molecular profiling to guide treatment. Even if a new biopsy is available for analysis, is one sample sufficient, if multiple lesions may have a very different molecular profile? How many areas of a single tumor need to be biopsied to obtain comprehensive information of the drivers of the tumor in a patient? Answers to these questions are urgently needed.
Given the emergence of resistance as described above, it is expected that future patients' tumors will need to be profiled at several times during treatment in order to determine best treatment options. Indeed, some academic cancer centers have repeat-biopsy programs in place, pursuing new biopsies when patients progress on targeted treatment, in particular with EGFR or ALK inhibitors. How realistic are multiple longitudinal biopsies for a majority of cancer patients across tumor types and do we urgently need to increase our efforts to further explore other technologies that may be able to assess tumor mutations without multiple longitudinal biopsies or parallel biopsies of different lesions?
To minimize invasive procedures, the analysis of cfDNA or CTCs should be explored further to evaluate if these technologies can replace multiple biopsies for molecular analysis, especially, as patients move through successive lines of treatment. Initial results are promising (50-52). Is genetic profiling really enough? How do we approach the many targets for which treatment options do not yet exist, including the ones that are presently considered un-druggable, like TP53 and KRAS (53-56). Do we need broader molecular profiling that includes other endpoints to strengthen scientific understanding and other selection methods, especially if they can be applied to samples other than tumor biopsies?
Serum proteins have been established as general prognostic factors, for example PSA levels in prostate cancer patients. CEA is measured as a tumor marker across multiple tumor types and high levels have been shown to correlate with poor prognosis (57-59). Attempts to identify serum proteins as predictors for response to targeted agents so far have had limited success (60, 61), but this has not been assessed broadly. Further serum marker studies in clinical trials should be encouraged, especially, if tumor samples at baseline of these trials are available and could be compared to serum assays.
Other areas presently explored are RNA profiling (mRNA and miRNA), epigenetic and immune profiling (62-64). RNA analyses will strengthen the understanding of de-regulated signaling pathways in cancer that genetic profiling alone may not reveal and may lead to novel hypotheses for targets. Immune profiling will become important as new therapies, especially immune check point therapies demonstrate striking activity is several cancer types.
While great progress has been made towards molecular profiling for personalized cancer therapy, it is expected that with rapidly developing technology, molecular determinants other than genetic aberrations may emerge that will contribute to treatment decisions. The next five years will decide if personalized medicine will become standard practice across all tumor types and if this will revolutionize treatment options for greater survival benefit for cancer patients.
Acknowledgments
Grant Support: This work was supported by the NIH (2P30CA016359-34; to J.P. Eder, through his institution) and the NIH/NCI (1R01CA155196; to R.S. Herbst).
Footnotes
Disclosure of Potential Conflicts of Interest: J.M. Jürgensmeier was an employee of and reports receiving a commercial research grant from AstraZeneca. R.S. Herbst reports receiving a commercial research grant from Genentech and is a consultant/advisory board member for Biothera, Diatech, Kolltan, and N-of-One. No potential conflicts of interest were disclosed by the other author.
