Abstract
Longitudinal electronic health records on 99,785 Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort individuals provided 1,342,814 systolic and diastolic blood pressure measurements for a genome-wide association study on long-term average systolic, diastolic, and pulse pressure. We identified 39 novel among 75 significant loci (P≤5×10−8), most replicating in the combined International Consortium for Blood Pressure (ICBP, n=69,396) and UK Biobank (UKB, n=152,081) studies. Combining GERA with ICBP yielded 36 additional novel loci, most replicating in UKB. Combining all three studies (n=321,262) yielded 241 additional genome-wide significant loci, although for these no replication sample was available. All associated loci explained 2.9%/2.5%/3.1% of systolic/diastolic/pulse pressure variation in GERA non-Hispanic whites. Using multiple BP measurements in GERA doubled the variance explained. A normalized risk score was associated with time-to-onset of hypertension (hazards ratio=1.18, P=10−44). Expression quantitative trait locus analysis of BP loci showed enrichment in aorta and tibial artery.
Keywords: blood pressure, hypertension, genome-wide association study, electronic health records
Blood pressure (BP) is an important cardiovascular risk factor1, with estimated 30-50% heritability2,3. Over the past several years, genome-wide association studies (GWAS) have identified 85 BP SNPs4–22. However, the heritability explained remains less than other quantitative cardiovascular traits, e.g., lipids23. Three strategies to identify additional variants are the use of: larger sample sizes, more precise measurements, and more extensive imputation panels. To date, all large studies have used measurements from research protocols rather than clinical records. There is little doubt that the phenotype observed in observational research or randomized trials is similar to a clinical encounter, but clinical measures may be influenced by somewhat different circumstances and measurements may be obtained under a less stringent protocol24. However, studies using clinical measurements from electronic health records (EHR) permit not only very large sample sizes, but also a long-term average of multiple independent clinical measurements from many different visits, yielding reduced phenotype variance (as shown by simulation and experimental data)7. We therefore reasoned a large-sample BP GWAS with longitudinal EHR-based measures would provide improved statistical power and understanding of BP genomic architecture, which we show theoretically (Online Methods) and through data application.
Results
GERA cohort
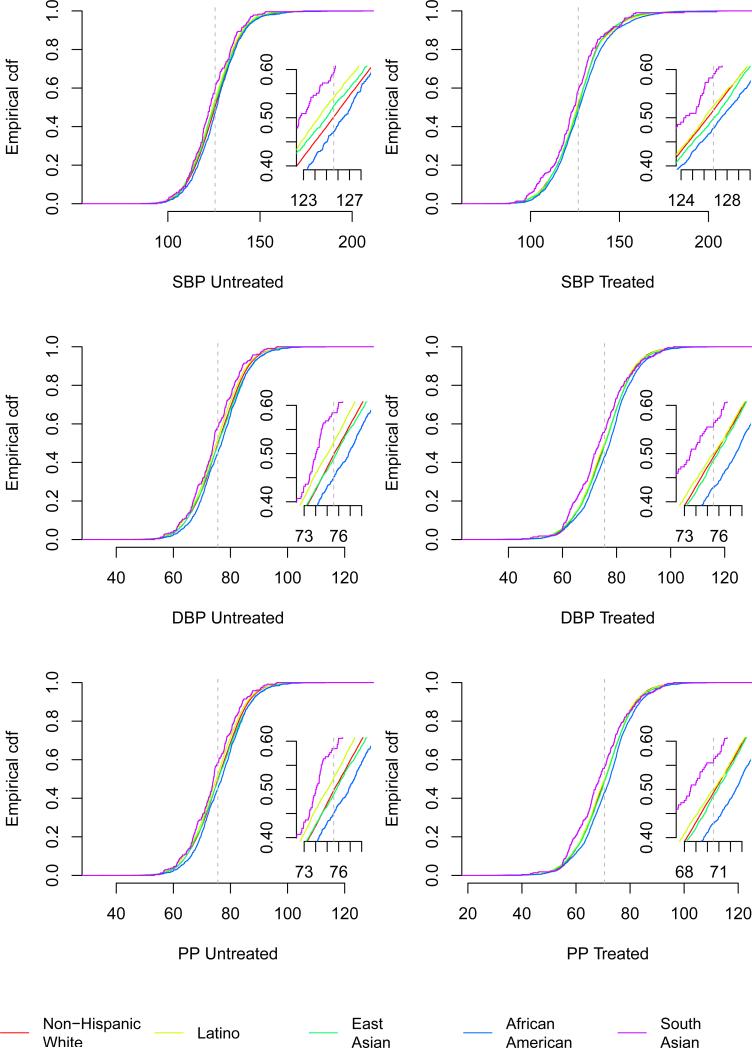
We conducted primary discovery in the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort (n=99,785 for this study) that is composed of non-Hispanic whites (81%; 80,792), Latinos (8%; 8,231), East Asians (7%; 7,243), African Americans (3%; 3,058), and South Asians (1%; 461) (Table 1). GERA is part of the Kaiser Permanente Research Program on Genes, Environment, and Health (RPGEH), whose participants are members of an integrated health care delivery system. The average follow-up time was 4 years, beginning at age 60.9, leading to high prevalence of hypertension and anti-hypertensive therapy. Figure 1 describes the EHR extraction and study design (Online Methods). Multiple BP measurements (1,342,814 total) were available for many participants: 46.4% had at least one untreated measurement and 62.6% had at least one treated measurement. We included all individuals who had at least one (untreated or treated) BP measurement. The multiple measurements enabled the use of a long-term average to increase accuracy7. There were differences in anthropometric and BP values at the first visit among the race/ethnicity groups (Table 1): African Americans and Latinos had the highest BMI, while South Asians had the lowest, although this group was on average the youngest. Untreated systolic blood pressure (SBP) and diastolic blood pressure (DBP) were highest in African Americans followed by non-Hispanic whites; South Asians had lower values (Figure 2). Untreated BPs were higher in males than females across groups, as also found previously25.
Table 1.
Characteristics of the GERA cohort
| Group | Non-Hispanic white | Latino | East Asian | African American | South Asian | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| N/Mean | %/SE | N/Mean | %/SE | N/Mean | %/SE | N/Mean | %/SE | N/Mean | %/SE | |
| N (% total) | 80792 | 81.0% | 8231 | 8.2% | 7243 | 7.3% | 3058 | 3.1% | 461 | 0.5% |
| N female (%) | 46771 | 57.9% | 4960 | 60.2% | 4190 | 57.9% | 1819 | 59.5% | 184 | 39.9% |
| Avg # meas | 13.6 | 13.2 | 11.1 | 15.6 | 10.7 | |||||
| N treated (%) | 51221 | 63.4% | 4741 | 57.6% | 3931 | 54.3% | 2276 | 74.4% | 247 | 53.6% |
| Avg # treated meas | 16.07 | 15.98 | 14.06 | 17.66 | 13.67 | |||||
| N untreated (%) | 36931 | 45.7% | 4261 | 51.8% | 3837 | 53.0% | 1008 | 33.0% | 248 | 53.8% |
| Avg # untreated meas | 7.5 | 7.7 | 6.5 | 7.5 | 6.3 | |||||
| Age (at first meas) | ||||||||||
| Male mean (SE) | 63.9 | 12.1 | 59.1 | 13.7 | 59.2 | 13.6 | 61.5 | 11.7 | 54.8 | 14.0 |
| Female mean (SE) | 60.6 | 13.5 | 53.3 | 14.8 | 53.9 | 14.6 | 56.8 | 14.1 | 48.5 | 13.7 |
| BMI (at first meas) | ||||||||||
| Male mean (SE) | 28.0 | 4.6 | 29.0 | 4.9 | 26.1 | 4.0 | 29.3 | 5.2 | 25.8 | 3.7 |
| Female mean (SE) | 27.3 | 6.0 | 28.6 | 6.3 | 24.5 | 4.5 | 30.8 | 6.9 | 25.1 | 4.2 |
| SBP (at first meas, mmHg) | ||||||||||
| Treated male mean (SE) | 128.2 | 15.2 | 128.2 | 15.5 | 127.4 | 14.8 | 130.8 | 15.4 | 125.0 | 16.2 |
| Untreated male mean (SE) | 125.2 | 13.0 | 124.7 | 13.0 | 122.4 | 12.8 | 127.0 | 14.5 | 120.4 | 12.5 |
| Treated female mean (SE) | 129.7 | 15.6 | 129.2 | 15.9 | 128.1 | 15.5 | 130.9 | 15.9 | 123.1 | 14.8 |
| Untreated female mean (SE) | 121.2 | 14.4 | 118.7 | 14.1 | 117.3 | 14.3 | 122.5 | 13.4 | 113.8 | 13.3 |
| DBP (at first meas, mmHg) | ||||||||||
| Treated male mean (SE) | 73.9 | 10.1 | 75.0 | 10.3 | 74.8 | 10.2 | 76.9 | 10.1 | 73.2 | 10.5 |
| Untreated male mean (SE) | 75.3 | 8.8 | 75.5 | 9.0 | 75.0 | 9.1 | 76.8 | 9.2 | 73.2 | 8.9 |
| Treated female mean (SE) | 73.8 | 9.9 | 74.5 | 10.1 | 74.4 | 10.5 | 75.9 | 10.4 | 73.3 | 9.8 |
| Untreated female mean (SE) | 72.4 | 9.2 | 72.0 | 9.3 | 71.2 | 9.5 | 75.0 | 8.8 | 69.7 | 8.8 |
| PP (at first meas, mmHg) | ||||||||||
| Treated male mean (SE) | 54.4 | 12.5 | 53.2 | 12.3 | 52.6 | 12.4 | 54.0 | 12.5 | 51.8 | 11.8 |
| Untreated male mean (SE) | 49.9 | 10.3 | 49.2 | 10.2 | 47.4 | 9.6 | 50.1 | 10.9 | 47.2 | 9.5 |
| Treated female mean (SE) | 55.9 | 13.8 | 54.8 | 13.6 | 53.7 | 13.2 | 55.0 | 13.7 | 49.8 | 11.4 |
| Untreated female mean (SE) | 54.0 | 17.4 | 51.8 | 17.3 | 52.1 | 18.0 | 51.6 | 17.3 | 43.0 | 14.6 |
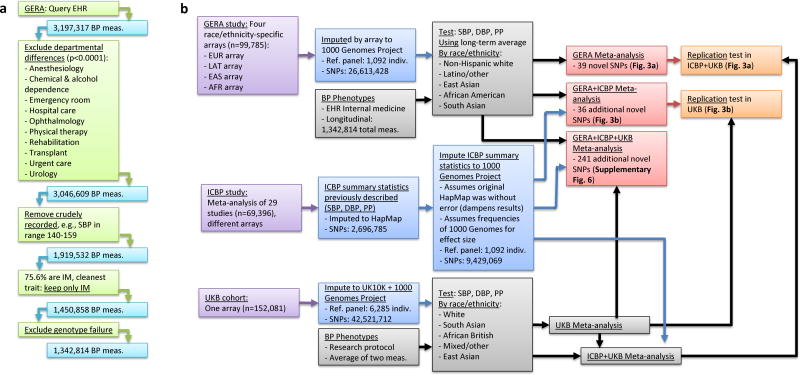
Figure 1.
