Abstract
Single-center studies have previously reported associations of MHC Class I Chain-Related Gene A (MICA) polymorphisms and donor-recipient MICA mismatching with graft-versus-host disease (GVHD) after unrelated donor hematopoietic cell transplantation (HCT). In this study, we investigated the association of MICA polymorphism (MICA-129, MM versus MV versus VV) and MICA mismatches after HCT with 10/10 HLA–matched (n = 552) or 9/10 (n = 161) unrelated donors. Included were adult patients with a first unrelated bone marrow or peripheral blood HCT for acute lymphoblastic leukemia, acute myeloid leukemia, or myelodysplastic syndrome that were reported to the Center for International Blood and Marrow Transplant Research between 1999 and 2011. Our results showed that neither MICA mismatch nor MICA-129 polymorphism were associated with any transplantation outcome (P < .01), with the exception of a higher relapse in recipients of MICA-mismatched HLA 10/10 donors (hazard ratio [HR], 1.7; P = .003). There was a suggestion of association between MICA mismatches and a higher risk of acute GVHD grades II to IV (HR, 1.4; P = .013) There were no significant interactions between MICA mismatches and HLA matching (9/10 versus 10/10). In conclusion, the findings in this cohort did not confirm prior studies reporting that MICA polymorphism and MICA mismatches were associated with HCT outcomes.
Keywords: MICA mismatch, HLA-B mismatch, MICA-129 polymorphism
INTRODUCTION
HLA mismatches are associated with worse clinical outcomes in unrelated hematopoietic cell transplantation (HCT) [1,2]. Reports of the clinical relevance of mismatches at other polymorphic genetic loci, such as the MHC Class I Chain-Related Gene A (MICA) gene have been contradictory. At least 2 single-center studies have reported significant associations between MICA mismatches and increased risk of severe acute graft-versus-host disease (GVHD; grades III and IV), whereas 1 registry study did not find an association [3–5]. It was also argued that because of strong linkage between MICA and HLA-B locus (D′.99484, P < .001), it is unlikely that an HLA-B–matched donor-recipient pair will be mismatched for MICA [5].
The MICA gene was described in 1994 and spans a 2 kb region within the MHC class I region of chromosome 6 [6]. It is a member of the MIC gene family, which consists of 7 members (MICA through MICG) that collectively comprise approximately 11 kb located about 46.4 kb centromeric to the HLA-B locus. MICA is considered a nonclassical MHC class I gene as opposed to classical MHC class I genes encoding HLA class I loci A, B, and C. The MICA gene is highly polymorphic, although it is less so than the HLA genes. One hundred five alleles of MICA have been reported (according to IMGT Release 3.24.0) [7]. The MICA gene encodes molecules with a domain structure similar to that of classical HLA class I molecules with 3 extracellular domains, (α1, α2, and α3), a transmembrane segment and a carboxy-terminal cytoplasmic tail [8]. MICA molecules are considered nonclassical MHC class I molecules because they are not expressed on the surface of human leukocytes and their putative peptide binding groove does not present a peptide. However, MICA molecules are expressed on endothelial cells, dendritic cells, fibroblasts, epithelial cells, and many tumors, where they are targets for both cellular and humoral immune responses [6,9]. At rest, most healthy tissues have low levels of MICA expression, but this can be significantly upregulated upon malignant transformation or under various stimuli [10,11]. Moreover, both activated CD4+ and CD8+ T cells express MICA via a nuclear factor kB-driven pathway [12].
MICA is a ligand for activating natural killer group 2, member D (NKG2D) receptor, which is expressed on the surface of natural killer (NK), NKT, CD8, and TCRɣδ T cells. NKG2D binding leads to the activation of NK cells via a Src-PI3 kinase signaling pathway that results in cytotoxicity and release of IFNγ. Allelic variants of MICA have been reported to exhibit large differences in binding affinity to NKG2D [13,14]. These MICA alleles are defined by a dimorphism of a single SNP (rs1051792 A > G) at position 454 in the third exon of the MICA gene, corresponding to amino acid 129 in the alpha-2 domain of the MICA protein [15]. MICA alleles with a methionine (M) or valine (V) have been classified as having strong or weak binding affinity for NKG2D, respectively. These variable affinities have been suggested to affect thresholds of NK cell triggering and T cell modulation and, consequently, influence clinical phenotypes in autoimmune disorders and malignancies [13,16]. It has been proposed that the influence of MICA mismatches on HCT outcomes could be due to differential strengths of binding of various MICA alleles to the cognate NKG2D activating receptor. These variable affinities may affect thresholds of NK cell triggering and T cell modulation [14]. The MICA-129 non-VV genotypes (VM/MM) were reported to be associated with chronic GVHD (cGVHD) [17].
In the present study, we investigate linkage disequilibrium (LD) using the normalized measure (D′) between MICA and the neighboring HLA-B locus, the incidence of MICA mismatches among 10/10- and HLA-B–mismatched 9/10 donor-recipient pairs, and the association between the outcomes of unrelated donor HCT and donor-recipient MICA matching as well as MICA 129 genotype.
METHODS
Study Design and Population
The study population included 713 unrelated donor HCT cases receiving a HLA-A, -B, -C, -DRB1, and DQB1–matched (10/10) (n = 552) or 9/10 HLA-B–mismatch only (n = 161) graft. We included adult and pediatric patients who were reported to the Center for International Blood and Marrow Transplant Research (CIBMTR) between 1999 and 2011 and had genomic DNA available at the CIBMTR research repository for MICA genotyping of recipients and their corresponding donors. MICA genotyping was performed and, subsequently, analysis of MICA matching between recipients and donors was conducted by Allogen Laboratories, Cleveland Clinic Foundation, using its standard MICA testing protocol. The study was limited to patients receiving a first myeloablative unrelated donor HCT for acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), and myelodysplastic syndrome (MDS). The 9/10-mismatched HCT were limited to HLA-B mismatches to maximize the rate of MICA mismatches, because of the strong LD between HLA-B and MICA loci reported in a number of populations [18–20]. All HLA typing was verified using DNA-based methods at high resolution, as previously described [21].
