Abstract
The most frequent recurring mutations in neurofibromatosis type 1 (NF1) are large deletions encompassing the NF1 gene and its flanking regions (NF1 microdeletions). The majority of these deletions encompass 1.4-Mb and are associated with the loss of 14 protein-coding genes and four microRNA genes. Patients with germline type-1 NF1 microdeletions frequently exhibit dysmorphic facial features, overgrowth/tall-for-age stature, significant delay in cognitive development, large hands and feet, hyperflexibility of joints and muscular hypotonia. Such patients also display significantly more cardiovascular anomalies as compared with patients without large deletions and often exhibit increased numbers of subcutaneous, plexiform and spinal neurofibromas as compared with the general NF1 population. Further, an extremely high burden of internal neurofibromas, characterised by >3000 ml tumour volume, is encountered significantly, more frequently, in non-mosaic NF1 microdeletion patients than in NF1 patients lacking such deletions. NF1 microdeletion patients also have an increased risk of malignant peripheral nerve sheath tumours (MPNSTs); their lifetime MPNST risk is 16–26%, rather higher than that of NF1 patients with intragenic NF1 mutations (8–13%). NF1 microdeletion patients, therefore, represent a high-risk group for the development of MPNSTs, tumours which are very aggressive and difficult to treat. Co-deletion of the SUZ12 gene in addition to NF1 further increases the MPNST risk in NF1 microdeletion patients. Here, we summarise current knowledge about genotype–phenotype relationships in NF1 microdeletion patients and discuss the potential role of the genes located within the NF1 microdeletion interval whose haploinsufficiency may contribute to the more severe clinical phenotype.
Introduction
Neurofibromatosis type 1 (NF1; MIM#162200) is a tumour predisposition syndrome with an incidence at birth of 1 in 2000–3000 (Crowe et al. 1956; Lammert et al. 2005; Uusitalo et al. 2015). The hallmark features of NF1 are café-au-lait spots (CALS) and the pathognomonic neurofibromas. The majority of NF1 patients are characterised by mutations residing within the boundaries of the NF1 gene, which spans 287-kilobases (kb) of chromosome 17q11.2 and comprises 57 constitutive and 3 alternatively spliced exons.
Only a few genotype–phenotype correlations in NF1 have been identified to date. One of these relates to spinal neurofibromatosis (SNF) which is characterised by bilateral neurofibromas located at all 38 spinal nerve roots. The risk of having SNF versus NF1 without spinal neurofibromas, or NF1 with neurofibromas affecting only some but not all spinal nerve roots, is significantly increased in individuals harbouring NF1 missense mutations (Ruggieri et al. 2015). Furthermore, the recurrent three base-pair in-frame deletion, c.2970-2972 delAAT, within exon 17 of the NF1 gene leads to the loss of a single amino acid (p.Met992del) and is associated with a relatively mild NF1 phenotype that is characterised by the occurrence of CALS and skinfold freckling but a lack of externally visible cutaneous or plexiform neurofibromas (Upadhyaya et al. 2007). The second well-established genotype–phenotype correlation in NF1 is associated with missense mutations affecting codon p.Arg1809. Individuals with these very specific missense NF1 mutations exhibit CALS (with or without freckling) and Lisch nodules, but no externally visible plexiform neurofibromas or cutaneous neurofibromas (Pinna et al. 2015; Rojnueangnit et al. 2015). Approximately, 25% of the individuals with missense mutations affecting codon p.Arg1809 have Noonan-like features including pulmonic stenosis and short stature whilst 50% of them exhibit developmental delay and/or learning disability (Rojnueangnit et al. 2015). However, missense mutations affecting codon p.Arg1809 appear to be quite rare, since they were observed in only 1.2% of the cohort of 7000 NF1 patients with identified mutations. In the same cohort of patients, the prevalence of the recurrent one amino acid deletion (p.Met992del) was 0.8% (Rojnueangnit et al. 2015).
The third genotype–phenotype relationship evident in NF1 is that associated with large NF1 deletions and is the topic of this review. An estimated 4.7–11% of all NF1 patients have large deletions encompassing the entire NF1 gene and its flanking regions at 17q11.2 (Cnossen et al. 1997; Rasmussen et al. 1998; Kluwe et al. 2004; Zhang et al. 2015). Large deletions of the NF1 gene and its flanking regions (generally termed ‘NF1 microdeletions’) are frequently associated with a severe clinical manifestation of NF1 as described below.
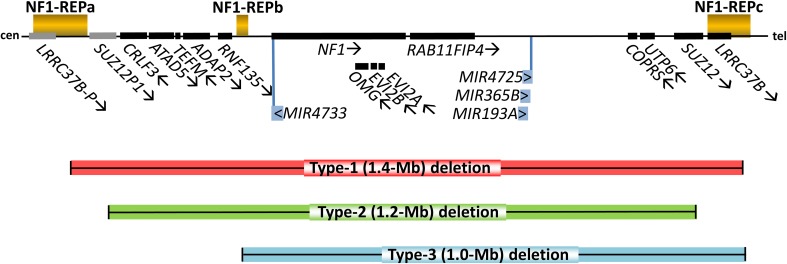
Altogether, four types of large NF1 deletion (type-1, 2, 3 and atypical) have been identified that are distinguishable in terms of their size and breakpoint location, by the number of genes located within the deletion region or by the frequency of somatic mosaicism with normal cells not harbouring the deletion. Most frequent are the type-1 NF1 deletions which encompass 1.4-Mb and include 14 protein-coding genes as well as four microRNA genes (Fig. 1) (Dorschner et al. 2000; Jenne et al. 2001; López-Correa et al. 2001). Type-1 deletions account for 70–80% of all large NF1 deletions and usually occur as germline deletions that are present in all cells of the affected patients (Messiaen et al. 2011). Most type-1 NF1 deletions are caused by interchromosomal non-allelic homologous recombination (NAHR) during maternal meiosis (López-Correa et al. 2000; Steinmann et al. 2008). The NAHR events causing type-1 NF1 deletions are mediated by the low-copy repeats, NF1-REPa and NF1-REPc. Within these low-copy repeats, recurrent breakpoints have been detected within two NAHR hotspots, termed paralogous recombination sites 1 and 2 (Forbes et al. 2004; De Raedt et al. 2006; Bengesser et al. 2014; Hillmer et al. 2016).
Fig. 1.
Schema of the genomic region at 17q11.2 harbouring the NF1 gene and its flanking genes included within the boundary of the type-1 NF1 deletion interval encompassing 1.4-Mb (red bar). The arrows given subsequent to the symbols of the genes denote their transcriptional orientation. SUZ12P1 and LRRC37B-P are non-functional pseudogenes. cen centromeric direction, tel telomeric direction
In contrast to type-1 NF1 deletions, type-2 deletions encompass only 1.2-Mb and are associated with hemizygosity for 13 protein-coding genes since the LRRC37B gene is absent from the deleted region (Fig. 1). At least 10% of large NF1 deletions are type-2 but this is very likely to be an underestimate (Messiaen et al. 2011). Type-2 deletions are also mediated by NAHR but in contrast to type-1 NF1 deletions, their breakpoints are located within SUZ12 and its highly homologous pseudogene SUZ12P1 which flank NF1-REPc and NF1-REPa, respectively (Fig. 1) (Petek et al. 2003; Vogt et al. 2012). Type-2 NF1 deletions are frequently of postzygotic origin, mediated by mitotic NAHR, and hence are associated with somatic mosaicism of normal cells lacking the deletion (Kehrer-Sawatzki et al. 2004; Steinmann et al. 2007; Roehl et al. 2010, 2012).
Type-3 NF1 deletions are very rare; these 1.0-Mb deletions occur in only 1-4% of all patients with gross NF1 deletions and are mediated by NAHR between NF1-REPb and NF1-REPc leading to hemizygosity for a total of nine protein-coding genes (Fig. 1) (Bengesser et al. 2010; Pasmant et al. 2010; Zickler et al. 2012). As their name suggests, atypical large NF1 deletions do not exhibit recurrent breakpoints and are quite heterogeneous in terms of their size and the number of genes located within the deleted region (Upadhyaya et al. 1996; Cnossen et al. 1997; Dorschner et al. 2000; Kehrer-Sawatzki et al. 2003, 2005, 2008; Venturin et al. 2004a; Gervasini et al. 2005; Mantripragada et al. 2006; Pasmant et al. 2008, 2009, 2010; Vogt et al. 2014). It has been estimated that 8–10% of all large NF1 deletions are atypical (Pasmant et al. 2010; Messiaen et al. 2011). Atypical NF1 deletions may occur as germline mutations but can also be of postzygotic origin and hence may be associated with somatic mosaicism with normal cells (Taylor Tavares et al. 2013; Vogt et al. 2014). Atypical NF1 deletions are not only highly heterogeneous in terms of their length but also in terms of their underlying mutational mechanisms which may involve aberrant DNA double strand break repair and/or replication and retrotransposon-mediated mechanisms (Vogt et al. 2014 and references therein). The architecture of the genomic regions flanking the NF1 gene in 17q11.2, characterised by low-copy repeats, predisposes to large deletions mediated by various different mutational mechanisms occurring in the germline of an unaffected parent or during mitotic postzygotic cell divisions.
NF1 microdeletions are important from the clinical standpoint because, as noted above, they are often associated with more severe manifestations of NF1 than those noted in patients with intragenic NF1 mutations. One interpretation of this observation is that some of those genes co-deleted with NF1 exert an influence on the clinical manifestation of the disease in patients with NF1 microdeletions. In the following, we shall review current knowledge about NF1 microdeletions in terms of potential genotype–phenotype relationships and the putative modifier role of genes located within the NF1 microdeletion interval.
Genotype–phenotype relationships in patients with NF1 microdeletions
The NF1 gene was identified more than 25 years ago (Viskochil et al. 1990; Wallace et al. 1990). Genotype–phenotype analyses suggested from very early on that patients with NF1 microdeletions often exhibit a more severe clinical phenotype than patients with intragenic NF1 mutations; the former are frequently characterised by dysmorphic facial features and severe developmental delay (Kayes et al. 1992, 1994; Wu et al. 1995, 1997, 1999; Riva et al. 1996, 2000; Upadhyaya et al. 1996, 1998; Leppig et al. 1997; Tonsgard et al. 1997; Valero et al. 1997; Rasmussen et al. 1998; Streubel et al. 1999; Dorschner et al. 2000; Kobayashi et al. 2012).
Subsequent follow-up studies confirmed that these deletions frequently lead to severe clinical manifestations of the disease including a high tumour load and cardiovascular anomalies (Venturin et al. 2004b; Mensink et al. 2006; Pasmant et al. 2010; Zhang et al. 2015). Although, as a group, NF1 microdeletion patients tend to exhibit a comparatively severe form of NF1, some variability of clinical symptoms has nevertheless been observed when comparing individuals with NF1 microdeletions. These clinical phenotypic differences may be influenced by the variable expressivity of NF1 characteristic of all NF1 patients, irrespective of the type of NF1 gene mutation involved (Sabbagh et al. 2009; reviewed by Pasmant et al. 2012). However, a certain proportion of the phenotypic variability exhibited by patients with NF1 microdeletions who were investigated in the above-mentioned studies may have been due to differences in deletion size and breakpoint location which together determine the number of genes included within the deletion interval.
Somatic mosaicism with cells not harbouring the NF1 microdeletion in question is likely to have a disproportionately large impact upon the manifestations of disease (Rasmussen et al. 1998; Tinschert et al. 2000; Maertens et al. 2007; Kehrer-Sawatzki and Cooper 2008; Roehl et al. 2012; Kehrer-Sawatzki et al. 2012). This is particularly relevant since some types of NF1 microdeletion, such as the type-2 and atypical NF1 deletions, are frequently of postzygotic origin and hence occur as mosaic deletions alongside normal cells in the body of an affected patient (Steinmann et al. 2007; Vogt et al. 2014). By contrast, type-1 NF1 deletions are only very rarely of postzygotic origin (Messiaen et al. 2011). This notwithstanding, in many of the reported studies of genotype–phenotype correlations in patients with NF1 microdeletions, neither deletion size nor somatic mosaicism has been specifically taken into consideration.
To refine the genotype–phenotype analysis of NF1 microdeletions, Mautner et al. (2010) investigated 29 NF1 patients with non-mosaic type-1 (1.4-Mb) NF1 deletions. A combination of breakpoint-spanning PCRs and/or polymorphic marker analysis confirmed that the deletion breakpoints were located within specific regions of the NF1-REPs. Thus, all 29 patients studied were hemizygous for the same number of genes at 17q11.2 (Fig. 1). The clinical analysis of these 29 patients with type-1 NF1 microdeletions served to confirm that several clinical phenotypic features were relatively frequent in patients with NF1 microdeletions but less common (or even not identifiable) in the general NF1 population (as may be concluded from the studies listed in Table 1). These features included facial dysmorphism, overgrowth/tall-for-age stature, significant delay in cognitive development, scoliosis, bone cysts, large hands and feet with excessive soft tissue, hyperflexibility of joints of hands and feet and pronounced muscular hypotonia. Further, patients with type-1 NF1 microdeletions exhibited a variety of features that were markedly more frequent than in the general NF1 population including intellectual disability, high numbers of subcutaneous and spinal neurofibromas, and the occurrence of plexiform neurofibromas (Table 1). An increased frequency of optic gliomas was however not observed in patients with type-1 NF1 deletions as compared to the general NF1 population. In the studies listed in Table 1 that compared the frequency of clinical features, NF1 patients were analysed irrespective of the type of NF1 mutation they harboured; these patients are therefore referred to as the ‘general NF1 population’. Such comparisons will tend to err on the conservative side since 4.7–11% of the individuals in the general NF1 population harbour NF1 microdeletions.
Table 1.
Frequency of clinical symptoms in patients with type-1 NF1 microdeletions investigated by Mautner et al. (2010) and in the general NF1 population
| Clinical features | Frequency in patients with type-1 NF1 deletions, (%) | Frequency in the general NF1 population (reference), (%) | |
|---|---|---|---|
| Facial dysmorphism | 90 | n.d. | |
| Hypertelorism | 86 | n.d. | |
| Facial asymmetry | 28 | 8 6 |
(Friedman and Birch 1997) (Sbidian et al. 2012) |
| Coarse face | 59 | n.d. | |
| Broad neck | 31 | n.d. | |
| Tall-for-age stature | 46 | n.d. | |
| Macrocephalya | 39 | 45 29 43 24 |
(Huson et al. 1988) (Riccardi 1992) (North 1993) (Sbidian et al. 2012) |
| Large hands and feet | 46 | n.d. | |
| Pes cavus | 17 | n.d. | |
| Café-au-lait spots | 93 | 87 95 86 99 |
(McGaughran et al. 1999) (Friedman 1999) (Duong et al. 2011) (Sbidian et al. 2012) |
| Axillary and inguinal freckling | 86 | 86 89 |
(Duong et al. 2011) (Plotkin et al. 2012) |
| Lisch nodules | 93 | 63 93 50 45 |
(McGaughran et al. 1999) (Huson et al. 1988) (Friedman 1999) (Sbidian et al. 2012) |
| Significant delay in cognitive development | 48 | 17 | (Klein-Tasman et al. 2014) |
| General learning difficulties | 45 | 45 47 32 39 31 |
(North et al. 1995) (Brewer et al. 1997)b (Hyman et al. 2006) (Krab et al. 2008)c (Plotkin et al. 2012) |
| IQ < 70 | 38 | 8 7 |
(Ferner et al. 1996) (Hyman et al. 2005) |
| Attention deficit hyperactivity disorder | 33 | 49 38 |
(Mautner et al. 2002) (Hyman et al. 2005) |
| Skeletal anomalies | 76 | 31 | (Plotkin et al. 2012) |
| Scoliosis | 43 | 26 12 10 20 25 28 |
(Friedman and Birch 1997) (McGaughran et al. 1999) (Riccardi 1999a) (North 2000) (Duong et al. 2011) (Plotkin et al. 2012) |
| Pectus excavatum | 31 | 50 12 |
(Riccardi 1999b) (Castle et al. 2003) |
| Bone cysts | 50 | 1 | (Plotkin et al. 2012) |
| Hyperflexibility of joints | 72 | n.d. | |
| Excess soft tissue in hands and feet | 50 | n.d. | |
| Congenital heart defects | 29 | 2 1.6–2 |
(Friedman and Birch 1997) (Lin et al. 2000) |
| Epilepsy | 7 | 7 4 13 4 |
(Huson et al. 1988) (North 1993) (Ferner et al. 1996) (Kulkantrakorn and Geller 1998) |
| Muscular hypotonia | 45 | 27 | (Wessel et al. 2013) |
| Speech difficulties | 48 | 25 20–55 |
(North 1999
n.d.) (Alivuotila et al. 2010)d |
| Subcutaneous neurofibromas | 76 | 48 | (Tucker et al. 2005) |
| Cutaneous neurofibromas | 86 | 38–44 59 85 76 84 |
(Friedman and Birch 1997) (McGaughran et al. 1999) (Tucker et al. 2005) (Duong et al. 2011) (Plotkin et al. 2012) |
| Plexiform neurofibromas | 76 | 15 44 50 30 54 |
(McGaughran et al. 1999) (Waggoner et al. 2000) (Ferner et al. 2007) (Duong et al. 2011) (Plotkin et al. 2012) |
| Malignant peripheral nerve sheath tumours | 21 | 2–5 7 |
(Ferner and Gutmann 2002) (Duong et al. 2011) |
| Spinal neurofibromas | 64 | 30 24 |
(Tucker et al. 2005) (Plotkin et al. 2012) |
| Optic pathway gliomas | 19 | 15 19 14 11 18 |
(Lewis et al. 1984) (Listernick et al. 1997) (Duong et al. 2011) (Plotkin et al. 2012) (Millichap 2015; Prada et al. 2015) |
| T2 hyperintensities | 45 | 34 77 79 71 |
(Ferner et al. 1993) (Itoh et al. 1994) (Sevick et al. 1992) (Hyman et al. 2007) |
n.d. not determined; these features are either absent or rare in the general NF1 population
aThe evaluation criteria in these studies included a definition of macrocephaly as an occipitofrontal circumference greater than the 98th centile or two standard deviations above the mean. Despite consistent evaluation criteria having been employed, a high degree of variability in terms of the frequency of macrocephaly has been observed
b39% of the children with NF1 analysed by Brewer et al. (1997) exhibited general learning disabilities whereas an additional 14% exhibited visuospatial-construction deficiencies (specific learning disabilities)
c39% of the children with NF1 investigated by Krab et al. (2008) had general learning disabilities whilst an additional 39% had specific learning disabilities
d Alivuotila et al. (2010) investigated the speech characteristics of 62 NF1 patients (40 adults and 22 children) and compared them with those observed in 24 control individuals. Patients with NF1 exhibited deviations in voice quality (35% of the adult NF1 patients and 55% of the children with NF1), problems in regulating pitch (53% of the adult NF1 patients and 55% of the children), deviant nasality (20% of the adult NF1 patients and 45% of the children) and disfluency (20% of the adult NF1 patients and 41% of the children)
In addition to the previously mentioned clinical features, type-1 NF1 microdeletion patients were found to have an increased risk of malignant peripheral nerve sheath tumours (MPNSTs) (De Raedt et al. 2003; Mautner et al. 2010) (Table 1). These findings confirmed that patients with non-mosaic type-1 NF1 deletions exhibit, as a group, a severe form of NF1. However, even within this group of patients that are hemizygous for the same number of genes, some variability in expression of the clinical symptoms has been detected (Mautner et al. 2010). Hence, the phenotype associated with NF1 microdeletions is likely to be influenced to a certain degree by the genetic background (e.g. the expression level of non-deleted genes), as well as by environmental factors.
It should be emphasised that many of the studies that have addressed the question of whether or not specific disease features occur disproportionately more frequently in patients with NF1 microdeletions have employed, for the purposes of comparative analysis, frequency values for these clinical features that were derived from the general NF1 population obtained in different studies. More appropriate would have been a methodical comparative analysis of a large number of age-matched patients with and without germline type-1 NF1 deletions investigated by standardised analytical tools. Although such comparative analyses have been attempted to assess differences in height (Ning et al. 2016), cognitive capability (Descheemaeker et al. 2004) or the frequency of cardiovascular anomalies (Nguyen et al. 2013), they have not as yet been performed methodically in the context of the number of neurofibromas and other NF1 microdeletion-associated clinical features.
In the following, the clinical features associated with type-1 NF1 microdeletions are explored in greater detail.