Project workflow. (a) EHR phenotype extraction for the GERA cohort. (b) GWAS analysis approaches.
Figure 2.
Empirical cumulative distribution functions of BP measures (mmHg), stratified by GERA race/ethnicity group and normalized to a 61 year old male with BMI 27kg/m2 for reference, indicated by the vertical dashed line. There were 80,792 non-Hispanic whites, 8,231 Latinos, 7,243 East Asians, 3,058 African Americans, and 461 South Asians.
To further investigate covariate effects, we assessed age, sex, BMI, and genetic ancestry on SBP, DBP, and pulse pressure (PP) within each race/ethnicity group (Supplementary Table 1). Age and age2 accounted for substantial SBP variation as expected, ranging from 10.6% (African Americans) to 29.0% (South Asians; although this large number may simply reflect small sample size). Age explained little DBP variance in any group. BMI explained moderate SBP variance, ranging from 2.1% (African Americans) to 5.4% (South Asians). While males had higher BPs than females across groups, sex contributed little to BP variance. Although statistically significant, ancestry principal components (PCs) explained little variance for any BP phenotype in any group (generally <0.1%), except European ancestry in African Americans (1% of SBP and DBP variance, decreased SBP, DBP, and PP with lower European ancestry).
Novel BP loci in GERA and meta-analyses with ICBP and UKB
The GERA GWAS discovery stage did not indicate significant genomic inflation with genomic-control (λ)26 values of 1.063, 1.058, and 1.065 for SBP, DBP, and PP, respectively (Supplementary Figures 1-4, Online Methods). In addition to the linear regression analytic approach used in previous GWAS13,14,17 we used a mixed model approach that yielded slightly smaller λ values, suggesting an improved population substructure and/or cryptic relatedness adjustment (Supplementary Table 2). We detected 75 independent, genome-wide significant (P≤5×10−8) loci associated with one or more BP phenotypes (Supplementary Figures 1-4, Supplementary Tables 1-5).
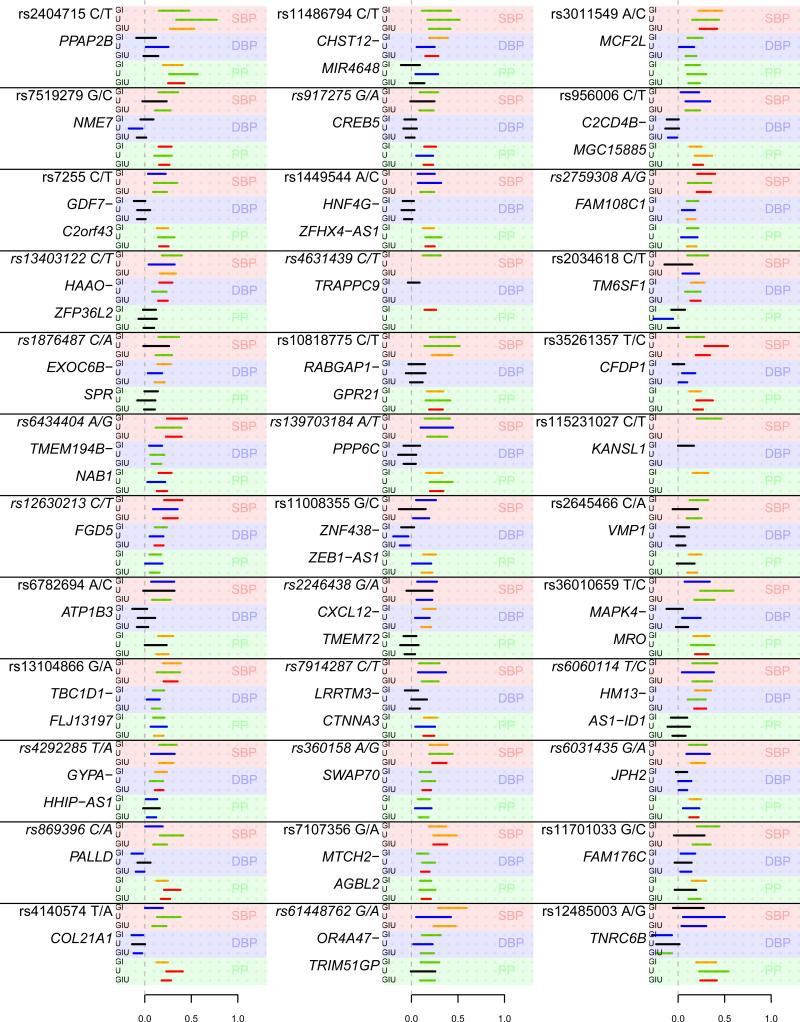
Of the 75 identified loci, 36 replicated previous GWAS findings. Of the remaining 39 novel loci (Figure 3), 25 were strictly replicated (P≤0.00067, Bonferroni correction for 75=39+36 SNPs – see Online Methods) in 221,477 individuals from the International Consortium on Blood Pressure (ICBP; HapMap summary statistics augmented to 1000 Genomes Project; Online Methods; Supplementary Figure 5)17 and UK Biobank (UKB; imputed additionally using UK10K)27). Among the remaining 14 loci, 8 had suggestive significance (P≤0.01), and one X chromosome SNP was unavailable for replication. All SNPs of at least suggestive significance (P≤0.05) had effects in the same direction as in GERA, and had no significant heterogeneity among the GERA race/ethnicity groups or between GERA and/or ICBP and/or UKB (Figure 3 and Supplementary Table 3), giving further credibility that these loci are also true-positive findings. Of note, ICBP alone poorly replicated novel SNPs (only 3 SNPs met Bonferroni correction in ICBP alone), although the SNPs were highly enriched for small P-values. These results emphasize the importance of large replication cohorts.
Figure 3.
Novel BP loci detected in GERA and tested for replication in the ICBP+UKB meta-analysis. The two SNPs rs76217164 and rs143118162 failed to impute in ICBP (owing to low allele frequency), and rs141216986 was on the X chromosome and not available in ICBP or UK. We used an additive model. The effect allele is the allele to the left (e.g., A in A/G). Effect sizes are indicated in mmHg. Each line represents the effect size and 95% confidence interval for each group, with the text on top of each line representing the group tested: G, GERA (n=99,785); IU, meta-analysis of the ICBP and UKB (n=221,477); and GIU, meta-analysis of GERA, ICBP, and the UKB (n=321,262). The color of each line indicates the statistical significance of the test: red, P≤10−9; orange, 10−9<P≤5×10−8; green, 5×10−8<P≤0.00067 (Bonferroni correction for 39+36=75 SNPs); blue, 0.00066<P≤0.05; black, P>0.05.
Expanding our discovery to a meta-analysis of GERA and ICBP also did not indicate significant inflation (average λ=1.042, Supplementary Table 2); this λ is slightly smaller than GERA alone, likely due to the slightly conservative nature of extending the ICBP summary statistics (Online Methods). Thirty-six additional new loci reached genome-wide significance for at least one BP phenotype. Using 152,081 individuals from UKB for replication, 22 loci replicated at P≤0.00067 (Bonferroni for 75 SNPs, see above), 7 were suggestive with P<0.01, and 2 reached nominal significance (P<0.05). As before, all SNPs at least of nominal significance (P<0.05) had the same effect direction in UKB, arguing for a low rate of false positive findings (Figure 4, Supplementary Table 3). We did not detect significant heterogeneity for any lead SNP.
Figure 4.
Novel BP loci identified in the GERA+ICBP meta-analysis, and tested for replication in the UKB. We used an additive model. The effect allele is the allele to the left (e.g., A in A/G). Effect sizes are indicated in mmHg. Each line represents the effect size and 95% confidence interval for each group, with the text on top of each line representing the group tested: GI, meta-analysis of GERA and ICBP (n=169,181); U, UK Biobank (n=152,081); and GIU, meta-analysis of GERA, ICBP, and the UKB (n=321,262). The color of each line indicates the statistical significance of the test: red, P≤10−9; orange, 10−9<P≤5×10−8; green, 5×10−8<P≤0.00067 (Bonferroni correction for 39+36=75 SNPs); blue, 0.00066<P≤0.05; black, P>0.05.
Finally, to maximize discovery power, we combined all three studies (GERA, ICBP, and UKB, n=321,262). Our genome-wide meta-analysis of SBP, DBP, and PP had λ=1.069, 1.076, and 1.076, respectively. We identified 241 additional novel genome-wide significant loci (Supplementary Figure 6, Supplementary Table 3), although replication was not possible. Only rs139491786 showed heterogeneity evidence (I2=88, P=1.5×10−5).
Conditional analysis
We first searched for additional genome-wide significant SNPs within a 1Mb window (±0.5Mb of the lead SNP) involving each previously-described or novel locus in GERA, testing for replication in UKB. We first identified an additional novel SNP, rs1322640, 129Kb from rs13197550 (lead GERA SNP), that replicated in UKB (P=8.3×10−6, Table 2a, Supplementary Table 5). We next identified a novel INDEL (chromosome=20, b37 position=10,573,001) located 396Kb from rs2104574 (lead GERA SNP), that replicated in UKB (P=0.012, Table 2a, Supplementary Table 5).
Table 2.