Outcome Definitions
The primary outcomes of interest in this study were acute GVHD (aGVHD; grades II to IV, III and IV, and gastrointestinal) and cGVHD (limited and extensive). Grades II to IV and III to IV a GVHD were defined by the Glucksberg scale, and cGVHD was defined as limited or extensive cGVHD according to the Seattle criteria [22,23]. Secondary outcomes analyzed included overall survival (OS), disease relapse, disease-free survival (DFS), treatment-related mortality (TRM), and neutrophil engraftment. OS was defined as time from HCT to death from any cause. Relapse and DFS were defined per CIBMTR criteria [24]. TRM was defined as death in continuous remission from the primary malignancy. Engraftment was defined as achieving an absolute neutrophil count of 500/mL for 3 consecutive days.
MICA Genotyping and Analysis of the Linkage between MICA and HLA-B loci
MICA genotyping was performed on genomic DNA samples using commercially available Luminex based reverse sequence–specific oligonucleotide probes kits (One Lambda Thermo Fisher Scientific, Canoga Park, CA). MICA genotypes were utilized to determine the MICA-129 M/V genotype and the donor-recipient MICA mismatches. Allele frequencies of MICA were calculated by direct gene counting. Deviations from Hardy-Weinberg equilibrium (HWE) proportions were assessed at the allele-family level (first nomenclature field) using a modified version of the Guo and Thompson algorithm as implemented in the software Pypop [25,26]. Two-loci MICA~HLA-B haplotype frequencies were estimated on self-reported European-Caucasian donors (n = 623), resolving phase and allelic ambiguity using an expectation-maximization algorithm designed to handle mixed resolution data [27]. LD was estimated between MICA and HLA-B alleles using the normalized measure D′ [28].
Statistical Analysis
Sample size
The target sample size was determined to be 700 donor-recipient pairs based on the desired statistical power of 80% and 2-sided log-rank test at significance level of .05. This calculation assumed overall cumulative 100-day incidence of aGVHD II to IV of 38% for MICA-matched and 51% for single MICA–mismatched and cumulative 100-day incidence of aGVHD III and IV of 26% for MICA-matched with a hazard ratio (HR) of ≥1.5 for MICA mismatched versus matched. It also assumed that MICA mismatches occurred in 10% of 10/10 pairs in the study population and higher in 9/10 pairs with mismatches at HLA-B locus. The power calculation for the MICA-129 M/V polymorphism was determined based on a previously published frequency of MICA-129 genotype VV of approximately 50% (versus 50% for VM/MM) in a study population with a 1% incidence of cGVHD for VV versus 26% for VM/MM 1 year after transplantation [17].
Variables analyzed
The main explanatory variables in our analysis are MICA mismatches and MICA-129 V/M dimorphism (donor and recipient MM versus MV versus VV) and different donor-recipient combinations. Patient-related variables included age at time of transplantation, gender, and Karnofsky score. Disease-related variables included disease (ALL, AML, or MDS) and disease status (early versus intermediate versus advanced versus others). Transplantation-related variables included source of hematopoietic cells (bone marrow versus peripheral blood stem cells), donor age, year of transplantation, gender match (male-male versus male-female versus female-male versus female-female), donor-recipient cytomegalovirus status (−/− versus −/+ versus +/− versus +/+ versus unknown), conditioning regimen (myeloablative versus reduced-intensity/nonmyeloablative), and GVHD prophylaxis (tacrolimus ± others versus cyclosporine A ± others versus others).
Descriptive statistics, univariable, and multivariable analyses
Descriptive statistics included medians and ranges for continuous variables and frequencies for categorical variables. Multivariable models were built for OS, DFS, relapse, TRM, aGVHD (grades II to IV and III and IV), and cGVHD using the Cox proportional hazards model. All the clinical variables were tested for the affirmation of the proportional hazards assumption (P < .01). Factors violating the proportional hazards assumption were adjusted through stratification. A stepwise variable selection procedure was then used to select adjusted clinical variables for each outcome with a threshold of .05 for both entry and retention in the model. Then the association of MICA polymorphism and number of MICA mismatches with clinical outcomes was tested with adjustments for the selected clinical variables. Interactions between MICA polymorphism and HLA mismatches were tested and no significant interactions were detected. For MICA polymorphism and mismatches, a P value of < .01 was considered significant to adjust for multiple testing. SAS version 9.4 (SAS Institute, Cary, NC) was used for all the analyses.
RESULTS
Study Population
A cohort of 713 patients was included in the analysis. The distribution of patient-related variables among the cohort split by MICA matched versus mismatched pairs is summarized in Table 1. The distribution of donor and transplantation-related variables among the cohort split by MICA-matched versus mismatched pairs is summarized in Table 2. The majority received peripheral blood stem cell grafts (75%) after myeloablative conditioning (70%) and were male (53%) and Caucasian (91%) with a median age of 49 years. AML (55%) was the most common transplantation indication, followed by MDS (29%) and ALL (16%), and most patients had early stage disease (70%). All patients received calcineurin inhibitor–based GVHD prophylaxis, with a tacrolimus-based regimen as the most common (76%).
Table 1.