High number of neurofibromas in patients with NF1 microdeletions
Several studies have suggested that NF1 microdeletion patients frequently exhibit a disproportionately high number of cutaneous and subcutaneous neurofibromas (Mensink et al. 2006 and references therein; Pasmant et al. 2010). Indeed, the 29 patients with type-1 NF1 microdeletions investigated by Mautner et al. (2010) exhibited significantly increased numbers of subcutaneous but also spinal neurofibromas by comparison with the general NF1 population as concluded from some of the studies listed in Table 1. Additionally, externally visible plexiform neurofibromas are significantly more frequent in patients with NF1 microdeletions than in the general NF1 population (Table 1; Mautner et al. 2010). Remarkably, 10 of 20 (50%) of the adult type-1 NF1 microdeletion patients investigated by Mautner et al. (2010) exhibited a very high number of cutaneous neurofibromas (N > 1000). Such a high burden of cutaneous neurofibromas may also be present in some patients with intragenic NF1 mutations but the precise frequency of this feature in this subgroup of NF1 patients is currently unknown. Plotkin et al. (2012) reported that 13% of 141 NF1 patients exhibited more than 500 cutaneous neurofibromas, but the precise proportions of patients harbouring either NF1 microdeletions or intragenic NF1 mutations among the 141 NF1 patients investigated had not been determined. A detailed comparison between both patient groups, involving the careful age-matching of patients, would be necessary to clarify whether a very high load of cutaneous neurofibromas (N > 1000) is encountered significantly more frequently in patients with type-1 NF1 microdeletions as compared to patients with intragenic NF1 mutations. In passing, it should be pointed out that counting neurofibromas one by one in NF1 patients who may have thousands of such tumours is likely to be subject to considerable intra- and inter-examiner variability, and hence the precise number of neurofibromas in each patient should always be regarded as a rough estimate. In an attempt to improve upon this state of affairs, Cunha et al. (2014) developed a new method using paper frames to quantify cutaneous neurofibromas. Combined with computerised analysis, this method could yet prove to be very useful for the comparative quantification of cutaneous neurofibromas in patients with NF1 microdeletions and those lacking such deletions.
Several studies have suggested that patients with NF1 microdeletions not only exhibit a high number of cutaneous neurofibromas but also an early (pre-pubertal) onset of cutaneuous neurofibroma growth (Kayes et al. 1992, 1994; Mensink et al. 2006; Leppig et al. 1997; Dorschner et al. 2000). However, a comprehensive analysis of age-matched children has not yet been performed to ascertain whether an early (pre-pubertal) onset in growth of multiple cutaneous neurofibromas is significantly more prevalent in children with NF1 microdeletions as compared to children with intragenic NF1 mutations.
In addition to tumours that are visible by external investigation, NF1 patients may also possess internal neurofibromas (mostly of the plexiform type) which would only be detectable by magnetic-resonance imaging (MRI). Kluwe et al. (2012) showed that an extremely high burden of internal neurofibromas, characterised by >3000 ml tumour volume as determined by whole-body MRI, was significantly more frequent in non-mosaic type-1 and type-2 NF1 microdeletion patients than in NF1 patients with intragenic lesions (13 vs. 1%). Consequently, a readily identifiable subgroup of NF1 patients with germline NF1 microdeletions is likely to exhibit an extremely high burden of internal tumours. These patients require special attention in terms of clinical care and surveillance since a strong association has been observed between the presence of internal neurofibromas and the occurrence of malignant peripheral nerve sheath tumours (MPNSTs) (Tucker et al. 2005; Mautner et al. 2008; Nguyen et al. 2014). MPNSTs are very aggressive, have a poor prognosis and frequently arise in pre-existing plexiform neurofibromas (Ferner and Gutmann 2002) which are often diagnosed before the age of 5, suggesting that they are congenital lesions (Waggoner et al. 2000). This postulate receives strong support from the observation that children with NF1 who do not exhibit plexiform neurofibromas upon first MRI examination are unlikely to develop new plexiform neurofibromas later in life (Nguyen et al. 2012). Hence, a high burden of internal neurofibromas may well be strongly associated with an increased MPNST risk and this should be taken into account when planning the clinical care of patients with NF1 microdeletions.
Increased risk of MPNSTs in patients with NF1 microdeletions
MPNSTs are rare soft tissue sarcomas occurring with an incidence of 0.001% in the overall (general) population (Ducatman et al. 1986) and are known to have an association with NF1. Thus, some 28–52% of patients with MPNSTs also have NF1 (Ducatman et al. 1986; Evans et al. 2002). The estimated lifetime risk of an MPNST in all NF1 patients is 8–13% (Evans et al. 2002, 2012) or 15.8% according to Uusitalo et al. (2016). However, individuals with NF1 microdeletions have an even higher lifetime MPNST risk, in the range of 16–26% (De Raedt et al. 2003; Mautner et al. 2010). Further, MPNSTs may occur significantly earlier in patients with NF1 microdeletions as compared with NF1 patients with intragenic mutations (De Raedt et al. 2003). These findings clearly indicate that patients with NF1 microdeletions constitute a high-risk group for the development of MPNSTs. Hence patients with NF1 microdeletions should be under regular surveillance from an early age.
MPNSTs are difficult to treat, particularly if they are at an advanced stage and have already metastasized. The complete surgical excision of non-metastatic MPNSTs represents the mainstay of an effective therapy but this is only possible if the tumour is detected at an early stage (reviewed by Karajannis and Ferner 2015). MPNSTs represent the biggest contributory factor to reduced life expectancy in NF1 (Evans et al. 2012). It may well be that an NF1 microdeletion and high internal tumour load are independent risk factors for MPNST. Thus, patients with an NF1 microdeletion and a high internal tumour load could represent an ultra-high risk group for MPNST. For this group of patients, long-term follow-up investigations using whole-body MRI and serial 18Fluorodeoxyglucose positron emission tomography (PET) scans are likely to be critically important to identify malignant transformation at an early stage (Salamon et al. 2015).
Intellectual disability in patients with NF1 microdeletions
An estimated, 4.8–8% of all NF1 patients are characterised by intellectual disability (mean full-scale IQ (FSIQ) <70), a somewhat higher proportion than the 2% observed in the normal population (Ferner et al. 1996; reviewed by North et al. 1997). Several studies have suggested that intellectual disability occurs disproportionately more frequently in NF1 microdeletion patients than in the general NF1 population (Kayes et al. 1994; Rasmussen et al. 1998; Korf et al. 1999; Venturin et al. 2004b; reviewed by Mensink et al. 2006). However, some of the early studies were biassed by including mostly patients with a particularly severe phenotype and uncharacterised deletion size. Only in two studies, the frequency of intellectual disability has been analysed systematically by including exclusively non-mosaic patients with NF1 microdeletions of the same size (type-1 deletions spanning 1.4-Mb) (Descheemaeker et al. 2004; Mautner et al. 2010). Intellectual disability was evident in eight of 21 patients (38%) with type-1 NF1 deletions analysed by Mautner et al. (2010) and in two of 11 patients with type-1 NF1 microdeletions (18%) investigated by Descheemaeker et al. (2004). Taken together, these findings indicate that intellectual disability is markedly more frequent in patients with type-1 NF1 microdeletions as compared to patients with intragenic NF1 mutations. Furthermore, borderline intellectual disability, characterised by a full-scale IQ (FSIQ) higher than 70 but lower than 85 (70 ≤ FSIQ ≤ 85), was noted in seven (33%) of the 21 type-1 NF1 microdeletion patients analysed by Mautner et al. (2010) and in nine of the 11 type-1 NF1 microdeletion patients (82%) investigated by Descheemaeker et al. (2004).
Patients with NF1, irrespective of their mutation type, have a mean FSIQ of 88-99 which is in the range of one standard deviation lower than the FSIQ of the general population (100 ± 15) (North et al. 1997; Krab et al. 2008). A mean FSIQ of 76.9 was documented in the 21 type-1 NF1 microdeletion patients investigated by Mautner et al. (2010). This is similar to the mean FSIQ of 76.0 ascertained in the 11 type-1 NF1 microdeletion patients analysed by Descheemaeker et al. (2004). By comparison, a mean FSIQ of 88.5 was determined in 106 NF1 patients without an NF1 deletion (Descheemaeker et al. 2004). Despite the relatively small numbers of individuals available in these studies, the tentative conclusion to be drawn from them is that the mean FSIQ in patients with type-1 NF1 microdeletions is markedly lower than the mean FSIQ in patients with intragenic NF1 mutations. However, Descheemaeker et al. (2004) noted a considerable overlap regarding the range of the FSIQ observed in patients with NF1 microdeletions (65–85) as compared with the range observed in patients without microdeletions (54–126), even though the average intelligence (as measured by FSIQ) of type-1 NF1 microdeletion patients is generally lower than that of NF1 patients without large microdeletions.
Overgrowth associated with NF1 microdeletions
Stature is reduced to some extent in virtually all patients with intragenic NF1 mutations. On average, adolescents and adults with NF1 are one standard deviation shorter than would be expected for their age and sex (Clementi et al. 1999; Szudek et al. 2000). Short stature, characterised by a height of more than two standard deviations below the predicted mean, is evident in 8–13% of children with NF1 (Szudek et al. 2000; Sbidian et al. 2012; Soucy et al. 2013) whilst a body height below the third percentile has been observed in 15% of children with NF1 (Clementi et al. 1999).
Short stature in children with NF1 and intragenic NF1 mutations has been suggested to be caused by a paucity of growth hormone as a consequence of abnormal hypothalamic–pituitary axis function (Vassilopoulou-Sellin et al. 2000). Support for this hypothesis comes from the phenotype observed in conditional knockout mice with specific Nf1 gene inactivation in neuroglial progenitor cells using the brain lipid-binding protein promoter (Hegedus et al. 2008). These mice exhibit significantly reduced body weight and anterior pituitary gland size caused by the loss of neurofibromin expression in the hypothalamus, leading to reduced production of growth hormone releasing hormone, pituitary growth hormone and liver-expressed insulin-like growth factor-1 (IGF1). Thus, it would appear that neurofibromin plays a critical role in hypothalamic–pituitary axis function and hence its loss may cause growth abnormalities in patients with NF1 (Hegedus et al. 2008).
In contrast to the reduced stature observed in most patients with intragenic NF1 mutations, tall stature in adults and childhood overgrowth has been reported to occur frequently in patients with NF1 microdeletions (van Asperen et al. 1998; Spiegel et al. 2005; Mensink et al. 2006; Pasmant et al. 2010). Tall-for-age stature, with height measurements at or above the 94th percentile, was noted in 46% of patients with germline type-1 NF1 deletions (Mautner et al. 2010). Since intragenic NF1 mutations lead to shorter height, and total loss of the NF1 gene plus flanking genes often results in tall-for-age stature, it may be concluded that haploinsufficiency of a gene or genes co-deleted in patients with NF1 microdeletions causes this overgrowth phenotype (Mautner et al. 2010). In a study of 21 patients with type-1 NF1 microdeletions, overgrowth was most evident in preschool children (2–6 years, n = 10) (Spiegel et al. 2005). These findings were confirmed by Ning et al. (2016) who performed longitudinal growth measurements in 56 NF patients with NF1 microdeletions and 226 NF1 patients with intragenic NF1 mutations. Most height measurements in 2–18-year-old boys and girls with NF1 microdeletions were greater than the median observed in non-deletion NF1 patients. However, extreme body height (more than three standard deviations above the mean) was still unusual among patients with NF1 microdeletions (Ning et al. 2016). These authors also showed that children with NF1 microdeletions were usually much taller than non-deletion NF1 patients after the age of 2 years. In early infancy, before the age of 2, the body length of microdeletion and non-deletion NF1 patients was found to be similar (Ning et al. 2016). The reasons for these differences in growth pattern are currently unknown.
Dysmorphic facial features
Patients with NF1 microdeletions frequently exhibit dysmorphic facial features not seen in patients with intragenic NF1 mutations. These features include broad neck, hypertelorism, downslanted palpebral fissures, a broad nasal bridge and a coarse, sometimes fleshy facial appearance (Mensink et al. 2006 and references therein; Venturin et al. 2004b; Pasmant et al. 2010; Mautner et al. 2010). A dysmorphic facial appearance is the most commonly observed feature in patients with NF1 microdeletions (Table 1) and is probably caused by haploinsufficiency for a gene or genes located within the deletion interval. The dysmorphic facial appearance as seen in patients with NF1 microdeletions is generally absent in patients with intragenic NF1 mutations. Using Face2Gene facial recognition software (http://www.fdna.com) may be helpful in characterising and quantifying the dysmorphic facial features observed in patients with NF1 microdeletions, particularly if comparison is made with family members lacking the NF1 microdeletion.
Cardiovascular malformations
Several studies have reported the occurrence of heart defects in a proportion of patients with NF1 microdeletions (Kayes et al. 1994; Tonsgard et al. 1997; Wu et al. 1997; Dorschner et al. 2000; Riva et al. 2000; Oktenli et al. 2003; Mensink et al. 2006; Venturin et al. 2004b; De Luca et al. 2007). However, the overall frequency of such defects in patients with NF1 microdeletions has remained unclear. In the study of Mautner et al. (2010), eight (29%) of the 28 type-1 NF1 deletion patients investigated had cardiovascular anomalies. The first detailed analysis of the prevalence of heart defects in patients harbouring NF1 microdeletions was performed by Nguyen et al. (2013) who observed major cardiac abnormalities in 6 of 16 NF1 microdeletion patients whereas none of 16 patients with intragenic NF1 mutations exhibited heart defects. Consequently, it would appear that heart defects are significantly more common in patients with NF1 microdeletions as compared to patients with intragenic NF1 mutations. However, the type and frequency of the heart defects observed in patients with NF1 microdeletions are quite heterogeneous, including pulmonic stenosis, ventricular septal defect, aortic stenosis, atrial septal defect, aortic stenosis, mitral valve prolapse or insufficiency, and hypertrophic cardiomyopathy (Table 2). Larger studies are clearly required to ascertain the type and the frequency of heart defects in patients with NF1 microdeletions with more precision.
Table 2.
Heart defects observed in patients with NF1 microdeletions
| Type of heart defect | Number of NF1 microdeletion patients exhibiting the heart defect (reference)a |
|---|---|
| Pulmonic stenosis | 1 (Tonsgard et al. 1997) 2 (Dorschner et al. 2000) 2 (Riva et al. 2000) |
| Ventricular septal defect | 1 (Tonsgard et al. 1997) 1 (Venturin et al. 2004b) 1 (Nguyen et al. 2013) |
| Atrial septal defect | 1 (Kayes et al. 1994) 1 (Dorschner et al. 2000) |
| Aortic stenosis | 1 (Nguyen et al. 2013) |
| Aortic dissection | 1 (Leppig et al. 1997) |
| Patent ductus arteriosus | 1 (Upadhyaya et al. 1996) |
| Mitral valve prolapse | 1 (Wu et al. 1997) 3 (Mensink et al. 2006) 1 (Venturin et al. 2004b) 1 (Oktenli et al. 2003) |
| Mitral valve insufficiency | 1 (Venturin et al. 2004b) 2 (Nguyen et al. 2013) |
| Aortic valve insufficiency | 1 (Nguyen et al. 2013) 1 (Dorschner et al. 2000) |
| Hypertrophic cardiomyopathy | 1 (Mensink et al. 2006) 1 (Venturin et al. 2004b) 3 (Nguyen et al. 2013) |
| Intracardiac neurofibromas | 2 (Nguyen et al. 2013) |
aIn the study of Nguyen et al. (2013), 6 of 16 NF1 microdeletion patients had major cardiac abnormalities. Two of these patients exhibited several heart abnormalities: Patient 5a had an aortic and a mitral valve insufficiency as well as a cardiac tumour whereas patient 8a had aortic stenosis, mitral valve insufficiency and hypertrophic cardiomyopathy
Co-deleted genes with the potential to influence the clinical phenotype in patients with NF1 microdeletions
In addition to the deletion of the NF1 gene, hemizygosity of any one of a number of genes located within the deletion interval at 17q11.2 may contribute to the clinical phenotype observed in patients with NF1 microdeletions. Some of these genes may have tumour suppressive functions and their hemizygosity could predispose to an increased tumour risk or might facilitate tumour progression. There is certainly good evidence that biallelic SUZ12 loss promotes MPNST progression (De Raedt et al. 2014; Lee et al. 2014; Zhang et al. 2014). Haploinsufficiency of other genes during early embryonic development and/or during later stages may, however, contribute to clinical sequelae such as overgrowth, reduced cognitive capabilities, heart defects and dysmorphic facial features. Thus, RNF135 haploinsufficiency is associated with dysmorphic facial features and overgrowth (Douglas et al. 2007). The consequences of the deletion of the other genes located within the 1.4-Mb NF1 microdeletion region (listed in Table 3) are less clear. This notwthstanding, the function, as well as the expression pattern of some of these genes, renders it highly likely that their loss impacts upon the NF1 microdeletion-associated phenotype. The haploinsufficiency of some of these genes may even synergize with the loss of the NF1 gene to aggravate the clinical manifestations of patients with NF1 microdeletions. To estimate the likely consequence of haploinsufficiency of genes located within the NF1 microdeletion region, the probability of loss-of-function (LoF) intolerance may be considered as calculated from the ExAC data set (Lek et al. 2016). The metric ‘probability of being LoF intolerant (pLI)’ separates genes into LoF intolerant (pLI ≥ 0.9) or LoF tolerant (pLI ≤ 0.1) categories. Importantly, ATAD5, RAB11FIP4, LRRC37B and SUZ12 reside in the category of LoF intolerant genes suggesting that their haploinsufficiency in patients with NF1 microdeletions is highly likely to have pathological consequences. By contrast, TEFM, ADAP2, RNF135, EVI2A and UTP6 are LoF tolerant (Table 3). It cannot, however, be excluded that the hemizygosity of these latter genes may still contribute in one way or another to the NF1 microdeletion phenotype.
Table 3.
Protein-coding and microRNA genes located within the NF1 microdeletion region at 17q11.2
| Official HGNC gene symbol | Alternative names | MIM# | Official gene name | Probability of loss of function intolerance (pLI) |
|---|---|---|---|---|
| CRLF3 | FRWS; CRLM9; p48.2;CYTOR4; CREME-9 | 614853 | Cytokine receptor-like factor 3 | 0.98 |
| ATAD5 | ELG1; FRAG1; C17orf41 | 609534 | ATPase family, AAA domain containing 5 | 1.00 |
| TEFM | C17orf42 | 616422 | Transcription elongation factor, mitochondrial | 0.04 |
| ADAP2 | CENTA2; Cent-b; HSA272195 | 608635 | ArfGAP with dual PH domains 2 | 0.00 |
| RNF135 | L13; MMFD; REUL; Riplet | 611358 | Ring finger protein 135p | 0.00 |
| MIR4733 | None | — | microRNA 4733 | |
| NF1 | WSS; NFNS; VRNF | 162200 | neurofibromin 1 | 1.00 |
| OMG | OMGP | 164345 | Oligodendrocyte myelin glycoprotein | 0.86 |
| EVI2B | EVDB; CD361; D17S376 | 158381 | Ecotropic viral integration site 2B | 0.18 |
| EVI2A | EVDA; EVI2; EVI-2A | 158380 | Ecotropic viral integration site 2A | 0.00 |
| RAB11FIP4 | FIP4-Rab11; RAB11-FIP4 | 611999 | RAB11 family-interacting protein 4 | 0.99 |
| MIR193A | MIRN193; MIRN193A; mir-193a | 614,733 | microRNA 193a | |
| MIR365B | MIR365-2; mir-365b; MIRN365-2;hsa-mir-365b | 614,733 | microRNA 365b | |
| MIR4725 | mir-4725 | — | microRNA 4725 | |
| COPRS | TTP1; COPR5; C17orf79; HSA272196 | 616477 | Coordinator of PRMT5 and differentiation stimulator | 0.83 |
| UTP6 | HCA66; C17orf40 | — | UTP6, small subunit processome component | 0.00 |
| SUZ12 | CHET9; JJAZ1 | 613675 | SUZ12 polycomb-repressive complex 2 subunit | 1.00 |
| LRRC37B | None | 616558 | Leucine-rich repeat containing 37B | 0.95 |
aThe ExAC browser (http://exac.broadinstitute.org/) provides the constraint metric termed “probability of loss of function” (pLI). To determine the pLI metric, the observed and expected variant counts for a given gene included in the ExAC dataset are considered. The closer the pLI value is to one, the more loss of function-intolerant the gene appears to be. A pLI value ≥ 0.9 is indicative of genes extremely intolerant of loss function variants
Our current knowledge of the genes located within the NF1 microdeletion region that have the potential to modify the clinical phenotype in patients with NF1 microdeletions, as concluded from previously published studies, is summarised below.
RNF135 may be involved in overgrowth and dysmorphic facial features
The RNF135 gene located upstream of NF1 represents a good candidate to account for the overgrowth phenotype observed in NF1 microdeletion patients. This conclusion derives directly from the findings of Douglas et al. (2007), who analysed a cohort of 245 individuals with overgrowth, learning disability, dysmorphic facial features and detected RNF135 mutations in 6 of them. The 245 patients investigated by Douglas et al. had been previously shown to be negative for NSD1 mutations, a frequent cause of Sotos syndrome characterised by overgrowth, dysmorphic facial features and learning disability (Tatton-Brown et al. 2005). Five of the six RNF135 mutation-positive patients identified by Douglas et al. harboured intragenic RNF135 mutations whereas the other patient exhibited a microdeletion resulting from an NAHR event between NF1-REPa and NF1-REPb which included RNF135 plus five other genes, but not NF1 (Douglas et al. 2007). Inactivating mutations or entire gene deletions of RNF135 do not appear to be frequent in patients with an overgrowth phenotype since RNF135 mutations were not detected in another cohort of 160 NSD1 mutation-negative patients with features of Sotos syndrome (Visser et al. 2009). Remarkably, among these 160 patients with suggested Sotos syndrome and overgrowth was a 4-year-old girl with dysmorphic facial features, two CALS and developmental delay who was found to have an NF1 microdeletion. This finding indicates that an NF1 microdeletion should be considered in the differential diagnosis of children with Sotos syndrome-associated features (Visser et al. 2009).