Conditional/stepwise regression models to test for additional independent BP SNPs at each locus. Effect sizes (mmHg) and P-values are given from a univariate fit of each SNP one-at-a-time (univariate) and a joint fit of all of the SNPs at each locus (joint).
| (a) SNPs discovered in GERA. The SNPs on chromosome 20 show an independent phenotype trait association for different SNPs. Rs2104574 is the lead GERA SNP near a previously-identified SNP. | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GERA Meta-analysis (n=99,785) | UKB Meta-analysis (n=152,081) | GERA + UKB Meta-analysis (n=251,866) | ||||||||||||||
| Univariate | Joint | Univariate | Joint | Univariate | Joint | |||||||||||
| Chr | Trait | SNP | Position | Allele | Eff | P | Eff | P | Eff | P | Eff | P | Eff | P | Eff | P |
| 6 | PP | rs1322640 | 169586887 | T/C | −0.27 | 8.9×10−11 | −0.28 | 4.7×10−11 | −0.40 | 4.0×10−13 | −0.41 | 1.3×10−13 | −0.32 | 1.1×10−21 | −0.33 | 2.1×10−22 |
| rs13197550 | 169716025 | C/A | −0.23 | 2.1×10−9 | −0.23 | 1.4×10−9 | −0.20 | 2.3×10−5 | −0.21 | 8.3×10−6 | −0.22 | 2.5×10−13 | −0.22 | 5.8×10−14 | ||
| 20 | PP | rs2104574 | 10968891 | C/T | −0.10 | 0.012 | −0.091 | 0.024 | −0.18 | 0.00025 | −0.18 | 0.0005 | −0.13 | 2.2×10−5 | −0.12 | 8.3×10−5 |
| 20:10573001:I | 10573001 | C/CG | 0.45 | 6.2×10−9 | 0.44 | 1.0×10−8 | 0.24 | 0.012 | 0.22 | 0.023 | 0.37 | 1.1×10−9 | 0.36 | 3.8×10−9 | ||
| 20 | DBP | rs2104574 | 10968891 | C/T | −0.25 | 2.6×10−10 | −0.25 | 2.4×10−10 | −0.21 | 2.2×10−7 | −0.22 | 7.1×10−8 | −0.23 | 4.0×10−16 | −0.23 | 1.1×10−16 |
| 20:10573001:I | 10573001 | C/CG | 0.0026 | 0.97 | −0.015 | 0.84 | −0.25 | 0.00089 | −0.28 | 0.00024 | −0.12 | 0.021 | −0.15 | 0.0067 | ||
| (b) SNPs discovered in the GERA and UKB meta-analysis. The SNP rs12770529 on chromosome 10 was associated with DBP. | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GERA Meta-analysis (n=99,785) | UKB Meta-analysis (n=152,081) | GERA + UKB Meta-analysis (n=251,866) | ||||||||||||||
| Univariate | Joint | Univariate | Joint | Univariate | Joint | |||||||||||
| Chr | Trait | SNP | Position | Allele | Eff | P | Eff | P | Eff | P | Eff | P | Eff | P | Eff | P |
| 2 | PP | rs13403122 | 43078758 | C/T | 0.22 | 1.3×10−7 | 0.21 | 4.5×10−7 | 0.16 | 8e-05 | 0.15 | 0.00018 | 0.19 | 7.2×10−11 | 0.18 | 5.2×10−10 |
| rs2115859 | 43386092 | T/C | 0.17 | 3.1×10−6 | 0.17 | 9.9×10−6 | 0.14 | 0.00027 | 0.13 | 0.0006 | 0.16 | 4.1×10−9 | 0.15 | 2.7×10−8 | ||
| 3 | PP | rs149240564 | 41867621 | A/G | 0.39 | 1.6×10−7 | −0.048 | 0.62 | 0.76 | 3.3×10−32 | 0.77 | 1.9×10−31 | 0.60 | 3.9×10−35 | 0.51 | 1.4×10−20 |
| rs9816560 | 41872527 | G/T | 0.43 | 2e×10−19 | 0.45 | 7.5×10−13 | 0.30 | 0.19 | −0.26 | 0.29 | 0.43 | 9.8×10−20 | 0.41 | 2.1×10−11 | ||
| 6 | PP | rs147384090 | 32013850 | C/T | −0.90 | 0.18 | −0.92 | 0.17 | −0.29 | 1.3×10−16 | −0.29 | 1.7×10−16 | −0.29 | 7.7×10−17 | −0.29 | 1.0×10−16 |
| rs3129927 | 32333827 | C/A | 0.23 | 0.00061 | 0.23 | 0.00059 | 0.35 | 5.4×10−7 | 0.34 | 7.3×10−7 | 0.29 | 2.6×10−9 | 0.29 | 3.2×10−9 | ||
| 10 | PP | 10:104957628:D | 104957628 | AT/A | 0.61 | 1.2×10−17 | 0.61 | 2.1×10−17 | 1.6 | 0.073 | 1.6 | 0.071 | 0.62 | 4.5×10−18 | 0.62 | 7.7×10−18 |
| rs12770529 | 105149645 | C/T | −0.02 | 0.71 | 0.032 | 0.55 | 0.062 | 0.36 | 0.066 | 0.33 | 0.012 | 0.78 | 0.046 | 0.28 | ||
Chr, chromosome; Position, b37 position; Eff, effect.
We further combined GERA and UKB in a discovery conditional meta-analysis, identifying an additional 4 independent signals (Table 2b, Supplementary Table 5). No replication was possible for these.
Replication of previous GWAS results
We also investigated replication of previously-described BP loci in GERA (Supplementary Table 6, which also reports the GERA lead SNP when it differs from the previously-described lead SNP at the locus)4–22. For the 85 previously-described lead SNPs (or an r2=1.00 proxy for one SNP), 62.4% (53/85) were significantly associated with at least one GERA BP phenotype at P<0.00059 (Bonferroni adjustment for 85 tests) and had the same direction of effect; 78.8% (67/85) were nominally significant; 95.3% (81/85) had effects in the same direction. Replication was stronger in UKB, with 77.6% (66/85) replicating at Bonferroni significance, 89.4% (76/85) at nominal significance, and 96.5% (82/85) in the same direction. The replication was further improved in meta-analysis of GERA and UKB, where 84.7% (72/85) met Bonferroni significance, 89.4% (76/85) were nominally significant, and 96.5% (82/85) had effects in the same direction.
In addition, testing an aggregate, weighted genetic risk score (GRS) using all 85 previously-described SNPs for each BP trait led to highly significant associations in all GERA groups with P<10−168 (whites), P<10−22 (Latinos), P<10−9 (East Asians), and P<0.002 (African Americans), and P<10−350 in UKB whites, for all BP traits, (Table 3). In GERA, Latinos had a larger mean SBP GRS than whites (P=0.053), while African Americans had a lower one (P=0.032). When GERA African Americans were stratified by European ancestry, SBP GRS were lower in individuals with 0%-50% European ancestry (coefficient=0.65, 95% CI=0.18-1.13) than in those with 50%-100% European ancestry (coefficient=1.04, 95% CI=0.56-1.51), although these confidence intervals overlap. The same trend appeared for DBP and PP (Table 3). There was also a very high degree of concordance of the estimated regression coefficients for SBP, DBP, and PP among the non-Hispanic whites in GERA, ICBP, and the UKB (Supplementary Figure 7).
Table 3.
Variance explained by BP genetic risk scores (standardized to a unit normal distribution). In GERA, there were 80,792 non-Hispanic whites, 8,231 Latinos, 7,243 East Asians, 3,058 African Americans, and 461 South Asians; there were 145,341 UKB whites.