Patient Variables Split by MICA-Matched versus MICA-Mismatched within 10/10 and 9/10 HLA Matching
| Variable | MICA Matched | MICA Mismatched | P Value* |
|---|---|---|---|
| No. of patients | 522 | 191 | |
| No. of centers | 99 | 63 | |
| Recipient age at transplantation, yr | .42 | ||
| 10–19 | 12 (2) | 8 (4) | |
| 20–29 | 84 (16) | 37 (19) | |
| 30–39 | 64 (12) | 20 (10) | |
| 40–49 | 111 (21) | 34 (18) | |
| 50–59 | 251 (48) | 92 (48) | |
| Median (range) | 49 (18–74) | 49 (18–75) | .29 |
| Recipient race/ethnicity | .36 | ||
| Caucasian, non-Hispanic | 481 (94) | 169 (90) | |
| African-American, non-Hispanic | 5 (1) | 5 (3) | |
| Asian, non-Hispanic | 4 (1) | 3 (2) | |
| Native American, non-Hispanic | 2 (<1) | 0 | |
| Hispanic, Caucasian | 18 (4) | 10 (5) | |
| Hispanic, African-American | 1 (<1) | 0 | |
| Hispanic, race unknown | 1 (<1) | 1 (1) | |
| Unknown | 10 (N/A) | 3 (N/A) | |
| Recipient sex | .64 | ||
| Male | 274 (52) | 104 (54) | |
| Female | 248 (48) | 87 (46) | |
| Karnofsky score | .30 | ||
| 10–80 | 177 (37) | 58 (32) | |
| 90–100 | 305 (63) | 121 (68) | |
| Unknown | 40 (N/A) | 12 (N/A) | |
| Disease at transplantation | .67 | ||
| AML | 293 (56) | 102 (53) | |
| ALL | 83 (16) | 29 (15) | |
| MDS | 146 (28) | 60 (31) | |
| Disease status at transplantation | .97 | ||
| Early | 365 (70) | 134 (70) | |
| Intermediate | 4 (1) | 2 (1) | |
| Advanced | 133 (25) | 47 (25) | |
| Other | 20 (4) | 8 (4) |
Data presented are n (%) unless otherwise indicated.
The Pearson chi-square test was used for comparing discrete variables; the Kruskal-Wallis test was used for comparing continuous variables.
Table 2.
Summary Donor and Transplantation Variables Distribution Split by MICA Matched versus MICA Mismatched within 10/10 and 9/10 HLA Matching
| Variable | MICA Matched | MICA Mismatched | P Value* |
|---|---|---|---|
| High-resolution HLA matches available out of 10 | <.001 | ||
| 9/10 (mismatch at HLA-B) | 54 (10) | 107 (56) | |
| 10/10 | 468 (90) | 84 (44) | |
| Graft type | .12 | ||
| Marrow | 139 (27) | 40 (21) | |
| PBSC | 383 (73) | 151 (79) | |
| Conditioning regimen | .68 | ||
| Myeloablative | 359 (69) | 140 (73) | |
| RIC | 115 (22) | 37 (19) | |
| Nonmyeloablative | 29 (6) | 9 (5) | |
| Other | 19 (4) | 5 (3) | |
| Donor age at donation, yr | .05 | ||
| 10–19 | 17 (3) | 7 (4) | |
| 20–29 | 184 (35) | 52 (27) | |
| 30–39 | 174 (33) | 57 (30) | |
| 40–49 | 110 (21) | 53 (28) | |
| ≥ 50 | 37 (7) | 22 (12) | |
| Median (range) | 33 (18–61) | 37 (19–60) | .003 |
| Donor/recipient CMV serostatus | .92 | ||
| Negative/negative | 144 (28) | 58 (30) | |
| Negative/positive | 180 (34) | 63 (33) | |
| Positive/negative | 56 (11) | 19 (10) | |
| Positive/positive | 130 (25) | 48 (25) | |
| Unknown | 12 (2) | 3 (2) | |
| GVHD Prophylaxis | .45 | ||
| Tacrolimus + MMF ± others | 96 (18) | 26 (14) | |
| Tacrolimus + MTX ± others (except MMF) | 303 (58) | 117 (61) | |
| CSA + MMF ± others (except tacrolimus) | 42 (8) | 14 (7) | |
| CSA + MTX ± others (except tacrolimus, MMF) | 81 (16) | 34 (18) | |
| Donor/recipient sex match | .74 | ||
| Male/male | 191 (37) | 72 (38) | |
| Male/female | 155 (30) | 49 (26) | |
| Female/male | 83 (16) | 32 (17) | |
| Female/female | 93 (18) | 38 (20) | |
| Year of transplantation | .88 | ||
| 2000 | 10 (2) | 2 (1) | |
| 2001 | 8 (2) | 6 (3) | |
| 2002 | 17 (3) | 5 (3) | |
| 2003 | 34 (7) | 9 (5) | |
| 2004 | 36 (7) | 16 (8) | |
| 2005 | 67 (13) | 30 (16) | |
| 2006 | 71 (14) | 25 (13) | |
| 2007 | 77 (15) | 29 (15) | |
| 2008 | 67 (13) | 25 (13) | |
| 2009 | 66 (13) | 18 (9) | |
| 2010 | 43 (8) | 16 (8) | |
| 2011 | 26 (5) | 10 (5) | |
| Follow-up among survivors, mo | |||
| No. evaluated | 215 | 68 | |
| Median (range) | 51.2 (3.3–147.6) | 60.7 (11.8–119.8) | .09 |
PBSC indicates peripheral blood stem cells, RIC, reduced-intensity conditioning; CMV, cytomegalovirus; MMF, mycophenolate mofetil; MTX, methotrexate; CSA, cyclosporin A.
The Pearson chi-square test was used for comparing discrete variables; the Kruskal-Wallis test was used for comparing continuous variables.
MICA/HLA-B Frequency Analysis and LD
Because of sample size constraints for minority subjects, frequency analysis was only run on the self-reported Caucasian donor subset of the study cohort (n = 623). Table 3 reports the results of deviations from HWE proportion test. Expectedly, both MICA and HLA-B deviated from HWE proportions in this study cohort since the criteria for random selection were not met. These results partly support prior assertions of tight LD [5].
Table 3.
Deviation from HWE Proportion Chi-Square Test
| Locus | Homozygotes
|
Heterozygotes
|
||||
|---|---|---|---|---|---|---|
| Observed | Expected | P Value | Observed | Expected | P Value | |
| MICA | 47 | 122.745 | .000000 | 576 | 500.255 | .000708 |
| HLA-B | 12 | 71 | .000000 | 610 | 551 | .011967 |
Haplotype Frequency
Among 623 Caucasian donors, 62 unique MICA-HLA-B haplotypes were identified (Supplementary Table S1). Haplotype frequency ranged from 23.29% to <.01%. The 4 most frequent haplotypes (MICA*002~HLA-B*35:01, 23.29%; MICA*009~HLA-B*51:01, 12:02%; MICA*008 ~ HLA-B*07:02, 8.88%; MICA*008 ~ HLA-B*08:01, 6.51%) were represented in >50% of the donors analyzed. The remaining haplotypes were represented at fairly low frequency (.05% to 4.97%) and 14 haplotypes were each observed in only 1 subject.