RNF135 encodes an E3 ubiquitin ligase with an N-terminal RING finger domain and C-terminal SPRY and PRY motifs. It is expressed in many different tissues (Oshiumi et al. 2009). RNF135 ubiquitinates RIG-I (retinoic acid-inducible gene-I protein) and promotes its signal transduction capacity so as to produce antiviral type-I interferon 1 (Oshiumi et al. 2010). Owing to its ring finger domain and the PRY motif, RNF135 is likely to bind numerous proteins, suggestive of a wide range of functions. The mechanism underlying the overgrowth phenotype mediated by the loss of one RNF135 copy is currently unclear and needs to be further investigated.
Importantly, patients with RNF135 mutations exhibit dysmorphic facial features including hypertelorism, down-slanting palpebral fissures and a broad nasal tip giving rise to a facial appearance similar to that observed in patients with NF1 microdeletions (Douglas et al. 2007). These findings suggest that RNF135 haploinsufficiency may be responsible for the dysmorphic facial features observed in patients with NF1 microdeletions. However, since only five patients with intragenic RNF135 mutations but lacking NF1 microdeletions have so far been reported, more extended genotype/phenotype studies are necessary to assess whether the dysmorphic facial features are indeed caused by RNF135 mutations.
SUZ12 and its role in MPNST development in patients with NF1 microdeletions
The increased risk of MPNSTs in patients with large NF1 microdeletions is probably associated with hemizygosity of the SUZ12 gene, located telomeric to NF1 within the NF1 microdeletion region (Fig. 1). SUZ12 is frequently bi-allelically inactivated in MPNSTs suggestive of a tumour suppressor function in this tumour type (De Raedt et al. 2014; Lee et al. 2014; Zhang et al. 2014). As a component of the Polycomb repressive complex 2 (PRC2), the SUZ12 protein is involved in the epigenetic silencing of many different genes by establishing di- and tri-methylation of histone H3 lysine 27 (reviewed by Di Croce and Helin 2013). Loss of histone H3 lysine 27 trimethylation has been observed in 50–70% of MPNSTs. By contrast, H3 lysine 27 trimethylation is retained in benign neurofibromas and hence serves as a diagnostic marker for malignant transformation (Asano et al. 2017; Cleven et al. 2016; Prieto-Granada et al. 2016; Schaefer et al. 2016; Röhrich et al. 2016). The genes targeted by PRC2 regulate cell cycle progression, stem cell self-renewal, cell fate decisions and cellular identity. The expression changes of some of these genes consequent to the loss of PRC2 function appear likely to contribute to tumorigenesis (reviewed by Conway et al. 2015; Laugesen et al. 2016). Thus, PRC2 loss has been shown to amplify Ras-driven gene expression through epigenetic changes (De Raedt et al. 2014). Somatic inactivating mutations of SUZ12 or other genes encoding PRC2 components have been detected in MPNSTs but not in benign neurofibromas and atypical neurofibromas. The latter are considered to be premalignant tumours with high potential to transform into MPNSTs (De Raedt et al. 2014; Lee et al. 2014; Zhang et al. 2014; Pemov et al. 2016). Consequently, loss of PRC2 function is important during malignant transformation and/or progression of MPNSTs. In patients with germline NF1 microdeletions, one SUZ12 allele is deleted in all cells and the probability of acquiring a somatic mutation of the remaining SUZ12 allele is clearly going to be higher than acquiring two independent somatic SUZ12 mutations (as would be necessary in NF1 patients with intragenic NF1 mutations, or in patients without NF1 who exhibit sporadic MPNSTs). Hence, the constitutional deletion of one SUZ12 allele represents a predisposing factor that contributes to the increased risk of MPNSTs in patients with NF1 microdeletions.
Other genes with known tumour suppressor function within the NF1 microdeletion region
In addition to the NF1 and SUZ12 genes, patients with NF1 microdeletions are also hemizygous for three other genes with putative tumour suppressor function: ATAD5 and the microRNA genes MIR193A and MIR365B. Whilst the importance of SUZ12 loss in MPNST progression has now been well documented, rather less is known about the co-deleted genes ATAD5, MIR193A, MIR365B and their involvement in MPNST pathogenesis. However, as deduced from their function, it is not unreasonable to suppose that haploinsufficiency of these genes could promote MPNST development in patients with NF1 microdeletions as discussed below.
ATAD5
Another tumour suppressor gene located within the NF1 microdeletion region is ATAD5 (ATPase family AAA domain–containing protein 5) (Fig. 1). The ATAD5 protein is involved in the stabilisation of stalled DNA replication forks by regulating proliferating cell nuclear antigen (PCNA) ubiquitination during DNA damage bypass, thereby promoting the exchange of a low-fidelity translesion polymerase back to a high-fidelity replication polymerase (Lee et al. 2010, 2013). Mice haploinsufficient for Atad5 (Atad5+/m) display high levels of genomic instability (Bell et al. 2011). Embryonic fibroblasts from Atad5+/m mice exhibit molecular defects in PCNA deubiquitination in response to DNA damage, as well as DNA damage hypersensitivity, high levels of genomic instability and aneuploidy. More than 90% of haploinsufficient Atad5+/m mice developed tumours such as sarcomas, carcinomas and adenocarcinomas that exhibited high levels of genomic instability (Bell et al. 2011). Furthermore, somatic ATAD5 mutations were identified in a subset of sporadic human endometrial tumours (Bell et al. 2011) as well as breast and ovarian tumour cell lines (Abaan et al. 2013). Hence, ATAD5 is regarded as a tumour suppressor gene (Bell et al. 2011; Kubota et al. 2013). Rare germline missense variants of ATAD5 with predicted pathogenicity have been reported to be enriched in patients with ovarian cancer as compared with controls (Maleva Kostovska et al. 2016). Targeted knockdown of ATAD5 expression in human cell lines has been shown to confer sensitivity to DNA damaging agents and cause severe genomic instability (Sikdar et al. 2009). Consequently, we may surmise that ATAD5 functions as an important regulator of genome instability (Gazy et al. 2015). Taken together, ATAD5 haploinsufficiency is likely to contribute to tumorigenesis in patients with NF1 microdeletions, in particular MPNST pathogenesis, since these tumours exhibit a high degree of genome instability including numerous copy number variants as well as chromosomal rearrangements (Mantripragada et al. 2009; Beert et al. 2011 and references therein).
MicroRNA genes
MicroRNAs (miRNAs) can play a critical role during tumorigenesis by directly interacting with the 3′UTRs of specific target mRNAs and inhibiting their translation (reviewed by Lovat et al. 2011). MicroRNAs have also been implicated in NF1-associated tumorigenesis (Sedani et al. 2012). Four microRNA genes are located within the 1.4-Mb NF1 microdeletion region, MIR193A, MIR365B, MIR4725 and MIR4733 (Fig. 1). One of these, MIR193A, encodes two mature miRNAs with well-known tumour suppressor functions; miR193a-3p and miR193a-5p are generated from the primary transcript by means of several maturation steps. The expression preference of miR193a-5p and miR193a-3p is likely to be determined by the Ago protein (reviewed by Tsai et al. 2016). According to the mirbase database (http://www.mirbase.org/), miR193a-3p is more abundantly expressed in human tissues than miR193a-5p. Several studies have indicated that miR193a-3p suppresses tumour development by silencing multiple target genes including SRSF2, HIC2, HOXC9, PSEN1, LOXL4, ING5, KIT, PLAU and MCL1 (Tsai et al. 2016). miR193a-3p has been found to be down-regulated by hypermethylation in oral squamous cell carcinoma cell lines (Kozaki et al. 2008), non-small cell lung cancer (Heller et al. 2012; Wang et al. 2013; Liang et al. 2015; Ren et al. 2015), bladder cancer (Deng et al. 2014; Lv et al. 2014; Li et al. 2015), hepatocellular carcinoma (Salvi et al. 2013), BRAF mutation-positive malignant melanoma (Caramuta et al. 2010), acute myeloid leukaemia (Xing et al. 2015) and pleural mesothelioma (Williams et al. 2015). However, it is unclear whether the down-regulation of miR193a-3p is a cause or a consequence of tumorigenesis. Decreased expression of miR193a-3p has been found to be correlated with metastasis, apoptosis and proliferation in breast cancer cell lines (Iliopoulos et al. 2011; Tsai et al. 2016) and ovarian tumour tissue (Nakano et al. 2013). Moreover, miR193a-5p is known to possess tumour suppressor functions since it inhibits the growth of breast cancer cells (Tsai et al. 2016) and endometrioid endometrial carcinoma cells by down-regulation of the transcription factor YY1 (Yang et al. 2013). Both miR193a-5p and miR193a-3p suppress lung cancer cell migration and invasion by co-regulating the ERBB4/PIK3R3/mTOR/S6K2 signalling pathway (Yu et al. 2015). These findings indicate that miR193a-3p and miR193a-5p play a tumour suppressor role in many different tumour types. In particular, the putative tumour suppressor function of miR193a-3p in breast cancer cell lines is noteworthy since breast cancer occurs at an increased frequency in patients with NF1 (Sharif et al. 2007; Seminog and Goldacre 2013, 2015; Uusitalo et al. 2016). However, it is unknown whether the breast cancer risk is higher in patients with NF1 microdeletions than in patients with intragenic NF1 mutations.
The MIR365B gene, located within the NF1 microdeletion region (Fig. 1), also encodes an miRNA with known tumour suppressor function, as evidenced by its ability to target specific transcription factors, such as NKX2-1 and TTF1, in non-small cell lung cancer (Qi et al. 2012; Kang et al. 2013; Sun et al. 2015). miR365 is down-regulated in colon cancer (Nie et al. 2012), cutaneous squamous cell carcinoma (Zhou et al. 2014, 2015), hepatocellular carcinoma (Chen et al. 2015), gastric cancer (Guo et al. 2013) and malignant melanoma (Bai et al. 2015a, b). By contrast, putative tumour suppressor functions of the other two miRNAs located within the NF1 microdeletion region, encoded by MIR4725 and MIR4733, respectively, have not so far been reported.
Taken together, the loss of the MIR193A and MIR365B genes in patients with NF1 microdeletions may well contribute to tumorigenesis in these patients. However, miRNAs are not only involved in tumour development; they also play a role as key regulators of metabolic homeostasis and tissue differentiation (reviewed by Vienberg et al. 2017). Additional studies are necessary to investigate the influence of the hemizygous loss of the miRNA genes on the clinical phenotype associated with NF1 microdeletions.
Other genes that may contribute to tumour development in patients with NF1 microdeletions
In addition to the deletion of SUZ12 and ATAD5, hemizygosity for the genes COPRS, UTP6 and RNF135 may also contribute to the increased tumour risk associated with NF1 microdeletions. Current knowledge about the function of these genes is summarised in the following paragraphs.
COPRS
COPRS is another gene located within the NF1 microdeletion interval that may well play a role in MPNST development. COPRS encodes an adaptor protein that binds strongly to protein-arginine methyltransferase 5 (PRTM5) and to histone H4. By these means, COPRS recruits PRMT5 to chromatin and also modulates PRMT5 substrate specificity since PRMT5 bound to COPRS preferentially methylates histone H4 instead of histone H3 (Lacroix et al. 2008). COPRS binding to PRMT5 is essential for myogenic differentiation, possibly through altered targeting of PRMT5 to specific gene promoters (Paul et al. 2012). These observations suggest that the COPRS–PRMT5 complex regulates cell differentiation, a process that is frequently perturbed during tumorigenesis. MPNSTs often exhibit regions of divergent differentiation possibly including rhabdomyosarcomatous, chondral, glandular, neuroendocrine, gangliocytic and liposarcomatous components. These regions of divergent differentiation may be focal on a background of typical spindle-shaped tumour cells. For example, in some cases, rhabdomyosarcomatous differentiation may become predominant rendering even differential diagnosis very difficult (Guo et al. 2012). Aberrant regulation of the COPRS–PRMT5 complex due to COPRS haploinsufficiency may contribute to these divergent differentiation patterns. Increased expression of PRMT5 has been noted in a wide variety of cancer types (reviewed by Stopa et al. 2015), but further studies are necessary to investigate how COPRS haploinsufficiency in patients with NF1 microdeletions could alter PRMT5 function, thereby contributing to tumorigenesis.
Analysis of the expression level of COPRS in MPNST cell lines has yielded inconsistent results both within and between studies (Bartelt-Kirbach et al. 2009; Pasmant et al. 2011). Overexpression of COPRS (five to 10-fold) was observed in two MPNST tissue samples from patients with intragenic NF1 mutations as compared with cutaneous neurofibroma tissue (Bartelt-Kirbach et al. 2009). However, by contrast, these authors detected low expression of COPRS in an MPNST cell line, which was as low as the COPRS expression level in neurofibroma-derived fibroblast cell cultures. The MPNST cell line analysed by these authors was derived from an NF1 patient but the germline NF1 mutation in this patient had not been determined. In similar vein, whilst Pasmant et al. (2011) observed high COPRS expression in a series of MPNST cell lines as compared with plexiform and cutaneous neurofibroma samples, in other MPNST cell lines, COPRS expression levels were as low as in plexiform and cutaneous neurofibromas (Pasmant et al. 2011). Unfortunately, these authors failed to specify the origin of the MPNST cell lines analysed, whether they were derived from NF1 patients and if so, whether they harboured NF1 microdeletions. The inconsistent findings regarding the COPRS expression level in MPNSTs reported by Bartelt-Kirbach et al. (2009) and Pasmant et al. (2011) are difficult to interpret. It may be that MPNST cell lines are an inappropriate system in which to investigate COPRS expression, perhaps because these cell lines have been subject to massive in vitro selection and hence may not be representative of key stages of MPNST development in vivo. Thus, the analysis of COPRS expression should be performed using primary MPNST samples (with high proportions of tumour cells) from patients with NF1 microdeletions to explore the role of COPRS during MPNST development or progression in NF1 microdeletion patients.
UTP6
Tumorigenesis in patients with NF1 microdeletions may also be influenced by haploinsufficiency of UTP6, located telomeric to NF1 within the NF1 microdeletion region (Fig. 1). The protein encoded by UTP6 is involved in apoptosome-dependent apoptosis and it has been suggested that UTP6 haploinsufficiency could render cells with NF1 microdeletions less susceptible to apoptosis (Piddubnyak et al. 2007). UTP6 is required for ribosome synthesis (Bonnart et al. 2012) and is, as a component of the centrosome, required for centriole duplication and the establishment of a bipolar spindle ensuring proper chromosome segregation during mitosis (Fant et al. 2009; Ferraro et al. 2011). Somatic loss of the remaining UTP6 allele in tumours of patients with NF1 microdeletions may contribute to malignant transformation by increasing chromosome instability and aneuploidy.
RNF135
In addition to its influence on the childhood overgrowth phenotype and the dysmorphic facial features observed in patients with NF1 microdeletions, RNF135 haploinsufficiency may also promote tumorigenesis. The RNF135 gene has been shown to be down-regulated in cells from malignant peripheral nerve sheath tumours (MPNSTs) and MPNST cell lines suggesting that RNF135 loss is involved in conferring the increased MPNST risk characteristic of NF1 microdeletion patients (Pasmant et al. 2011). In glioblastomas, however, RNF135 has been found to be upregulated and promotes the proliferation of human glioblastoma cells in vivo and in vitro via the ERK pathway (Liu et al. 2016). Furthermore, RNF135 would appear to promote the expression of PTEN and TP53 in tongue cancer SCC25 cells and RNF135 overexpression inhibits the viability, proliferation, and invasion of these cells (Jin et al. 2016). Taken together, these findings suggest that changes in the dosage of RNF135 might contribute to tumorigenesis although further studies are necessary to clarify this postulate.
Haploinsufficiency of genes in NF1 microdeletion patients and intellectual disability
OMG
Hemizygosity of a gene (or several genes) located within the NF1 microdeletion region may contribute to the intellectual disability noted in patients with large NF1 deletions (Venturin et al. 2006). A good candidate is the OMG gene which encodes the oligodendrocyte myelin glycoprotein (OMgp) involved in the regulation of synaptic plasticity (reviewed by Mironova and Giger 2013). Synaptic plasticity and structural changes of the synapse have been suggested to cause cognitive and functional defects observed in intellectual disability, autism spectrum disorders and schizophrenia (Bernardinelli et al. 2014).
OMgp is anchored to the myelin membrane through a glycosylphosphatidyl inositol lipid molecule and is expressed in neurons as well as oligodendrocytes (Habib et al. 1998; Raiker et al. 2010 and references therein). OMgp belongs to the group of myelin-associated inhibitor proteins (MAIPs) which act as central nervous system (CNS) regeneration inhibitors by preventing injured axons from regenerating beyond the injury site. Prototypical MAIPs, including OMgp, are expressed in the healthy as well as the injured brain and bind to the Nogo-66 receptor (NgR1) and the paired Ig-like receptor B (PirB) which appear to inhibit neurite outgrowth in the adult CNS (Atwal et al. 2008; Akbik et al. 2012; Geoffroy and Zheng 2014; Baldwin and Giger 2015). However, in the adult CNS, OMgp and other MAIPs also regulate neuronal morphology, dendritic spine shape and activity-driven synaptic plasticity by binding to their receptors as determined both by NgR1 and PirB knockout mouse models and human cell lines (McGee et al. 2005; Syken et al. 2006; Lee et al. 2008; Raiker et al. 2010; reviewed by Mironova and Giger 2013).
In addition to its function in the adult CNS, OMgp plays important roles during early stages of brain development before the onset of myelination, possibly by regulating neurogenesis (Martin et al. 2009). During normal mouse development, neuronal OMgp is expressed, from embryonic day E14 on, in growing axons during axonal tract formation following the maturation of cortical connexions (Gil et al. 2010). In primary hippocampal cultures of adult normal mice, OMgp is present in the neuronal membrane, synaptosomal fractions and axonal varicosities (Gil et al. 2010). OMgp-null mice show impaired myelination and thalamo-cortical projection (Gil et al. 2010) as well as hypomyelination of the spinal cord that correlates with lower propagation of ascending and descending electrical impulses (Lee et al. 2011). Even although OMgp-null mice may not represent a wholly appropriate model with which to ascertain the consequences of OMG haploinsufficiency in humans, data derived from this system are consistent with the view that OMgp plays a key role in axonal target specification and synaptic plasticity.
Many studies have indicated that dysfunction of synapse formation, shape or density and synaptic plasticity cause intellectual disability and neuropsychiatric disorders (reviewed by Pittenger 2013; Srivastava and Schwartz 2014). Since MAIPs and their receptors play important roles in regulating synapse formation and plasticity, altered expression or function of these proteins may contribute to intellectual disability and other brain disorders (Sinibaldi et al. 2004; Budel et al. 2008; Tews et al. 2013; Llorens et al. 2011; Willi and Schwab, 2013; Petrasek et al. 2014). Consequently, OMgp haploinsufficiency may well contribute to the intellectual disability observed in patients with NF1 microdeletions. The negative effects of OMgp haploinsufficiency on synaptic plasticity could be additive in relation to the consequences of the loss of the NF1 gene product neurofibromin, an important regulator of Ras signalling in the brain. At least 50% of the patients with intragenic NF1 mutations suffer from intellectual disabilities manifesting as cognitive slowing, memory disturbances, difficulties in solving strategic problems, visuospatial impairment and deficits in motor coordination (Diggs-Andrews and Gutmann 2013; Violante et al. 2013). These symptoms are further aggravated in patients with NF1 microdeletions who exhibit a significantly lower mean FSIQ than patients with intragenic NF1 mutations (Descheemaeker et al. 2004).
Similar to the phenotype observed in patients with NF1, behavioural studies in Nf1-deficient mouse models indicated deficits in spatial learning and motor coordination (Shilyansky et al. 2010a, b; van der Vaart et al. 2011). These mouse models also revealed that increased Ras/MAPK (mitogen-activated protein kinases) signalling results in higher GABA (gamma-aminobutyric acid) release during learning causing deficits in hippocampal long-term potentiation (LTP) that could account for the spatial learning and memory deficits of these mutant mice (Costa et al. 2002; Cui et al. 2008). Hence, neurofibromin is an important Ras regulator in interneurons influencing hippocampal-dependent learning. Ras signalling in dendritic spines of pyramidal neurons is required for many forms of synaptic plasticity, including LTP, spine structural plasticity, and new spine formation (reviewed by Oliveira and Yasuda 2014). Consequently, NF1 and OMG haploinsufficiency are likely to exert additive negative effects that are causally associated with the cognitive deficit evident in many patients with NF1 microdeletions. Loss-of-function mutations in the OMG gene have not, however, been observed in patients with idiopathic intellectual disability (Venturin et al. 2006).