| Previously identified | Previously identified, GERA | Previously identified, GERA, GERA+ICBP | Previously identified, GERA, GERA+ICBP, GERA+ICBP+UKB | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | Trait | Eff/HR | P | Rsq | Eff/HR | P | Rsq | Eff/HR | P | Rsq | Eff/HR | P | Rsq |
| GERA Non-Hispanic white | SBP | 1.26 | 1.1 ×10−191 | 0.011 | 1.46 | 8.9×10−262 | 0.015 | 1.63 | 10−327 | 0.018 | 2.03 | 10−521 | 0.029 |
| GERA Latino | SBP | 1.52 | 2.8×10−30 | 0.016 | 1.71 | 1.5×10−38 | 0.020 | 1.86 | 2.4×10−45 | 0.024 | 2.21 | 5.0×10−64 | 0.034 |
| GERA East Asian | SBP | 1.22 | 1.4×10−14 | 0.008 | 1.43 | 1.3×10−19 | 0.011 | 1.62 | 1.0×10−23 | 0.014 | 2.11 | 1.9×10−39 | 0.024 |
| GERA African American | SBP | 0.80 | 0.00054 | 0.004 | 0.98 | 1.6×10−5 | 0.006 | 1.22 | 1.1×10−7 | 0.009 | 1.78 | 3.2×10−15 | 0.020 |
| GERA South Asian | SBP | 0.90 | 0.11 | 0.006 | 1.33 | 0.021 | 0.012 | 1.06 | 0.073 | 0.007 | 1.58 | 0.0066 | 0.017 |
| UKB white | SBP | 1.93 | 10−373 | 0.012 | 2.03 | 10−417 | 0.013 | 2.17 | 10−477 | 0.015 | 2.91 | 10−869 | 0.027 |
| UKB South Asian | SBP | 1.31 | 0.00035 | 0.006 | 1.44 | 7.8×10−5 | 0.007 | 1.63 | 7.6×10−6 | 0.009 | 2.27 | 1.5×10−9 | 0.016 |
| UKB African British | SBP | 1.80 | 4.3 ×10−5 | 0.008 | 1.82 | 3.5×10−5 | 0.008 | 1.77 | 7.9×10−5 | 0.008 | 2.47 | 3.0×10−8 | 0.015 |
| UKB Mixed | SBP | 1.60 | 5.5×10−5 | 0.008 | 1.87 | 3.2×10−6 | 0.011 | 1.91 | 1.8×10−6 | 0.012 | 2.60 | 7.1×10−11 | 0.022 |
| UKB East Asian | SBP | 4.65 | 2.2 ×10−7 | 0.058 | 4.67 | 1.7×10−7 | 0.058 | 4.16 | 5.7×10−6 | 0.044 | 3.72 | 4.9×10−5 | 0.036 |
| GERA Non-Hispanic white | DBP | 0.80 | 1.9 ×10−186 | 0.01 | 0.91 | 1.3×10−247 | 0.014 | 0.99 | 2.8×10−285 | 0.016 | 1.25 | 10−453 | 0.025 |
| GERA Latino | DBP | 0.87 | 2.4 ×10−23 | 0.012 | 0.97 | 1.4×10−28 | 0.015 | 1.05 | 2.3 ×10−33 | 0.018 | 1.31 | 2.4×10−49 | 0.026 |
| GERA East Asian | DBP | 0.68 | 5.2 ×10−10 | 0.005 | 0.79 | 2.5×10−13 | 0.007 | 0.88 | 1.4×10−15 | 0.009 | 1.20 | 3.4×10−28 | 0.017 |
| GERA African American | DBP | 0.49 | 0.0014 | 0.003 | 0.54 | 0.00035 | 0.004 | 0.62 | 4.2×10−5 | 0.006 | 0.96 | 1.5×10−10 | 0.013 |
| GERA South Asian | DBP | 0.54 | 0.17 | 0.004 | 0.77 | 0.047 | 0.009 | 0.73 | 0.068 | 0.008 | 0.81 | 0.046 | 0.009 |
| UKB white | DBP | 1.08 | 10−378 | 0.012 | 1.14 | 10−422 | 0.013 | 1.17 | 10−450 | 0.014 | 1.57 | 10−815 | 0.025 |
| UKB South Asian | DBP | 0.89 | 2.6 ×10−5 | 0.008 | 1.01 | 1.3×10−6 | 0.010 | 0.94 | 7.4×10−6 | 0.009 | 1.32 | 5.1×10−10 | 0.017 |
| UKB African British | DBP | 0.97 | 0.00027 | 0.007 | 1.06 | 5.9×10−5 | 0.008 | 1.08 | 5.2×10−5 | 0.008 | 1.29 | 1.6×10−6 | 0.011 |
| UKB Mixed | DBP | 0.82 | 0.00026 | 0.007 | 0.89 | 7.9×10−5 | 0.008 | 1.01 | 5.7×10−6 | 0.011 | 1.41 | 1.1×10−10 | 0.021 |
| UKB East Asian | DBP | 2.27 | 2.0 ×10−5 | 0.039 | 1.93 | 0.00026 | 0.029 | 1.75 | 0.0013 | 0.023 | 2.12 | 5.3×10−5 | 0.035 |
| GERA Non-Hispanic white | PP | 0.81 | 2.6 ×10−169 | 0.01 | 1.02 | 3.6×10−268 | 0.015 | 1.18 | 10−352 | 0.020 | 1.44 | 10−561 | 0.031 |
| GERA Latino | PP | 0.96 | 2.8 ×10−28 | 0.015 | 1.17 | 5.6×10−41 | 0.022 | 1.28 | 6.2×10−48 | 0.026 | 1.50 | 9.9×10−68 | 0.036 |
| GERA East Asian | PP | 0.76 | 1.7 ×10−15 | 0.009 | 0.96 | 2.5×10−24 | 0.014 | 1.12 | 1.5×10−30 | 0.018 | 1.31 | 1.5×10−42 | 0.026 |
| GERA African American | PP | 0.53 | 0.00037 | 0.004 | 0.72 | 1.3×10−6 | 0.008 | 0.88 | 2.2×10−8 | 0.010 | 1.23 | 1.3×10−15 | 0.021 |
| GERA South Asian | PP | 0.36 | 0.28 | 0.003 | 0.72 | 0.042 | 0.009 | 0.64 | 0.082 | 0.007 | 0.96 | 0.0075 | 0.016 |
| UKB white | PP | 1.32 | 10−351 | 0.011 | 1.52 | 10−467 | 0.015 | 1.64 | 10−542 | 0.017 | 2.16 | 10−960 | 0.030 |
| UKB South Asian | PP | 0.84 | 0.00037 | 0.006 | 0.95 | 7.1×10−5 | 0.007 | 1.18 | 1.0×10−6 | 0.011 | 1.47 | 4.3×10−9 | 0.015 |
| UKB African British | PP | 1.08 | 5.7 ×10−5 | 0.008 | 1.12 | 3.8×10−5 | 0.008 | 1.15 | 5.5×10−5 | 0.008 | 1.71 | 1.3×10−9 | 0.018 |
| UKB Mixed | PP | 1.30 | 8.4 ×10−7 | 0.012 | 1.59 | 2.9×10−9 | 0.018 | 1.57 | 2.0×10−9 | 0.018 | 1.91 | 2.0×10−12 | 0.025 |
| UKB East Asian | PP | 2.03 | 0.00021 | 0.03 | 2.51 | 2.6×10−6 | 0.048 | 2.54 | 4.1×10−6 | 0.046 | 2.26 | 5.9×10−5 | 0.035 |
| GERA Non-Hispanic white | HTN by SBP | 1.11 | 4.0×10−18 | 1.13 | 10−24 | 1.14 | 10−29 | 1.18 | 10−44 | ||||
| GERA Latino | HTN by SBP | 1.11 | 0.0032 | 1.13 | 0.00059 | 1.14 | 0.00020 | 1.19 | 1.4×10−6 | ||||
| GERA East Asian | HTN by SBP | 1.14 | 0.0053 | 1.14 | 0.0052 | 1.14 | 0.0088 | 1.16 | 0.0021 | ||||
| GERA African American | HTN by SBP | 1.20 | 0.019 | 1.18 | 0.030 | 1.26 | 0.0031 | 1.32 | 0.00024 | ||||
| GERA Non-Hispanic white | HTN by DBP | 1.09 | 2.3×10−14 | 1.11 | 10−18 | 1.11 | 10−20 | 1.14 | 10−30 | ||||
| GERA Latino | HTN by DBP | 1.06 | 0.087 | 1.08 | 0.037 | 1.08 | 0.033 | 1.11 | 0.0052 | ||||
| GERA East Asian | HTN by DBP | 1.13 | 0.012 | 1.12 | 0.026 | 1.15 | 0.0074 | 1.17 | 0.0022 | ||||
| GERA African American | HTN by DBP | 1.15 | 0.057 | 1.15 | 0.060 | 1.23 | 0.0066 | 1.30 | 0.00037 | ||||
| GERA Non-Hispanic white | HTN by PP | 1.10 | 1.4×10−14 | 1.11 | 10−19 | 1.13 | 10−23 | 1.15 | 10−33 | ||||
| GERA Latino | HTN by PP | 1.15 | 9.4×10−5 | 1.17 | 1.4×10−5 | 1.19 | 2.4×10−6 | 1.24 | 5.3×10−9 | ||||
| GERA East Asian | HTN by PP | 1.11 | 0.0021 | 1.12 | 0.015 | 1.08 | 0.11 | 1.11 | 0.042 | ||||
| GERA African American | HTN by PP | 1.16 | 0.040 | 1.13 | 0.086 | 1.18 | 0.031 | 1.20 | 0.015 | ||||
HTN, hypertension; Eff, effect size (for SBP/DBP/PP linear regression); HR, hazards ratio (for HTN time-to-onset analysis).
Examining the effects of individual SNPs, for those discovered in ICBP the effects are typically weaker in GERA, likely due to the winner's curse28. The opposite is also the case: SNPs discovered in GERA have weaker effects in ICBP. UKB comparisons are similar, with the discovery cohort (GERA or GERA+ICBP) having stronger effect sizes than the replication cohort (UKB). Seven SNPs exhibited significant heterogeneity among studies (P<0.00059, Bonferroni correction for 85 SNPs) at the lead trait (Supplementary Table 6).
Variance explained and gain using multiple BP measurements
The variance explained in an additive linear model by the 75 genome-wide significant loci identified in our GERA discovery cohort was 1.4%/1.2%/1.8% for SBP/DBP/PP in GERA non-Hispanic whites; note that the same individuals were used for discovery and testing, but with the independent ICBP estimated effect size. The results for the other GERA groups were: 2.0%/1.6/2.4% in Latinos, 0.9%/0.7%/1.4% in East Asians, 1.3%/0.6%/1.6% in African Americans, and 1.7%/1.7%/0.7% in South Asians. Including the remaining of the 85 previously-described SNPs not genome-wide significant in GERA and the 36 novel SNPs from the GERA and ICBP meta-analysis modestly increased variance explained (Table 3). All previously-described and novel loci explain 2.9%/2.5%/3.1% of SBP/DBP/PP variation in GERA non-Hispanic whites, with an estimated greater (but not significantly different) variance in Latinos (3.4%/2.6%/3.6%) and less in East Asians (2.4%/1.7%/2.6%) and African Americans (2.0%/1.3%/2.1%), who similarly have the lowest GRS; UKB results were generally slightly lower than GERA, e.g., 2.7%/2.5%/3.0% for UKB whites. Adding dominance terms to the linear regression model did not increase variance explained (none significant after multiple comparison correction).
We subsequently investigated the impact of multiple BP measurements in an analysis restricted to individuals who had ≥5 measurements (Supplementary Figure 8). Using all measurements, compared to just one, reduced the regression coefficient standard error (SE) by 25%; the regression coefficient estimate itself did not change significantly. With a large number of measurements, the GRS approximately doubled variance explained for SBP and DBP, but was over 3-fold greater for PP, due to the latter's greater measurement error (Supplementary Table 7). The BP variance due to measurement error was estimated (Online Methods) as 56.5% (SBP), 47.5% (DBP) and 71.5% (PP). Lastly, the number of genome-wide significant variants that would have been found when using 1/2/3/4/all measurements (in a fixed subset of non-Hispanic white individuals with ≥5 measurements and using genotyped SNPs only) was 2/3/3/7/7 SBP, 2/4/7/7/11 DBP, and 4/7/15/14/23 PP, demonstrating a large increase with more measurements included. However, when not fixing the sample size, and using all individuals with at least 1/2/3/4/5+ measurements, we found 12/10/11/10/7 genome-wide SBP, 14/14/14/13/11 DBP, and 20/21/23/21/23 PP significant loci, using a total of 80,792/78,372/75,446/71,834/67,547 individuals, reflecting the loss of statistical power with decreasing sample size. Consequently, it is difficult to determine the optimal minimum number of measurements for subject inclusion, due to the precision vs. sample size tradeoff.