LD
Figure 1 represents a heat map of the relative LD for different MICA~HLA-B haplotypes. Only 6 MICA~HLA-B haplotypes were observed at D′ > .40 (MICA*002~HLA-B*35:01, MICA*008 ~ HLA-B*35:01, MICA*009 ~ HLA-B*51:01, MICA*002 ~ HLA-B*51:01, MICA*008 ~ HLA-B*51:01 and MICA*008 ~ HLA-B*07:02) and cumulatively represented 44.7% of the overall haplotype frequency in the used donor cohort. Sixteen MICA~HLA-B haplotypes were observed at D′ <.10. These results are not consistent with universal tight LD between alleles of the HLA-B and MICA loci.
Figure 1.
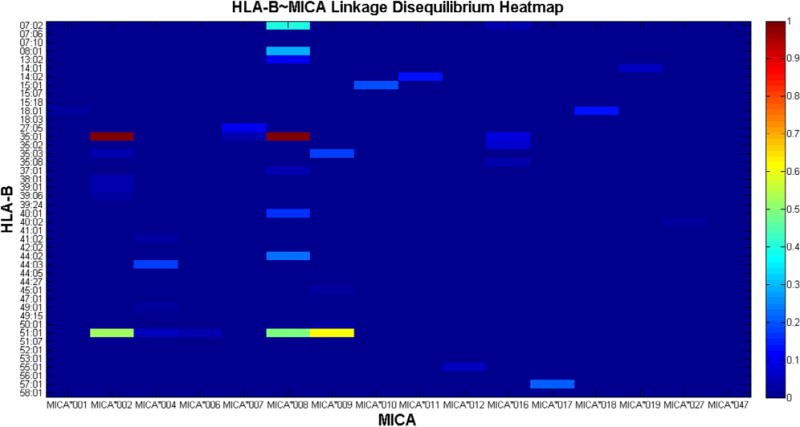
Heat map of MICA~HLA-B haplotype linkage disequilibrium (LD) expressed in terms of the D′ measure. The abscissa and ordinate represent MICA and HLA-B alleles respectively. The right color-bar represents the range of D′ (0–1). Cool colors represent lower values of D′ while warm colors demonstrate high haplotype LD. (This figure is available in color online at www.bbmt.org.)
Donor-Recipient MICA Mismatches
MICA mismatches occurred in 191 out of the 713 (27%) of the pairs, of which 182 (26%) pairs had single MICA mismatch (83 were 10/10 and 99 were 9/10) and 9 pairs (1%) had double MICA mismatches (1 10/10 and 8 9/10). In the 10/10 HLA–matched pairs(n = 552), MICA mismatches were significantly less frequent than in the 9/10 HLA–matched pairs (n = 161) (15% versus 66%, respectively).
Association between Donor-Recipient MICA Mismatches and Clinical Outcomes
The incidence of aGVHD grades III and IV in the MICA-matched patients in this cohort was 16%. In univariable analysis, there was no significant association (at level of P < .01) between MICA mismatches and 1-year, 3-year, or 5-year OS, DFS, or TRM (Table 4). There was also no association with neutrophil engraftment, 100-day incidence of GVHD (II to IV or III and IV), or cGVHD (at 1 or 2 years). Table 5 shows that in multivariable analysis, MICA mismatches remained nonsignificant for all these outcomes with the exception of an unexpected finding of significantly higher relapse in association with MICA mismatches in 10/10 patients (HR, 1.7; 95% confidence interval [CI], 1.2 to 2.4; P = .003) but not in 9/10 patients (Figure 2). There was a suggestion of an association between MICA mismatches and aGVHD grades II to IV (HR, 1.4; 95% CI, 1.1 to 1.9; P = .013) but not with grades III and IV (Figure 3). A post-hoc power calculation determined that based on the aGVHD rates in the study cohort, the power to detect the MICA effect for aGVHD grades II to IV and III and IV were 85% and 11%, respectively. There was also a suggestion of an association between MICA mismatches and cGVHD in 7/8 HLA–matched transplantations (HR, 1.8; 95% CI, 1.0 to 3.1; P = .04) (Table 6). There was no evidence of significant interactions between MICA mismatches and HLA matching (9/10 versus 10/10).
Table 4.