RNF135
RNF135 haploinsufficiency may also contribute to the reduced cognitive capabilities observed in patients with NF1 microdeletions. As mentioned earlier, RNF135 encodes an E3 ubiquitin ligase; other ubiquitin ligase genes have already been implicated in the development of intellectual disability and autism (Tenorio et al. 2014; reviewed by Tastet et al. 2015).
A significantly increased frequency of genotypes carrying the rare allele of the RNF135 missense variant rs111902263 (p.R115 K) has been observed in patients with autism as compared with healthy controls (P = 0.0019, odds ratio: 4.23, 95% confidence interval: 1.87–9.57) (Tastet et al. 2015). These authors also showed that the RNF135 gene is expressed in the cerebral cortex of humans and mice. The RNF135-encoded protein, termed ‘Riplet’, regulates the cytosolic viral RNA receptors RIG-I by ubiquitination (Oshiumi et al. 2009, 2010, 2013). RIG-I and other RIG-I-like receptors contribute to innate antiviral immunity by inducing antiviral responses such as the production of type I interferons (IFNs) and proinflammatory cytokines (reviewed by Yoneyama et al. 2015). RIG-1 is upregulated in neurons upon viral infection and is an important component of the intrinsic antiviral pathways in the CNS (Nazmi et al. 2011). RIG-I knockdown in a mouse model was associated with reduced neural stem/progenitor cell proliferation (Mukherjee et al. 2015), suggesting that the RNF135 protein plays a role in neurogenesis. Further studies will be necessary to determine the role of RNF135/Riplet in neural stem/progenitor cells and during brain development, roles which may yet prove to be relevant in the context of RNF135 haploinsufficiency and cognitive disability in patients with NF1 microdeletions.
Intriguingly, the ADAP2 gene, which is also located within the NF1 microdeletion region, encodes another key regulator of RIG-I signalling. ADAP2, an ADP-ribosylation factor GTPase-activating protein (ArfGAP) with dual PH domains 2, plays an important role as a scaffold protein that couples different modules of RIG-I signalling, leading to the up-regulation of type-I interferon gene transcription in response to viral infection (Bist et al. 2016). The potential role of ADAP2 in the aetiology of cardiovascular malformations is discussed below.
ADAP2 and cardiovascular malformations in patients with NF1 microdeletions
Hemizygosity of the ADAP2 gene may contribute to the cardiovascular malformations observed in patients with NF1 microdeletions. This conclusion is drawn from the observation that ADAP2 is highly expressed during early stages of heart development in both mouse and human (Venturin et al. 2005, 2014). In zebrafish, ADAP2 loss of function leads to circulatory deficiencies and heart shape defects or defective valvulogenesis (Venturin et al. 2014). The ADAP2-encoded protein acts as a GTPase-activating protein (GAP) of the ADP-ribosylation factor 6 (ARF6), a small GTPase involved in actin cytoskeleton remodelling. ADAP2 (centaurin-alpha2) is located in the cytoplasm but after EGF stimulation, it binds to the plasma membrane via phosphatidylinositols. Plasma membrane association of ADAP2 prevents ARF6 translocation to the plasma membrane. By these means, ADAP2 negatively regulates ARF6-mediated actin cytoskeleton reorganisation (Venkateswarlu et al. 2007). ADAP2 also interacts with beta-tubulin and stabilises microtubules (Zuccotti et al. 2012). As a microtubulin-associated protein expressed during early embryonic development in the central nervous system and in the heart, ADAP2 is likely to mediate microtubule cytoskeleton reorganisation during cell differentiation and migration. It is well known that the interaction between microtubules and the actin cytoskeleton in association with membrane-associated proteins regulates cell shape and cellular remodelling (reviewed by Basu and Chang 2007; Bezanilla et al. 2015). Since ADAP2 interacts with the microtubule/actin cytoskeleton, it may function as a cytoskeleton cross-talker that increases microtubule stability and modulates actin reorganisation and hence cellular morphology (Zucotti et al. 2012). Disturbances of the cytoskeletal organisation in myocytes during embryonal development may be responsible for the cardiovascular malformations observed in patients with NF1 microdeletions. This postulate was reinforced by the findings of Venturin et al. (2014) who showed that in zebrafish, ADAP2 is required for normal cardiac morphogenesis.
The protein product of the NF1 gene, neurofibromin, is essential for embryonic cardiac valve formation and the study of mouse models has indicated that neurofibromin loss leads to cardiovascular lethality during early embryonic development; Nf1 regulation of Ras in the developing endothelium is required for regular development of endocardial cushions and the ventricular myocardium (Gitler et al. 2003; Ismat et al. 2006; Xu et al. 2009; Bajaj et al. 2012; Yzaguirre et al. 2015). Haploinsufficiency for both NF1 and ADAP2 may contribute either cooperatively or additively to the increased frequency of heart defects in patients with NF1 microdeletions. Further, SUZ12 and UTP6 are highly expressed during the development of the human heart; it follows that their haploinsufficiency may also contribute to the increased prevalence of congenital heart defects in patients with NF1 microdeletions (Venturin et al. 2005).
Clinical phenotype in patients with NF1 microdeletions: influence of mosaicism and deletion size
The presence of normal cells not harbouring an NF1 microdeletion exerts a major influence on disease severity in patients with mosaic large NF1 deletions. Depending upon the proportion of cells harbouring the deletion, the clinical phenotype can be very mild or may affect only certain body segments (Tinschert et al. 2000; Maertens et al. 2007). The frequency of somatic mosaicism is strongly associated with the type of NF1 microdeletion. Type-2 NF1 deletions, caused by NAHR between SUZ12 and SUZ12P1, are frequently of postzygotic origin. Patients with postzygotic type-2 NF1 deletions exhibit somatic mosaicism of cells with the deletion and normal cells not harbouring the deletion (Kehrer-Sawatzki et al. 2004; Steinmann et al. 2007; Vogt et al. 2012). It has been estimated that at least 63% of all type-2 NF1 deletions are associated with somatic mosaicism (Vogt et al. 2012). Atypical NF1 deletions are also frequently mosaic; among the 17 atypical NF1 deletion patients investigated by Vogt et al. (2014), 10 patients (59%) exhibited somatic mosaicism with normal cells. By contrast, only a very low proportion (2–4%) of type-1 NF1 microdeletions is associated with somatic mosaicism (Messiaen et al. 2011). Remarkably, patients with type-2 deletions exhibit tissue-specific differences in the proportion of cells with the deletion (termed del(+/−) cells), whereas the proportion of del(+/−) cells is very high (94–99%) in the blood of these patients, and much lower proportions of del(+/−) cells are evident in urine samples (24–82%) (Roehl et al. 2012). Since mosaic type-2 NF1 microdeletions occur in most instances during early embryonic development, the tissue-specific differences in the proportion of del(+/−) cells should result from cell type-specific selection.
Genotype–phenotype correlations in patients with mosaic NF1 microdeletions are difficult to perform because the variable proportion of normal cells in different tissues is likely to influence the expression of clinical symptoms. The proportion of normal cells is difficult to assess and may vary from tissue to tissue and from patient to patient. Unfortunately, only a small number of patients with mosaic NF1 deletions have been analysed in any detail. None of the eight patients with mosaic type-2 NF1 microdeletions exhibited facial dysmorphism, nor was there any evidence of delayed cognitive development and/or learning disabilities, cognitive impairment, congenital heart disease, hyperflexibility of joints, large hands and feet, muscular hypotonia or bone cysts, all of which are frequently observed in patients with germline type-1 NF1 microdeletions (Table 1). Furthermore, externally visible and internal plexiform neurofibromas were significantly less prevalent in patients with mosaic type-2 NF1 microdeletions as compared with patients carrying constitutional (germline) type-1 NF1 microdeletions (Kehrer-Sawatzki et al. 2012). These differences in clinical phenotype are unlikely to be caused by the differing extent of type-1 and type-2 deletions. Even although only two patients with non-mosaic type-2 NF1 microdeletions have so far been analysed in terms of their clinical phenotype (Vogt et al. 2011), it may be concluded that patients with non-mosaic type-2 deletions exhibit most of the clinical features that have been reported in individuals with germline type-1 NF1 deletions. Thus, a severe disease manifestation is not confined to patients with type-1 NF1 deletions but may also occur in individuals with non-mosaic type-2 NF1 deletions. The loss of the LRRC37B gene, associated with type-1 microdeletions but not with type-2 microdeletions, is unlikely to exert a major influence on the clinical phenotype. We conclude that the less severe clinical phenotype observed in patients with mosaic type-2 NF1 microdeletions is unrelated to the extent of the deletion but is instead associated with the presence of normal cells that lack the microdeletion. Nevertheless, an increased risk of MPNSTs may also exist for patients with mosaic type-2 NF1 microdeletions and plexiform neurofibromas, since most MPNSTs develop from pre-existing plexiform neurofibromas (Tucker et al. 2005) and the concomitant loss of NF1 and SUZ12 in plexiform neurofibroma cells harbouring the type-2 NF1 microdeletion increases the likelihood of malignant transformation.
The extent of the deletion may nevertheless be important in the context of genotype-phenotype relationships in patients with NF1 microdeletions. It has been noted that patients with very large atypical NF1 deletions that encompass several Mb, much larger than the classical 1.4-Mb spanning type-1 NF1 deletions, exhibit very severe disease manifestations associated with many additional clinical features that are not generally found to be associated with type-1 NF1 deletions (Upadhyaya et al. 1996; Cnossen et al. 1997; Dorschner et al. 2000; Kehrer-Sawatzki et al. 2003; Pasmant et al. 2008). However, these deletions were very heterogeneous in size and hence genotype–phenotype analyses are scarcely feasible. More interesting in this regard are shorter deletions with recurrent breakpoints such as type-3 NF1 microdeletions. These deletions encompass only 1-Mb and do not include the five functional genes (CRLF3, ATAD5, TEFM, ADAP2 and RNF135) located centromeric to NF1-REPb (Fig. 1). However, only eight type-3 NF1 deletions have so far been identified by means of high-resolution breakpoint analysis (Bengesser et al. 2010; Pasmant et al. 2010; Messiaen et al. 2011). Unfortunately, the clinical data from these eight patients are far from comprehensive or completely unavailable. Intellectual disability or cognitive impairment was observed in four of these eight patients with type-3 NF1 microdeletions. Consequently, a gene (or genes) influencing the cognitive capabilities in these patients is located either within the NF1 gene itself (e.g. OMG) or located telomeric to NF1; OMG is probably the best candidate for such an influence, by virtue of its function. Remarkably, dysmorphic facial features were observed in six patients from whom clinical phenotypic data were available (Bengesser et al. 2010; Pasmant et al. 2010). Since the RNF135 gene was not deleted in these patients, it would appear that RNF135 haploinsufficiency cannot be held responsible for the dysmorphic facial features in these individuals. The RNF135 gene is located 46-kb upstream of the centromeric breakpoint of type-3 NF1 deletions. However, it cannot be unequivocally excluded that a regulatory element which influences RNF135 expression has been deleted in patients with type-3 NF1 deletions. The deletion of such a regulatory element could have impaired RNF135 expression in those patients with type-3 deletions, a postulate which remains to be investigated. Unfortunately, since it is not yet known if patients with type-3 NF1 deletions are affected by childhood overgrowth or tall stature as adults, no further conclusions can be drawn concerning RN135 haploinsufficieny and its role in height determination in patients with NF1 deletions. Detailed clinical characterisation of a larger number of patients with type-3 NF1 deletions would be necessary to assess the contribution of the genes listed in Table 1 to the clinical phenotype associated with large NF1 deletions.
The clinical phenotype in NF1 microdeletion vs. NF1 microduplication patients
For many disease-associated microdeletions encompassing several hundred kb, the reciprocal microduplications have been identified. In most instances, microdeletions and the reciprocal microduplications differ in terms of the associated clinical phenotype (reviewed by Vissers and Stankiewicz 2012; Weise et al. 2012). Microduplications reciprocal to NF1 microdeletions are not associated with a classical NF1 phenotype but instead with developmental delay and learning disabilities as the major clinical features. So far, 29 NF1 microduplication carriers have been reported, 18 of them were unrelated cases (Lu et al. 2007; Grisart et al. 2008; Moles et al. 2012; Coe et al. 2014; Kehrer-Sawatzki et al. 2014). None of the individuals with an NF1 microduplication so far reported exhibited neurofibromas or other NF1-associated tumours. Only two of the 29 NF1 microduplication carriers had CALS. However, these CALS were atypical, with irregular borders and nonhomogeneous pigmentation which is not generally characteristic of those CALS typically seen in patients with NF1 (Kehrer-Sawatzki et al. 2014). One of the two NF1 microduplication patients with CALS fulfilled the diagnostic criteria for NF1 because he not only had ten CALS but also Lisch nodules. Since only blood cells from this patients were available for investigation, it could not be excluded that the NF1 microduplication observed in this patient was of postzygotic origin and that the patient might also harbour cells in his body that contained the reciprocal NF1 microdeletion (rather than the NF1 microduplication). The potential co-occurrence of cells with the reciprocal NF1 microdeletion could have been responsible for the CALS and Lisch nodules observed in this patient. Furthermore, it could not be excluded that somatically acquired NF1 mutations in melanocyte progenitor cells contributed to the occurrence of CALS and Lisch nodules in this individual. Since melanocytes from CALS of this patient could not be investigated, his clinical manifestations are difficult to interpret with regard to the underlying mutation (Kehrer-Sawatzki et al. 2014). Nevertheless, the analysis of the other 28 NF1 microduplication carriers reported to date implies that these duplications do not cause a classical NF1 phenotype.
Importantly, the clinical phenotype associated with NF1 microdeletions is fully penetrant; clinically unaffected individuals with germline NF1 microdeletions have not been reported. By contrast, three carriers of familial NF1 microduplications have been observed who, according to the authors, do not exhibit any obvious clinical signs (Grisart et al. 2008; Moles et al. 2012). However, NF1 microduplications are very unlikely to be frequent neutral copy number variants since they have not been observed in a total of 30,134 control individuals (Shaikh et al. 2009; Cooper et al. 2011; Moles et al. 2012; Coe et al. 2014). In a disease context, NF1 microduplications have been observed in 14 unrelated individuals identified from among a total of 77,902 patients who were investigated by array CGH due to developmental delay (Moles et al. 2012; Coe et al. 2014).
NF1 microdeletions are estimated to occur with a frequency of approximately 1 in 60,000 individuals, calculated on the basis that large NF1 deletions are observed in ~5% of all patients with NF1 which occurs with an incidence of ~1 in 3000. NF1 microduplications were not observed in 30,134 control individuals analysed by array CGH (Shaikh et al. 2009; Cooper et al. 2011; Moles et al. 2012; Coe et al. 2014), but this number of individuals is still too low to estimate the frequency of NF1 microduplications in the general population or to assess whether NF1 microdeletions are more frequent than NF1 microduplications.
Conclusions
NF1 microdeletions are often associated with a severe clinical phenotype characterised by features not observed at all (or at significantly lower frequency) in patients with intragenic NF1 mutations. Although many published studies have described the NF1 microdeletion-associated phenotype, what is lacking are large studies comparing NF1 patients with and without NF1 microdeletions according to standardised evaluation criteria to ensure that the same analytical methods are identically applied in the investigation of both patient groups. Ideally, such studies should be performed by comparing patients with NF1 microdeletions and patients with intragenic loss-of-function NF1 mutations to minimise the effects of mutation severity. Further, these studies should include sufficiently high numbers of patients to perform meaningful statistical analysis whilst the age of the patients should be matched since many NF1-associated features are age-related. This would be best performed as a multicenter collaborative study to collect large numbers of patients with different NF1 microdeletion types to analyse the different deletion types separately. Comprehensive comparative analyses of this kind could help to answer several open questions that have not so far been systematically addressed, e.g., whether an early (pre-pubertal) onset of growth of multiple cutaneous neurofibromas is significantly more prevalent in children with NF1 microdeletions as compared to children with intragenic NF1 mutations. In addition, a comparative analysis including a large number of age-matched adult patients is urgently required to ascertain whether high numbers of cutaneous neurofibromas (N > 1000) occur significantly more often in patients with NF1 microdeletions than in patients with intragenic mutations. Clearly, these and other analyses of the genotype–phenotype relationship should be performed so as to include patients with non-mosaic NF1 microdeletions with well characterised deletion breakpoints to assess the number and identity of the co-deleted genes. The most frequently occurring type-1 NF1 deletions are important with regard to extended genotype-phenotype correlations since they are the easiest group in which to discern such correlations; however, the less frequent but recurrent type-3 NF1 deletions are also of interest. Analyses of a larger number of patients with type-3 NF1 microdeletions would be necessary to determine the influence of genes such as RNF135 on the overgrowth and dysmorphic facial features, or the influence on the deletion-associated phenotype of other genes not rendered hemizygous by the type-3 NF1 deletions (Fig. 1). Although the deletion of SUZ12 may well predispose patients with NF1 microdeletions to malignancy, in particular to the development of MPNSTs, the reasons for the disproportionately higher frequency of benign plexiform, subcutaneous and spinal neurofibromas in patients with NF1 microdeletions is still unclear. It is possible that deleted genes other than NF1 may also promote tumorigenesis and such additive effects could be investigated by the targeted knockout of individual genes located within the NF1 microdeletion interval using the CRISPR/Cas9 system in human cells. Similar experiments might also be performed with mouse cells but it should be appreciated that the genomic region on mouse chromosome 11 orthologous to the NF1 microdeletion region exhibits differences in both the number and arrangement of genes as compared with the human genomic region at 17q11.2 (Jenne et al. 2003).
Although NF1 microdeletion patients as a group exhibit a more severe clinical phenotype than that generally exhibited by patients with NF1 intragenic lesions, a certain degree of variability in terms of the expression of clinical symptoms is observed when individual patients with NF1 microdeletions are compared, even in cases where their germline deletions are identical. In patients with intragenic NF1 mutations, the level of expression of the wild-type NF1 allele has been suggested to impact upon the clinical phenotype since skewed allele-specific expression of the NF1 gene has been observed in healthy individuals (Hoffmeyer et al. 1995; Cowley et al. 1998; Jentarra et al. 2012). It would be important to investigate whether the phenotypic variability observed between patients with NF1 microdeletions might also be caused by differences in expression of the wild-type alleles of the genes that are present in only one copy owing to the large NF1 deletion. Such analyses, as well as an extended comparative analysis of the clinical phenotype of NF1 patients with and without NF1 microdeletions, would be necessary to improve our understanding of the mechanisms involved as well as to characterise the deletion-associated phenotype in a more systematic and comprehensive manner.