BP risk scores and onset of hypertension
We tested the association of the GRS (described above for SBP, DBP, and PP) with time-to-onset of hypertension. Predictive value of the GRS increased with the number of BP SNPs included (Table 3), as expected. Including SNPs from the meta-analysis of all three cohorts, the SBP GRS was the strongest hypertension predictor with a non-Hispanic white hazards ratio (HR)=1.18 (P=10−44); the DBP GRS was slightly less significant with HR=1.14 (P=10−30), as was that for PP with HR=1.15 (P=10−33). The GRS were also predictive in other groups; e.g., for SBP GRS, Latino P=1.4×10−6, East Asian P=0.0021, and African American P=0.00024.
Sex Differences
We tested SNP effect size differences by sex (heterogeneity test, Supplementary Table 8; coefficients plot, Supplementary Figure 9). After Bonferroni correction (α=0.00013, all 386 novel and previously-described SNPs), none was significantly different. However, 25 SNPs were nominally significant (P<0.05) at the lead trait, which is in slight excess of the 19.3 expected; of those in the same effect direction in males and females, 17/20 (85.0%, 95% CI=61.1%-96.0%) had stronger magnitude in females than males.
Differences in SBP, DBP, and PP effects
We tested whether the normalized effect size of each SNP was greater in SBP or DBP (Online Methods, Supplementary Table 9); 26.2% of the SNPs had significantly different normalized effect sizes for between SBP and DBP (P<0.00013, Bonferroni correction for 386 SNPs); of these, for 57.4% the normalized effect was greater for SBP than DBP.
Heritability from all Genotyped and Imputed SNPs
Array heritability estimates derived from genotyped SNPs based on PC-Relate kinship estimates29, to account for population stratification in the kinship estimate, using GEAR30 in the non-Hispanic whites was 15.5% (95% CI=13.9%-17.1%) for SBP, 15.1% (95% CI=13.5%-16.7%) for DBP, and 14.5% (95% CI=12.7%-16.2%) for PP, increasing only modestly when adding imputed SNPs to 16.1% (95% CI=14.5-17.7%) for SBP, 17.0% (95% CI=15.6%-18.4%) for DBP, and 15.6% (95%CI=14.0%-17.2%) for PP. These estimates were similar to estimates not accounting for population stratification in the kinship estimates but adjusting for it in the phenotype model instead using GCTA31 (SBP h2=16.8%, 95% CI=15.1%-18.6%); this may be because the ancestry effect in non-Hispanic whites is modest. Sample sizes were too small to evaluate other GERA groups.
eQTL analysis in different tissues
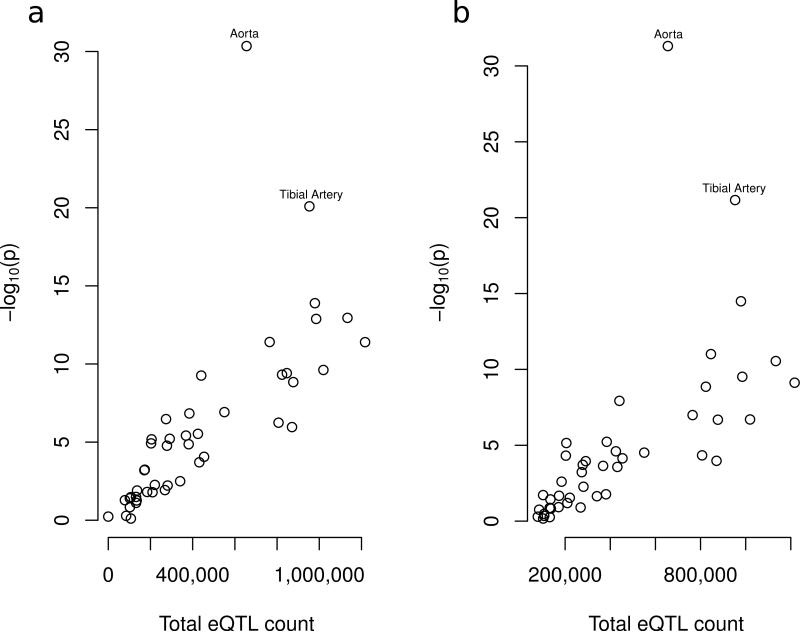
We investigated whether the previously-identified and all novel loci co-localized with Expression Quantitative Trait Loci (eQTLs). We used eQTLs from 44 Genotype-Tissue Expression (GTEx) tissues and kidney 32,33. Across all tissues, 186 of 367 sentinel SNPs were eQTLs in at least one tissue; at least one SNP in 213 of the same 367 loci was an eQTL. We determined for each tissue whether the number of eQTLs (either by sentinel SNP or by locus) was greater than expected by chance, where expectation was derived from a random sampling of SNPs and loci (Online Methods). We ranked the tissues by eQTL P-value, both for the sentinel SNP and locus analysis. We generally expect tissues with more eQTLs to overlap more SNP sets, and enrichment to be greater simply because of chance GWAS set overlap, especially when eQTLs in tissues relevant to the phenotype are also found in these tissues. To observe whether the enrichment visible for a given tissue is greater than expected relative to the total number of eQTLs it contains, we examined the relationship between P-value and total eQTL count per tissue (Figure 5). The aorta and tibial artery are clear outliers compared to other tissues, even accounting for total number of eQTLs.
Figure 5.
Tissue specific expression quantitative trait loci (eQTL) analysis of 51 tissues. The two outlier tissues, accounting for total eQTL count, are labeled. Tissue total eQTL counts vs P-values at (a) the locus and (b) the sentinel variant.
Enrichment analysis for functional elements
We subsequently investigated whether genes near sentinel variants were enriched for certain functional pathways. We included genes within ±0.5Mb of the 390 sentinel variants with a significant eQTL in either tissue identified above (aorta and tibial artery). We identified 2,013 genes near all 390 sentinel variants (Online Methods) and tested for functional annotation enrichment. Using DAVID 6.834,35, 1,480 had annotations, producing 26 significant annotation terms (Benjamini-Hochberg P<0.05, Supplementary Table 10), without a clear functional pathway emerging.
Discussion
In this large, ethnically-diverse GERA cohort with EHR-derived BP measures, we discovered 39 novel genome-wide significant BP loci, most replicating in ICBP and UKB. Merging GERA and ICBP identified 36 additional novel genome-wide significant loci, most replicating in UKB. Finally, merging all three cohorts identified 241 additional genome-wide significant loci, although no replication was available. Conversely, we were able to replicate almost all 85 previously-described BP SNPs. We also showed that using multiple EHR BP measurements almost doubled variance explained, although the total variance explained remains small (e.g., 2.9% for SBP in non-Hispanic whites). We also showed that BP signals are enriched in two large arteries, aorta and tibial.
Our study used a large general population sample with EHR-derived data for the first time in BP GWAS. The consistency and generalizability of BP genomics findings from one-time research-protocol-based assessments to purely clinical measures recorded in an EHR has been questioned36. We were able to replicate most previously-identified loci from many cohorts using research-based assessments, demonstrating BP genetic findings are not significantly different between studies using research assessments and those using clinical, EHR-derived ones. This is important because clinical measures recorded in the EHR are the basis for clinical decisions in general, real-world, clinical practice. Moreover, this extends GWAS reach to numerous clinical samples.
EHR-based studies offer additional benefits. Our identification of new variants takes advantage of multiple independent measurements in the EHR to increase statistical power7. Our study increased the standardization and reduced the variability of the EHR-derived BP measures by excluding measures obtained in clinical settings with increased measurement variability, e.g., emergency rooms, retaining measures obtained in visits to primary care/Internal Medicine departments.
The new BP SNPs identified have similar genomic context to those previously-described, which were located 8.2%/20.0%/32.9%/38.8% in exon/UTR/intron/intergenic regions while novel SNPs identified in GERA were distributed 2.6%/23.1%/33.3%/41.0%, respectively, those from the GERA+ICBP meta-analysis 0%/2.8%/55.6%/41.7%, and those from the GERA+ICBP+UKB meta-analysis 2.5%/14.9%/41.5%/41.1% (Supplementary Table 4)37. Frequencies and variant types of lead SNPs are also similar to those previously described; for European ancestry, 85.9% of previously-described SNPs have minor allele frequencies (MAF)>0.10, compared to 89.7% of GERA-identified SNPs; 94.4% of GERA+ICBP SNPs; and 82.2% of GERA+ICBP+UKB SNPs. Comparing results across traits within GERA, the leading trait locus was more often PP for novel loci than before (24.7% PP for previously-described SNPs, versus 59.0%, 58.3%, and 41.9% PP for GERA, GERA+ICBP, and GERA+ICBP+UKB, respectively); this may reflect that earlier BP studies tested SBP/DBP, but not PP. We additionally demonstrated the significant effect of the summary BP SNP scores on time-to-onset of hypertension, enabled by GERA longitudinal EHR data. We note that a GERA hypertension GWAS produced no additional novel results (and results much less significant than for the continuous BP traits, as expected).
One limitation was that 1000 Genomes imputed results were unavailable in ICBP; however, the much larger UKB replication did not have this limitation. For ICBP, we therefore relied on summary test statistic imputation from HapMap. The use of these approximated results, and the fact that all test statistics from ICBP were based on SNP results imperfectly imputed to HapMap, likely led to diminished effect sizes in ICBP. Overall, we needed a very large number of individuals for replication, both to replicate our novel GERA results, which improved greatly when adding UKB to ICBP, and to replicate previously-described results, which improved when adding UKB to GERA.
Another advantage of a single large cohort, such as GERA, is the ability to directly assess additional local SNPs by conditional analysis. The absence of individual-level data requires LD assumptions from other studies. Nevertheless, we only found two additional variants in GERA that were ultimately not explained by nearby previously-described SNPs, and an additional four when combining GERA and UKB. We further note these additional conditional hits were located at a substantial distance from the locus sentinel SNP, likely indicating an independent gene and/or mechanism involved. The lack of identification of additional SNPs close to sentinel SNPs is quite distinct from what is observed for serum lipids, for example38, and suggests that lower frequency variants with larger effects within the same loci identified here are uncommon. A similar conclusion was recently obtained in a sequencing study of type 2 diabetes39.