Univariate Results: MICA Mismatching Univariate Probability of Outcomes after Checking Bidirectional MICA Mismatch Status
| Outcome | MICA Matched
|
Single MICA MM
|
Double MICA MM
|
Pointwise
|
|||
|---|---|---|---|---|---|---|---|
| n | Prob (95% CI) | n | Prob (95% CI) | n | Prob (95% CI) | P Value | |
| Survival | 522 | 182 | 9 | 0.21* | |||
| 1 Yr | 57 (53–61) | 52 (45–59) | 67 (35–92) | .44 | |||
| 3 Yr | 44 (39–48) | 39 (32–46) | 67 (35–92) | .15 | |||
| 5 Yr | 38 (34–43) | 33 (26–41) | 67 (35–92) | .09 | |||
| DFS | 519 | 182 | 9 | 0.13* | |||
| 1 Yr | 47 (43–51) | 43 (36–50) | 67 (35–92) | .27 | |||
| 3 Yr | 38 (34–42) | 33 (26–40) | 67 (35–92) | .08 | |||
| 5 Yr | 32 (27–36) | 28 (21–35) | 67 (35–92) | .05 | |||
| Relapse | 519 | 182 | 9 | ||||
| 1 Yr | 31 (28–36) | 32 (26–39) | 0 (.–.) | ||||
| 3 Yr | 35 (31–40) | 36 (30–44) | 0 (.–.) | ||||
| 5 Yr | 38 (34–43) | 39 (32–46) | 0 (.–.) | ||||
| TRM | 519 | 182 | 9 | ||||
| 1 Yr | 22 (18–25) | 25 (19–31) | 33 (8–65) | .55 | |||
| 3 Yr | 27 (23–31) | 31 (24–38) | 33 (8–65) | .57 | |||
| 5 Yr | 30 (26–34) | 34 (27–41) | 33 (8–65) | .70 | |||
| Neutrophil engraftment | 521 | 182 | 9 | ||||
| 28 D | 93 (91–95) | 94 (90–97) | 100 (.–.) | ||||
| 42 D | 95 (94–97) | 95 (91–98) | 100 (.–.) | ||||
| Grades II–IV aGVHD | 520 | 182 | 9 | ||||
| 100 D | 38 (33–42) | 51 (43–58) | 44 (15–76) | .01 | |||
| Grades III and IV aGVHD | 521 | 182 | 9 | ||||
| 100 D | 16 (13–19) | 18 (13–24) | 33 (8–65) | .49 | |||
| cGVHD | 515 | 181 | 8 | ||||
| 1 Yr | 46 (42–51) | 54 (46–61) | 50 (18–82) | .21 | |||
| 2 Yr | 52 (48–57) | 58 (51–66) | 75 (42–97) | .15 | |||
Prob indicates probability (%).
Log-rank P-value.
Table 5.
Multivariate Results: MICA Mismatching
| Outcome | MICA Match (ref)
|
MICA Mismatch
|
|||
|---|---|---|---|---|---|
| n | HR (ref) | N | HR (95% CI) | P Value | |
| OS | 518 | 1.00 | 180 | 1.14 (.92–1.41) | .23 |
| DFS | 515 | 1.00 | 180 | 1.13 (.93–1.40) | .22 |
| Relapse | 515 | 1.00 | 180 | 1.10 (.83–1.46) | .51 |
| TRM | 515 | 1.00 | 180 | .91 (.63–1.30) | .59 |
| aGVHD II–IV | 535 | 1.00 | 163 | 1.44 (1.08-1.91) | .01 |
| aGVHD III–IV | 541 | 1.00 | 164 | 1.04 (.66–1.63) | .88 |
| cGVHD | 535 | 1.00 | 163 | 1.10 (.86–1.4) | .47 |
| Neutrophil engraftment | 534 | 1.00 | 167 | 1.02 (.85–1.22) | .88 |
Ref indicates reference.
Figure 2.
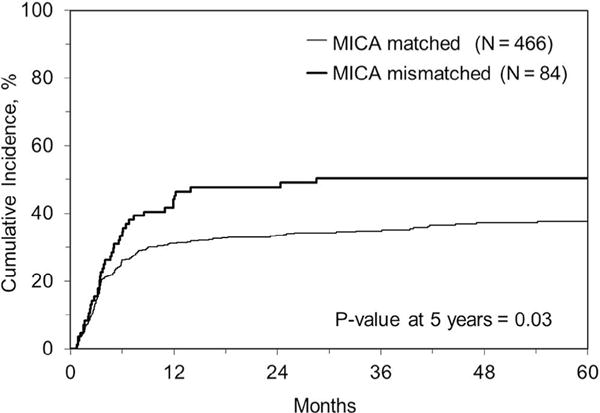
Cumulative incidence of relapse in MICA-matched versus mismatched cases in the HLA 10/10–matched subset.
Figure 3.
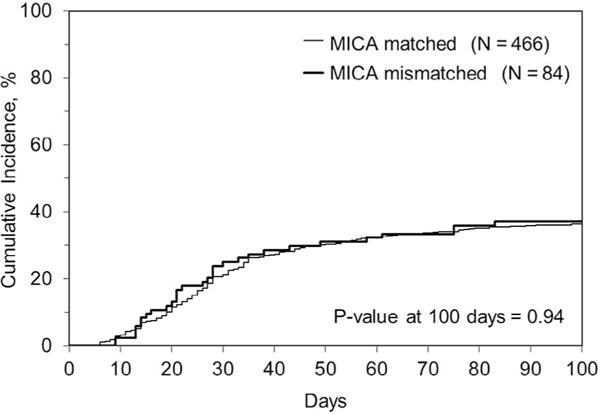
Cumulative incidence of acute GVHD grades II to IV in MICA-matched versus mismatched cases in the HLA 10/10–matched subset.
Table 6.
Association between MICA Mismatches for cGVHD in the Subset of 7/8 HLA–Matched Transplantations
| Factor | Level | Count | Event | HR | HR_Low | HR_Up | P Value |
|---|---|---|---|---|---|---|---|
| *Overall | . | . | . | . | . | .0361 | |
| MICA matches | 0 | 66 | 27 | 1.000 | . | . | . |
| MICA mismatches | 1 | 92 | 54 | 1.797 | 1.039 | 3.108 | .0361 |
Donor and Recipient MICA-129 M/V Genotypes
The distribution of MICA-129 M/V genotypes was comparable among donors and recipients. Of the recipients, 101 were MICA-129 MM (14%), 363 were MV (52%), and 239 were VV (34%). Of the donors, 106 were MICA-129 MM (15%), 375 were MV (53%), and 229 were VV (32%). The distribution of VV genotypes differed in this population compared with those reported in prior publications at 34% and 50%, respectively [16].
Association between MICA-129 Polymorphism and Clinical Outcomes
On univariable analysis, there were no significant associations between recipient (Supplemental Table S1) or donor (Supplemental Table S2) MICA-129 M/V genotype and 1-year, 3-year, or 5-year OS, DFS, or TRM. There was also no association with neutrophil engraftment, aGVHD (II to IV or III and IV), or cGVHD (at 1 or 2 years). The incidences of aGVHD III and IV were 16% for VV and 17% for VM/MM. There were also no significant associations found in multivariable analyses between any outcome and MICA-129 polymorphism (data not shown). There was a suggestion of an association between donor MICA-129 non-VV genotypes and slower platelet engraftment with HR of 1.4 (95% CI, 1.109 to 1.985; P = .02).