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Abaan OD, Polley EC, Davis SR, Zhu YJ, Bilke S, Walker RL, Pineda M, Gindin Y, Jiang Y, Reinhold WC, Holbeck SL, Simon RM, Doroshow JH, Pommier Y, Meltzer PS. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res. 2013;73:4372–4382. doi: 10.1158/0008-5472.CAN-12-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbik F, Cafferty WB, Strittmatter SM. Myelin associated inhibitors: a link between injury-induced and experience-dependent plasticity. Exp Neurol. 2012;235:43–52. doi: 10.1016/j.expneurol.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alivuotila L, Hakokari J, Visnapuu V, Korpijaakko-Huuhka AM, Aaltonen O, Happonen RP, Peltonen S, Peltonen J. Speech characteristics in neurofibromatosis type 1. Am J Med Genet A. 2010;152A:42–51. doi: 10.1002/ajmg.a.33178. [DOI] [PubMed] [Google Scholar]
- Asano N, Yoshida A, Ichikawa H, Mori T, Nakamura M, Kawai A, Hiraoka N. Immunohistochemistry for trimethylated H3K27 in the diagnosis of malignant peripheral nerve sheath tumours. Histopathology. 2017;70(30):385–393. doi: 10.1111/his.13072. [DOI] [PubMed] [Google Scholar]
- Atwal JK, Pinkston-Gosse J, Syken J, Stawicki S, Wu Y, Shatz C, Tessier-Lavigne M. PirB is a functional receptor for myelin inhibitors of axonal regeneration. Science. 2008;322:967–970. doi: 10.1126/science.1161151. [DOI] [PubMed] [Google Scholar]
- Bai J, Zhang Z, Li X, Liu H. MicroRNA-365 inhibits growth, invasion and metastasis of malignant melanoma by targeting NRP1 expression. Int J Clin Exp Pathol. 2015;8:4913–4922. [PMC free article] [PubMed] [Google Scholar]
- Bai J, Zhang Z, Li X, Liu H. MicroRNA-365 inhibits growth, invasion and metastasis of malignant melanoma by targeting NRP1 expression. Cancer Biomark. 2015;15:599–608. doi: 10.3233/CBM-150500. [DOI] [PubMed] [Google Scholar]
- Bajaj A, Li QF, Zheng Q, Pumiglia K. Loss of NF1 expression in human endothelial cells promotes autonomous proliferation and altered vascular morphogenesis. PLoS One. 2012;7:e49222. doi: 10.1371/journal.pone.0049222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin KT, Giger RJ. Insights into the physiological role of CNS regeneration inhibitors. Front Mol Neurosci. 2015;8:23. doi: 10.3389/fnmol.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartelt-Kirbach B, Wuepping M, Dodrimont-Lattke M, Kaufmann D. Expression analysis of genes lying in the NF1 microdeletion interval points to four candidate modifiers for neurofibroma formation. Neurogenetics. 2009;10:79–85. doi: 10.1007/s10048-008-0154-0. [DOI] [PubMed] [Google Scholar]
- Basu R, Chang F. Shaping the actin cytoskeleton using microtubule tips. Curr Opin Cell Biol. 2007;19:88–94. doi: 10.1016/j.ceb.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Beert E, Brems H, Daniëls B, De Wever I, Van Calenbergh F, Schoenaers J, Debiec-Rychter M, Gevaert O, De Raedt T, Van Den Bruel A, de Ravel T, Cichowski K, Kluwe L, Mautner V, Sciot R, Legius E. Atypical neurofibromas in neurofibromatosis type 1 are premalignant tumors. Genes Chromosomes Cancer. 2011;50:1021–1032. doi: 10.1002/gcc.20921. [DOI] [PubMed] [Google Scholar]
- Bell DW, Sikdar N, Lee KY, Price JC, Chatterjee R, Park HD, Fox J, Ishiai M, Rudd ML, Pollock LM, Fogoros SK, Mohamed H, Hanigan CL, Zhang S, Cruz P, Renaud G, Hansen NF, Cherukuri PF, Borate B, McManus KJ, Stoepel J, Sipahimalani P, Godwin AK, Sgroi DC, Merino MJ, Elliot G, Elkahloun A, Vinson C, Takata M, Mullikin JC, Wolfsberg TG, Hieter P, Lim DS, Myung K, NISC Comparative Sequencing Program Predisposition to cancer caused by genetic and functional defects of mammalian Atad5. PLoS Genet. 2011;7:e1002245. doi: 10.1371/journal.pgen.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengesser K, Cooper DN, Steinmann K, Kluwe L, Chuzhanova NA, Wimmer K, Tatagiba M, Tinschert S, Mautner VF, Kehrer-Sawatzki H. A novel third type of recurrent NF1 microdeletion mediated by nonallelic homologous recombination between LRRC37B-containing low-copy repeats in 17q11.2. Hum Mutat. 2010;31:742–751. doi: 10.1002/humu.21254. [DOI] [PubMed] [Google Scholar]
- Bengesser K, Vogt J, Mussotter T, Mautner VF, Messiaen L, Cooper DN, Kehrer-Sawatzki H. Analysis of crossover breakpoints yields new insights into the nature of the gene conversion events associated with large NF1 deletions mediated by nonallelic homologous recombination. Hum Mutat. 2014;35:215–226. doi: 10.1002/humu.22473. [DOI] [PubMed] [Google Scholar]
- Bernardinelli Y, Nikonenko I, Muller D. Structural plasticity: mechanisms and contribution to developmental psychiatric disorders. Front Neuroanat. 2014;8:123. doi: 10.3389/fnana.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Gladfelter AS, Kovar DR, Lee WL. Cytoskeletal dynamics: a view from the membrane. J Cell Biol. 2015;209:329–337. doi: 10.1083/jcb.201502062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bist P, Kim SY, Pulloor NK, McCaffrey K, Nair SK, Liu Y, Lin R, Krishnan MN (2016) ArfGAP domain containing protein 2 (ADAP2) integrates upstream and downstream modules of RIG-I signaling, and facilitates Type I interferon production. Mol Cell Biol Dec 12. pii: MCB.00537-16. doi: 10.1128/MCB.00537-16 (Epub ahead of print) [DOI] [PMC free article] [PubMed]
- Bonnart C, Gérus M, Hoareau-Aveilla C, Kiss T, Caizergues-Ferrer M, Henry Y, Henras AK. Mammalian HCA66 protein is required for both ribosome synthesis and centriole duplication. Nucleic Acids Res. 2012;40:6270–6289. doi: 10.1093/nar/gks234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer VR, Moore BD, 3rd, Hiscock M. Learning disability subtypes in children with neurofibromatosis. J Learn Disabil. 1997;30:521–533. doi: 10.1177/002221949703000508. [DOI] [PubMed] [Google Scholar]
- Budel S, Padukkavidana T, Liu BP, Feng Z, Hu F, Johnson S, Lauren J, Park JH, McGee AW, Liao J, Stillman A, Kim JE, Yang BZ, Sodi S, Gelernter J, Zhao H, Hisama F, Arnsten AF, Strittmatter SM. Genetic variants of Nogo-66 receptor with possible association to schizophrenia block myelin inhibition of axon growth. J Neurosci. 2008;28:13161–13172. doi: 10.1523/JNEUROSCI.3828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramuta S, Egyházi S, Rodolfo M, Witten D, Hansson J, Larsson C, Lui WO. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130:2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- Castle B, Baser ME, Huson SM, Cooper DN, Upadhyaya M. Evaluation of genotype/phenotype correlations in neurofibromatosis type 1. J Med Genet. 2003;40:e109. doi: 10.1136/jmg.40.10.e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Huang Z, Ye Q, Ming Y, Zhang S, Zhao Y, Liu L, Wang Q, Cheng K. Prognostic significance and anti-proliferation effect of microRNA-365 in hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:1705–1711. [PMC free article] [PubMed] [Google Scholar]
- Clementi M, Milani S, Mammi I, Boni S, Monciotti C, Tenconi R. Neurofibromatosis type 1 growth charts. Am J Med Genet. 1999;87:317–323. doi: 10.1002/(SICI)1096-8628(19991203)87:4<317::AID-AJMG7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Cleven AH, Sannaa GA, Briaire-de Bruijn I, Ingram DR, van de Rijn M, Rubin BP, de Vries MW, Watson KL, Torres KE, Wang WL, van Duinen SG, Hogendoorn PC, Lazar AJ, Bovée JV. Loss of H3K27 tri-methylation is a diagnostic marker for malignant peripheral nerve sheath tumors and an indicator for an inferior survival. Mod Pathol. 2016;29:582–590. doi: 10.1038/modpathol.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnossen MH, van der Est MN, Breuning MH, van Asperen CJ, Breslau-Siderius EJ, van der Ploeg AT, de Goede-Bolder A, van den Ouweland AM, Halley DJ, Niermeijer MF. Deletions spanning the neurofibromatosis type 1 gene: implications for genotype-phenotype correlations in neurofibromatosis type 1? Hum Mutat. 1997;9:458–464. doi: 10.1002/(SICI)1098-1004(1997)9:5<458::AID-HUMU13>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Coe BP, Witherspoon K, Rosenfeld JA, van Bon BW, Vulto-van Silfhout AT, Bosco P, Friend KL, Baker C, Buono S, Vissers LE, Schuurs-Hoeijmakers JH, Hoischen A, Pfundt R, Krumm N, Carvill GL, Li D, Amaral D, Brown N, Lockhart PJ, Scheffer IE, Alberti A, Shaw M, Pettinato R, Tervo R, de Leeuw N, Reijnders MR, Torchia BS, Peeters H, O’Roak BJ, Fichera M, Hehir-Kwa JY, Shendure J, Mefford HC, Haan E, Gécz J, de Vries BB, Romano C, Eichler EE. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway E, Healy E, Bracken AP. PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr Opin Cell Biol. 2015;37:42–48. doi: 10.1016/j.ceb.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H, Bader P, McCracken E, Niyazov D, Leppig K, Thiese H, Hummel M, Alexander N, Gorski J, Kussmann J, Shashi V, Johnson K, Rehder C, Ballif BC, Shaffer LG, Eichler EE. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RM, Federov NB, Kogan JH, Murphy GG, Stern J, Ohno M, Kucherlapati R, Jacks T, Silva AJ. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- Cowley GS, Murthy AE, Parry DM, Schneider G, Korf B, Upadhyaya M, Harper P, MacCollin M, Bernards A, Gusella JF. Genetic variation in the 3′ untranslated region of the neurofibromatosis 1 gene: application to unequal allelic expression. Somat Cell Mol Genet. 1998;24:107–119. doi: 10.1023/B:SCAM.0000007113.28381.53. [DOI] [PubMed] [Google Scholar]
- Crowe FW, Schull WJ, Neel JV. A clinical, pathological and genetic study of multiple neurofibromatosis. Springfield: Charles C Thomas; 1956. [Google Scholar]
- Cui Y, Costa RM, Murphy GG, Elgersma Y, Zhu Y, Gutmann DH, Parada LF, Mody I, Silva AJ. Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell. 2008;135:549–560. doi: 10.1016/j.cell.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha KS, Rozza-de-Menezes RE, Andrade RM, Theos A, Luiz RR, Korf B, Geller M. Validity and interexaminer reliability of a new method to quantify skin neurofibromas of neurofibromatosis 1 using paper frames. Orphanet J Rare Dis. 2014;9:202. doi: 10.1186/s13023-014-0202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca A, Bottillo I, Dasdia MC, Morella A, Lanari V, Bernardini L, Divona L, Giustini S, Sinibaldi L, Novelli A, Torrente I, Schirinzi A, Dallapiccola B. Deletions of NF1 gene and exons detected by multiplex ligation-dependent probe amplification. J Med Genet. 2007;44:800–808. doi: 10.1136/jmg.2007.053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Raedt T, Brems H, Wolkenstein P, Vidaud D, Pilotti S, Perrone F, Mautner V, Frahm S, Sciot R, Legius E. Elevated risk for MPNST in NF1 microdeletion patients. Am J Hum Genet. 2003;72:1288–1292. doi: 10.1086/374821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Raedt T, Stephens M, Heyns I, Brems H, Thijs D, Messiaen L, Stephens K, Lazaro C, Wimmer K, Kehrer-Sawatzki H, Vidaud D, Kluwe L, Marynen P, Legius E. Conservation of hotspots for recombination in low-copy repeats associated with the NF1 microdeletion. Nat Genet. 2006;38:1419–1423. doi: 10.1038/ng1920. [DOI] [PubMed] [Google Scholar]
- De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, Helin K, Hornick JL, Mautner V, Kehrer-Sawatzki H, Clapp W, Bradner J, Vidaud M, Upadhyaya M, Legius E, Cichowski K. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014;514:247–251. doi: 10.1038/nature13561. [DOI] [PubMed] [Google Scholar]
- Deng H, Lv L, Li Y, Zhang C, Meng F, Pu Y, Xiao J, Qian L, Zhao W, Liu Q, Zhang D, Wang Y, Zhang H, He Y, Zhu J. miR-193a-3p regulates the multi-drug resistance of bladder cancer by targeting the LOXL4 gene and the oxidative stress pathway. Mol Cancer. 2014;13:234. doi: 10.1186/1476-4598-13-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descheemaeker MJ, Roelandts K, De Raedt T, Brems H, Fryns JP, Legius E. Intelligence in individuals with a neurofibromatosis type 1 microdeletion. Am J Med Genet A. 2004;131:325–326. doi: 10.1002/ajmg.a.30346. [DOI] [PubMed] [Google Scholar]
- Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- Diggs-Andrews KA, Gutmann DH. Modeling cognitive dysfunction in neurofibromatosis-1. Trends Neurosci. 2013;36:237–247. doi: 10.1016/j.tins.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner MO, Sybert VP, Weaver M, Pletcher BA, Stephens K. NF1 microdeletion breakpoints are clustered at flanking repetitive sequences. Hum Mol Genet. 2000;9:35–46. doi: 10.1093/hmg/9.1.35. [DOI] [PubMed] [Google Scholar]
- Douglas J, Cilliers D, Coleman K, Tatton-Brown K, Barker K, Bernhard B, Burn J, Huson S, Josifova D, Lacombe D, Malik M, Mansour S, Reid E, Cormier-Daire V, Cole T, Collaboration Childhood Overgrowth, Rahman N. Mutations in RNF135, a gene within the NF1 microdeletion region, cause phenotypic abnormalities including overgrowth. Nat Genet. 2007;39:963–965. doi: 10.1038/ng2083. [DOI] [PubMed] [Google Scholar]
- Ducatman BS, Scheithauer BW, Piepgras DG, Reiman HM, Ilstrup DM. Malignant peripheral nerve sheath tumors. A clinicopathologic study of 120 cases. Cancer. 1986;57:2006–2021. doi: 10.1002/1097-0142(19860515)57:10<2006::AID-CNCR2820571022>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Duong TA, Sbidian E, Valeyrie-Allanore L, Vialette C, Ferkal S, Hadj-Rabia S, Glorion C, Lyonnet S, Zerah M, Kemlin I, Rodriguez D, Bastuji-Garin S, Wolkenstein P. Mortality associated with neurofibromatosis 1: a cohort study of 1895 patients in 1980-2006 in France. Orphanet J Rare Dis. 2011;6:18. doi: 10.1186/1750-1172-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Baser ME, McGaughran J, Sharif S, Howard E, Moran A. Malignant peripheral nerve sheath tumours in neurofibromatosis 1. J Med Genet. 2002;39:311–314. doi: 10.1136/jmg.39.5.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Huson SM, Birch JM. Malignant peripheral nerve sheath tumours in inherited disease. Clin Sarcoma Res. 2012;2:17. doi: 10.1186/2045-3329-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fant X, Gnadt N, Haren L, Merdes A. Stability of the small gamma-tubulin complex requires HCA66, a protein of the centrosome and the nucleolus. J Cell Sci. 2009;122:1134–1144. doi: 10.1242/jcs.035238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferner RE, Gutmann DH. International consensus statement on malignant peripheral nerve sheath tumors in neurofibromatosis. Cancer Res. 2002;62:1573–1577. [PubMed] [Google Scholar]
- Ferner RE, Chaudhuri R, Bingham J, Cox T, Hughes RA. MRI in neurofibromatosis 1. The nature and evolution of increased intensity T2 weighted lesions and their relationship to intellectual impairment. J Neurol Neurosurg Psychiatry. 1993;56:492–495. doi: 10.1136/jnnp.56.5.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferner RE, Hughes RA, Weinman J. Intellectual impairment in neurofibromatosis 1. J Neurol Sci. 1996;138:125–133. doi: 10.1016/0022-510X(96)00022-6. [DOI] [PubMed] [Google Scholar]
- Ferner RE, Huson SM, Thomas N, Moss C, Willshaw H, Evans DG, Upadhyaya M, Towers R, Gleeson M, Steiger C, Kirby A. Guidelines for the diagnosis and management of individuals with neurofibromatosis 1. J Med Genet. 2007;44:81–88. doi: 10.1136/jmg.2006.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro E, Pesaresi MG, De Zio D, Cencioni MT, Gortat A, Cozzolino M, Berghella L, Salvatore AM, Oettinghaus B, Scorrano L, Pérez-Payà E, Cecconi F. Apaf1 plays a pro-survival role by regulating centrosome morphology and function. J Cell Sci. 2011;124:3450–3463. doi: 10.1242/jcs.086298. [DOI] [PubMed] [Google Scholar]
- Forbes SH, Dorschner MO, Le R, Stephens K. Genomic context of paralogous recombination hotspots mediating recurrent NF1 region microdeletion. Genes Chromosomes Cancer. 2004;41:12–15. doi: 10.1002/gcc.20065. [DOI] [PubMed] [Google Scholar]
- Friedman JM. Evaluation and management. In: Friedman J, Gutmann D, MacCollin M, Riccardi VM, editors. Neurofibromatosis: phenotype, natural history and pathogenesis. 3. Baltimore: Johns Hopkins University Press; 1999. pp. 90–91. [Google Scholar]
- Friedman JM, Birch PH. Type 1 neurofibromatosis: a descriptive analysis of the disorder in 1,728 patients. Am J Med Genet. 1997;70:138–143. doi: 10.1002/(SICI)1096-8628(19970516)70:2<138::AID-AJMG7>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Gazy I, Liefshitz B, Parnas O, Kupiec M. Elg1, a central player in genome stability. Mutat Res Rev Mutat Res. 2015;763:267–279. doi: 10.1016/j.mrrev.2014.11.007. [DOI] [PubMed] [Google Scholar]
- Geoffroy CG, Zheng B. Myelin-associated inhibitors in axonal growth after CNS injury. Curr Opin Neurobiol. 2014;27:31–38. doi: 10.1016/j.conb.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervasini C, Venturin M, Orzan F, Friso A, Clementi M, Tenconi R, Larizza L, Riva P. Uncommon Alu-mediated NF1 microdeletion with a breakpoint inside the NF1 gene. Genomics. 2005;85:273–279. doi: 10.1016/j.ygeno.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Gil V, Bichler Z, Lee JK, Seira O, Llorens F, Bribian A, Morales R, Claverol-Tinture E, Soriano E, Sumoy L, Zheng B, Del Río JA. Developmental expression of the oligodendrocyte myelin glycoprotein in the mouse telencephalon. Cereb Cortex. 2010;20:1769–1779. doi: 10.1093/cercor/bhp246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler AD, Zhu Y, Ismat FA, Lu MM, Yamauchi Y, Parada LF, Epstein JA. Nf1 has an essential role in endothelial cells. Nat Genet. 2003;33:75–79. doi: 10.1038/ng1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisart B, Rack K, Vidrequin S, Hilbert P, Deltenre P, Verellen-Dumoulin C, Destrée A. NF1 microduplication first clinical report: association with mild mental retardation, early onset of baldness and dental enamel hypoplasia? Eur J Hum Genet. 2008;16:305–311. doi: 10.1038/sj.ejhg.5201978. [DOI] [PubMed] [Google Scholar]
- Guo A, Liu A, Wei L, Song X. Malignant peripheral nerve sheath tumors: differentiation patterns and immunohistochemical features—a mini-review and our new findings. J Cancer. 2012;3:303–309. doi: 10.7150/jca.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SL, Ye H, Teng Y, Wang YL, Yang G, Li XB, Zhang C, Yang X, Yang ZZ, Yang X. Akt-p53-miR-365-cyclin D1/cdc25A axis contributes to gastric tumorigenesis induced by PTEN deficiency. Nat Commun. 2013;4:2544. doi: 10.1038/ncomms3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib AA, Marton LS, Allwardt B, Gulcher JR, Mikol DD, Högnason T, Chattopadhyay N, Stefansson K. Expression of the oligodendrocyte-myelin glycoprotein by neurons in the mouse central nervous system. J Neurochem. 1998;70:1704–1711. doi: 10.1046/j.1471-4159.1998.70041704.x. [DOI] [PubMed] [Google Scholar]
- Hegedus B, Yeh TH, Lee DY, Emnett RJ, Li J, Gutmann DH. Neurofibromin regulates somatic growth through the hypothalamic-pituitary axis. Hum Mol Genet. 2008;17:2956–2966. doi: 10.1093/hmg/ddn194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller G, Weinzierl M, Noll C, Babinsky V, Ziegler B, Altenberger C, Minichsdorfer C, Lang G, Döme B, End-Pfützenreuter A, Arns BM, Grin Y, Klepetko W, Zielinski CC, Zöchbauer-Müller S. Genome-wide miRNA expression profiling identifies miR-9-3 and miR-193a as targets for DNA methylation in non-small cell lung cancers. Clin Cancer Res. 2012;18:1619–1629. doi: 10.1158/1078-0432.CCR-11-2450. [DOI] [PubMed] [Google Scholar]