References
- 1.MacConaill LE. Existing and emerging technologies for tumor genomic profiling. J Clin Oncol. 2013;31:1815–1824. doi: 10.1200/JCO.2012.46.5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran B, Dancey JE, Kamel-Reid S, et al. Cancer genomics: technology, discovery, and translation. J Clin Oncol. 2012;30:647–660. doi: 10.1200/JCO.2011.39.2316. [DOI] [PubMed] [Google Scholar]
- 3.Baselga J, Tripathy D, Mendelsohn J, et al. Phase II study of weekly intravenous recombinant humanized anti-p185HER2 monoclonal antibody in patients with HER2/neu-overexpressing metastatic breast cancer. J Clin Oncol. 1996;14:737–744. doi: 10.1200/JCO.1996.14.3.737. [DOI] [PubMed] [Google Scholar]
- 4.Figueroa-Magalhaes MC, Jelovac D, Connolly RM, Wolff AC. Treatment of HER2-positive breast cancer. Breast. 2013 doi: 10.1016/j.breast.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Mello RA, Marques AM, Araujo A. HER2 therapies and gastric cancer: a step forward. World J Gastroenterol. 2013;19:6165–6169. doi: 10.3748/wjg.v19.i37.6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thambi P, Sausville EA. STI571 (imatinib mesylate): the tale of a targeted therapy. Anticancer Drugs. 2002;13:111–114. doi: 10.1097/00001813-200202000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Ashman LK, Griffith R. Therapeutic targeting of c-KIT in cancer. Expert Opin Investig Drugs. 2013;22:103–115. doi: 10.1517/13543784.2013.740010. [DOI] [PubMed] [Google Scholar]
- 8.Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer. 2011;11:865–878. doi: 10.1038/nrc3143. [DOI] [PubMed] [Google Scholar]
- 9.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 12.Adelstein BA, Dobbins TA, Harris CA, et al. A systematic review and meta-analysis of KRAS status as the determinant of response to anti-EGFR antibodies and the impact of partner chemotherapy in metastatic colorectal cancer. Eur J Cancer. 2011;47:1343–1354. doi: 10.1016/j.ejca.2011.03.031. [DOI] [PubMed] [Google Scholar]
- 13.Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 14.Jimeno A, Messersmith WA, Hirsch FR, et al. KRAS mutations and sensitivity to epidermal growth factor receptor inhibitors in colorectal cancer: practical application of patient selection. J Clin Oncol. 2009;27:1130–1136. doi: 10.1200/JCO.2008.19.8168. [DOI] [PubMed] [Google Scholar]
- 15.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 16.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 17.Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol. 2012;13:1011–1019. doi: 10.1016/S1470-2045(12)70344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist. 2013;18:865–875. doi: 10.1634/theoncologist.2013-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohno T, Tsuta K, Tsuchihara K, et al. RET fusion gene: translation to personalized lung cancer therapy. Cancer Sci. 2013;104:1396–1400. doi: 10.1111/cas.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drilon A, Wang L, Hasanovic A, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013;3:630–635. doi: 10.1158/2159-8290.CD-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gainor JF, Shaw AT. The new kid on the block: RET in lung cancer. Cancer Discov. 2013;3:604–606. doi: 10.1158/2159-8290.CD-13-0174. [DOI] [PubMed] [Google Scholar]
- 25.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gainor JF, Shaw AT. Emerging paradigms in the development of resistance to tyrosine kinase inhibitors in lung cancer. J Clin Oncol. 2013;31:3987–3996. doi: 10.1200/JCO.2012.45.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solomon B, Wilner KD, Shaw AT. Current status of targeted therapy for anaplastic lymphoma kinase-rearranged non-small cell lung cancer. Clin Pharmacol Ther. 2013;95:15–23. doi: 10.1038/clpt.2013.200. [DOI] [PubMed] [Google Scholar]
- 28.Sakamoto H, Tsukaguchi T, Hiroshima S, et al. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell. 2011;19:679–690. doi: 10.1016/j.ccr.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Seto T, Kiura K, Nishio M, et al. CH5424802 (RO5424802) for patients with ALK-rearranged advanced non-small-cell lung cancer (AF-001JP study): a single-arm, open-label, phase 1-2 study. Lancet Oncol. 2013;14:590–598. doi: 10.1016/S1470-2045(13)70142-6. [DOI] [PubMed] [Google Scholar]
- 30.Ranson MPW, Planchard D, Ohe Y, Kim SW, Kim DW, Felip E, Ghiorghiu S, Cantarini M, Cross D, Janne P. AZD9291; an irreversible potent and selective tyrosine kinase inhibitor of activating (EGFRm+) and resistance (EGFRm+/T790M+) mutations in NSCLC. J Thorac Oncol. 2013;8:S389. abs MO321.312. [Google Scholar]