DISCUSSION
The impact of donor-recipient matching at HLA loci A, B, C, DRB1, DQB1, and DPB1 on clinical outcomes of unrelated HCT has been established by numerous studies [1,2,29,30]. However, the effect of MICA donor-recipient mismatches remains controversial. In the univariate and multivariate analyses, there is a trend of association between MICA and grade II to IV aGVHD; this association may be due to a weaker effect in determining risk for GVHD. In addition, the clinical relevance of matching for MICA in the context of HLA-matched HCT donor-recipient pairs has been questioned based on the putative strong LD between MICA and HLA-B [5] reported in different populations [18–20]. Although the global LD between the 2 loci is significant, it was reported that a limited number of HLA-B alleles showed significant LD with a specific MICA allele [19]. In the present study, some of the estimated MICA~HLA-B haplotypes showed significant LD (>D′ = .4), but these haplotypes represented approximately 45% of the total haplotype frequency in the analyzed cohort. In contrast, for haplotypes composing approximately 20% of the total haplotype frequencies, there was no appreciable LD between MICA and HLA-B (D′ <.1). Therefore, the notion that HCT donor-recipient matching for HLA-B alleles limits the odds of mismatches at MICA alleles due to the putative strong LD was not supported by the findings of our study. These results are also supported by 2 prior studies of the impact of MICA mismatches on GVHD after unrelated donor HCT, in which the incidence of MICA mismatches were 12.7% and 8.5% [3,4]. The rate of MICA mismatches in the present study and the 2 previous studies contrasts with the significantly lower mismatch rate of 2.6% seen in another CIBMTR study [5]. This remarkably low rate of mismatch may be at least partially explained by the selection criteria of that study limiting the analysis to only 38 transplantation pairs that were 12/12 HLA–matched (matched at loci HLA-A, B, C, DRB1, DQB1, and DPB1). It is estimated that only about 10% to 15 % of 8/8 HLA–matched donors are matched at HLA-DPB1 because of weak LD between HLA-DPB1 and the other HLA class II loci [31]. Therefore, limiting inclusion to a subset of remarkably matched pairs would result in a higher than expected level of HLA haplotype homogeneity with lower MICA mismatch rate.
In our study population, the incidence aGVHD III and IV was 16% for VV and 17% for VM/MM, compared with 1% and 26% reported in the prior publication [16]. In the current cohort, the largest studied to date, none of the investigated outcomes were significantly associated with MICA mismatching and the MICA-129 polymorphism VV, with the exception of an unexpected finding of significantly higher relapse in association with MICA mismatches in 10/10 patients. We observed a suggestion of an association between MICA mismatches and aGVHD grades II to IV and cGVHD, but these findings were not previously observed and may be spurious. The lack of association between MICA mismatches and grade III and IV acute GVHD in this registry analysis is discordant with prior reports from single center studies [3,4]. These results, however, are consistent with those of a prior smaller CIBMTR study that showed no association between MICA mismatches and any clinical outcome [5].
There are a number of possible explanations for the discordant results. The current study is more homogeneous and includes only patients with ALL, AML, and MDS compared with prior studies that included all diagnoses. The effect of MICA mismatching may be more pronounced in the diagnoses not included in the current study. Another possible explanation is the heterogeneity introduced by inclusion of many centers in the current CIBMTR study compared to single-center studies, particularly in the scoring of aGVHD among different centers compared to being assigned more consistently in the single-center studies. Finally, the unexpected lower incident rate of GVHD in the study population compared to what was assumed for the power calculation may have rendered the study underpowered to detect statistically significant associations.
The finding of a significantly higher relapse rate in association with MICA mismatches in 10/10 patients is unexpected, as we hypothesized that MICA mismatches would lead to higher rates of GVHD, correlating with better relapse control due to an enhanced graft-versus-leukemia effect. The lack of an association between MICA mismatches and GVHD may suggest that MICA-related alloreactivity may be insufficient to cause clinically relevant GVHD or relapse control. However, our results contradict a recently published study that showed a lower risk for relapse in MICA-mismatched 10/10 allele–matched HCT pairs of multiple diagnoses suggesting a possible graft-versus-leukemia effect [32]. In vitro studies have suggested a mechanism by which overexpression of MICA/B molecules on chronic myeloid leukemia CD34+ cells and their ability to bind the NK activation ligand NKG2D to mediate leukemia-specific cytotoxicity [33]. Further, in vivo mechanistic studies are needed to elucidate the contribution of MICA mismatches to graft-versus-leukemia effect.
It is noteworthy that opposite effects on outcome after unrelated donor transplantation depending on the degree of other HLA allele matching has been reported previously. In 1 study, HLA-DPB1 matching was significantly associated with improved survival in 10/10 HLA–matched patients but worse survival in 9/10-matched cases [34]. Similarly, Pidala et al. reported HLA-DPB1 mismatch was associated with increased risk for aGVHD and decreased relapse compared with HLA-DPB1 allele–matched cases among 8/8 HL–matched cases; however, among 7/8-matched cases, no significant effects of HLA-DPB1 mismatch were observed [2]. There was no evidence of significant interactions between MICA mismatches and the degree of HLA matching (9/10 versus 10/10) in our study or mismatches at HLA-DPB1 locus.
In our study, the observed distribution of MICA-129 genotypes MM, MV, and VV was comparable in recipients and donors. These distributions are not significantly different from what was reported previously by Boukouaci et al. [17]. In the previous study, recipient homozygosity of V alleles (VV) was associated with cGVHD. In the present study, neither donor nor recipient MICA-129 genotypes were significantly associated with any analyzed outcome except for a suggestion of an association between MICA- 129 non-VV genotypes (MM and MV) and slower platelet engraftment with HR of 1.4 (95% CI, 1.109 to 1.985; P = .02). Although the observed association was not significant, one could hypothesize that MM and MV alleles with stronger interactions with NKG2D receptors would lead to increased donor NK cell activation and production of granulocyte macrophage colony–stimulating factor with increased granulocyte macrophage and faster engraftment [35].