- Hillmer M, Wagner D, Summerer A, Daiber M, Mautner VF, Messiaen L, Cooper DN, Kehrer-Sawatzki H. Fine mapping of meiotic NAHR-associated crossovers causing large NF1 deletions. Hum Mol Genet. 2016;25:484–496. doi: 10.1093/hmg/ddv487. [DOI] [PubMed] [Google Scholar]
- Hoffmeyer S, Assum G, Griesser J, Kaufmann D, Nürnberg P, Krone W. On unequal allelic expression of the neurofibromin gene in neurofibromatosis type 1. Hum Mol Genet. 1995;4:1267–1272. doi: 10.1093/hmg/4.8.1267. [DOI] [PubMed] [Google Scholar]
- Huson SM, Harper PS, Compston DAS. von Recklinghausen neurofibromatosis: a clinical and population study in South East Wales. Brain. 1988;111:1355–1381. doi: 10.1093/brain/111.6.1355. [DOI] [PubMed] [Google Scholar]
- Hyman SL, Shores A, North KN. The nature and frequency of cognitive deficits in children with neurofibromatosis type 1. Neurology. 2005;65:1037–1044. doi: 10.1212/01.wnl.0000179303.72345.ce. [DOI] [PubMed] [Google Scholar]
- Hyman SL, Arthur Shores E, North KN. Learning disabilities in children with neurofibromatosis type 1: subtypes, cognitive profile, and attention-deficit-hyperactivity disorder. Dev Med Child Neurol. 2006;48:973–977. doi: 10.1017/S0012162206002131. [DOI] [PubMed] [Google Scholar]
- Hyman SL, Gill DS, Shores EA, Steinberg A, North KN. T2 hyperintensities in children with neurofibromatosis type 1 and their relationship to cognitive functioning. J Neurol Neurosurg Psychiatry. 2007;78:1088–1091. doi: 10.1136/jnnp.2006.108134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Rotem A, Struhl K. Inhibition of miR-193a expression by Max and RXRα activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer Res. 2011;71:5144–5153. doi: 10.1158/0008-5472.CAN-11-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismat FA, Xu J, Lu MM, Epstein JA. The neurofibromin GAP-related domain rescues endothelial but not neural crest development in Nf1 mice. J Clin Invest. 2006;116:2378–2384. doi: 10.1172/JCI28341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Magnaldi S, White RM, Denckla MB, Hofman K, Naidu S, Bryan RN. Neurofibromatosis type 1: the evolution of deep gray and white matter MR abnormalities. Am J Neuroradiol. 1994;15:1513–1519. [PMC free article] [PubMed] [Google Scholar]
- Jenne DE, Tinschert S, Reimann H, Lasinger W, Thiel G, Hameister H, Kehrer-Sawatzki H. Molecular characterization and gene content of breakpoint boundaries in patients with neurofibromatosis type 1 with 17q11.2 microdeletions. Am J Hum Genet. 2001;69:516–527. doi: 10.1086/323043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenne DE, Tinschert S, Dorschner MO, Hameister H, Stephens K, Kehrer-Sawatzki H. Complete physical map and gene content of the human NF1 tumor suppressor region in human and mouse. Genes Chromosomes Cancer. 2003;37:111–120. doi: 10.1002/gcc.10206. [DOI] [PubMed] [Google Scholar]
- Jentarra GM, Rice SG, Olfers S, Rajan C, Saffen DM, Narayanan V. Skewed allele-specific expression of the NF1 gene in normal subjects: a possible mechanism for phenotypic variability in neurofibromatosis type 1. J Child Neurol. 2012;27:695–702. doi: 10.1177/0883073811423439. [DOI] [PubMed] [Google Scholar]
- Jin J, Zhao L, Li Z. The E3 ubiquitin ligase RNF135 regulates the tumorigenesis activity of tongue cancer SCC25 cells. Cancer Med. 2016;5:3140–3146. doi: 10.1002/cam4.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SM, Lee HJ, Cho JY. MicroRNA-365 regulates NKX2-1, a key mediator of lung cancer. Cancer Lett. 2013;335:487–494. doi: 10.1016/j.canlet.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Karajannis MA, Ferner RE. Neurofibromatosis-related tumors: emerging biology and therapies. Curr Opin Pediatr. 2015;27:26–33. doi: 10.1097/MOP.0000000000000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayes LM, Riccardi VM, Burke W, Bennett RL, Stephens K. Large de novo DNA deletion in a patient with sporadic neurofibromatosis 1, mental retardation, and dysmorphism. J Med Genet. 1992;29:686–690. doi: 10.1136/jmg.29.10.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayes LM, Burke W, Riccardi VM, Bennett R, Ehrlich P, Rubenstein A, Stephens K. Deletions spanning the neurofibromatosis 1 gene: identification and phenotype of five patients. Am J Hum Genet. 1994;54:424–436. [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Cooper DN. Mosaicism in sporadic neurofibromatosis type 1: variations on a theme common to other hereditary cancer syndromes? J Med Genet. 2008;45:622–631. doi: 10.1136/jmg.2008.059329. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Tinschert S, Jenne DE. Heterogeneity of breakpoints in non-LCR-mediated large constitutional deletions of the 17q11.2 NF1 tumor suppressor region. J Med Genet. 2003;40:E116. doi: 10.1136/jmg.40.10.e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Kluwe L, Sandig C, Kohn M, Wimmer K, Krammer U, Peyrl A, Jenne DE, Hansmann I, Mautner VF. High frequency of mosaicism among patients with neurofibromatosis type 1 (NF1) with microdeletions caused by somatic recombination of the JJAZ1 gene. Am J Hum Genet. 2004;75:410–423. doi: 10.1086/423624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Kluwe L, Fünsterer C, Mautner VF. Extensively high load of internal tumors determined by whole body MRI scanning in a patient with neurofibromatosis type 1 and a non-LCR-mediated 2-Mb deletion in 17q11.2. Hum Genet. 2005;116:466–475. doi: 10.1007/s00439-005-1265-4. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Schmid E, Fünsterer C, Kluwe L, Mautner VF. Absence of cutaneous neurofibromas in an NF1 patient with an atypical deletion partially overlapping the common 1.4 Mb microdeleted region. Am J Med Genet A. 2008;146A:691–699. doi: 10.1002/ajmg.a.32045. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Vogt J, Mußotter T, Kluwe L, Cooper DN, Mautner VF. Dissecting the clinical phenotype associated with mosaic type-2 NF1 microdeletions. Neurogenetics. 2012;13:229–236. doi: 10.1007/s10048-012-0332-y. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Bengesser K, Callens T, Mikhail F, Fu C, Hillmer M, Walker ME, Saal HM, Lacassie Y, Cooper DN, Messiaen L. Identification of large NF1 duplications reciprocal to NAHR-mediated type-1 NF1 deletions. Hum Mutat. 2014;35:1469–1475. doi: 10.1002/humu.22692. [DOI] [PubMed] [Google Scholar]
- Klein-Tasman BP, Janke KM, Luo W, Casnar CL, Hunter SJ, Tonsgard J, Trapane P, van der Fluit F, Kais LA. Cognitive and psychosocial phenotype of young children with neurofibromatosis-1. J Int Neuropsychol Soc. 2014;20:88–98. doi: 10.1017/S1355617713001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluwe L, Siebert R, Gesk S, Friedrich RE, Tinschert S, Kehrer-Sawatzki H, Mautner VF. Screening 500 unselected neurofibromatosis 1 patients for deletions of the NF1 gene. Hum Mutat. 2004;23:111–116. doi: 10.1002/humu.10299. [DOI] [PubMed] [Google Scholar]
- Kluwe L, Nguyen R, Vogt J, Bengesser K, Mussotter T, Friedrich RE, Jett K, Kehrer-Sawatzki H, Mautner VF. Internal tumor burden in neurofibromatosis type I patients with large NF1 deletions. Genes Chromosomes Cancer. 2012;51:447–451. doi: 10.1002/gcc.21931. [DOI] [PubMed] [Google Scholar]
- Kobayashi R, Matsune K, Ohashi H. Fused teeth, macrodontia and increased caries are characteristic features of neurofibromatosis type 1 patients with NF1 gene microdeletion. J Pediatr Genet. 2012;1:25–31. doi: 10.3233/PGE-2012-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf BR, Schneider G, Poussaint TY. Structural anomalies revealed by neuroimaging studies in the brains of patients with neurofibromatosis type 1 and large deletions. Genet Med. 1999;1:136–140. doi: 10.1097/00125817-199905000-00004. [DOI] [PubMed] [Google Scholar]
- Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- Krab LC, Aarsen FK, de Goede-Bolder A, Catsman-Berrevoets CE, Arts WF, Moll HA, Elgersma Y. Impact of neurofibromatosis type 1 on school performance. J Child Neurol. 2008;23:1002–1010. doi: 10.1177/0883073808316366. [DOI] [PubMed] [Google Scholar]
- Kubota T, Myung K, Donaldson AD. Is PCNA unloading the central function of the Elg1/ATAD5 replication factor C-like complex? Cell Cycle. 2013;12:2570–2579. doi: 10.4161/cc.25626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkantrakorn K, Geller TJ. Seizures in neurofibromatosis 1. Pediatr Neurol. 1998;19:347–350. doi: 10.1016/S0887-8994(98)00075-7. [DOI] [PubMed] [Google Scholar]
- Lacroix M, El Messaoudi S, Rodier G, Le Cam A, Sardet C, Fabbrizio E. The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Rep. 2008;9:452–458. doi: 10.1038/embor.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammert M, Friedman JM, Kluwe L, Mautner VF. Prevalence of neurofibromatosis 1 in German children at elementary school enrolment. Arch Dermatol. 2005;141:71–74. doi: 10.1001/archderm.141.1.71. [DOI] [PubMed] [Google Scholar]
- Laugesen A, Højfeldt JW, Helin K. Role of the polycomb repressive complex 2 (PRC2) in transcriptional regulation and cancer. Cold Spring Harb Perspect Med. 2016;6:a026575. doi: 10.1101/cshperspect.a026575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Raiker SJ, Venkatesh K, Geary R, Robak LA, Zhang Y, Yeh HH, Shrager P, Giger RJ. Synaptic function for the Nogo-66 receptor NgR1: regulation of dendritic spine morphology and activity-dependent synaptic strength. J Neurosci. 2008;28:2753–2765. doi: 10.1523/JNEUROSCI.5586-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Yang K, Cohn MA, Sikdar N, D’Andrea AD, Myung K. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through its interactions with PCNA and USP1. J Biol Chem. 2010;285:10362–10369. doi: 10.1074/jbc.M109.092544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee X, Hu Y, Zhang Y, Yang Z, Shao Z, Qiu M, Pepinsky B, Miller RH, Mi S. Oligodendrocyte differentiation and myelination defects in OMgp null mice. Mol Cell Neurosci. 2011;46:752–761. doi: 10.1016/j.mcn.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Lee KY, Fu H, Aladjem MI, Myung K. ATAD5 regulates the lifespan of DNA replication factories by modulating PCNA level on the chromatin. J Cell Biol. 2013;200:31–44. doi: 10.1083/jcb.201206084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Teckie S, Wiesner T, Ran L, Prieto Granada CN, Lin M, Zhu S, Cao Z, Liang Y, Sboner A, Tap WD, Fletcher JA, Huberman KH, Qin LX, Viale A, Singer S, Zheng D, Berger MF, Chen Y, Antonescu CR, Chi P. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46:1227–1232. doi: 10.1038/ng.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppig KA, Kaplan P, Viskochil D, Weaver M, Ortenberg J, Stephens K. Familial neurofibromatosis 1 microdeletions: cosegregation with distinct facial phenotype and early onset of cutaneous neurofibromata. Am J Med Genet. 1997;73:197–204. doi: 10.1002/(SICI)1096-8628(1997)73:2<197::AID-AJMG17>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Lewis RA, Gerson LP, Axelson KA, Riccardi VM, Whitford RP. von Recklinghausen neurofibromatosis II. Incidence of optic gliomata. Ophthalmology. 1984;91:929–935. doi: 10.1016/S0161-6420(84)34217-8. [DOI] [PubMed] [Google Scholar]
- Li Y, Deng H, Lv L, Zhang C, Qian L, Xiao J, Zhao W, Liu Q, Zhang D, Wang Y, Yan J, Zhang H, He Y, Zhu J. The miR-193a-3p-regulated ING5 gene activates the DNA damage response pathway and inhibits multi-chemoresistance in bladder cancer. Oncotarget. 2015;6:10195–10206. doi: 10.18632/oncotarget.3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Liu M, Yan X, Zhou Y, Wang W, Wang X, Fu Z, Wang N, Zhang S, Wang Y, Zen K, Zhang CY, Hou D, Li J, Chen X. miR-193a-3p functions as a tumor suppressor in lung cancer by down-regulating ERBB4. J Biol Chem. 2015;290:926–940. doi: 10.1074/jbc.M114.621409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AE, Birch PH, Korf BR, Tenconi R, Niimura M, Poyhonen M, Armfield Uhas K, Sigorini M, Virdis R, Romano C, Bonioli E, Wolkenstein P, Pivnick EK, Lawrence M, Friedman JM. Cardiovascular malformations and other cardiovascular abnormalities in neurofibromatosis 1. Am J Med Genet. 2000;95:108–117. doi: 10.1002/1096-8628(20001113)95:2<108::AID-AJMG4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Listernick R, Louis DN, Packer RJ, Gutmann DH. Optic pathway gliomas in children with neurofibromatosis 1: consensus statement from the NF1 optic pathway glioma task force. Ann Neurol. 1997;41:143–149. doi: 10.1002/ana.410410204. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang F, Liu Y, Yao Y, Lv X, Dong B, Li J, Ren S, Yao Y, Xu Y. RNF135, RING finger protein, promotes the proliferation of human glioblastoma cells in vivo and in vitro via the ERK pathway. Sci Rep. 2016;6:20642. doi: 10.1038/srep20642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens F, Gil V, del Río JA. Emerging functions of myelin-associated proteins during development, neuronal plasticity, and neurodegeneration. FASEB J. 2011;25:463–475. doi: 10.1096/fj.10-162792. [DOI] [PubMed] [Google Scholar]
- López-Correa C, Brems H, Lázaro C, Marynen P, Legius E. Unequal meiotic crossover: a frequent cause of NF1 microdeletions. Am J Hum Genet. 2000;66:1969–1974. doi: 10.1086/302920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Correa C, Dorschner M, Brems H, Lázaro C, Clementi M, Upadhyaya M, Dooijes D, Moog U, Kehrer-Sawatzki H, Rutkowski JL, Fryns JP, Marynen P, Stephens K, Legius E. Recombination hotspot in NF1 microdeletion patients. Hum Mol Genet. 2001;10:1387–1392. doi: 10.1093/hmg/10.13.1387. [DOI] [PubMed] [Google Scholar]
- Lovat F, Valeri N, Croce CM. MicroRNAs in the pathogenesis of cancer. Semin Oncol. 2011;38:724–733. doi: 10.1053/j.seminoncol.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Lu X, Shaw CA, Patel A, Li J, Cooper ML, Wells WR, Sullivan CM, Sahoo T, Yatsenko SA, Bacino CA, Stankiewicz P, Ou Z, Chinault AC, Beaudet AL, Lupski JR, Cheung SW, Ward PA. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS One. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Deng H, Li Y, Zhang C, Liu X, Liu Q, Zhang D, Wang L, Pu Y, Zhang H, He Y, Wang Y, Yu Y, Yu T, Zhu J. The DNA methylation-regulated miR-193a-3p dictates the multi-chemoresistance of bladder cancer via repression of SRSF2/PLAU/HIC2 expression. Cell Death Dis. 2014;5:e1402. doi: 10.1038/cddis.2014.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens O, De Schepper S, Vandesompele J, Brems H, Heyns I, Janssens S, Speleman F, Legius E, Messiaen L. Molecular dissection of isolated disease features in mosaic neurofibromatosis type 1. Am J Hum Genet. 2007;81:243–251. doi: 10.1086/519562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleva Kostovska I, Wang J, Bogdanova N, Schürmann P, Bhuju S, Geffers R, Dürst M, Liebrich C, Klapdor R, Christiansen H, Park-Simon TW, Hillemanns P, Plaseska-Karanfilska D, Dörk T. Rare ATAD5 missense variants in breast and ovarian cancer patients. Cancer Lett. 2016;376:173–177. doi: 10.1016/j.canlet.2016.03.048. [DOI] [PubMed] [Google Scholar]
- Mantripragada KK, Thuresson AC, Piotrowski A, Díaz de Ståhl T, Menzel U, Grigelionis G, Ferner RE, Griffiths S, Bolund L, Mautner V, Nordling M, Legius E, Vetrie D, Dahl N, Messiaen L, Upadhyaya M, Bruder CE, Dumanski JP. Identification of novel deletion breakpoints bordered by segmental duplications in the NF1 locus using high resolution array-CGH. J Med Genet. 2006;43:28–38. doi: 10.1136/jmg.2005.033795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantripragada KK, Díaz de Ståhl T, Patridge C, Menzel U, Andersson R, Chuzhanova N, Kluwe L, Guha A, Mautner V, Dumanski JP, Upadhyaya M. Genome-wide high-resolution analysis of DNA copy number alterations in NF1-associated malignant peripheral nerve sheath tumors using 32 K BAC array. Genes Chromosomes Cancer. 2009;48:897–907. doi: 10.1002/gcc.20695. [DOI] [PubMed] [Google Scholar]
- Martin I, Andres CR, Védrine S, Tabagh R, Michelle C, Jourdan ML, Heuze-Vourc’h N, Corcia P, Duittoz A, Vourc’h P. Effect of the oligodendrocyte myelin glycoprotein (OMgp) on the expansion and neuronal differentiation of rat neural stem cells. Brain Res. 2009;1284:22–30. doi: 10.1016/j.brainres.2009.05.070. [DOI] [PubMed] [Google Scholar]
- Mautner VF, Kluwe L, Thakker SD, Leark RA. Treatment of ADHD in neurofibromatosis type 1. Dev Med Child Neurol. 2002;44:164–170. doi: 10.1017/S0012162201001876. [DOI] [PubMed] [Google Scholar]
- Mautner VF, Asuagbor FA, Dombi E, Fünsterer C, Kluwe L, Wenzel R, Widemann BC, Friedman JM. Assessment of benign tumor burden by whole-body MRI in patients with neurofibromatosis 1. Neuro Oncol. 2008;10:593–598. doi: 10.1215/15228517-2008-011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mautner VF, Kluwe L, Friedrich RE, Roehl AC, Bammert S, Högel J, Spöri H, Cooper DN, Kehrer-Sawatzki H. Clinical characterisation of 29 neurofibromatosis type-1 patients with molecularly ascertained 1.4 Mb type-1 NF1 deletions. J Med Genet. 2010;47:623–630. doi: 10.1136/jmg.2009.075937. [DOI] [PubMed] [Google Scholar]
- McGaughran JM, Harris DI, Donnai D, Teare D, MacLeod R, Westerbeek R, Kingston H, Super M, Harris R, Evans DG. A clinical study of type 1 neurofibromatosis in north west England. J Med Genet. 1999;36:197–203. [PMC free article] [PubMed] [Google Scholar]
- McGee AW, Yang Y, Fischer QS, Daw NW, Strittmatter SM. Experience-driven plasticity of visual cortex limited by myelin and Nogo receptor. Science. 2005;309:2222–2226. doi: 10.1126/science.1114362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensink KA, Ketterling RP, Flynn HC, Knudson RA, Lindor NM, Heese BA, Spinner RJ, Babovic-Vuksanovic D. Connective tissue dysplasia in five new patients with NF1 microdeletions: further expansion of phenotype and review of the literature. J Med Genet. 2006;43:e8. doi: 10.1136/jmg.2005.034256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messiaen L, Vogt J, Bengesser K, Fu C, Mikhail F, Serra E, Garcia-Linares C, Cooper DN, Lazaro C, Kehrer-Sawatzki H. Mosaic type-1 NF1 microdeletions as a cause of both generalized and segmental neurofibromatosis type-1 (NF1) Hum Mutat. 2011;32:213–219. doi: 10.1002/humu.21418. [DOI] [PubMed] [Google Scholar]
- Millichap JG. MRI Screening for optic gliomas in neurofibromatosis type 1. Pediatr Neurol Briefs. 2015;29:72. doi: 10.15844/pedneurbriefs-29-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironova YA, Giger RJ. Where no synapses go: gatekeepers of circuit remodeling and synaptic strength. Trends Neurosci. 2013;36:363–373. doi: 10.1016/j.tins.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moles KJ, Gowans GC, Gedela S, Beversdorf D, Yu A, Seaver LH, Schultz RA, Rosenfeld JA, Torchia BS, Shaffer LG. NF1 microduplications: identification of seven nonrelated individuals provides further characterization of the phenotype. Genet Med. 2012;14:508–514. doi: 10.1038/gim.2011.46. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Ghosh S, Nazmi A, Basu A. RIG-I knockdown impedes neurogenesis in a murine model of Japanese encephalitis. Cell Biol Int. 2015;39:224–229. doi: 10.1002/cbin.10354. [DOI] [PubMed] [Google Scholar]