While our sample sizes were smaller in the other race/ethnicity groups than for the non-Hispanic whites, we noticed that Latinos had the highest standardized GRS, followed closely by non-Hispanic whites, and then by East Asians and African Americans. In African Americans, European ancestry was associated with lower BP, but individuals with more European ancestry had higher BP standardized GRS (created from previously-described SNPs); this is counter-intuitive, but may reflect the fact that the GWAS discovery occurred primarily in European ancestry individuals, and suggests there may be other SNPs in African Americans remaining to be identified.
We also looked for a pattern in terms of which loci replicated. Logically, the largest replication indicator was discovery P-value, as stronger associations likely require a smaller sample size for replication than weaker ones. In GERA, loci with P≤1×10−9 replicated at a Bonferroni level at a rate of 76.5% (13/17) vs. 54.5% (12/22) for those with 5×10−8≤P<1×10−9; all of the ICBP SNPs with P≤1×10−9 replicated at a Bonferroni level in GERA+UKB; however, the pattern was not seen in the GERA and ICBP meta-analysis with 57.1% (4/7) with P≤1×10−9 vs. 62.1% (18/29) with 5×10−8≤P<1×10−9 although numbers were small. Perhaps also of note, the two SNPs in GERA with MAF<0.001 failed to replicate in UKB (P>0.05).
We also searched for eQTL enrichment in a variety of tissues. Both the aorta and tibial arteries were clear outliers compared to other tissues, suggesting genetic factors influencing vascular elasticity and/or stiffness are important determinants of BP and hypertension.
There are several reasons for the enhanced discovery in our study: an increased sample size, multiple BP measures (reducing phenotype variability), better designed arrays with increased genomic coverage40,41, and larger imputation reference panels (reducing error and providing additional imputed SNPs). We showed a 25% SNP effect se reduction using multiple BP measurements. In addition, 15 SNPs not present in 1000 Genomes were genome-wide significant in the UKB data alone (6.2% of the 241 novel SNPs), while none of the SNPs in 1000 Genomes surrounding them met genome-wide significance.
After completion of our analyses, three additional large-scale BP/hypertension GWAS have been published42–44, including, as in our study, hundreds of thousands of individuals in discovery and replication phases. Notable among the findings were an enrichment of SNPs also involved in cardiometabolic traits42 and the implication of genetic variation in vascular function42,44, as we also found. Two of the studies42,43 also focused on rare variation, and identified a few larger-effect rare missense and nonsense variants in eight distinct genes. These studies collectively identified 71 distinct novel genome-wide significant loci. Using a broad definition of overlap (r2>0.3), a cursory examination suggests that 16 of these overlap with our 316 novel hits (2 of the 39 from GERA alone, 4 of 36 from GERA+ICBP, and 10 of 241 from GERA+ICBP+UKB). These studies, along with ours, demonstrate the enhanced power of both gene discovery and characterization afforded by expanded sample sizes.
In summary, the current study demonstrates the utility of a large general cohort with EHR-derived multiple independent measurements for studying BP genetics; it is reassuring that the same BP loci found in research-based cohorts are captured with high significance, and also that the longitudinal data typical for EHRs provide important opportunities for novel SNP discovery. The new SNPs found here may provide novel mechanistic insight into the control and treatment of hypertension, ultimately preventing a variety of clinical sequelae.
Online methods
All statistical tests were two-sided.
Participants, phenotype, and genotyping
Our primary analysis used individuals from the RPGEH GERA cohort, which has been described45,46. We used three trait outcomes: SBP, DBP, and PP, where PP=SBP-DBP. We began with 3,197,317 GERA EHR BP measurements. In KPNC, BP is measured and recorded in the EHR at the beginning of each clinic visit, regardless of the visit reason. Examination of mean BP measurements by medical specialty showed that, compared to Internal Medicine (IM), average BP measurements obtained in the following departments were significantly higher (p<0.0001): anesthesiology, chemical and alcohol dependency, health education, emergency room, hospital care, ophthalmology, physical therapy, rehabilitation, transplant, urgent care, and urology. Higher average BP measurements in these specialties likely indicated effects of acute illnesses or other effects on BP, and we excluded all BP measurements obtained in these specialty visits; 3,046,609 BP measurements (95%) remained after these exclusions. We further excluded 1,127,077 measurements recorded as binned into 5 systolic and 7 diastolic BP ranges (e.g., systolic BP recorded in the range 140-159); this was an early recording method prior to the full EHR implementation in 2006. After noting that 75.6% of the 1,919,532 remaining measurements were from IM visits, we excluded the 188,173 OB/GYN and 280,501 other departmental measurements to obtain the most homogeneous BP phenotype, resulting in 1,450,858 measurements from IM visits on 107,196 individuals. Finally, after excluding those failing genotyping, 1,342,814 independent SBP and DBP IM visit measurements from different days (345,031 untreated and 997,783 treated) on 99,785 individuals obtained from the beginning of 2006 to the end of 2011 remained for analysis. Anti-hypertensive medication treatment was assessed via EHR prescription filling information; once an individual started a drug, they were considered treated on all subsequent measurements. We added 15mmHg to treated SBP values and 10mmHg to treated DBP values,47 similar to previous BP GWAS,17 to correct for treatment effect.
Individuals were genotyped at over 650,000 SNPs on one of four race/ethnicity-specific Affymetrix Axiom arrays optimized for individuals of European (EUR), Latino (LAT), East Asian (EAS), and African American (AFR) race/ethnicity40,41. We analyzed 80,792 non-Hispanic whites, 8,231 Latinos/other, 7,243 East Asians, 3,058 African Americans, and 461 South Asians (genotyped on the EUR array). The Kaiser Foundation Research Institute and University of California San Francisco Institutional Review Boards approved this project. Written informed consent was obtained from all subjects.
Genotype quality control and imputation
Initial genotype quality control was performed per race/ethnicity-specific array, as described46. In addition, we required an array per-SNP call-rate ≥90%, resulting in 665,350 (EUR), 777,927 (LAT), 704,105 (EAS), 864,905 (AFR), and 663,783 South Asian (SAS) SNPs. We excluded SNPs with a minor allele count (MAC)<20, resulting in an MAF cutoff of 0.0001 (EUR), 0.001 (LAT), 0.001 (EAS), 0.003 (AFR), and 0.02 (SAS) and a total number of 662,517, 758,681, 700,291, 855,429, and 568,707 SNPs, respectively.
Imputation was performed on an array-wise basis. We first pre-phased the genotypes with Shape-it v2.r7271948. We then imputed variants from the 1000 Genomes Project (phase I integrated release, March 2012, with Aug 2012 chromosome X update, a cosmopolitan reference panel with singletons removed) with Impute2 v2.3049–51. The estimated quality control metric rinfo2 used in this study is the info metric from Impute2, which is an estimate of the imputed genotype correlation to the true genotype52. Poorly imputed (rinfo2<0.3) and MAC<20 SNPs were removed, resulting in 24,149,855 (EUR), 20,828,585 (LAT), 15,248,462 (EAS), 21,485,958 (AFR), and 8,607,429 (SAS) SNPs (28,613,428 unique SNPs) available for analyses.
GWAS analysis and covariate adjustment
We first analyzed each of the five race/ethnicity groups separately. Data from each SNP were modeled using additive dosages accounting for imputation uncertainty53. For each quantitative trait (treatment adjusted SBP, DBP, and PP), for computational efficiency, we first ran a mixed model of the BP measurement adjusted for age, age2, BMI, and sex using all BP measurements for each individual. We then constructed a long-term average residual for each individual as the dependent variable in a linear mixed model using estimated kinship matrices with leave-one-chromosome-out (LOCO) to account for population substructure and cryptic relatedness with Bolt-LMM54. Finally, we undertook a fixed-effects meta-analysis to combine the results of the five groups using Metasoft v2.055. We considered as novel loci that were at a physical distance >0.5Mb from any previously-described locus (and visual inspection for longer LD stretches, see below).
To find additional independent SNPs at each locus, we ran a conditional stepwise regression analysis at all SNPs with rinfo2>0.8 in the GERA meta-analysis, around each previously-described and novel GERA SNP. We looked for additional genome-wide significant SNPs within a 1Mb window (±0.5Mb) of the lead SNP. While this generally worked well, certain portions of the genome have stronger LD (we noted particularly at ends of chromosomes and centromeres, where recombination is suppressed), which we assessed via visual inspection of the Manhattan plots to form an expanded window size, and repeated the stepwise regression on the expanded window. In these analyses we adjusted for ancestry PCs (see below) instead of the mixed model approach, both for simplicity and computational efficiency.
To adjust for genetic ancestry/population stratification when not using Bolt-LMM LOCO, we performed a PC analysis, as described45. The first 10 eigenvectors for non-Hispanic whites and the first 6 eigenvectors for all other race/ethnicity groups were included as covariates in the regression model described above. When we tested European vs. African ancestry percentages in African Americans, we used PC1 as a European admixture surrogate45.
Replication of novel GERA SNPs using ICBP and UKB
To test the 39 novel GERA genome-wide significant SNPs for replication, we evaluated the associations utilizing a fixed effects meta-analysis of ICBP and UKB. We also tested the 36 novel SNPs found in the meta-analysis of GERA and ICBP for replication in UKB. We report associations that replicate at a strict Bonferroni threshold (P<0.00067, to account for a total of 75 novel SNPs tested), as well as suggestive (P<0.01) and nominally suggestive (P<0.05) findings with effects in the same direction as the original.
ICBP
ICBP GWAS summary statistics from 69,396 individuals at 2,696,785 SNPs were obtained from dbGaP17. As only summary statistics were available, we did not use these data to replicate conditional SNPs.