In conclusion, the study results show that neither MICA-129 polymorphism nor MICA mismatches was associated with HCT outcomes at a level that reached the predetermined level of statistical significance. This registry analysis in patients with ALL, AML, and MDS was discordant with prior reports of single-center studies of patients with mixed diagnoses.
Supplementary Material
Acknowledgments
This study was partially supported by grant #IRG-91-022-18 to the Case Comprehensive Cancer Center from the American Cancer Society, Allogen Laboratories, Cleveland Clinic Foundation and funds from the US National Marrow Donor Program (NMDP) Immunobiology Research Grant Program. The authors acknowledge Michael George from the National Marrow Donor Program Bioinformatics Research Department for his valuable input related to the haplotype frequency analysis. The CIBMTR is supported by Public Health Service Grant/Cooperative Agreement 5U24-CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); a grant/cooperative agreement 5U10HL069294 from NHLBI and NCI; a contract HHSH250201200016C with Health Resources and Services Administration (HRSA/DHHS); 2 grants N00014-15-1-0848 and N00014-16-1-2020 from the Office of Naval Research; and grants from Alexion; *Amgen, Inc.; anonymous donation to the Medical College of Wisconsin; Astellas Pharma US; AstraZeneca; Be the Match Foundation; *Bluebird Bio, Inc.; *Bristol Myers Squibb Oncology; *Celgene Corporation; Cellular Dynamics International, Inc.; *Chimerix, Inc.; Fred Hutchinson Cancer Research Center; Gamida Cell Ltd.; Genentech, Inc.; Genzyme Corporation; *Gilead Sciences, Inc.; Health Research, Inc. Roswell Park Cancer Institute; HistoGenetics, Inc.; Incyte Corporation; Janssen Scientific Affairs, LLC; *Jazz Pharmaceuticals, Inc.; Jeff Gordon Children’s Foundation; The Leukemia & Lymphoma Society; Medac, GmbH; MedImmune; The Medical College of Wisconsin; *Merck & Co, Inc.; Mesoblast; MesoScale Diagnostics, Inc.; *Miltenyi Biotec, Inc.; National Marrow Donor Program; Neovii Biotech NA, Inc.; Novartis Pharmaceuticals Corporation; Onyx Pharmaceuticals; Optum Healthcare Solutions, Inc.; Otsuka America Pharmaceutical, Inc.; Otsuka Pharmaceutical Co, Ltd. – Japan; PCORI; Perkin Elmer, Inc.; Pfizer, Inc; *Sanofi US; *Seattle Genetics; *Spectrum Pharmaceuticals, Inc.; St. Baldrick’s Foundation; *Sunesis Pharmaceuticals, Inc.; Swedish Orphan Biovitrum, Inc.; Takeda Oncology; Telomere Diagnostics, Inc.; University of Minnesota; and *Wellpoint, Inc. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, Health Resources and Services Administration or any other agency of the U. Government.
*Corporate Members.
APPENDIX. SUPPLEMENTARY DATA
Supplementary data related to this article can be found online at doi:10.1016/j.bbmt.2016.11.021.
Footnotes
Conflict of interest statement: There are no conflicts of interest to report.
Authorship statement: MA, R.S., T.W., M.H., S.J.L., and S.R.S. drafted the research plan; N.M., A.M., K.F., K.H., M.V., and M.F.V. critically revised the research plan; D.T. and A.Z. performed MICA genetic typing on samples; T.W. and M.H. performed statistical analysis; M.A., R.S., S.J.L., and S.R.S. analyzed and interpreted data; M.A., R.S., T.W., M.H., S.J.L., and S.R.S. drafted the paper; and N.M., A.M., D.T., A.Z., K.F., K.H., M.V., and M.F.V. critically revised the paper.
References
- 1.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110:4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 2.Pidala J, Lee SJ, Ahn KW, et al. Nonpermissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood. 2014;124:2596–2606. doi: 10.1182/blood-2014-05-576041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Askar M, Sun Y, Rybicki L, et al. Synergistic effect of major histocompatibility complex class I-related chain A and human leukocyte antigen-DPB1 mismatches in association with acute graft-versus-host disease after unrelated donor hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2014;20:1835–1840. doi: 10.1016/j.bbmt.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 4.Parmar S, Del Lima M, Zou Y, et al. Donor-recipient mismatches in MHC class I chain-related gene A in unrelated donor transplantation lead to increased incidence of acute graft-versus-host disease. Blood. 2009;114:2884–2887. doi: 10.1182/blood-2009-05-223172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson E, Grzywacz B, Wang H, et al. Limited role of MHC class I chain-related gene A (MICA) typing in assessing graft-versus-host disease risk after fully human leukocyte antigen-matched unrelated donor transplantation. Blood. 2009;114:4753–4754. doi: 10.1182/blood-2009-08-239301. author reply 4754–4755. [DOI] [PubMed] [Google Scholar]
- 6.Bahram S, Bresnahan M, Geraghty DE, Spies T. A second lineage of mammalian major histocompatibility complex class I genes. Proc Natl Acad Sci USA. 1994;91:6259–6263. doi: 10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43:D423–D431. doi: 10.1093/nar/gku1161. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahram S. MIC genes: from genetics to biology. Adv Immunol. 2000;76:1–60. doi: 10.1016/s0065-2776(01)76018-x. [DOI] [PubMed] [Google Scholar]
- 9.Zou Y, Stastny P. Role of MICA in the immune response to transplants. Tissue Antigens. 2010;76:171–176. doi: 10.1111/j.1399-0039.2010.01527.x. [DOI] [PubMed] [Google Scholar]
- 10.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schilling D, Kuhnel A, Tetzlaff F, Konrad S, Multhoff G. NZ28-induced inhibition of HSF1, SP1 and NF-kappaB triggers the loss of the natural killer cell-activating ligands MICA/B on human tumor cells. Cancer Immunol Immunother. 2015;64:599–608. doi: 10.1007/s00262-015-1665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molinero LL, Fuertes MB, Girart MV, et al. NF-kappa B regulates expression of the MHC class I-related chain A gene in activated T lymphocytes. J Immunol. 2004;173:5583–5590. doi: 10.4049/jimmunol.173.9.5583. [DOI] [PubMed] [Google Scholar]