- 31.Yu HA, Pao W. Targeted therapies: Afatinib--new therapy option for EGFR-mutant lung cancer. Nat Rev Clin Oncol. 2013;10:551–552. doi: 10.1038/nrclinonc.2013.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berardi R, Santoni M, Morgese F, et al. Novel small molecule EGFR inhibitors as candidate drugs in non-small cell lung cancer. Onco Targets Ther. 2013;6:563–576. doi: 10.2147/OTT.S28155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 35.Janku F, Wheler JJ, Westin SN, et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 2012;30:777–782. doi: 10.1200/JCO.2011.36.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Von Hoff DD, Stephenson JJ, Jr, Rosen P, et al. Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol. 2010;28:4877–4883. doi: 10.1200/JCO.2009.26.5983. [DOI] [PubMed] [Google Scholar]
- 37.Meric-Bernstam F, Farhangfar C, Mendelsohn J, Mills GB. Building a personalized medicine infrastructure at a major cancer center. J Clin Oncol. 2013;31:1849–1857. doi: 10.1200/JCO.2012.45.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim ES, Herbst RS, Wistuba II, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Tourneau C, Kamal M, Tredan O, et al. Designs and challenges for personalized medicine studies in oncology: focus on the SHIVA trial. Target Oncol. 2012;7:253–265. doi: 10.1007/s11523-012-0237-6. [DOI] [PubMed] [Google Scholar]
- 40.Tsimberidou AM, Iskander NG, Hong DS, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res. 2012;18:6373–6383. doi: 10.1158/1078-0432.CCR-12-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnedos M, Andre F, Farace F, et al. The challenge to bring personalized cancer medicine from clinical trials into routine clinical practice: the case of the Institut Gustave Roussy. Mol Oncol. 2012;6:204–210. doi: 10.1016/j.molonc.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.ClinicalTrials.gov. Novartis Pharmaceuticals, NCT01831726, NCT 01833169, NCT 01885195, NCT 01981187, NCT 02002689. ClinicalTrials.gov.
- 43.Conley BA. Precision Cancer Medicine; Exceptional Responders; NCI-MATCH. http://deainfo.nci.nih.gov/advisory/ctac/1113/PrecisionCancerMedicine.pdf.
- 44.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 45.Awada A, Aftimos PG. Targeted therapies of solid cancers: new options, new challenges. Curr Opin Oncol. 2013;25:296–304. doi: 10.1097/CCO.0b013e32835ff318. [DOI] [PubMed] [Google Scholar]
- 46.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vakiani E, Janakiraman M, Shen R, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol. 2012;30:2956–2962. doi: 10.1200/JCO.2011.38.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jakobsen JN, Sorensen JB. Intratumor heterogeneity and chemotherapy-induced changes in EGFR status in non-small cell lung cancer. Cancer Chemother Pharmacol. 2012;69:289–299. doi: 10.1007/s00280-011-1791-9. [DOI] [PubMed] [Google Scholar]
- 50.Narayan A, Carriero NJ, Gettinger SN, et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012;72:3492–3498. doi: 10.1158/0008-5472.CAN-11-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perkins G, Yap TA, Pope L, et al. Multi-purpose utility of circulating plasma DNA testing in patients with advanced cancers. PLoS One. 2012;7:e47020. doi: 10.1371/journal.pone.0047020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu M, Stott S, Toner M, et al. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shangary S, Wang S. Targeting the MDM2-p53 interaction for cancer therapy. Clin Cancer Res. 2008;14:5318–5324. doi: 10.1158/1078-0432.CCR-07-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maurer T, Garrenton LS, Oh A, et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci U S A. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ostrem JM, Peters U, Sos ML, et al. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebeling FG, Stieber P, Untch M, et al. Serum CEA and CA 15-3 as prognostic factors in primary breast cancer. Br J Cancer. 2002;86:1217–1222. doi: 10.1038/sj.bjc.6600248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okamoto T, Nakamura T, Ikeda J, et al. Serum carcinoembryonic antigen as a predictive marker for sensitivity to gefitinib in advanced non-small cell lung cancer. Eur J Cancer. 2005;41:1286–1290. doi: 10.1016/j.ejca.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 59.Jürgensmeier JM, Schmoll HJ, Robertson JD, et al. Prognostic and predictive value of VEGF, sVEGFR-2 and CEA in mCRC studies comparing cediranib, bevacizumab and chemotherapy. Br J Cancer. 2013;108:1316–1323. doi: 10.1038/bjc.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanrahan EO, Ryan AJ, Mann H, et al. Baseline vascular endothelial growth factor concentration as a potential predictive marker of benefit from vandetanib in non-small cell lung cancer. Clin Cancer Res. 2009;15:3600–3609. doi: 10.1158/1078-0432.CCR-08-2568. [DOI] [PubMed] [Google Scholar]