As the ICBP has been imputed to HapMap v22, a smaller reference panel than used here for GERA, we used ImpG v1.0.156 to estimate the summary statistics for the 1000 Genomes Project reference panel SNPs used for the GERA imputation. To solve for the effect size βj of the additive coded genotype Xij (i indexing N individuals, j indexing SNPs) from the summary statistics imputed to 1000 Genomes from ImpG, we assumed that the ICBP had the same allele frequency as in the 1000 Genomes European ancestry individuals and Hardy-Weinberg Equilibrium (HWE). Let qj be the MAF, and pj=1-qj. Assuming HWE, Npj2 individuals have Xij=0, N2pjqj have Xij=1, and Nqj2 have Xij=2. It is known that SE(βj)=∑irij2/sqrt(sxx,j), where rij is the residual of the phenotype regressed on the SNP genotype Xj and sxx,j= ∑i(Xij-mean(X.j))2. It can be shown that sxx,i=2npjqj. Although ∑rij2 is unknown in ICBP, a reasonable approximation is obtained by assuming that individually each SNP explains very little of the trait variance and thus ∑rij2 is constant and does not depend on j, i.e., ∑rij2=∑ri2, and solve for this quantity using the existing effect size estimate of βj from the available HapMap SNPs. Using ImpG assumes all HapMap SNPs were imputed without error; such error likely dampens the results.
UKB
The UKB cohort has been previously-described27. Of note, genotypes were imputed using a larger number of individuals from the UK10K combined with 1000 Genomes Project as a reference panel (n=6,285). SBP measures were taken from manual (variable 93.0-2.0-1) and automatic readings (4080.0-2.0-1), as were DBP (94.0-2.0-1 and 4079.0-2.0-1, respectively). Age was reported as the age at measurement (34.0.0). Anti-hypertensive use was assessed by self-report (6153.0-1.0 and 6177.0-2.0), and BPs were corrected as in GERA. BMI was calculated from measured weight and height (21001.0.0). Sex was determined genetically (22001.0.0). Analysis was done as in GERA, a meta-analysis of each self-reported race/ethnicity group (21000.0-2.0): we identified 145,341 individuals who reported any white race/ethnicity group and with global ancestry PC1≤50 and PC2≤50, where global PC1 and PC2 were calculated from the entire cohort (22009.0.1-2), including 2,274 South Asians, 2,029 African British, 1,979 mixed/other, and 458 East Asians, totaling 152,081 individuals. Ancestry PCs within whites were calculated using 50,000 random white individuals with the remaining subjects projected, which has been shown to work well45, and then within each other group. We analyzed 35,893,267, 12,078,001, 19,866,667, 15,820,020, and 7,298,789 SNPs with rinfo2≥0.3 and MAF≥0.0001, 0.005, 0.005, 0.005, and 0.025, in whites, South Asians, African-European, mixed/other, and East Asians, respectively (42,521,712 unique SNPs).
GERA meta-analysis with ICBP, and with UKB
We additionally performed meta-analysis of the GERA and ICBP results for genome-wide discovery using a fixed-effects meta-analysis, using UKB for replication. We further performed a discovery meta-analysis of GERA, ICBP and UKB for maximal discovery size, but with no replication sample available. In this analysis we reviewed the locus plots, manually merging the ±0.5Mb windows when necessary. Specifically, after assessing SNPs in the GERA+ICB+UKB meta-analysis, we checked if the SNPs appeared independent in a meta-analysis of GERA and UKB, as both had individual level data. Most regions were either obviously correlated with high r2, or obviously not with r2<0.05; however, to formalize the conditional analysis and retain a SNP as independent, we required that the reduction in p-values from univariate to joint in the GERA+UKB meta-analysis be less than 10-fold, and additionally that translating an equivalent reduction in p-values to the GERA+ICBP+UKB meta-analysis still led to a genome-wide significant result (i.e., if we assumed that Pjoint,GERA+ICBP+UKB/Punivariate,GERA+ICBP +UKB=Pjoint,G ERA+UKB/Punivariate,GERA+UKB, the approximated Pjoint,GERA+ICBP+UKB would still need to be genome-wide significant). This may have been slightly conservative.
Replication analysis of previously-described SNPs in GERA
To determine how many of the 85 previously-described loci from ICBP and other GWAS replicated in this study, we tested the sentinel SNPs from those studies in our dataset4–22. Frequently, multiple BP phenotypes are reported for the same loci. We used a Bonferroni correction for replication (85 SNPs, α=0.00059). The SNP rs2446849 was not in our reference panel, so we used the closest proxy, rs2513758, at a physical distance of 876bp and r2=1.00 in Europeans.
GRS construction
We constructed a GRS for each of the three BP traits for each individual by summing the additive coding of each set of SNPs associated with the particular BP trait weighted by the previously-described effect size from ICBP (phs000585.v1.p1), and then standardized the distribution of all groups simultaneously by the mean and standard deviation (i.e., to a standard normal distribution) for interpretability. We used the leading SNP from each locus.
Multiple Measurements
To assess the impact of multiple BP measurements, we compared the P-value and effect size estimates for the previously-described GWAS significant SNPs using one, two, three, four, and all measurements from each individual. We used a set of 67,547 non-Hispanic white individuals, all with ≥5 BP measurements available for this analysis, to keep the sample size identical among comparisons. We also examined the variance explained by a GRS of the previously-described hits assuming previous effect sizes as a function of number of BP measurements.
From this analysis, we can also estimate both the variance due to measurement error and variance explained by the GRS in the absence of measurement error, as follows. Let B = observed BP measurement, G = the GRS, E = residual genetic and environmental effect on BP, M = component of BP due to measurement error, and k = number of BP measurements. We assume that the measurement error is independent across multiple measures within an individual, and the additive model B=G+E+Mk for the average of k BP measurements. Let VB=Var(B), VG=Var(G), VE=Var(E), and VM=Var(M). For k BP measurements with independent measurement error, VMk=VM/k. The proportion H of BP variance attributable to the GRS is VG/(VG+VE+VM/k). Then 1/H = (1+VE/VG)+(VM/VG)/k=α+β(1/k) where α=1+VE/VG and β=VM/VG. We thus have a linear model of 1/H in terms of 1/k, and 1/α=VG/(VG+VE) is the proportion of variance due to the GRS in the absence of measurement error, and β/(α+β) is the proportion of variance in BP due to measurement error. Fitting a linear regression model to 1/H as a function of 1/k, we can then use the estimated intercept (α) and regression coefficient (β) to estimate the error variance and variance due to the GRS in the absence of measurement error.
BP risk scores and onset of hypertension
We additionally tested GRS constructed by weighting different subsets of identified BP-associated SNPs (i.e., identified for SBP, for DBP, and for PP, constructed as described above). Hypertension onset here was defined as the first hypertension treatment time, or the first time either SBP≥140 or DBP≥90 occurred in an individual and was maintained for the next subsequent BP measurement. Individuals were left censored at their first measurement (and not included if already meeting the hypertension diagnosis criterion), and right censored at their latest measurement if not hypertensive.
Differences in SBP, DBP, and PP effects
We also tested if the normalized effect size of each SNP was different for SBP versus DBP. Suppose that Y is SBP normalized to a standard normal (mean centered, then divided by the standard deviation) and Z is normalized DBP, and X is the SNP dosage. Then we model Y=aX+E and Z=bX+F, where a is the regression coefficient for Y on X and similarly b for Z; E and F are the residual errors, respectively. Since Var(Y)=Var(Z)=1, assuming a and b have the same sign (which is generally the case since the phenotypes are correlated), testing the equality of a and b is also a test of effect difference between SBP and DBP. Now, consider the difference Y-Z=(a-b)X+(E-F). Regressing Y-Z on X tests the difference between a and b; in this analysis, we additionally adjust for the same covariates as discussed previously.
GWAS Heritability from all Measured SNPs
We estimated the additive array heritability of each individual's long-term average age and BMI-adjusted BP residuals using GEAR v0.7.730. Array heritability estimates may be more sensitive to artifacts than GWAS results57, so we restricted our analysis to the largest group of individuals, non-Hispanic whites, that were run with the same reagent kit and type of microarray (n=73,133)46. We used only autosomal data, a common practice in array heritability estimation, and also LD-filtered our data so no two pairwise SNPs had r2>0.8 with a standard greedy algorithm in plink v1.0758. This resulted in 547,922 genotyped SNPs, and 3,796,606 imputed SNPs restricted to rinfo2>0.8. Because of population stratification, we used PC-Relate29 to estimate kinship coefficients rather than the standard GCTA estimates31 which assume a homogeneous population; we also compared the results to those obtained using the standard GCTA kinship estimates with PC adjustment. We used GEAR rather than GCTA to estimate heritability since the PC-Relate kinship matrix estimate was not positive definite; this can be explained by the fact that the matrix entries are computed based on different allele frequencies, i.e., those depending on ancestry from the PC analysis. In all analyses we removed individuals so that no two remaining individuals had a kinship estimate >0.025; sample size was maximized with Plink v1.959, leaving us with 62,133 individuals.
eQTL enrichment analysis
To carry out tissue-specific eQTL enrichment analysis, we used 44 tissue types with at least 70 samples available from GTex32 in addition to seven kidney eQTLs33. We used 367 sentinel variants from previously-identified SNPs and the three discovery stages presented here with MAF>0.001 and in eQTL databases. Next, 100 sets of 367 random pseudo-sentinel variants were selected matching the MAF to the original 367 (within ±0.5%). Within each set, the selection was done without replacement; the match for each variant was selected one-at-a-time, and selection of the subsequent variant excluded all previously-selected variants, as well as all variants within ±0.5 Mb of all previously-selected variants.
Enrichment was tested at both the sentinel SNP level and locus level, conceptually similar to Nicolae et al.60. At the sentinel SNP level, the number of variants that were also eQTLs in any of the 45 tissues was counted. At the locus level, variants in high LD (r2>0.8) with any of the 367 sentinel variants were examined for overlap with eQTLs, and if at least one variant within the locus was also an eQTL, the locus was counted. Subsequently, this was repeated for 100 randomly generated sets to observe if an eQTL enrichment was visible in the GWAS set. In order to assess which of the 45 tissues were driving the enrichment, counts were also computed per tissue. For each tissue, an upper-tailed p-value for enrichment of the GWAS count was calculated with a Z-score computed using the mean and standard deviation of the null distribution for that tissue.