- 13.Douik H, Ben Chaaben A, Attia Romdhane N, et al. Association of MICA-129 polymorphism with nasopharyngeal cancer risk in a Tunisian population. Hum Immunol. 2009;70:45–48. doi: 10.1016/j.humimm.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Steinle A, Li P, Morris DL, et al. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics. 2001;53:279–287. doi: 10.1007/s002510100325. [DOI] [PubMed] [Google Scholar]
- 15.Stephens HA. MICA and MICB genes: can the enigma of their polymorphism be resolved? Trends Immunol. 2001;22:378–385. doi: 10.1016/s1471-4906(01)01960-3. [DOI] [PubMed] [Google Scholar]
- 16.Amroun H, Djoudi H, Busson M, et al. Early-onset ankylosing spondylitis is associated with a functional MICA polymorphism. Hum Immunol. 2005;66:1057–1061. doi: 10.1016/j.humimm.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Boukouaci W, Busson M, Peffault de Latour R, et al. MICA-129 genotype, soluble MICA, and anti-MICA antibodies as biomarkers of chronic graft-versus-host disease. Blood. 2009;114:5216–5224. doi: 10.1182/blood-2009-04-217430. [DOI] [PubMed] [Google Scholar]
- 18.Gao X, Single RM, Karacki P, Marti D, O’Brien SJ, Carrington M. Diversity of MICA and linkage disequilibrium with HLA-B in two North American populations. Hum Immunol. 2006;67:152–158. doi: 10.1016/j.humimm.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Wenda S, Fae I, Sanchez-Mazas A, Nunes JM, Mayr WR, Fischer GF. The distribution of MICA alleles in an Austrian population: evidence for increasing polymorphism. Hum Immunol. 2013;74:1295–1299. doi: 10.1016/j.humimm.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Tian W, Cai J, Liu X. MICA genetic polymorphism and HLA-A,C,B,MICA and DRB1 haplotypic variation in a southern Chinese Han population: identification of two new MICA alleles, MICA*060 and MICA*062. Hum Immunol. 2011;72:510–515. doi: 10.1016/j.humimm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Spellman S, Setterholm M, Maiers M, et al. Advances in the selection of HLA-compatible donors: refinements in HLA typing and matching over the first 20 years of the National Marrow Donor Program Registry. Biol Blood Marrow Transplant. 2008;14(9 suppl):37–44. doi: 10.1016/j.bbmt.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Glucksberg H, Storb R, Fefer A, et al. Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation. 1974;18:295–304. doi: 10.1097/00007890-197410000-00001. [DOI] [PubMed] [Google Scholar]
- 23.Shulman HM, Sullivan KM, Weiden PL, et al. Chronic graft-versus-host syndrome in man. A long-term clinicopathologic study of 20 Seattle patients. Am J Med. 1980;69:204–217. doi: 10.1016/0002-9343(80)90380-0. [DOI] [PubMed] [Google Scholar]
- 24.Flomenberg N, Baxter-Lowe LA, Confer D, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104:1923–1930. doi: 10.1182/blood-2004-03-0803. [DOI] [PubMed] [Google Scholar]
- 25.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- 26.Lancaster AK, Single RM, Solberg OD, Nelson MP, Thomson G. PyPop update–a software pipeline for large-scale multilocus population genomics. Tissue Antigens. 2007;69(suppl 1):192–197. doi: 10.1111/j.1399-0039.2006.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kollman C, Maiers M, Gragert L, et al. Estimation of HLA-A, -B, -DRB1 haplotype frequencies using mixed resolution data from a National Registry with selective retyping of volunteers. Hum Immunol. 2007;68:950–958. doi: 10.1016/j.humimm.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Lewontin RC. The interaction of selection and linkage. I. general considerations; heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciurea SO, Saliba RM, Rondon G, et al. Outcomes of patients with myeloid malignancies treated with allogeneic hematopoietic stem cell transplantation from matched unrelated donors compared with one human leukocyte antigen mismatched related donors using HLA typing at 10 loci. Biol Blood Marrow Transplant. 2011;17:923–929. doi: 10.1016/j.bbmt.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleischhauer K, Shaw BE, Gooley T, et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13:366–374. doi: 10.1016/S1470-2045(12)70004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurley CK, Baxter-Lowe LA, Begovich AB, et al. The extent of HLA class II allele level disparity in unrelated bone marrow transplantation: analysis of 1259 National Marrow Donor Program donor-recipient pairs. Bone Marrow Transplant. 2000;25:385–393. doi: 10.1038/sj.bmt.1702161. [DOI] [PubMed] [Google Scholar]
- 32.Carapito R, Jung N, Kwemou M, et al. Matching for the nonconventional MHC-I MICA gene significantly reduces the incidence of acute and chronic GVHD. Blood. 2016;128:1979–1986. doi: 10.1182/blood-2016-05-719070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sconocchia G, Lau M, Provenzano M, et al. The antileukemia effect of HLA-matched NK and NK-T cells in chronic myelogenous leukemia involves NKG2D-target-cell interactions. Blood. 2005;106:3666–3672. doi: 10.1182/blood-2005-02-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw BE, Mayor NP, Russell NH, et al. Diverging effects of HLA-DPB1 matching status on outcome following unrelated donor transplantation depending on disease stage and the degree of matching for other HLA alleles. Leukemia. 2009;24:58–65. doi: 10.1038/leu.2009.239. [DOI] [PubMed] [Google Scholar]
- 35.Muller JR, Waldmann TA, Dubois S. Loss of cytotoxicity and gain of cytokine production in murine tumor-activated NK cells. PLoS ONE. 2014;9:e102793. doi: 10.1371/journal.pone.0102793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