- 61.Spencer SK, Pommier AJ, Morgan SR, et al. Prognostic/predictive value of 207 serum factors in colorectal cancer treated with cediranib and/or chemotherapy. Br J Cancer. 2013;109:2765–2773. doi: 10.1038/bjc.2013.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costa V, Aprile M, Esposito R, Ciccodicola A. RNA-Seq and human complex diseases: recent accomplishments and future perspectives. Eur J Hum Genet. 2013;21:134–142. doi: 10.1038/ejhg.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655–4669. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weichenhan D, Plass C. The evolving epigenome. Hum Mol Genet. 2013;22:R1–6. doi: 10.1093/hmg/ddt348. [DOI] [PubMed] [Google Scholar]
- 65.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 66.Chen Y, Takita J, Choi YL, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 67.Kohno T, Tsuta K, Tsuchihara K, et al. RET fusion gene: Translation to personalized lung cancer therapy. Cancer Sci. 2013 doi: 10.1111/cas.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Day E, Waters B, Spiegel K, et al. Inhibition of collagen-induced discoidin domain receptor 1 and 2 activation by imatinib, nilotinib and dasatinib. Eur J Pharmacol. 2008;599:44–53. doi: 10.1016/j.ejphar.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 69.Hammerman PS, Sos ML, Ramos AH, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov. 2011;1:78–89. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 71.Kelleher FC, O'Sullivan H, Smyth E, et al. Fibroblast growth factor receptors, developmental corruption and malignant disease. Carcinogenesis. 2013;34:2198–2205. doi: 10.1093/carcin/bgt254. [DOI] [PubMed] [Google Scholar]
- 72.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 73.Sierra JR, Tsao MS. c-MET as a potential therapeutic target and biomarker in cancer. Ther Adv Med Oncol. 2011;3:S21–35. doi: 10.1177/1758834011422557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ghiso E, Giordano S. Targeting MET: why, where and how? Curr Opin Pharmacol. 2013;13:511–518. doi: 10.1016/j.coph.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 75.Board R, Jayson GC. Platelet-derived growth factor receptor (PDGFR): a target for anticancer therapeutics. Drug Resist Updat. 2005;8:75–83. doi: 10.1016/j.drup.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 76.Neuzillet C, Tijeras-Raballand A, de Mestier L, et al. MEK in cancer and cancer therapy. Pharmacol Ther. 2014;141:160–171. doi: 10.1016/j.pharmthera.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 77.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 78.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Polivka J, Jr, Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Ther. 2013 doi: 10.1016/j.pharmthera.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 80.Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 81.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sato T, Nakashima A, Guo L, et al. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene. 2010;29:2746–2752. doi: 10.1038/onc.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hardt M, Chantaravisoot N, Tamanoi F. Activating mutations of TOR (target of rapamycin) Genes Cells. 2011;16:141–151. doi: 10.1111/j.1365-2443.2010.01482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chan JA, Zhang H, Roberts PS, et al. Pathogenesis of tuberous sclerosis subependymal giant cell astrocytomas: biallelic inactivation of TSC1 or TSC2 leads to mTOR activation. J Neuropathol Exp Neurol. 2004;63:1236–1242. doi: 10.1093/jnen/63.12.1236. [DOI] [PubMed] [Google Scholar]
- 85.Xie J, Johnson RL, Zhang X, et al. Mutations of the PATCHED gene in several types of sporadic extracutaneous tumors. Cancer Res. 1997;57:2369–2372. [PubMed] [Google Scholar]
- 86.Xie J, Murone M, Luoh SM, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 87.Takebe N, Nguyen D, Yang SX. Targeting Notch signaling pathway in cancer: Clinical development advances and challenges. Pharmacol Ther. 2014;141:140–149. doi: 10.1016/j.pharmthera.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shih Ie M, Wang TL. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res. 2007;67:1879–1882. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- 89.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8:547–566. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- 90.Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- 91.Fokas E, Prevo R, Hammond EM, et al. Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat Rev. 2014;40:109–117. doi: 10.1016/j.ctrv.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 92.Rosen EM, Pishvaian MJ. Targeting the BRCA1/2 Tumor Suppressors. Curr Drug Targets. 2014;15:17–31. doi: 10.2174/1389450114666140106095432. [DOI] [PubMed] [Google Scholar]
- 93.Hennessy BT, Timms KM, Carey MS, et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J Clin Oncol. 2010;28:3570–3576. doi: 10.1200/JCO.2009.27.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]