- Nakano H, Yamada Y, Miyazawa T, Yoshida T. Gain-of-function microRNA screens identify miR-193a regulating proliferation and apoptosis in epithelial ovarian cancer cells. Int J Oncol. 2013;42:1875–1882. doi: 10.3892/ijo.2013.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazmi A, Dutta K, Basu A. RIG-I mediates innate immune response in mouse neurons following Japanese encephalitis virus infection. PLoS One. 2011;6:e21761. doi: 10.1371/journal.pone.0021761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen R, Dombi E, Widemann BC, Solomon J, Fuensterer C, Kluwe L, Friedman JM, Mautner VF. Growth dynamics of plexiform neurofibromas: a retrospective cohort study of 201 patients with neurofibromatosis 1. Orphanet J Rare Dis. 2012;7:75. doi: 10.1186/1750-1172-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen R, Mir TS, Kluwe L, Jett K, Kentsch M, Mueller G, Kehrer-Sawatzki H, Friedman JM, Mautner VF. Cardiac characterization of 16 patients with large NF1 gene deletions. Clin Genet. 2013;84:344–349. doi: 10.1111/cge.12072. [DOI] [PubMed] [Google Scholar]
- Nguyen R, Jett K, Harris GJ, Cai W, Friedman JM, Mautner VF. Benign whole body tumor volume is a risk factor for malignant peripheral nerve sheath tumors in neurofibromatosis type 1. J Neurooncol. 2014;116:307–313. doi: 10.1007/s11060-013-1293-1. [DOI] [PubMed] [Google Scholar]
- Nie J, Liu L, Zheng W, Chen L, Wu X, Xu Y, Du X, Han W. microRNA-365, down-regulated in colon cancer, inhibits cell cycle progression and promotes apoptosis of colon cancer cells by probably targeting cyclin D1 and Bcl-2. Carcinogenesis. 2012;33:220–225. doi: 10.1093/carcin/bgr245. [DOI] [PubMed] [Google Scholar]
- Ning X, Farschtschi S, Jones A, Kehrer-Sawatzki H, Mautner VF, Friedman JM. Growth in neurofibromatosis 1 microdeletion patients. Clin Genet. 2016;89:351–354. doi: 10.1111/cge.12632. [DOI] [PubMed] [Google Scholar]
- North KN. Neurofibromatosis type 1: review of the first 200 patients in an Australian clinic. J Child Neurol. 1993;8:395–402. doi: 10.1177/088307389300800421. [DOI] [PubMed] [Google Scholar]
- North KN. Cognitive function and academic performance. In: Friedman J, Gutmann D, MacCollin M, Riccardi VM, editors. Neurofibromatosis: phenotype, natural history and pathogenesis. 3. Baltimore: Johns Hopkins University Press; 1999. p. 168. [Google Scholar]
- North K. Neurofibromatosis type 1. Am J Med Genet. 2000;97:119–127. doi: 10.1002/1096-8628(200022)97:2<119::AID-AJMG3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- North K, Joy P, Yuille D, Cocks N, Hutchins P. Cognitive function and academic performance in children with neurofibromatosis type 1. Dev Med Child Neurol. 1995;37:427–436. doi: 10.1111/j.1469-8749.1995.tb12026.x. [DOI] [PubMed] [Google Scholar]
- North KN, Riccardi V, Samango-Sprouse C, Ferner R, Moore B, Legius E, Ratner N, Denckla MB. Cognitive function and academic performance in neurofibromatosis 1: consensus statement from the NF1 cognitive disorders task force. Neurology. 1997;48:1121–1127. doi: 10.1212/WNL.48.4.1121. [DOI] [PubMed] [Google Scholar]
- Oktenli C, Saglam M, Demirbas S, Thompson P, Upadhyaya M, Consoli C, Ulucan H, Koz C, Durukan AH, Bozkurt A, Koc B, Kocar IH, Gul D. A large deletion (1.5 Mb) encompassing the neurofibromatosis type 1 gene in a patient with sporadic NF1 associated with dysmorphism, mental retardation, and unusual ocular and skeletal features. Clin Dysmorphol. 2003;12:199–201. doi: 10.1097/01.mcd.0000077565.66911.43. [DOI] [PubMed] [Google Scholar]
- Oliveira AF, Yasuda R. Neurofibromin is the major ras inactivator in dendritic spines. J Neurosci. 2014;34:776–783. doi: 10.1523/JNEUROSCI.3096-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem. 2009;284:807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- Oshiumi H, Miyashita M, Inoue N, Okabe M, Matsumoto M, Seya T. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe. 2010;8:496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Oshiumi H, Miyashita M, Matsumoto M, Seya T. A distinct role of Riplet-mediated K63-Linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 2013;9:e1003533. doi: 10.1371/journal.ppat.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmant E, de Saint-Trivier A, Laurendeau I, Dieux-Coeslier A, Parfait B, Vidaud M, Vidaud D, Bièche I. Characterization of a 7.6-Mb germline deletion encompassing the NF1 locus and about a hundred genes in an NF1 contiguous gene syndrome patient. Eur J Hum Genet. 2008;16:1459–1466. doi: 10.1038/ejhg.2008.134. [DOI] [PubMed] [Google Scholar]
- Pasmant E, Sabbagh A, Masliah-Planchon J, Haddad V, Hamel MJ, Laurendeau I, Soulier J, Parfait B, Wolkenstein P, Bièche I, Vidaud M, Vidaud D. Detection and characterization of NF1 microdeletions by custom high resolution array CGH. J Mol Diagn. 2009;11:524–529. doi: 10.2353/jmoldx.2009.090064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmant E, Sabbagh A, Spurlock G, Laurendeau I, Grillo E, Hamel MJ, Martin L, Barbarot S, Leheup B, Rodriguez D, Lacombe D, Dollfus H, Pasquier L, Isidor B, Ferkal S, Soulier J, Sanson M, Dieux-Coeslier A, Bièche I, Parfait B, Vidaud M, Wolkenstein P, Upadhyaya M, Vidaud D, Members of the NF France Network NF1 microdeletions in neurofibromatosis type 1: from genotype to phenotype. Hum Mutat. 2010;31:E1506–1518. doi: 10.1002/humu.21271. [DOI] [PubMed] [Google Scholar]
- Pasmant E, Masliah-Planchon J, Lévy P, Laurendeau I, Ortonne N, Parfait B, Valeyrie-Allanore L, Leroy K, Wolkenstein P, Vidaud M, Vidaud D, Bièche I. Identification of genes potentially involved in the increased risk of malignancy in NF1-microdeleted patients. Mol Med. 2011;17:79–87. doi: 10.2119/molmed.2010.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmant E, Vidaud M, Vidaud D, Wolkenstein P. Neurofibromatosis type 1: from genotype to phenotype. J Med Genet. 2012;49:483–489. doi: 10.1136/jmedgenet-2012-100978. [DOI] [PubMed] [Google Scholar]
- Paul C, Sardet C, Fabbrizio E. The histone- and PRMT5-associated protein COPR5 is required for myogenic differentiation. Cell Death Differ. 2012;19:900–908. doi: 10.1038/cdd.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemov A, HansenNH, Patidar R, Boland JF, Chandrasekharappa S, Mullikin JC,Wallace M, Khan J, Legius E, Widemann B, Stewart DR (2016) Comparative genomic analysis of NF1-associated atypical neurofibromas (ANF) and malignant peripheral nerve sheath tumors (MPNST). In: American Society of Human Genetics 66th Annual Meeting, 18–22 October 2016 Vancouver
- Petek E, Jenne DE, Smolle J, Binder B, Lasinger W, Windpassinger C, Wagner K, Kroisel PM, Kehrer-Sawatzki H. Mitotic recombination mediated by the JJAZF1 (KIAA0160) gene causing somatic mosaicism and a new type of constitutional NF1 microdeletion in two children of a mosaic female with only few manifestations. J Med Genet. 2003;40:520–525. doi: 10.1136/jmg.40.7.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek T, Prokopova I, Sladek M, Weissova K, Vojtechova I, Bahnik S, Zemanova A, Schönig K, Berger S, Tews B, Bartsch D, Schwab ME, Sumova A, Stuchlik A. Nogo-A-deficient transgenic rats show deficits in higher cognitive functions, decreased anxiety, and altered circadian activity patterns. Front Behav Neurosci. 2014;8:90. doi: 10.3389/fnbeh.2014.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddubnyak V, Rigou P, Michel L, Rain JC, Geneste O, Wolkenstein P, Vidaud D, Hickman JA, Mauviel A, Poyet JL. Positive regulation of apoptosis by HCA66, a new Apaf-1 interacting protein, and its putative role in the physiopathology of NF1 microdeletion syndrome patients. Cell Death Differ. 2007;14:1222–1233. doi: 10.1038/sj.cdd.4402122. [DOI] [PubMed] [Google Scholar]
- Pinna V, Lanari V, Daniele P, Consoli F, Agolini E, Margiotti K, Bottillo I, Torrente I, Bruselles A, Fusilli C, Ficcadenti A, Bargiacchi S, Trevisson E, Forzan M, Giustini S, Leoni C, Zampino G, Digilio MC, Dallapiccola B, Clementi M, Tartaglia M, De Luca A. p.Arg1809Cys substitution in neurofibromin is associated with a distinctive NF1 phenotype without neurofibromas. Eur J Hum Genet. 2015;23:1068–1071. doi: 10.1038/ejhg.2014.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger C. Disorders of memory and plasticity in psychiatric disease. Dialogues Clin Neurosci. 2013;15:455–463. doi: 10.31887/DCNS.2013.15.4/cpittenger. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin SR, Bredella MA, Cai W, Kassarjian A, Harris GJ, Esparza S, Merker VL, Munn LL, Muzikansky A, Askenazi M, Nguyen R, Wenzel R, Mautner VF. Quantitative assessment of whole-body tumor burden in adult patients with neurofibromatosis. PLoS One. 2012;7:e35711. doi: 10.1371/journal.pone.0035711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada CE, Hufnagel RB, Hummel TR, Lovell AM, Hopkin RJ, Saal HM, Schorry EK. The use of magnetic resonance imaging screening for optic pathway gliomas in children with neurofibromatosis type 1. J Pediatr. 2015;167(851–856):e1. doi: 10.1016/j.jpeds.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Granada CN, Wiesner T, Messina JL, Jungbluth AA, Chi P, Antonescu CR. Loss of H3K27me3 expression is a highly sensitive marker for sporadic and radiation-induced MPNST. Am J Surg Pathol. 2016;40:479–489. doi: 10.1097/PAS.0000000000000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Rice SJ, Salzberg AC, Runkle EA, Liao J, Zander DS, Mu D. MiR-365 regulates lung cancer and developmental gene thyroid transcription factor 1. Cell Cycle. 2012;11:177–186. doi: 10.4161/cc.11.1.18576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiker SJ, Lee H, Baldwin KT, Duan Y, Shrager P, Giger RJ. Oligodendrocyte-myelin glycoprotein and Nogo negatively regulate activity-dependent synaptic plasticity. J Neurosci. 2010;30:12432–12445. doi: 10.1523/JNEUROSCI.0895-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SA, Colman SD, Ho VT, Abernathy CR, Arn PH, Weiss L, Schwartz C, Saul RA, Wallace MR. Constitutional and mosaic large NF1 gene deletions in neurofibromatosis type 1. J Med Genet. 1998;35:468–471. doi: 10.1136/jmg.35.6.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Ding H, Huang S, Wang H, Wu M, Luo D, Dang Y, Yang L, Chen G. Expression and clinicopathological significance of miR-193a-3p and its potential target astrocyte elevated gene-1 in non-small lung cancer tissues. Cancer Cell Int. 2015;15:80. doi: 10.1186/s12935-015-0227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardi VM. Neurofibromatosis: phenotype, natural history and pathogenesis. 2. Baltimore: Johns Hopkins University Press; 1992. pp. 165–166. [Google Scholar]
- Riccardi VM. Skeletal system. In: Friedman J, Gutmann D, MacCollin M, Riccardi VM, editors. Neurofibromatosis: phenotype, natural history and pathogenesis. 3. Baltimore: Johns Hopkins University Press; 1999. pp. 262–263. [Google Scholar]
- Riccardi VM. Skeletal system. In: Friedman J, Gutmann D, MacCollin M, Riccardi VM, editors. Neurofibromatosis: phenotype, natural history and pathogenesis. 3. Baltimore: Johns Hopkins University Press; 1999. p. 260. [Google Scholar]
- Riva P, Castorina P, Manoukian S, Dalprà L, Doneda L, Marini G, den Dunnen J, Larizza L. Characterization of a cytogenetic 17q11.2 deletion in an NF1 patient with a contiguous gene syndrome. Hum Genet. 1996;98:646–650. doi: 10.1007/s004390050277. [DOI] [PubMed] [Google Scholar]
- Riva P, Corrado L, Natacci F, Castorina P, Wu BL, Schneider GH, Clementi M, Tenconi R, Korf BR, Larizza L. NF1 microdeletion syndrome: refined FISH characterization of sporadic and familial deletions with locus-specific probes. Am J Hum Genet. 2000;66:100–109. doi: 10.1086/302709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehl AC, Vogt J, Mussotter T, Zickler AN, Spöti H, Högel J, Chuzhanova NA, Wimmer K, Kluwe L, Mautner VF, Cooper DN, Kehrer-Sawatzki H. Intrachromosomal mitotic nonallelic homologous recombination is the major molecular mechanism underlying type-2 NF1 deletions. Hum Mutat. 2010;31:1163–1173. doi: 10.1002/humu.21340. [DOI] [PubMed] [Google Scholar]
- Roehl AC, Mussotter T, Cooper DN, Kluwe L, Wimmer K, Högel J, Zetzmann M, Vogt J, Mautner VF, Kehrer-Sawatzki H. Tissue-specific differences in the proportion of mosaic large NF1 deletions are suggestive of a selective growth advantage of hematopoietic del(±) stem cells. Hum Mutat. 2012;33:541–550. doi: 10.1002/humu.22013. [DOI] [PubMed] [Google Scholar]
- Röhrich M, Koelsche C, Schrimpf D, Capper D, Sahm F, Kratz A, Reuss J, Hovestadt V, Jones DT, Bewerunge-Hudler M, Becker A, Weis J, Mawrin C, Mittelbronn M, Perry A, Mautner VF, Mechtersheimer G, Hartmann C, Okuducu AF, Arp M, Seiz-Rosenhagen M, Hänggi D, Heim S, Paulus W, Schittenhelm J, Ahmadi R, Herold-Mende C, Unterberg A, Pfister SM, von Deimling A, Reuss DE. Methylation-based classification of benign and malignant peripheral nerve sheath tumors. Acta Neuropathol. 2016;131:877–887. doi: 10.1007/s00401-016-1540-6. [DOI] [PubMed] [Google Scholar]
- Rojnueangnit K, Xie J, Gomes A, Sharp A, Callens T, Chen Y, Liu Y, Cochran M, Abbott MA, Atkin J, Babovic-Vuksanovic D, Barnett CP, Crenshaw M, Bartholomew DW, Basel L, Bellus G, Ben-Shachar S, Bialer MG, Bick D, Blumberg B, Cortes F, David KL, Destree A, Duat-Rodriguez A, Earl D, Escobar L, Eswara M, Ezquieta B, Frayling IM, Frydman M, Gardner K, Gripp KW, Hernández-Chico C, Heyrman K, Ibrahim J, Janssens S, Keena BA, Llano-Rivas I, Leppig K, McDonald M, Misra VK, Mulbury J, Narayanan V, Orenstein N, Galvin-Parton P, Pedro H, Pivnick EK, Powell CM, Randolph L, Raskin S, Rosell J, Rubin K, Seashore M, Schaaf CP, Scheuerle A, Schultz M, Schorry E, Schnur R, Siqveland E, Tkachuk A, Tonsgard J, Upadhyaya M, Verma IC, Wallace S, Williams C, Zackai E, Zonana J, Lazaro C, Claes K, Korf B, Martin Y, Legius E, Messiaen L. High incidence of Noonan syndrome features including short stature and pulmonic stenosis in patients carrying NF1 missense mutations affecting p.Arg1809: genotype-phenotype correlation. Hum Mutat. 2015;36:1052–1063. doi: 10.1002/humu.22832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggieri M, Polizzi A, Spalice A, Salpietro V, Caltabiano R, D’Orazi V, Pavone P, Pirrone C, Magro G, Platania N, Cavallaro S, Muglia M, Nicita F. The natural history of spinal neurofibromatosis: a critical review of clinical and genetic features. Clin Genet. 2015;87:401–410. doi: 10.1111/cge.12498. [DOI] [PubMed] [Google Scholar]
- Sabbagh A, Pasmant E, Laurendeau I, Parfait B, Barbarot S, Guillot B, Combemale P, Ferkal S, Vidaud M, Aubourg P, Vidaud D, Wolkenstein P, Members of the NF France Network Unravelling the genetic basis of variable clinical expression in neurofibromatosis 1. Hum Mol Genet. 2009;18:2768–2778. doi: 10.1093/hmg/ddp212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamon J, Mautner VF, Adam G, Derlin T. Multimodal imaging in neurofibromatosis type 1-associated nerve sheath tumors. Rofo. 2015;187:1084–1092. doi: 10.1055/s-0035-1551330. [DOI] [PubMed] [Google Scholar]
- Salvi A, Conde I, Abeni E, Arici B, Grossi I, Specchia C, Portolani N, Barlati S, De Petro G. Effects of miR-193a and sorafenib on hepatocellular carcinoma cells. Mol Cancer. 2013;12:162. doi: 10.1186/1476-4598-12-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbidian E, Hadj-Rabia S, Riccardi VM, Valeyrie-Allanore LL, Barbarot S, Chosidow O, Ferkal S, Rodriguez D, Wolkenstein P, Bastuji-Garin S. Clinical characteristics predicting internal neurofibromas in 357 children with neurofibromatosis-1: results from a cross-selectional study. Orphanet J Rare Dis. 2012;7:62. doi: 10.1186/1750-1172-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer IM, Fletcher CD, Hornick JL. Loss of H3K27 trimethylation distinguishes malignant peripheral nerve sheath tumors from histologic mimics. Mod Pathol. 2016;29:4–13. doi: 10.1038/modpathol.2015.134. [DOI] [PubMed] [Google Scholar]
- Sedani A, Cooper DN, Upadhyaya M. An emerging role for microRNAs in NF1 tumorigenesis. Hum Genom. 2012;6:23. doi: 10.1186/1479-7364-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seminog OO, Goldacre MJ. Risk of benign tumours of nervous system, and of malignant neoplasms, in people with neurofibromatosis: population-based record-linkage study. Br J Cancer. 2013;108:193–198. doi: 10.1038/bjc.2012.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seminog OO, Goldacre MJ. Age-specific risk of breast cancer in women with neurofibromatosis type 1. Br J Cancer. 2015;112:1546–1548. doi: 10.1038/bjc.2015.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevick RJ, Barkovich AJ, Edwards MS, Koch T, Berg B, Lempert T. Evolution of white matter lesions in neurofibromatosis type 1: MR findings. Am J Roentgenol. 1992;159:171–175. doi: 10.2214/ajr.159.1.1609692. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Gai X, Perin JC, Glessner JT, Xie H, Murphy K, O’Hara R, Casalunovo T, Conlin LK, D’Arcy M, Frackelton EC, Geiger EA, Haldeman-Englert C, Imielinski M, Kim CE, Medne L, Annaiah K, Bradfield JP, Dabaghyan E, Eckert A, Onyiah CC, Ostapenko S, Otieno FG, Santa E, Shaner JL, Skraban R, Smith RM, Elia J, Goldmuntz E, Spinner NB, Zackai EH, Chiavacci RM, Grundmeier R, Rappaport EF, Grant SF, White PS, Hakonarson H. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif S, Moran A, Huson SM, Iddenden R, Shenton A, Howard E, Evans DG. Women with neurofibromatosis 1 are at a moderately increased risk of developing breast cancer and should be considered for early screening. J Med Genet. 2007;44:481–484. doi: 10.1136/jmg.2007.049346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilyansky C, Lee YS, Silva AJ. Molecular and cellular mechanisms of learning disabilities: a focus on NF1. Annu Rev Neurosci. 2010;33:221–243. doi: 10.1146/annurev-neuro-060909-153215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilyansky C, Karlsgodt KH, Cummings DM, Sidiropoulou K, Hardt M, James AS, Ehninger D, Bearden CE, Poirazi P, Jentsch JD, Cannon TD, Levine MS, Silva AJ. Neurofibromin regulates corticostriatal inhibitory networks during working memory performance. Proc Natl Acad Sci USA. 2010;107:13141–13146. doi: 10.1073/pnas.1004829107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikdar N, Banerjee S, Lee KY, Wincovitch S, Pak E, Nakanishi K, Jasin M, Dutra A, Myung K. DNA damage responses by human ELG1 in S phase are important to maintain genomic integrity. Cell Cycle. 2009;8:3199–3207. doi: 10.4161/cc.8.19.9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinibaldi L, De Luca A, Bellacchio E, Conti E, Pasini A, Paloscia C, Spalletta G, Caltagirone C, Pizzuti A, Dallapiccola B. Mutations of the Nogo-66 receptor (RTN4R) gene in schizophrenia. Hum Mutat. 2004;24:534–535. doi: 10.1002/humu.9292. [DOI] [PubMed] [Google Scholar]