DAVID analysis
Annotation of genes surrounding sentinel variants was conducted with DAVID 6.8 beta (non-beta was 6 years old)34,35. Genes within a ±0.5Mb window of each of the 390 sentinel variants were selected, as defined by GENCODE v19 GTF61. Subsequently, those with at least one significant eQTL in tissues identified from the previous enrichment analysis were included in the final list for analysis. Functional annotation analysis was run on the Homo sapiens background with default annotations in the categories of disease, functional categories, gene ontology, pathways, and protein domains, as well as with default parameters, retaining terms with at least two assigned genes. Annotation terms meeting Benjamini-Hochberg P<0.05 (adjusting for the number of terms) were considered significant.
Data availability
Data, including all genotype data and information on hypertension status, are available on approximately 78% of GERA participants from dbGaP under accession code phs000674.v1.p1 . This includes individuals who consented to having their data shared with dbGaP. The complete GERA data are available upon application to the KP Research Bank Portal, http://researchbank.kaiserpermanente.org/for-researchers/. The ICBP summary statistics are available from dbGaP under accession code phs000585.v1.p1. The UK Biobank data are available upon application to the UK Biobank, www.biobank.ac.uk.
Supplementary Material
Acknowledgements
We are grateful to the Kaiser Permanente Northern California members who have generously agreed to participate in the Kaiser Permanente Research Program on Genes, Environment, and Health. Support for participant enrollment, survey completion, and biospecimen collection for the RPGEH was provided by the Robert Wood Johnson Foundation, the Wayne and Gladys Valley Foundation, the Ellison Medical Foundation, and Kaiser Permanente Community Benefit Programs. Genotyping of the GERA cohort was funded by a grant from the National Institute on Aging, National Institute of Mental Health, and the National Institute of Health Common Fund (RC2 AG036607 to CAS and NJR). This research has been conducted using the UK Biobank Resource. This research has also been conducted using access-controlled ICBP data from dbGaP. We thank our colleagues for making these data available. Data analyses were facilitated by NHLBI grant R01 HL128782 (to AC and NJR). GE receives support from Geneva University Hospitals and The Foundation of Medical Researchers, Geneva. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflicts of interest: No conflicts of interest were disclosed.
URL section. UK Biobank, www.biobank.ac.uk; KP Research Bank Portal, http://researchbank.kaiserpermanente.org/for-researchers/; dbGaP, www.ncbi.nlm.nih.gov.
Accession codes. GERA dbGaP, phs000674.v1.p1; ICBP dbGaP, phs000585.v1.p1.
Author contributions. T.J.H., G.B.E., C.I., A.C., and N.R. conceived and designed the study. P.-Y.K. supervised the creation of genotype data. D.R., in collaboration with C.I., C.S., T.J.H. and N.R., extracted phenotype data from the EHR. T.J.H., P.N., D.R., and N.R. performed the statistical analyses. T.J.H., G.B.E., P.N., C.I., A.C., and N.R interpreted the results of analyses. All authors contributed to the drafting and critical review of the manuscript.
References (for main text)
- 1.Yang Q, Cogswell ME, Flanders W, et al. TRends in cardiovascular health metrics and associations with all-cause and cvd mortality among us adults. JAMA. 2012;307:1273–1283. doi: 10.1001/jama.2012.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy D, et al. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med. Genet. 2007;8(Suppl 1):S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miall WE, Oldham PD. The Hereditary Factor in Arterial Blood-pressure. Br. Med. J. 1963;1:75–80. doi: 10.1136/bmj.1.5323.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox ER, et al. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum. Mol. Genet. 2011;20:2273–2284. doi: 10.1093/hmg/ddr092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franceschini N, et al. Genome-wide Association Analysis of Blood-Pressure Traits in African-Ancestry Individuals Reveals Common Associated Genes in African and Non-African Populations. Am. J. Hum. Genet. 2013;93:545–554. doi: 10.1016/j.ajhg.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganesh SK, et al. Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum. Mol. Genet. 2013;22:1663–1678. doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganesh SK, et al. Effects of Long-Term Averaging of Quantitative Blood Pressure Traits on the Detection of Genetic Associations. Am. J. Hum. Genet. 2014;95:49–65. doi: 10.1016/j.ajhg.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho JE, et al. Discovery and replication of novel blood pressure genetic loci in the Women's Genome Health Study. J. Hypertens. 2011;29:62–69. doi: 10.1097/HJH.0b013e3283406927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson T, et al. Blood Pressure Loci Identified with a Gene-Centric Array. Am. J. Hum. Genet. 2011;89:688–700. doi: 10.1016/j.ajhg.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato N, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat. Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato N, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat. Genet. 2015;47:1282–1293. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly TN, et al. Genome-Wide Association Study Meta-Analysis Reveals Transethnic Replication of Mean Arterial and Pulse Pressure Loci. Hypertension. 2013;62:853–859. doi: 10.1161/HYPERTENSIONAHA.113.01148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy D, et al. Genome-wide association study of blood pressure and hypertension. Nat. Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newton-Cheh C, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padmanabhan S, et al. Genome-Wide Association Study of Blood Pressure Extremes Identifies Variant near UMOD Associated with Hypertension. PLoS Genet. 2010;6:e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simino J, et al. Gene-Age Interactions in Blood Pressure Regulation: A Large-Scale Investigation with the CHARGE, Global BPgen, and ICBP Consortia. Am. J. Hum. Genet. 2014;95:24–38. doi: 10.1016/j.ajhg.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehret GB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeuchi F, et al. Blood Pressure and Hypertension Are Associated With 7 Loci in the Japanese Population. Circulation. 2010;121:2302–2309. doi: 10.1161/CIRCULATIONAHA.109.904664. [DOI] [PubMed] [Google Scholar]
- 19.Tragante V, et al. Gene-centric Meta-analysis in 87,736 Individuals of European Ancestry Identifies Multiple Blood-Pressure-Related Loci. Am. J. Hum. Genet. 2014;94:349–360. doi: 10.1016/j.ajhg.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wain LV, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat. Genet. 2011;43:1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, et al. Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc. Natl. Acad. Sci. 2009;106:226–231. doi: 10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu X, et al. Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. Am. J. Hum. Genet. 2015;96:21–36. doi: 10.1016/j.ajhg.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Global Lipids Genetics Consortium Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sewell K, et al. Blood Pressure Measurement Biases in Clinical Settings, Alabama, 2010–2011. Prev. Chronic. Dis. 2016;13 doi: 10.5888/pcd13.150348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nwankwo T, Yoon SS, Burt V, Gu Q. Hypertension among adults in the United States: National Health and Nutrition Examination Survey, 2011-2012. NCHS Data Brief. 2013;1–8 [PubMed] [Google Scholar]
- 26.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 2004;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 27.Sudlow C, et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLOS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraft P. Curses—Winner's and Otherwise—in Genetic Epidemiology. Epidemiology. 2008;19:649–651. doi: 10.1097/EDE.0b013e318181b865. [DOI] [PubMed] [Google Scholar]
- 29.Conomos MP, Reiner AP, Weir BS, Thornton TA. Model-free Estimation of Recent Genetic Relatedness. Am. J. Hum. Genet. 2016;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen G-B. Estimating heritability of complex traits from genome-wide association studies using IBS-based Haseman–Elston regression. Stat. Genet. Methodol. 2014;5:107. doi: 10.3389/fgene.2014.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Consortium T. Gte. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, et al. Synthesis of 53 tissue and cell line expression QTL datasets reveals master eQTLs. BMC Genomics. 2014;15:532. doi: 10.1186/1471-2164-15-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kho AN, et al. Electronic Medical Records for Genetic Research: Results of the eMERGE Consortium. Sci. Transl. Med. 2011;3:79re1–79re1. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tada H, et al. Multiple Associated Variants Increase the Heritability Explained for Plasma Lipids and Coronary Artery Disease. Circ. Cardiovasc. Genet. 2014;7:583–587. doi: 10.1161/CIRCGENETICS.113.000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuchsberger C, et al. The genetic architecture of type 2 diabetes. Nature advance online publication. 2016 [Google Scholar]
- 40.Hoffmann TJ, et al. Next generation genome-wide association tool: Design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011;98:79–89. doi: 10.1016/j.ygeno.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann TJ, et al. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics. 2011;98:422–430. doi: 10.1016/j.ygeno.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu C, et al. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat. Genet. advance online publication. 2016 doi: 10.1038/ng.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Surendran P, et al. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat. Genet. advance online publication. 2016 doi: 10.1038/ng.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehret GB, et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat. Genet. advance online publication. 2016 doi: 10.1038/ng.3667. References (Methods-only) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banda Y, et al. Characterizing Race/Ethnicity and Genetic Ancestry for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200:1285–1295. doi: 10.1534/genetics.115.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kvale MN, et al. Genotyping Informatics and Quality Control for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200:1051–1060. doi: 10.1534/genetics.115.178905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat. Med. 2005;24:2911–2935. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 48.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 49.Howie B, Marchini J, Stephens M. Genotype Imputation with Thousands of Genomes. G3 Genes Genomes Genet. 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howie BN, Donnelly P, Marchini J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 53.Huang L, Wang C, Rosenberg NA. The Relationship between Imputation Error and Statistical Power in Genetic Association Studies in Diverse Populations. Am. J. Hum. Genet. 2009;85:692–698. doi: 10.1016/j.ajhg.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loh P-R, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han B, Eskin E. Random-Effects Model Aimed at Discovering Associations in Meta-Analysis of Genome-wide Association Studies. Am. J. Hum. Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pasaniuc B, et al. Fast and accurate imputation of summary statistics enhances evidence of functional enrichment. Bioinformatics. 2014;30:2906–2914. doi: 10.1093/bioinformatics/btu416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating Missing Heritability for Disease from Genome-wide Association Studies. Am. J. Hum. Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Purcell S, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4 doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nicolae DL, et al. Trait-Associated SNPs Are More Likely to Be eQTLs: Annotation to Enhance Discovery from GWAS. PLOS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harrow J, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data, including all genotype data and information on hypertension status, are available on approximately 78% of GERA participants from dbGaP under accession code phs000674.v1.p1 . This includes individuals who consented to having their data shared with dbGaP. The complete GERA data are available upon application to the KP Research Bank Portal, http://researchbank.kaiserpermanente.org/for-researchers/. The ICBP summary statistics are available from dbGaP under accession code phs000585.v1.p1. The UK Biobank data are available upon application to the UK Biobank, www.biobank.ac.uk.