- Soucy EA, van Oppen D, Nejedly NL, Gao F, Gutmann DH, Hollander AS. Height assessments in children with neurofibromatosis type 1. J Child Neurol. 2013;28:303–307. doi: 10.1177/0883073812446310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M, Oexle K, Horn D, Windt E, Buske A, Albrecht B, Prott EC, Seemanová E, Seidel J, Rosenbaum T, Jenne D, Kehrer-Sawatzki H, Tinschert S. Childhood overgrowth in patients with common NF1 microdeletions. Eur J Hum Genet. 2005;13:883–888. doi: 10.1038/sj.ejhg.5201419. [DOI] [PubMed] [Google Scholar]
- Srivastava AK, Schwartz CE. Intellectual disability and autism spectrum disorders: causal genes and molecular mechanisms. Neurosci Biobehav Rev. 2014;2:161–174. doi: 10.1016/j.neubiorev.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann K, Cooper DN, Kluwe L, Chuzhanova NA, Senger C, Serra E, Lazaro C, Gilaberte M, Wimmer K, Mautner VF, Kehrer-Sawatzki H. Type 2 NF1 deletions are highly unusual by virtue of the absence of nonallelic homologous recombination hotspots and an apparent preference for female mitotic recombination. Am J Hum Genet. 2007;81:1201–1220. doi: 10.1086/522089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann K, Kluwe L, Cooper DN, Brems H, De Raedt T, Legius E, Mautner VF, Kehrer-Sawatzki H. Copy number variations in the NF1 gene region are infrequent and do not predispose to recurrent type-1 deletions. Eur J Hum Genet. 2008;16:572–580. doi: 10.1038/sj.ejhg.5202002. [DOI] [PubMed] [Google Scholar]
- Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci. 2015;72:2041–2059. doi: 10.1007/s00018-015-1847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel B, Latta E, Kehrer-Sawatzki H, Hoffmann GF, Fonatsch C, Rehder H. Somatic mosaicism of a greater than 1.7-Mb deletion of genomic DNA involving the entire NF1 gene as verified by FISH: further evidence for a contiguous gene syndrome in 17q11.2. Am J Med Genet. 1999;87:12–16. doi: 10.1002/(SICI)1096-8628(19991105)87:1<12::AID-AJMG3>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Sun R, Liu Z, Ma G, Lv W, Zhao X, Lei G, Xu C. Associations of deregulation of mir-365 and its target mRNA TTF-1 and survival in patients with NSCLC. Int J Clin Exp Pathol. 2015;8:2392–2399. [PMC free article] [PubMed] [Google Scholar]
- Syken J, Grandpre T, Kanold PO, Shatz CJ. PirB restricts ocular-dominance plasticity in visual cortex. Science. 2006;313:1795–1800. doi: 10.1126/science.1128232. [DOI] [PubMed] [Google Scholar]
- Szudek J, Birch P, Friedman JM. Growth in North American white children with neurofibromatosis 1 (NF1) J Med Genet. 2000;37:933–938. doi: 10.1136/jmg.37.12.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tastet J, Decalonne L, Marouillat S, Malvy J, Thépault RA, Toutain A, Paubel A, Tabagh R, Bénédetti H, Laumonnier F, Barthélémy C, Bonnet-Brilhault F, Andres CR, Vourc’h P. Mutation screening of the ubiquitin ligase gene RNF135 in French patients with autism. Psychiatr Genet. 2015;25:263–267. doi: 10.1097/YPG.0000000000000100. [DOI] [PubMed] [Google Scholar]
- Tatton-Brown K, Douglas J, Coleman K, Baujat G, Cole TR, Das S, Horn D, Hughes HE, Temple IK, Faravelli F, Waggoner D, Turkmen S, Cormier-Daire V, Irrthum A, Rahman N, Collaboration Childhood Overgrowth. Genotype-phenotype associations in Sotos syndrome: an analysis of 266 individuals with NSD1 aberrations. Am J Hum Genet. 2005;77:193–204. doi: 10.1086/432082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor Tavares AL, Willatt L, Armstrong R, Simonic I, Park SM. Mosaic deletion of the NF1 gene in a patient with cognitive disability and dysmorphic features but without diagnostic features of NF1. Am J Med Genet A. 2013;161A:1185–1188. doi: 10.1002/ajmg.a.35853. [DOI] [PubMed] [Google Scholar]
- Tenorio J, Mansilla A, Valencia M, Martínez-Glez V, Romanelli V, Arias P, Castrejón N, Poletta F, Guillén-Navarro E, Gordo G, Mansilla E, García-Santiago F, González-Casado I, Vallespín E, Palomares M, Mori MA, Santos-Simarro F, García-Miñaur S, Fernández L, Mena R, Benito-Sanz S, del Pozo Á, Silla JC, Ibañez K, López-Granados E, Martín-Trujillo A, Montaner D, Heath KE, Campos-Barros Á, Dopazo J, Nevado J, Monk D, Ruiz-Pérez VL, Lapunzina P, SOGRI Consortium A new overgrowth syndrome is due to mutations in RNF125. Hum Mutat. 2014;35:1436–1441. doi: 10.1002/humu.22689. [DOI] [PubMed] [Google Scholar]
- Tews B, Schönig K, Arzt ME, Clementi S, Rioult-Pedotti MS, Zemmar A, Berger SM, Schneider M, Enkel T, Weinmann O, Kasper H, Schwab ME, Bartsch D. Synthetic microRNA-mediated downregulation of Nogo-A in transgenic rats reveals its role as regulator of synaptic plasticity and cognitive function. Proc Natl Acad Sci USA. 2013;110:6583–6588. doi: 10.1073/pnas.1217665110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinschert S, Naumann I, Stegmann E, Buske A, Kaufmann D, Thiel G, Jenne DE. Segmental neurofibromatosis is caused by somatic mutation of the neurofibromatosis type 1 (NF1) gene. Eur J Hum Genet. 2000;8:455–459. doi: 10.1038/sj.ejhg.5200493. [DOI] [PubMed] [Google Scholar]
- Tonsgard JH, Yelavarthi KK, Cushner S, Short MP, Lindgren V. Do NF1 gene deletions result in a characteristic phenotype? Am J Med Genet. 1997;73:80–86. doi: 10.1002/(SICI)1096-8628(19971128)73:1<80::AID-AJMG16>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Tsai KW, Leung CM, Lo YH, Chen TW, Chan WC, Yu SY, Tu YT, Lam HC, Li SC, Ger LP, Liu WS, Chang HT. Arm selection preference of microRNA-193a varies in breast cancer. Sci Rep. 2016;6:28176. doi: 10.1038/srep28176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker T, Wolkenstein P, Revuz J, Zeller J, Friedman JM. Association between benign and malignant peripheral nerve sheath tumors in NF1. Neurology. 2005;65:205–211. doi: 10.1212/01.wnl.0000168830.79997.13. [DOI] [PubMed] [Google Scholar]
- Upadhyaya M, Roberts SH, Maynard J, Sorour E, Thompson PW, Vaughan M, Wilkie AO, Hughes HE. A cytogenetic deletion, del(17)(q11.22q21.1), in a patient with sporadic neurofibromatosis type 1 (NF1) associated with dysmorphism and developmental delay. J Med Genet. 1996;33:148–152. doi: 10.1136/jmg.33.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya M, Ruggieri M, Maynard J, Osborn M, Hartog C, Mudd S, Penttinen M, Cordeiro I, Ponder M, Ponder BA, Krawczak M, Cooper DN. Gross deletions of the neurofibromatosis type 1 (NF1) gene are predominantly of maternal origin and commonly associated with a learning disability, dysmorphic features and developmental delay. Hum Genet. 1998;102:591–597. doi: 10.1007/s004390050746. [DOI] [PubMed] [Google Scholar]
- Upadhyaya M, Huson SM, Davies M, Thomas N, Chuzhanova N, Giovannini S, Evans DG, Howard E, Kerr B, Griffiths S, Consoli C, Side L, Adams D, Pierpont M, Hachen R, Barnicoat A, Li H, Wallace P, Van Biervliet JP, Stevenson D, Viskochil D, Baralle D, Haan E, Riccardi V, Turnpenny P, Lazaro C, Messiaen L. An absence of cutaneous neurofibromas associated with a 3-bp inframe deletion in exon 17 of the NF1 gene (c.2970-2972 delAAT): evidence of a clinically significant NF1 genotype-phenotype correlation. Am J Hum Genet. 2007;80:140–151. doi: 10.1086/510781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uusitalo E, Leppävirta J, Koffert A, Suominen S, Vahtera J, Vahlberg T, Pöyhönen M, Peltonen J, Peltonen S. Incidence and mortality of neurofibromatosis: a total population study in Finland. J Invest Dermatol. 2015;135:904–906. doi: 10.1038/jid.2014.465. [DOI] [PubMed] [Google Scholar]
- Uusitalo E, Rantanen M, Kallionpää RA, Pöyhönen M, Leppävirta J, Ylä-Outinen H, Riccardi VM, Pukkala E, Pitkäniemi J, Peltonen S, Peltonen J. Distinctive cancer associations in patients with neurofibromatosis type 1. J Clin Oncol. 2016;34:1978–1986. doi: 10.1200/JCO.2015.65.3576. [DOI] [PubMed] [Google Scholar]
- Valero MC, Pascual-Castroviejo I, Velasco E, Moreno F, Hernández-Chico C. Identification of de novo deletions at the NF1 gene: no preferential paternal origin and phenotypic analysis of patients. Hum Genet. 1997;99:720–726. doi: 10.1007/s004390050438. [DOI] [PubMed] [Google Scholar]
- van Asperen CJ, Overweg-Plandsoen WC, Cnossen MH, van Tijn DA, Hennekam RC. Familial neurofibromatosis type 1 associated with an overgrowth syndrome resembling Weaver syndrome. J Med Genet. 1998;35:323–327. doi: 10.1136/jmg.35.4.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart T, van Woerden GM, Elgersma Y, de Zeeuw CI, Schonewille M. Motor deficits in neurofibromatosis type 1 mice: the role of the cerebellum. Genes Brain Behav. 2011;10:404–409. doi: 10.1111/j.1601-183X.2011.00685.x. [DOI] [PubMed] [Google Scholar]
- Vassilopoulou-Sellin R, Klein MJ, Slopis JK. Growth hormone deficiency in children with neurofibromatosis type 1 without suprasellar lesions. Pediatr Neurol. 2000;22:355–358. doi: 10.1016/S0887-8994(00)00123-5. [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K, Brandom KG, Yun H. PI-3-kinase-dependent membrane recruitment of centaurin-alpha2 is essential for its effect on ARF6-mediated actin cytoskeleton reorganisation. J Cell Sci. 2007;120:792–801. doi: 10.1242/jcs.03373. [DOI] [PubMed] [Google Scholar]
- Venturin M, Gervasini C, Orzan F, Bentivegna A, Corrado L, Colapietro P, Friso A, Tenconi R, Upadhyaya M, Larizza L, Riva P. Evidence for non-homologous end joining and nonallelic homologous recombination in atypical NF1 microdeletions. Hum Genet. 2004;115:69–80. doi: 10.1007/s00439-004-1101-2. [DOI] [PubMed] [Google Scholar]
- Venturin M, Guarnieri P, Natacci F, Stabile M, Tenconi R, Clementi M, Hernandez C, Thompson P, Upadhyaya M, Larizza L, Riva P. Mental retardation and cardiovascular malformations in NF1 microdeleted patients point to candidate genes in 17q11.2. J Med Genet. 2004;41:35–41. doi: 10.1136/jmg.2003.014761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturin M, Bentivegna A, Moroni R, Larizza L, Riva P. Evidence by expression analysis of candidate genes for congenital heart defects in the NF1 microdeletion interval. Ann Hum Genet. 2005;69:508–516. doi: 10.1111/j.1529-8817.2005.00203.x. [DOI] [PubMed] [Google Scholar]
- Venturin M, Moncini S, Villa V, Russo S, Bonati MT, Larizza L, Riva P. Mutations and novel polymorphisms in coding regions and UTRs of CDK5R1 and OMG genes in patients with non-syndromic mental retardation. Neurogenetics. 2006;7:59–66. doi: 10.1007/s10048-005-0026-9. [DOI] [PubMed] [Google Scholar]
- Venturin M, Carra S, Gaudenzi G, Brunelli S, Gallo GR, Moncini S, Cotelli F, Riva P. ADAP2 in heart development: a candidate gene for the occurrence of cardiovascular malformations in NF1 microdeletion syndrome. J Med Genet. 2014;51:436–443. doi: 10.1136/jmedgenet-2013-102240. [DOI] [PubMed] [Google Scholar]
- Vienberg S, Geiger J, Madsen S, Dalgaard LT. MicroRNAs in metabolism. Acta Physiol (Oxf) 2017;219:346–361. doi: 10.1111/apha.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violante IR, Ribeiro MJ, Silva ED, Castelo-Branco M. Gyrification, cortical and subcortical morphometry in neurofibromatosis type 1: an uneven profile of developmental abnormalities. J Neurodev Disord. 2013;5:3. doi: 10.1186/1866-1955-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viskochil D, Buchberg AM, Xu G, Cawthon RM, Stevens J, Wolff RK, Culver M, Carey JC, Copeland NG, Jenkins NA, et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–192. doi: 10.1016/0092-8674(90)90252-A. [DOI] [PubMed] [Google Scholar]
- Visser R, Koelma N, Vijfhuizen L, van der Wielen MJ, Kant SG, Breuning MH, Wit JM, Losekoot M. RNF135 mutations are not present in patients with Sotos syndrome-like features. Am J Med Genet A. 2009;149A:806–808. doi: 10.1002/ajmg.a.32694. [DOI] [PubMed] [Google Scholar]
- Vissers LE, Stankiewicz P. Microdeletion and microduplication syndromes. Methods Mol Biol. 2012;838:29–75. doi: 10.1007/978-1-61779-507-7_2. [DOI] [PubMed] [Google Scholar]
- Vogt J, Nguyen R, Kluwe L, Schuhmann M, Roehl AC, Mußotter T, Cooper DN, Mautner VF, Kehrer-Sawatzki H. Delineation of the clinical phenotype associated with non-mosaic type-2 NF1 deletions: two case reports. J Med Case Rep. 2011;5:577. doi: 10.1186/1752-1947-5-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt J, Mussotter T, Bengesser K, Claes K, Högel J, Chuzhanova N, Fu C, van den Ende J, Mautner VF, Cooper DN, Messiaen L, Kehrer-Sawatzki H. Identification of recurrent type-2 NF1 microdeletions reveals a mitotic nonallelic homologous recombination hotspot underlying a human genomic disorder. Hum Mutat. 2012;33:1599–1609. doi: 10.1002/humu.22171. [DOI] [PubMed] [Google Scholar]
- Vogt J, Bengesser K, Claes KB, Wimmer K, Mautner VF, van Minkelen R, Legius E, Brems H, Upadhyaya M, Högel J, Lazaro C, Rosenbaum T, Bammert S, Messiaen L, Cooper DN, Kehrer-Sawatzki H. SVA retrotransposon insertion-associated deletion represents a novel mutational mechanism underlying large genomic copy number changes with non-recurrent breakpoints. Genome Biol. 2014;15:R80. doi: 10.1186/gb-2014-15-6-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner DJ, Towbin J, Gottesman G, Gutmann DH. A clinic-based study of plexiform neurofibromas in neurofibromatosis 1. Am J Med Genet. 2000;92:132–135. doi: 10.1002/(SICI)1096-8628(20000515)92:2<132::AID-AJMG10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Wallace MR, Andersen LB, Fountain JW, Odeh HM, Viskochil D, Marchuk DA, O’Connell P, White R, Collins FS. A chromosome jump crosses a translocation breakpoint in the von Recklinghausen neurofibromatosis region. Genes Chromosomes Cancer. 1990;2:271–277. doi: 10.1002/gcc.2870020404. [DOI] [PubMed] [Google Scholar]
- Wang J, Yang B, Han L, Li X, Tao H, Zhang S, Hu Y. Demethylation of miR-9-3 and miR-193a genes suppresses proliferation and promotes apoptosis in non-small cell lung cancer cell lines. Cell Physiol Biochem. 2013;32:1707–1719. doi: 10.1159/000356605. [DOI] [PubMed] [Google Scholar]
- Weise A, Mrasek K, Klein E, Mulatinho M, Llerena JC, Jr, Hardekopf D, Pekova S, Bhatt S, Kosyakova N, Liehr T. Microdeletion and microduplication syndromes. J Histochem Cytochem. 2012;60:346–358. doi: 10.1369/0022155412440001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel LE, Albers AC, Gutmann DH, Dunn CM. The association between hypotonia and brain tumors in children with neurofibromatosis type 1. J Child Neurol. 2013;28:1664–1667. doi: 10.1177/0883073812460918. [DOI] [PubMed] [Google Scholar]
- Willi R, Schwab ME. Nogo and Nogo receptor: relevance to schizophrenia? Neurobiol Dis. 2013;54:150–157. doi: 10.1016/j.nbd.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Williams M, Kirschner MB, Cheng YY, Hanh J, Weiss J, Mugridge N, Wright CM, Linton A, Kao SC, Edelman JJ, Vallely MP, McCaughan BC, Cooper W, Klebe S, Lin RC, Brahmbhatt H, MacDiarmid J, van Zandwijk N, Reid G. miR-193a-3p is a potential tumor suppressor in malignant pleural mesothelioma. Oncotarget. 2015;6:23480–23495. doi: 10.18632/oncotarget.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BL, Austin MA, Schneider GH, Boles RG, Korf BR. Deletion of the entire NF1 gene detected by the FISH: four deletion patients associated with severe manifestations. Am J Med Genet. 1995;59:528–535. doi: 10.1002/ajmg.1320590427. [DOI] [PubMed] [Google Scholar]
- Wu BL, Schneider GH, Korf BR. Deletion of the entire NF1 gene causing distinct manifestations in a family. Am J Med Genet. 1997;69:98–101. doi: 10.1002/(SICI)1096-8628(19970303)69:1<98::AID-AJMG19>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Wu R, López-Correa C, Rutkowski JL, Baumbach LL, Glover TW, Legius E. Germline mutations in NF1 patients with malignancies. Genes Chromosomes Cancer. 1999;26:376–380. doi: 10.1002/(SICI)1098-2264(199912)26:4<376::AID-GCC13>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Xing CY, Hu XQ, Xie FY, Yu ZJ, Li HY, Bin-Zhou WuJB, Tang LY, Gao SM. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 2015;589:1981–1987. doi: 10.1016/j.febslet.2015.04.061. [DOI] [PubMed] [Google Scholar]
- Xu J, Ismat FA, Wang T, Lu MM, Antonucci N, Epstein JA. Cardiomyocyte-specific loss of neurofibromin promotes cardiac hypertrophy and dysfunction. Circ Res. 2009;105:304–311. doi: 10.1161/CIRCRESAHA.109.201509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhou L, Lu L, Wang L, Li X, Jiang P, Chan LK, Zhang T, Yu J, Kwong J, Cheung TH, Chung T, Mak K, Sun H, Wang H. A novel miR-193a-5p-YY1-APC regulatory axis in human endometrioid endometrial adenocarcinoma. Oncogene. 2013;32:3432–3442. doi: 10.1038/onc.2012.360. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. Viral RNA detection by RIG-I-like receptors. Curr Opin Immunol. 2015;32:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Yu T, Li J, Yan M, Liu L, Lin H, Zhao F, Sun L, Zhang Y, Cui Y, Zhang F, Li J, He X, Yao M. MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene. 2015;34:413–423. doi: 10.1038/onc.2013.574. [DOI] [PubMed] [Google Scholar]
- Yzaguirre AD, Padmanabhan A, de Groh ED, Engleka KA, Li J, Speck NA, Epstein JA. Loss of neurofibromin Ras-GAP activity enhances the formation of cardiac blood islands in murine embryos. Elife. 2015;4:e07780. doi: 10.7554/eLife.07780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang Y, Jones S, Sausen M, McMahon K, Sharma R, Wang Q, Belzberg AJ, Chaichana K, Gallia GL, Gokaslan ZL, Riggins GJ, Wolinksy JP, Wood LD, Montgomery EA, Hruban RH, Kinzler KW, Papadopoulos N, Vogelstein B, Bettegowda C. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46:1170–1172. doi: 10.1038/ng.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tong H, Fu X, Zhang Y, Liu J, Cheng R, Liang J, Peng J, Sun Z, Liu H, Zhang F, Lu W, Li M, Yao Z. Molecular characterization of NF1 and neurofibromatosis type 1 genotype-phenotype correlations in a Chinese population. Sci Rep. 2015;5:11291. doi: 10.1038/srep11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Zhou L, Zheng L, Guo L, Wang Y, Liu H, Ou Ding CZ. miR-365 promotes cutaneous squamous cell carcinoma (CSCC) through targeting nuclear factor I/B (NFIB) PLoS One. 2014;9:e100620. doi: 10.1371/journal.pone.0100620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Wang Y, Ou C, Lin Z, Wang J, Liu H, Zhou M, Ding Z. microRNA-365-targeted nuclear factor I/B transcriptionally represses cyclin-dependent kinase 6 and 4 to inhibit the progression of cutaneous squamous cell carcinoma. Int J Biochem Cell Biol. 2015;65:182–191. doi: 10.1016/j.biocel.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Zickler AM, Hampp S, Messiaen L, Bengesser K, Mussotter T, Roehl AC, Wimmer K, Mautner VF, Kluwe L, Upadhyaya M, Pasmant E, Chuzhanova N, Kestler HA, Högel J, Legius E, Claes K, Cooper DN, Kehrer-Sawatzki H. Characterization of the nonallelic homologous recombination hotspot PRS3 associated with type-3 NF1 deletions. Hum Mutat. 2012;33:372–383. doi: 10.1002/humu.21644. [DOI] [PubMed] [Google Scholar]
- Zuccotti P, Cartelli D, Stroppi M, Pandini V, Venturin M, Aliverti A, Battaglioli E, Cappelletti G, Riva P. Centaurin-α2 interacts with β-tubulin and stabilizes microtubules. PLoS One. 2012;7:e52867. doi: 10.1371/journal.pone.0052867. [DOI] [PMC free article] [PubMed] [Google Scholar]